Generation of an Inbred Miniature Pig Model of Retinitis Pigmentosa (original) (raw)
Utilizing somatic cell nuclear transfer, the authors demonstrate the generation of a transgenic miniature pig model of retinitis pigmentosa. These pigs demonstrate a clinical phenotype similar to that observed in humans and may represent a novel tool for study of hereditary retinal degeneration.
Abstract
Purpose.
The Pro23His (P23H) rhodopsin (RHO) mutation underlies the most common form of human autosomal dominant retinitis pigmentosa (adRP). The objective of this investigation was to establish a transgenic miniature swine model of RP using the human P23H RHO gene.
Methods.
Somatic cell nuclear transfer (SCNT) was used to create transgenic miniature pigs that expressed the human P23H RHO mutation. From these experiments, six transgenic founders were identified whose retinal function was studied with full-field electroretinography (ffERG) from 3 months through 2 years. Progeny from one founder were generated and genotyped to determine transgene inheritance pattern. Retinal mRNA was isolated, and the ratio of P23H to wild-type pig RHO was measured.
Results.
A single transgene integration site was observed for five of the six founders. All founders had abnormal scotopic and photopic ffERGs after 3 months. The severity of the ffERG phenotype was grouped into moderately and severely affected groups. Offspring of one founder inherited the transgene as an autosomal dominant mutation. mRNA analyses demonstrated that approximately 80% of total RHO was mutant P23H.
Conclusions.
Expression of the human RHO P23H transgene in the retina creates a miniature swine model with an inheritance pattern and retinal function that mimics adRP. This large-animal model can serve as a novel tool for the study of the pathogenesis and therapeutic intervention in the most common form of adRP.
Retinitis pigmentosa (RP) is the most frequent cause of hereditary visual loss among adults. It is a group of inherited retinal diseases characterized by the onset of night blindness, the early loss of the peripheral visual field, and the late loss of central vision.1 Recently, important therapeutic advances have been made using gene therapy in a human form of the disease in children, Leber's congenital amaurosis.2 The great mutational heterogeneity in monogenic forms of RP, with over 200 mutations in 45 genes (www.sph.uth.tmc.edu/RetNet/home.htm, University of Texas Houston Health Science Center, Houston, TX), makes generalized gene therapy of limited utility in adult RP. In contrast, regenerative medicine may have a greater universal therapeutic potential and has elicited very promising results in a murine model of RP.3 Before embarking on human clinical trials using induced pluripotent stem cells (iPSCs) or other potential progenitor cells, it is imperative to demonstrate both efficacy and safety in a large-animal model of RP. The domestic pig is ideal for preclinical studies of regenerative medicine since its eye is similar to the human eye in size and structure, containing a rod photoreceptor dominant periphery and a cone photoreceptor dominant visual streak.4 Unfortunately, the large body size of the domestic pig limits its utility and restricts its widespread use in experimental studies.
All three modes of Mendelian inheritance occur in RP, although autosomal dominant RP is the most common accounting for 25% to 30% of all cases.5 Mutations in the rhodopsin gene alone (RHO; OMIM ID +180380; National Center for Biotechnology Information, Bethesda, MD) account for approximately 25% of the dominant forms of RP inheritance and a small percentage of recessively inherited cases. The most common form of autosomal dominant RP in humans is a proline–histidine substitution at the 23rd amino acid residue of rhodopsin (P23H), accounting for about one third of the dominant rhodopsin mutations or 8.5% of all RP cases.6 The objective of this project was to create a miniature pig model of RP that replicates the human phenotype. To this end, we used somatic cell nuclear transfer (SCNT) and produced inbred miniature transgenic pig lines that express the most common form of human autosomal dominant RP.
