Co-habiting amphibian species harbor unique skin bacterial communities in wild populations (original) (raw)
Abstract
Although all plant and animal species harbor microbial symbionts, we know surprisingly little about the specificity of microbial communities to their hosts. Few studies have compared the microbiomes of different species of animals, and fewer still have examined animals in the wild. We sampled four pond habitats in Colorado, USA, where multiple amphibian species were present. In total, 32 amphibian individuals were sampled from three different species including northern leopard frogs (Lithobates pipiens), western chorus frogs (Pseudacris triseriata) and tiger salamanders (Ambystoma tigrinum). We compared the diversity and composition of the bacterial communities on the skin of the collected individuals via barcoded pyrosequencing of the 16S rRNA gene. Dominant bacterial phyla included Acidobacteria, Actinobacteria, Bacteriodetes, Cyanobacteria, Firmicutes and Proteobacteria. In total, we found members of 18 bacterial phyla, comparable to the taxonomic diversity typically found on human skin. Levels of bacterial diversity varied strongly across species: L. pipiens had the highest diversity; A. tigrinum the lowest. Host species was a highly significant predictor of bacterial community similarity, and co-habitation within the same pond was not significant, highlighting that the skin-associated bacterial communities do not simply reflect those bacterial communities found in their surrounding environments. Innate species differences thus appear to regulate the structure of skin bacterial communities on amphibians. In light of recent discoveries that some bacteria on amphibian skin have antifungal activity, our finding suggests that host-specific bacteria may have a role in the species-specific resistance to fungal pathogens.
Keywords: amphibian, skin, bacteria, host specific, microbiome
Introduction
All species of plants and animals harbor assemblages of microbes and these symbionts often have vital roles in the health of their hosts. However, there have been few comprehensive studies assessing whether different species harbor distinct microbial symbionts. Work on mammals (Ley et al., 2008), primates (Yildirim et al., 2010) and plants (Redford et al., 2010) suggests that there is some degree of host specificity of microbial symbionts across taxa. However, such studies have not yet been performed in wild populations, nor have these studies been designed to distinguish environmental factors from innate host factors. For different species in the wild, the question of host-specific microbial assemblages becomes difficult to address because species often occupy different habitat niches, and different environmental conditions may confound questions of host specificity. For several reasons, we have identified pond-dwelling larval amphibian communities as a model system to address whether different, co-habiting species harbor unique microbial communities. First, populations of larval amphibians observed before metamorphosis (for example, tadpoles), develop within a single pond and therefore have a known environmental origin. Second, ponds can harbor several species of co-habiting larval amphibians that occupy the same environmental niche during their development. Third, different ponds that harbor multiple species of larval amphibians can serve as replicates to examine patterns of host-associated microbial community assembly across different sites.
We focused specifically on skin-associated bacterial communities of amphibians as these communities likely provide the first line of defense against pathogens and, for co-habiting species, are exposed to the same environmental sources of microbes. One specific skin pathogen of global significance is the fungus Batrachochytrium dendrobatidis (Bd), which has a key role in amphibian declines worldwide (Skerratt et al., 2007; Fisher et al., 2009). Amphibian species, and some populations within species, vary in their susceptibility to disease caused by this fungal pathogen (Kilpatrick et al., 2010). While there are many factors that may have a role in amphibian disease resistance including host genetics, immunology (Carey et al., 1999), skin peptides (Rollins-Smith 2009) and environmental factors (Lips et al., 2008), microbial communities living on the skin of amphibians may also have a crucial role in host defense mechanisms. Certain bacteria isolated from amphibian skin are known to produce fungicides and can inhibit growth of Bd (Harris et al., 2006) and even reduce amphibian mortality under controlled experimental conditions (Harris et al., 2009b). However, it is not known if different species of wild amphibians can select unique skin microbial assemblages from exposure to the same microbial background. If specific microbial assemblages are involved in resistance to pathogens (such as Bd), and amphibian species vary in their susceptibility to disease, then we would expect that unique skin microbial composition across species or populations might provide a mechanism underlying this observed pattern of differences in susceptibility. Our goal in the present study was to use co-habiting, pond-dwelling larval amphibians to determine whether different species of amphibians harbor unique communities of bacteria on their skin. If they do, it suggests that species-specific bacterial communities have the potential to contribute to differences in the resistance of individual species to skin pathogens.
Teasing apart how symbiotic microbial communities are influenced by factors such as the environment, diet or innate host traits can now be addressed with the use of newer high-throughput survey techniques such as barcoded pyrosequencing of the 16S rRNA gene. These methods allow for observing the composition and relative abundance of microbes with greater accuracy than previous methods used to estimate bacterial community structure (Costello et al., 2009). By applying these newer techniques in pond-dwelling amphibians, we hope to gain insight on the host-specific nature of skin microbes independent of environmental confounds as well as build a foundation to understand the potential for these microbes to have a role in host resistance against pathogens.