Materials and Methods
Cell Line Modification and Production
The miniature pigs were managed according to the guidelines of the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Ear snips were collected from 5-week-old male and female NIH Miniature pigs (SLAC/C haplotype), washed three times with PBS containing 10 ng/mL gentamicin, minced, and then digested with collagenase (200 U/mL) and DNaseI (25 Kunitz units/mL) in DMEM+15% FBS for 5 hours at 38.5°C. The digested cell solution was pelleted via centrifugation at 600_g_ for 5 minutes, washed in DMEM with 15% FBS, resuspended in DMEM with 15% FBS, and cultured until confluence at 38.5°C. Fibroblasts were then harvested via 0.05% trypsin-EDTA, aliquotted, frozen in FBS with 10% dimethyl sulfoxide (DMSO), and stored in liquid nitrogen. These fibroblast cells were used for SCNT, the reconstructed embryos were transferred to surrogates, and fetuses from the established pregnancies were collected on day 35 of gestation. Fetal fibroblast cells were collected using the above described methods after the preparation of the fetuses as previously described.7
DNA Constructs
An aliquot of the bacteriophage EMBL3 containing the human rhodopsin gene with the P23H mutation with 4.2 and 8.4 kb of sequence upstream and downstream from the coding sequence, respectively,8 were kindly provided by Thaddeus Dryja (Novartis, Cambridge, MA). The entire insert, including _Sal_I restriction enzyme sites, was subcloned into the Puc19 vector and amplified in DH10B electrocompetent Escherichia coli (Invitrogen, Carlsbad, CA). Human P23H RHO genomic (g)DNA was then isolated by digestion with _Sal_I, followed by size separation by agarose gel electrophoresis. The linearized human RHO fragment was purified and electroporated with one of two selectable marker constructs. The first selectable marker used was a linearized 1831-bp fragment of the neomycin resistance gene flanked by the PGK promoter and PGK polyA-tail. The second was a supercoiled vector expressing enhanced GFP, as previously described.9
Transfection and Selection
Exogenous DNA (12.5 μg/mL linearized human P23H RHO genomic construct) was delivered to the fibroblast cells using published electroporation conditions.10 Two strategies were used to create fetal fibroblast cell lines containing stably integrated human P23H RHO gene. In the first, passage-1 fetal fibroblasts were co-transfected via electroporation with the linearized human P23H rhodopsin fragment and a fragment containing the neomycin resistance gene flanked by the phosphoglycerate kinase promoter and a polyA-tail (10:1 molar ratio), respectively. Cells (final concentration of 1 × 106/mL) were transfected in 75% cytosalts (consisting in mM of 120 KCl, 0.15 CaCl2, 10 K2HPO4 [pH 7.6], and 5 MgCl2)11 and 25% growth factor formulation (Opti-MEM; Invitrogen). Two hundred microliters of cells was electroporated with three consecutive 250-V, 1-ms square wave pulses in a 2-mm gap cuvette in the presence of 12.5 μg/mL linearized human P23H RHO. Immediately after electroporation, the cells were resuspended in DMEM supplemented with 15% FBS and 2.5 ng/mL FGF. After 24 hours, cultured cells were selected for the presence of the neomycin resistance gene by the addition of geneticin (G418; 400 μg/mL) for 14 days. DNA from clonal colonies was used to verify the presence of the human P23H RHO gene by PCR. Cell lines were then used in SCNT and gave rise to four of the transgenic founders that were characterized: 51-1, 53-1, 62-1, and 64-1.
In the second strategy, we used exactly the same transfection techniques but fetal fibroblasts were co-transfected with linearized human P23H rhodopsin (12.5 μg/mL) and a supercoiled, enhanced green fluorescent protein vector9 (10:1 molar ratio). Twenty-four hours after electroporation, the cells were sorted by fluorescence activation (FACS), and single GFP-expressing cells were transferred into 96-well plates in DMEM with 15% FBS and cultured for 7 days. Colonies were then passaged into 24-well plates, and 2 days later, one third of the colony was genotyped for the presence of the human P23H RHO fragment; the rest were retained and frozen. Of 112 clonal colonies genotyped, 5 were verified positive for the rhodopsin gene via PCR. Two different cell lines were used to create one pregnancy that gave rise to two transgenic founders, 63-1 and 63-2.
Preparation of Donor Cells
Transgenic fetal fibroblast cells were thawed at room temperature, washed in culture medium (Gibco DMEM with 1000 mg/L glucose [Invitrogen], supplemented with 15% FBS [Hyclone, Logan, UT] and 2.5 ng/mL basic fibroblast growth factor [Sigma-Aldrich, St. Louis, MO]) and cultured for 1 to 2 days. The medium was removed, and the attached cells were washed with PBS followed by visually monitored dispersion of cells with 0.05% trypsin/EDTA on a heat plate at 37°C. Dispersed cells were resuspended in culture medium, centrifuged at 600_g_ for 5 minutes, and resuspended in micromanipulation medium (25 mM HEPES TCM 199, [Invitrogen] and 0.3% BSA).
Oocyte Maturation, SCNT, and Embryo Reconstruction
Oocyte-cumulus cell complexes (OCCs) were received in maturation medium from ART Inc. (Madison, WI) approximately 24 hours after harvest, primarily from ovaries of multiparous sows. Oocytes were transferred into fresh maturation medium in four-well dishes and allowed to mature for 42 to 44 hours in a humidified atmosphere of 5% CO2 at 38.5°C. After in vitro maturation, cumulus cells were removed from OCCs in 0.5 mg/mL hyaluronidase with gentle vortexing. After removal of cumulus cells, oocytes of good or excellent morphology with a polar body, indicative of metaphase II, were selected and placed in fresh manipulation medium containing 7.5 μg/mL cytochalasin B.
SCNT was conducted according to our published procedure.12 Briefly, the denuded oocyte was pierced with a microinjection pipette, and its polar body and adjacent cytoplasm containing the metaphase II chromosomes were removed. After enucleation, a donor fetal fibroblast cell was inserted into the oocyte and strategically placed beneath the zona pellucida, with care taken to maximize the amount of cell membrane contact between the donor fibroblast and the oocyte. The two cells of the nuclear transfer complex were fused by and activated using one of two methods. The first method included two direct-current pulses of 1.2 kV/cm for 30 μs, with an electroporator (Electro-cell Manipulator 2001; BTX Harvard Apparatus, Holliston, MA) in low-calcium fusion medium (0.3 M mannitol, 0.1 mM CaCl2 · 2H2O, 0.1 mM MgCl2 · 6H2O, and 0.5 mM HEPES). At least 30 minutes after fusion, reconstructed embryos were activated in the dark with 200 μM thimerosal for 10 minutes and rinsed and treated with 8 mM dithiothreitol (DTT) for 30 minutes.13 Trace amounts of DTT were removed from the reconstructed embryos by rinsing followed by washing three times in PZM3 solution with 0.3% BSA for 30 minutes. In the second method, we used two direct-current pulses of 1.2 kV/cm for 30 μs in high-calcium fusion medium (0.3 M mannitol, 1.0 mM CaCl2 · 2H2O, 0.1 mM MgCl2 · 6H2O, and 0.5 mM HEPES).14 After fusion and activation, the reconstructed embryos were cultured in PZM3 with 0.3% BSA for 15 to 21 hours, until surgically transferred into the surrogates.
Surrogates and Embryo Transfer
Landrace gilts exhibiting at least one natural estrous cycle were used on day 0 or day 1 of estrus for the transfer of reconstructed embryos. Briefly, anesthesia of the surrogates was induced using 1 g of pentothal sodium and then maintained via closed circuit inhalation of isoflurane (5%) mixed with oxygen (1.5 L/min). A skin incision of approximately 20 to 40 cm was made on the abdominal midline and extended through the muscle layer to the peritoneum. The peritoneum was carefully incised and one oviduct to the tip of the conjoined uterine horn was exteriorized. A minimum of 100 reconstructed embryos was transferred via a Tom cat catheter approximately midway between the uterotubal and the ampullary–isthmic junctions. The surrogates were monitored bi-weekly via ultrasound to monitor pregnancy. Piglets from surrogates that did not farrow naturally by day 115 of gestation were delivered via cesarean section.
Southern Blot Analysis
Fibroblast cells cultured from each of the founders and the original wild-type miniature pig were used for gDNA extraction (DNeasy Blood and Tissue Kit; Qiagen, Inc., Valencia, CA). Southern blot analysis of six founders was conducted by using a P32-labeled probe that was 635 bp, amplified by the PCR primers. The probe recognition site is between two _Nsi_I digestion sites that are specific to the human rhodopsin gene. gDNA from the original source of the fetal fibroblasts (18-1) was used as a negative control. Sample digestion, probe labeling, hybridization, and detection were conducted by Lofstrand Laboratories Limited (Gaithersburg, MD).
Fluorescent In Situ Hybridization
Lymphocytes were cultured with RPMI 1640 growth medium (HyClone), supplemented with 20% FBS (Atlanta Biologicals, Lawrenceville, GA) and 0.5% cell culture medium (Glutamax-1; Invitrogen). Chromosome preparations were made using standard cytogenetic protocols for thymidine synchronization. FISH probes were prepared by labeling the linearized P23H RHO gDNA transgene (Nick Translation Kit; Vysis, Abbott Park, IL) with a fluorophore (Spectrum Green; Vysis). Humid overnight incubation at 37°C followed co-denaturation at 77.5°C for 6 minutes. Observation was performed with a light microscope (Axioscope; Carl Zeiss Meditec, Dublin, CA) and documented on an imaging system (Metasystems, Altlussheim, Germany).