Materials and methods
Amphibian species and field collections
Four pond habitats in Colorado were identified in 2008 that each harbored at least two amphibian species that are common in Colorado and co-habit in ponds during the summer breeding season. Amphibian species sampled included Lithobates (=Rana) pipiens (northern leopard frog), Pseudacris triseriata (western chorus frog) and Ambystoma tigrinum (tiger salamander). Each pond was less than one hectare in surface area and located in a separate watershed. Site characteristics and the number of individual amphibians sampled per species per pond are described in Table 1. Amphibians were sampled for skin bacteria during August 2008 and all individuals from a given pond were sampled on the same day. Pre-metamorphic, larval amphibians were targeted because they develop over one season within a single pond, therefore, we can ensure that our cross-species comparisons only include individuals that shared the same environment and microbial inocula during their development. All three species named above are pond-breeders that lay eggs in early summer, which hatch larval swimming stages that develop over the course of the summer into metamorphic stages (Hammerson, 1999). This system allows us to control for confounding environmental conditions and isolate the effect of host species.
Table 1. Amphibian sampling scheme.
| Pond name | # Leopard frogs sampled | # Chorus frogs sampled | # Tiger salamanders sampled | Pond elevation (m) | Pond location in Colorado | Latitude longitude (UTM) |
|---|---|---|---|---|---|---|
| Monster bull | 4 | 2 | 4 | 2357 | Meeker | 40.010359–107.625487 |
| Chive | 0 | 4 | 4 | 2598 | Nederland | 39.951877–105.463940 |
| Horse | 0 | 4 | 4 | 1702 | Southwest Boulder | 39.946662–105.241441 |
| Eggleston3 | 3 | 4 | 0 | 1754 | Southeast Boulder | 39.927857–105.20637 |
Larval amphibians were captured from the water using hand-held dipnets or a blocking net seine. Within a given species, larval individuals of a similar size were selected for skin microbe sampling. Larval amphibians were held temporarily (<10 min) in plastic containers with pond water before sampling. In total, 32 individual amphibians were sampled (7_L. pipiens_, 14 P. triseriata and 12 A. tigrinum; see Table 1) from four ponds. Fresh latex gloves were used for every individual amphibian handled. Before sampling, each individual was held and rinsed with 100 ml of sterile water three times to ensure that the skin sample primarily included skin-associated microbes rather than pond-associated material, including pond water, sediment and transient microbes (Culp et al., 2007; Lauer et al., 2007). Prior studies by Culp et al. (2007) and Lauer et al. (2007) demonstrate that the composition of bacteria obtained from amphibian skin versus rinse water mostly differ and represent unique assemblages, thus, we have some precedent to assume that most of the bacteria observed in this study are likely associated with the amphibians rather than transient bacteria from the environment. Immediately following rinsing, each amphibian was sampled using a sterile cotton-tipped swab brushed over the entire body of the amphibian for 30 s. The small size of larval amphibians (3–12 cm, depending on species) prevented us from swabbing a specific site on the body, so we opted to uniformly swab the entire body (head, mouth, tail, dorsal and ventral surfaces) in an effort to capture the total skin microbiome of each amphibian. All individuals were swabbed according to a standard protocol such that the swab was brushed in the direction from the head to the tail and the swab was turned slightly with each move to a new body surface (for example, dorsal stroke, rotate swab, then lateral body stroke) in order to capture each body surface on the swab without saturating the whole swab with one body part; this technique was manageable given the small size of the animals. Each swab was placed in a sterile vial and stored on dry ice. Within a few hours, each sample was then transferred to a −20 °C freezer for storage until DNA extraction. All animals were handled and released according to an approved Institutional Animal Care and Use Committee protocol.
DNA extraction, bacterial 16S rRNA PCR amplification and pyrosequencing
Bacterial DNA was extracted from each swab using the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA). For each of the 32 samples, the swab was loaded into a bead tube from the extraction kit and the remaining procedures were performed according to the manufacturer's protocol. Bacterial community composition was determined using barcoded pyrosequencing, following the procedure described in detail in Fierer et al. (2008), which includes both the PCR conditions and error-correcting barcoded primer sequences. The primer set used was the 27F/388R that targets the V2 region of the 16S small subunit ribosomal gene. No template and template from blank filters were included as negative controls throughout the entire process from DNA extraction to PCR amplification to check for contamination. PCR amplicons were pooled from each sample to equimolar ratio in a single tube, and sequenced at the University of South Carolina Environmental Genomics Core Facility on a 454 Life Sciences Sequence GS FLX instrument (Roche, Florence, SC, USA). We obtained an average of 1220 sequences per sample (range 780–1510), with read lengths averaging 258 bp in length. Previous work has shown that reads of this length over the V2 region are sufficient for accurate taxonomic identification to at least the family level and for community characterization (Liu et al., 2007).
A portion of the extracted DNA was used to screen samples for Bd, the chytrid fungal pathogen. Frozen DNA was shipped to Pisces Molecular LLC (Boulder, CO, USA) for analysis using _Bd_-specific primers (Annis et al., 2004), PCR amplification and gel electrophoresis to examine for the presence or absence of bands.
Sequence analysis
All sequence analyses were conducted using the Quantitative Insights Into Microbial Ecology (QIIME) pipeline (Caporaso et al., 2010). In brief, the QIIME pipeline removed low-quality sequences (that is, those sequences <200 bp in length, had a quality score <25, contained ambiguous characters, having an unreadable barcode or did not contain the primer sequence). Remaining sequences were clustered using CD-HIT (Li and Godzik, 2006), using a minimum coverage of 99% and minimum sequence identity of 97%. A representative sequence was chosen for each phylotype by selecting the longest sequence with the highest number of hits to other sequences in the same phylotype. The representative sequences were aligned using PyNAST (Caporaso et al., 2010) and taxonomy was assigned using the RDP classifier (Wang et al., 2007). A phylogenetic tree of all aligned sequences was constructed with FastTree (Price et al., 2009) and used in all downstream phylogenetic community comparisons. Community similarity (that is, β-diversity) analyses were performed using the taxon-based Bray–Curtis dissimilarity metric, and the phylogenetic UniFrac metric: the Bray–Curtis metric examines taxon overlap, whereas the UniFrac metric is a phylogenetic approach based on the fraction of shared branch length between two communities within a phylogenetic tree (Lozupone et al., 2007). Phylogenetic trees were constructed using FastTree (Price et al., 2009). All samples were rarified to 750 sequences per sample to remove sample heterogeneity, which impacts α- and β-diversity metrics (Lozupone et al., 2011).
Statistical analyses
To address whether bacterial phylotype richness varied among species and/or among pond sites, we conducted an analysis of covariance. Phylotype richness per frog served as the response variable (_n_=32) and frog species and pond site served as two categorical covariables. This analysis was performed using the statistical software JMP (SAS version 8, Cary, NC, USA). The data were normally distributed as determined by a Shapiro–Wilk W goodness-of-fit test.
To determine the differences in bacterial community composition across individuals (β-diversity), we used two different distance metrics: a Bray–Curtis similarity index (a taxonomic metric) and the UniFrac algorithm, which provides a measure of phylogenetic distance between communities from individual samples (Lozupone et al., 2007). β-Diversity patterns were visualized using a MDS (non-metric multidimensional scaling) ordination approach (Clarke and Warwick, 2001). To test for significant differences in community composition, we used a two-way nested ANOSIM (analysis of similarity) to examine whether host amphibian species and/or pond site were significant predictors of variability in bacterial communities across individual amphibian samples (PRIMER-E). We ran two separate analyses, one with Bray–Curtis similarity as the response variable and one with unweighted UniFrac distances as the response variable.
Lastly, to visualize the relative abundances of the most common bacterial groups across amphibian species per pond, we constructed a heat map figure. By removing all sequences that comprised ⩽5% of the total number of classified sequences across all samples, 19 phylotypes remained. By reducing the data set to the 19 most abundant phylotypes, visualization of the compositional differences was tractable and issues related to sequencing error are avoided (Quince et al., 2009; Huse et al., 2010; Kunin et al., 2010).
Results
From the rarefied 750 sequences per sample (_n_=32), we found between 15 and 345 unique phylotypes per sample representing 18 different bacterial phyla. Diversity results thus apply only to the dominant members of the community, and it is possible that sequences scored as absent are in fact present at lower abundance; however, the communities are substantially different from one another when sampled at this depth. The dominant phyla across all samples were Acidobacteria, Actinobacteria, Bacteriodetes, Cyanobacteria, Firmicutes and Proteobacteria. A list of all phylotypes and their taxonomic affiliation is provided in Supplementary Information (Supplementary Table S1).