Genotyping Assays
Genotyping was conducted by using 10 ng of gDNA for each 25 μL PCR reaction. PCR was conducted with a commercial real-time thermal cycler (Mastercycler Pro; Eppendorf, Hauppauge, NY). Taq polymerase (either Biolase; Bioline, Taunton, MA, or GoTaq; Promega, Madison, WI) was used as directed by the manufacturers. Detection of the presence of the human transgene was conducted with PCR primers specific to human genomic rhodopsin synthesized to recognize and amplify a 635 bp amplicon between exons 2 and 3, as described in Figure 1A (forward: 5′-ACCTTGTATGTCTCCTGGCCCAAA-3′ and reverse: 5′-AGGACGTGGCAATGGTTCCTGATA-3′). PCR cycling conditions were as follows: 95°C for 3 minutes followed by 32 cycles at 95°C for 15 seconds, 64°C for 30 seconds, and 72°C for 45 seconds. Porcine-specific primers (forward: 5′-ACGTCCTTGTAGGCACAGCTAACA-3′ and reverse: 5′-GTCACTGTTTGTTTGGTGCCGTGA-3′) amplified a 425-bp amplicon to serve as a positive control reaction for pig DNA samples using the following PCR conditions: 95°C for 3 minutes followed by 32 cycles at 95°C for 15 seconds, 59°C for 30 seconds, and 72°C for 45 seconds.
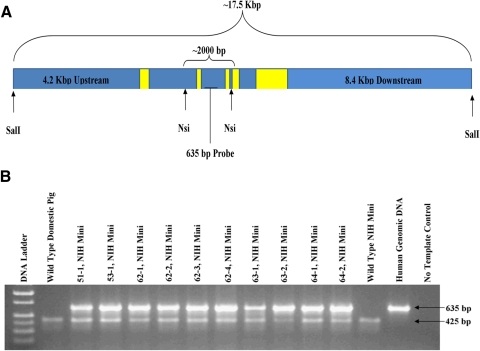
Figure 1.
Generation of P23H miniature pig RP model. (A) The 17.5-kb construct provided by Thaddeus Dryja.8 The construct was linearized with _Sal_I before electroporation into the cells. Yellow boxes: approximate location and size of the five RHO exons. _Nsi_I was used for Southern blot analysis detected by a 635-bp radiolabeled probe. (B) A total of 10 piglets were born alive, all PCR positive for the genomic human RHO construct (635-bp amplicon), while endogenous porcine gDNA (425-bp amplicon) was amplified in all pig samples.
P23H mRNA Expression in the Pig Retina
All offspring from a cross of one founder (53-1) to a domestic, wild-type sow were euthanized. The retina from one eye was removed, and total RNA was isolated (TRIzol; Invitrogen). After extraction and quantification, cDNA was reverse transcribed (RT) from approximately 1 mg total RNA from each offspring, using transcription reagent (Superscript II; Invitrogen) according to manufacturer recommendations for a gene-specific primer (GSP). One microliter of each RT reaction was used for PCR amplification (GoTaq polymerase; Promega). The GSP was used as the reverse primer for the RT reaction and represented a region of the RHO mRNA that is 100% homologous between human and pig (5′-TTCTCCCCGAAGCGGAAGTTGCTCAT-3′). The forward PCR primer (5′-GGCCCaAACTTCTACGTGCC-3′) had a single-nucleotide difference between human and pig (indicated by lowercase base). Together, these primers produce a 437-bp amplicon containing both swine and human rhodopsin cDNA (Supplementary Fig. S7, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8784/-/DCSupplemental) that is distinguishable based on the lack of sequence homology between the primer sites. After amplification, the 437-bp PCR product was visualized on an agarose gel, excised, purified, and used for cloning into the pGEMT easy vector (Promega). After transformation into JM109 chemically competent E. coli were plated on Luria broth (LB) agar and incubated overnight at 37°C. Individual colonies were selected and propagated in LB broth, and the plasmid DNA was purified and sequenced at the Iowa State University DNA Core facility, using the M13 reverse-sequencing primer. Sequence data (46 sequence reads representing the five transgenic piglets and 34 reads representing the four wild-type piglets) were used to perform BLASTn and BLAST alignments.15 Using this sequence information, we identified the proportion of the RHO mRNA expressed from the porcine genome and the P23H transgene to assess their relative representation in the retina at post natal day 1.