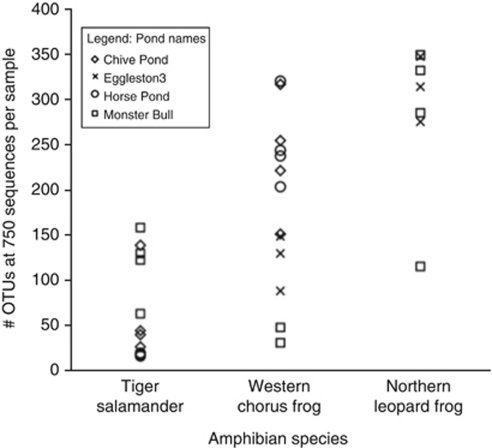
Phylotype richness varied between the amphibian species (Figure 1). Northern leopard frogs had the highest bacterial phylotype richness relative to the first 750 sequences sampled (mean=287 phylotypes per sample), western chorus frogs had a moderate richness (mean=185) and tiger salamanders had the lowest (mean=66). These phylotype richness patterns varied significantly among species (P<0.0001, _r_2=0.60) but were not significantly different among ponds (_P_=0.47), as determined by an analysis of covariance. We cannot effectively estimate absolute levels of diversity because sequencing errors may lead to an inflation of phylotype numbers (Quince et al., 2009) and because deeper sequencing depths would undoubtedly increase the number of phylotypes identified. However, since all samples were compared at an equivalent sequencing depth, these results do suggest that there are strong differences in relative levels of bacterial diversity across the amphibian species sampled here.
Figure 1.
Phylotype richness per amphibian species. The number of unique phylotypes per individual amphibian, estimated from a sampling depth of 750 sequences per sample. Each symbol represents the estimate of unique phylotypes or operational taxonomic units (OTUs) per amphibian individuals at a given site. Different symbols correspond to different pond locations.
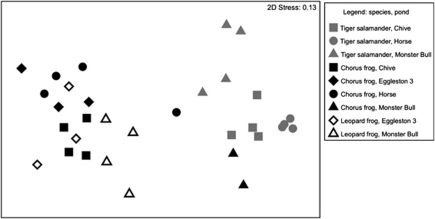
There were strong differences in the composition of the skin-associated bacterial communities found on the different amphibian species, a pattern evident in the ordination plot (Figure 2). These patterns are confirmed by the ANOSIM analyses; with both the taxon-based Bray–Curtis distance and the unweighted UniFrac phylogenetic distance metrics, host species identity was a highly significant predictor of skin bacterial community uniqueness (P<0.001; Table 2). Host species explained a large proportion of the variance in the skin bacterial communities (86% using Bray–Curtis and 91% using unweighted UniFrac). Different pond sites were not significant predictors of skin bacterial communities on amphibians (Table 2), although variation among sites within species does exist, as evident from Figure 2.
Figure 2.
Ordination plot. β-Diversity patterns were visualized using a MDS ordination approach with skin-associated bacterial community differences represented as Bray–Curtis distances. The 2D stress (or measure of distortion) in the MDS configuration was relatively low (0.13), so the 2D distance between points in the ordination plot is a good representation of the degree of similarity between each sample's bacterial community. Each point represents the skin bacterial community of an individual amphibian; color indicates species and shape indicates pond location.
Table 2. Results of the two-way nested analysis of similarity (ANOSIM).
| | Bray–Curtis similarity | Unweighted UniFrac distance | | | | | --------------------------- | ----------------------------- | ---------- | -------------- | ------ | | | Global R | Significance | Global R | Significance | | | Between amphibian species | 0.868 | <0.001 | 0.914 | <0.001 | | Between pond sites | 0.089 | 0.391 | −0.178 | 0.714 |
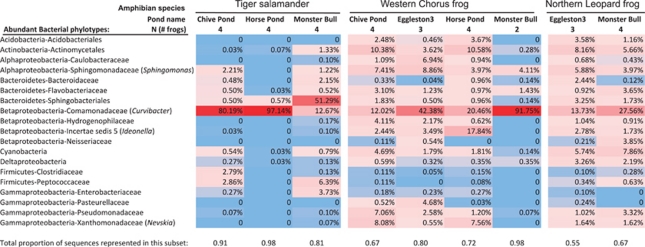
The dominant phylotypes across all amphibian samples and their relative abundances are presented in Figure 3, with further details provided in Supplementary Table S1. The most dominant phylotype across all three amphibian species was classified as belonging to the genus Curvibacter (Betaproteobacteria: Comamonadaceae), which accounted for 12–97% of the sequences from individual samples, depending on the species and pond location (Figure 3). A Bacteroidetes phylotype belonging to the group Sphingobacteriales was dominant on tiger salamanders from one pond, comprising 51% of the community, but otherwise, the remaining phylotypes observed on salamanders comprised ⩽6% of the bacterial sequences. A species of Ideonella (Betaproteobacteria) was abundant on chorus frogs from one pond, comprising 18% of the community and another phylotype belonging to the Actinobacteria (Actinomycetales) was also abundant on chorus frogs at two of the pond sites. Leopard frogs had many phylotypes present at lower abundances. For instance, several phylotypes comprised between 3% and 8% of the leopard frog skin communities, and these phylotypes belonged to a range of different phyla and subphyla.
Figure 3.
Heat map. A color-scale heat map demonstrates the relative abundance of the 19 most common bacterial phylotypes across species and ponds; red cells indicate higher proportional abundance and blue cells indicate lower proportional abundance. The relative abundance of each phylotype was converted to a proportion of the total number of classified sequences. The total number of unclassified sequences summed together for each amphibian sample comprised on average only 5.7% (±s.d.=0.06) of the sequences per sample and were removed from the table. In order to condense the bacterial groups and present the most common phylotypes in the heat map, we further removed all phylotypes that comprised ⩽5% of the total number of classified sequences across all samples. The proportion of sequences that are represented in the heat map for each species in each pond site varied between 55–98%. Nineteen unique phylotypes remained and the average proportion was calculated per amphibian species per pond.
One individual western chorus frog (P. triseriata) from Chive Pond was found to be positive for Bd, the chytrid fungal skin pathogen. Due to a lack of other positive individuals in this data set, we cannot draw any conclusions about the possible interactions of chytrid infection with the skin bacterial communities.
Discussion
Our understanding of the microbes associated with plants and animals, including humans, is in a phase of rapid discovery owing, in part, to the development of high-throughput sequence-based approaches that allow us to document the structure and composition of numerous complex bacterial communities simultaneously. With improved community-level data, we can explore questions regarding how microbiomes compare across individuals, species or environments, yet there is a paucity of studies comparing microbiomes across animal species. Ley et al. (2008) compared gut microbiomes of 60 different mammal species and discovered that both host phylogeny and diet appear to influence the bacterial communities. Roughly half of the samples they observed were from animals in the wild, but from populations that did not overlap in their geographic range, and the other half were from captive zoo animals. Yildirim et al. (2010) compared the gut microbiome of three different primate species from wild troops whose home ranges overlapped. While the authors found highly significant differences in the gut bacteria among the primate species, it is difficult to completely disentangle the role of social interactions (for example, grooming, mating, aggressive interactions) and close physical contact. It is possible that the similarity of the gut communities of primate individuals is, in part, due to closeness and physical contact within a troop rather than phylogenetic host differences. Thus, many wild animal species may not directly overlap in their habitat use or level of social contact with other species, and these factors may confound our understanding of species-specific differences of their microbiomes.
As the skin microbiome is in more constant contact with the external environment than the gut microbiome, we might expect this to obscure patterns of phylogenetic host specificity. For example, we may expect the skin microbiome to consist of many exogenous bacteria originating from the environment (Belden and Harris, 2007), rather than bacteria that are host specific. Although the human skin microbiome and its spatial variability has been reasonably well studied (Costello et al., 2009; Grice et al., 2009), patterns of microbial assemblages living on the skin of other animals is largely unknown. Without fur or feathers, amphibians provide an excellent model system for studying skin-associated microbial communities. Pre-metamorphic amphibians in pond habitats provide a rare example of multispecies communities that co-exist temporally and spatially in aquatic habitats. We can be confident that all larval individuals present in a given pond during a breeding season originated there and are exposed to the same microbial inocula over the course of their larval development. If there are no differences in skin bacterial communities across species that co-habit in a pond, but there are differences across pond sites, then we can argue that the composition of skin-associated bacterial communities is most strongly influenced by the pond environment. Conversely, if we observe that the skin communities are unique to different amphibian species and that those patterns are consistent across multiple sites, then we can assert that the skin bacterial communities are demonstrating evidence for host specificity. Our results indicate strong support for the latter case, skin bacteria of amphibians appear to be strongly species specific across different sites, even when species co-exist in the same pond (Table 2).
The number of unique skin bacterial phyla varied between 10 and 18 phyla depending upon amphibian species. Using similar techniques, Costello et al. (2009) found the human skin microbiome is comprised of 18 different bacterial phyla. Therefore, the phylum-level bacterial richness found on the skin of amphibians appears high and on par with that of human skin. Previous studies have reported a diversity of bacteria from amphibian skin using different methods. Using a PCR-based fingerprinting technique (denaturing gradient gel electrophoresis) of 16S rRNA fragments, Lauer et al. (2007) reported up to 19 unique bacteria from eastern red-backed salamanders (Plethodon cinereus), representing four different phyla. Likewise, Woodhams et al. (2007b) used a culture-based approach and found 40 unique bacterial isolates, representing three different phyla, from 70 frog (Rana muscosa) individuals (mean of eight isolates per frog individual). As in other environments, culture-based approaches likely capture only a small fraction of the bacterial diversity present on amphibian skin (Pace, 1997). While culture-based approaches allow one to determine the specific attributes of individual bacterial taxa, such as their ability to limit fungal growth (Lauer et al., 2007, 2008), combining molecular with culture-based approaches is clearly important for understanding the role of bacterial communities on their amphibian hosts.
All of the phyla reported from previous studies of bacteria on amphibian skin that employed different methods, including Firmicutes, Actinobacteria, Proteobacteria and Bacteroidetes (Culp et al., 2007; Lauer et al., 2007, 2008; Woodhams et al., 2007a; Lam et al., 2010), were also found in the present study and comprised some of the more abundant groups observed. Given that the barcoded pyrosequencing method allowed us to observe 14 additional phyla, we can examine community-level patterns and composition to a far greater degree. Closer examination of the composition of the bacterial communities revealed that, for several amphibians sampled in this study, their skin bacterial communities were dominated by a few phylotypes (Figure 3). For example, on tiger salamanders and chorus frogs from four different pond sites, between 42–97% of the bacterial community was dominated by members of the genus Curvibacter, a Betaproteobacteria in the family Comamonadaceae. This was by far the most abundant phylotype across all species in all sites. This genus and its close relatives are commonly found in a wide variety of natural aquatic systems (Shaw et al., 2008; Mueller-Spitz et al., 2009; Hutalle-Schmelzer et al., 2010) and they appear to be primarily freshwater organisms with a wide range of temperature tolerance (9–40 °C) and pH tolerance (5.5–8.5) for growth (Ding and Yokota, 2004). Due to the lack of comparable studies, we do not know if members of this genus are commonly associated with other species of amphibians found at other locations. Another dominant phylotype was identified as being related to Ideonella (Betaproteobacteria) and was particularly abundant on western chorus frogs. The sequence matches that of a symbiont found on the epithelium of Hydra, a small freshwater cnidarian (Fraune et al., 2010). A few of the other more abundant phylotypes found on the amphibians also match sequences of bacterial strains that are previously known as symbionts living with hosts. These include a strain of Sphingomonas (Alphaproteobacteria) reported to be a symbiont with plant hopper insects (Tang et al., 2010), a strain of Hydrogenophilus (Betaproteobacteria) reported from the human oral microbiome (Dewhirst et al., 2010) and a strain of Caulobacter (Alphaproteobacteria) reported from the human gut (Frank et al., 2007).
Overall, several bacterial types that are related to those found in freshwater systems (for example, Nevskia; Pladdies et al., 2004) are abundant on amphibian skin, as well as several that are related to known symbionts of other organisms. While we used sterile water rinsing of amphibians specifically to remove transient microbes before swab sampling (sensu Culp et al., 2007), it is possible that not all of the bacteria obtained from the swab are actual epibiotic symbionts of the amphibians. However, since we found that amphibian species inhabiting the same pond harbored distinct bacterial communities, it is likely that most of the taxa were epibiotic and not simply transient microbes from the pond environment. Furthermore, we do not know whether transient microbes potentially interact with the epibiotic community on amphibians or simply exist temporarily in proximity to the skin microbiome. The mechanism by which amphibian species acquire unique skin communities is not known. Sources of microbial inocula may include vertical transmission (from parent to offspring), horizontal transmission (between individuals) and a variety of environmental sources (water, sediment, algae, invertebrates, organic matter, etc.). While some terrestrial species of amphibians exhibit parental care of egg nests and young (Banning et al., 2008), the aquatic species studied in this system do not display parental care; thus, vertical transmission seems unlikely in this system. However, Fraune et al. (2010) provide evidence that freshwater cnidarian (_Hydr_a) embryos obtain maternally derived peptides that drive the assembly of the epithelial bacterial community, indicating that it may be possible for organisms without extended parental care to receive vertical transmission of peptides that select for the type of community that resides on the offspring. In any case, the assembly of microbial communities that live on amphibians is an intriguing area that can be examined with both field observations and experimental approaches.
While Curvibacter is clearly the most abundant bacteria observed on amphibians in this data set, the presence and relative abundance of other phylotypes largely account for the species-specific patterns observed (Figure 3). Pond sites were not significant in determining the differences among individual amphibians (Table 2); however, we can see that there is some variation across sites; for example, chorus frogs from four different sites have variation in their communities (Figure 3). Three of the Colorado sites occur on the Front Range and one is on the Western Slope (∼200 miles away), yet several dominant phylotypes are still found in both regions (for example, Curvibacter, a Sphingobacteriales phylotype, a Bacteroidaceae phylotype and Sphingomonas; see Figure 3). It is possible to study larger-scale regional variation (for example, continental scale) for some wider ranging species such as leopard frogs, to see if dominant groups shift at different spatial scales; however, these patterns are completely unknown.
Skin-associated bacteria have the potential to influence susceptibility to infectious diseases that affect amphibians. Several studies have cultured bacteria from various amphibians and demonstrated that certain isolates can inhibit the growth of fungi, including Bd (Harris et al., 2006, 2009b; Lauer et al., 2007, 2008; Woodhams et al., 2007b). Harris et al. (2009a) used an anti-Bd isolate (Janthinobacterium lividum) in a bioaugmentation experiment and demonstrated success in reducing mortality rates due to Bd for susceptible amphibians in the laboratory. The same bacterial strain and other anti-Bd bacteria were found to be more abundant naturally on endangered mountain yellow-legged frog (Rana sierrae) populations that have co-existed with Bd relative to populations that have been extirpated due to Bd (Woodhams et al., 2007b; Lam et al., 2010). Just as some populations of amphibians differ with respect to their tolerance of Bd, different species of amphibians are known to vary in their ability to tolerate and resist infection with Bd (Garner et al., 2006; Kilpatrick et al., 2010). Our finding that amphibians have species-specific bacterial communities on their skin further suggests that these skin bacterial communities have the potential to influence species differences in susceptibility to Bd. Amphibians that harbor more antifungal bacterial species are likely to be better at tolerating or resisting infection with Bd. Broadly, how microbiotas influence the susceptibility to infectious disease is a next frontier with many potential applications relevant to human medicine (Stecher et al., 2010) and wildlife conservation (Harris et al., 2009b; Lam et al., 2010).
Acknowledgments
We thank Anna Peterson, Ana Lisette Arellano and August Jensen for assistance in the field. We thank Donna Berg-Lyons for assistance in preparing samples for pyrosequencing. The Colorado Division of Wildlife and the City of Boulder Open Space and Mountain Parks provided permission to access some of the wetlands to obtain amphibian samples. This work was supported in part by the National Institutes of Health and by the Howard Hughes Medical Institute.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
Supplementary Table S1
References
- Annis SL, Dastoor FP, Ziel H, Daszak P, Longcore JE. A DNA-based assay identifies Batrachochytrium dendrobatidis in amphibians. J Wildl Dis. 2004;40:420–428. doi: 10.7589/0090-3558-40.3.420. [DOI] [PubMed] [Google Scholar]
- Banning JL, Weddle AL, Wahl GW, Simon MA, Lauer A, Walters RL, et al. Antifungal skin bacteria, embryonic survival, and communal nesting in four-toed salamanders, Hemidactylium scutatum. Oecologia. 2008;156:423–429. doi: 10.1007/s00442-008-1002-5. [DOI] [PubMed] [Google Scholar]
- Belden LK, Harris RN. Infectious diseases in wildlife: the community ecology context. Front Ecol Environ. 2007;5:533–539. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C, Cohen N, Rollins-Smith L. Amphibian declines: an immunological perspective. Dev Comp Immunol. 1999;23:459–472. doi: 10.1016/s0145-305x(99)00028-2. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Warwick RM.2001Change in Marine Communities: an Approach to Statistical Analysis and Interpretation2nd edn.PRIMER-E: Plymouth [Google Scholar]
- Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culp CE, Falkinham JO, Belden LK. Identification of the natural bacterial microflora on the skin of eastern newts, bullfrog tadpoles and redback salamanders. Herpetologica. 2007;63:66–71. [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding LX, Yokota A. Proposals of Curvibacter gracilis gen. nov., sp nov and Herbaspirillum putei sp. nov. for bacterial strains isolated from well water and reclassification of Pseudomonas huttiensis, Pseudomonas lanceolata, Aquaspirillum delicatum and Aquaspirillum autotrophicum as Herbaspirillum huttiense comb. nov., Curvibacter lanceolatus comb. nov., Curvibacter delicatus comb. nov and Herbaspirillum autotrophicum comb. nov. Int J Syst Evol Microbiol. 2004;54:2223–2230. doi: 10.1099/ijs.0.02975-0. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA. 2008;105:17994–17999. doi: 10.1073/pnas.0807920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher MC, Garner TWJ, Walker SF. Global emergence of Batrachochytrium dendrobatidis and amphibian Chytridiomycosis in space, time, and host. Ann Rev Microbiol. 2009;63:291–310. doi: 10.1146/annurev.micro.091208.073435. [DOI] [PubMed] [Google Scholar]
- Frank DN, Amand ALS, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune S, Augustin R, Anton-Erxleben F, Wittlieb J, Gelhaus C, Klimovich VB, et al. In an early branching metazoan, bacterial colonization of the embryo is controlled by maternal antimicrobial peptides. Proc Natl Acad Sci USA. 2010;107:18067–18072. doi: 10.1073/pnas.1008573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner TWJ, Perkins MW, Govindarajulu P, Seglie D, Walker S, Cunningham AA, et al. The emerging amphibian pathogen Batrachochytrium dendrobatidis globally infects introduced populations of the North American bullfrog, Rana catesbeiana. Biol Lett. 2006;2:455–459. doi: 10.1098/rsbl.2006.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerson GA. Amphibians and Reptiles in Colorado: a Colorado Field Guide. University Press of Colorado: Niwot, CO; 1999. [Google Scholar]
- Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, et al. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 2009a;3:818–824. doi: 10.1038/ismej.2009.27. [DOI] [PubMed] [Google Scholar]
- Harris RN, James TY, Lauer A, Simon MA, Patel A. Amphibian pathogen Batrachochytrium dendrobatidis is inhibited by the cutaneous bacteria of Amphibian species. Ecohealth. 2006;3:53–56. [Google Scholar]
- Harris RN, Lauer A, Simon MA, Banning JL, Alford RA. Addition of antifungal skin bacteria to salamanders ameliorates the effects of chytridiomycosis. Dis Aquat Organ. 2009b;83:11–16. doi: 10.3354/dao02004. [DOI] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutalle-Schmelzer KML, Zwirnmann E, Kruger A, Grossart HP. Changes in pelagic bacteria communities due to leaf litter addition. Microb Ecol. 2010;60:462–475. doi: 10.1007/s00248-010-9639-0. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Briggs CJ, Daszak P. The ecology and impact of chytridiomycosis: an emerging disease of amphibians. Trends Ecol Evol. 2010;25:109–118. doi: 10.1016/j.tree.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol. 2010;12:118–123. doi: 10.1111/j.1462-2920.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Lam BA, Walke JB, Vredenburg VT, Harris RN. Proportion of individuals with anti-Batrachochytrium dendrobatidis skin bacteria is associated with population persistence in the frog Rana muscosa. Biol Conserv. 2010;143:529–531. [Google Scholar]
- Lauer A, Simon MA, Banning JL, Andre E, Duncan K, Harris RN. Common cutaneous bacteria from the eastern red-backed salamander can inhibit pathogenic fungi. Copeia. 2007;3:630–640. [Google Scholar]
- Lauer A, Simon MA, Banning JL, Lam BA, Harris RN. Diversity of cutaneous bacteria with antifungal activity isolated from female four-toed salamanders. ISME J. 2008;2:145–157. doi: 10.1038/ismej.2007.110. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Lips KR, Diffendorfer J, Mendelson JR, Sears MW. Riding the wave: reconciling the roles of disease and climate change in amphibian declines. PLoS Biol. 2008;6:441–454. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007;35:e120. doi: 10.1093/nar/gkm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73:1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Spitz SR, Goetz GW, McLellan SL. Temporal and spatial variability in nearshore bacterioplankton communities of Lake Michigan. FEMS Microbiol Ecol. 2009;67:511–522. doi: 10.1111/j.1574-6941.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Pladdies T, Babenzien HD, Cypionka H. Distribution of Nevskia ramosa and other rosette-forming neustonic bacteria. Microb Ecol. 2004;47:218–223. doi: 10.1007/s00248-003-1070-3. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, et al. Accurate determination of microbial diversity from 454 pyrosequencing data. Nat Methods. 2009;6:639–641. doi: 10.1038/nmeth.1361. [DOI] [PubMed] [Google Scholar]
- Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Env Microbiol. 2010;12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins-Smith LA. The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim Biophys Acta Biomembr. 2009;1788:1593–1599. doi: 10.1016/j.bbamem.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JBH. It's all relative: ranking the diversity of aquatic bacterial communities. Env Microbiol. 2008;10:2200–2210. doi: 10.1111/j.1462-2920.2008.01626.x. [DOI] [PubMed] [Google Scholar]
- Skerratt LF, Berger L, Speare R, Cashins S, McDonald KR, Phillott AD, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. Ecohealth. 2007;4:125–134. [Google Scholar]
- Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Lv L, Jing SL, Zhu LL, He GC. Bacterial symbionts of the Brown Planthopper, Nilaparvata lugens (Homoptera: Delphacidae) Appl Environ Microbiol. 2010;76:1740–1745. doi: 10.1128/AEM.02240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams DC, Rollins-Smith LA, Alford RA, Simon MA, Harris RN. Innate immune defenses of amphibian skin: antimicrobial peptides and more. Animal Conserv. 2007a;10:425–428. [Google Scholar]
- Woodhams DC, Vredenburg VT, Simon MA, Billheimer D, Shakhtour B, Shyr Y, et al. Symbiotic bacteria contribute to innate immune defenses of the threatened mountain yellow-legged frog, Rana muscosa. Biol Conserv. 2007b;138:390–398. [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One. 2010;5:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1