Clinical Examination
At each time point, a ffERG was obtained under inhalation anesthesia, and slit lamp biomicroscopy and indirect ophthalmoscopy were performed through a dilated pupil. The anterior and posterior segments of the eye were observed for cataract formation in the lens, as well as the absence of optic nerve pallor, attenuation of the retinal arterial vasculature, and intraretinal migration of pigment.
Full-field ERG
Animals were sedated (Telazol 10 mg/kg; Fort Dodge Animal Health, Fort Dodge, IA), and anesthesia was maintained on isoflurane (1.5%–2.5%). Heart rate and respiration were continuously monitored. Unilateral ffERGs were recorded using JET electrodes and a portable ERG unit (HMsERG model 1000, RetVet Corp., Columbia, MO), with a mini-Ganzfeld dome positioned approximately 1 cm from the eye. Each ERG session consisted of a series of scotopic and photopic flashes as recommended by the International Society of Electrophysiology and Vision (ISCEV) for ERGs in human subjects. After 20 minutes of dark adaptation, scotopic low-intensity rod responses were elicited at a stimulus intensity of 0.01 cd · s · m−2; averaged responses to 10 flashes, given at 2-second interval, were recorded. After 10 minutes of light adaptation with a background luminance of 30 cd · m−2, photopic single-flash responses were recorded using 3 cd · s · m−2 flash stimuli, averaging 32 flashes at an interval of 0.5 seconds, followed by evaluation of the 30-Hz flicker response at the same light intensity. The b-wave was used as the metric of photoreceptor function, and its amplitude was measured from baseline to peak (scotopic) or from trough of the a-wave to peak of the b-wave (photopic).
Histologic Analyses
After the last ffERG experiment, anesthetized pigs were euthanized and their eyes enucleated. Isolated eyes were hemisected and immersion fixed overnight in 1% paraformaldehyde, 2.5% glutaraldehyde, 3% sucrose, and 0.01% CaCl2, in 0.1 M phosphate buffer (pH 7.4). Tissues were dehydrated in graded methanol and acetone, embedded in epoxy resin, serial sectioned at 200 nm, and exposed to 4′,6-diamidino-2-phenylindole (DAPI). Images were captured digitally, logically inverted, and linearly stretched to enhance detail (Photoshop CS5; Adobe Systems, San Jose, CA).
Results
Somatic Cell Nuclear Transfer
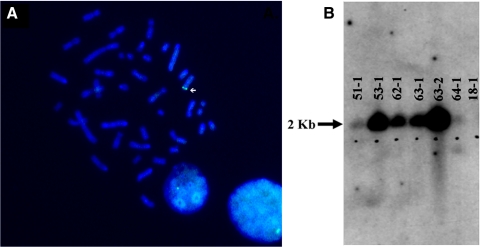
To generate a miniature swine model of RP, we stably transfected a 17.5-kb transgene (Fig. 1A) containing human P23H RHO into NIH miniature pig fetal fibroblasts with a defined swine leukocyte antigen haplotype (SLAC/C) and used them for SCNT. A total of 10 male piglets were produced, and all were positive for the human P23H RHO transgene (Fig. 1B). Six founder piglets (Fig. 2) survived and were examined using Southern blot analysis and fluorescence in situ hybridization (FISH) to assess transgene integration sites. Using FISH (Fig. 3A; Supplementary Figs. S1–S6, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8784/-/DCSupplemental), we detected a single chromosomal integration site in five of the six founders, but were unable to detect an integration site for founder 51-1 (Supplementary Fig. S1, http://www.iovs.org/lookup/suppl/doi:10.1167/iovs.11-8784/-/DCSupplemental). Southern blot analysis of the founders indicated variable intensities of the _Nsi_I fragment containing the transgene (Fig. 3B), suggesting that for some of the founders, concatamerization of the transgene before genome integration most likely occurred.
Figure 2.
The first of six founder piglets, 51-1, born May 25, 2008.
Figure 3.
(A) A representative FISH image (founder 53-1) indicating the chromosomal integration site of the human RHO transgene. All founder pigs demonstrated a single integration site, except 51-1 for which a signal was not detected. (B) Southern blot of six living founders. A total of 10 μg digested with _Nsi_I was loaded per lane. Pig 18-1 was the original source of the fetal fibroblasts used to create the transgenic founders. Signal for all transgenic pigs was detected, while 51-1 and 64-1 demonstrated the least signal. Pigs 53-1 and 63-2 most likely had a greater number of human RHO copies integrated into the genome.
P23H Founders Show Progressive Rod and Cone Photoreceptor Dysfunction and Degeneration
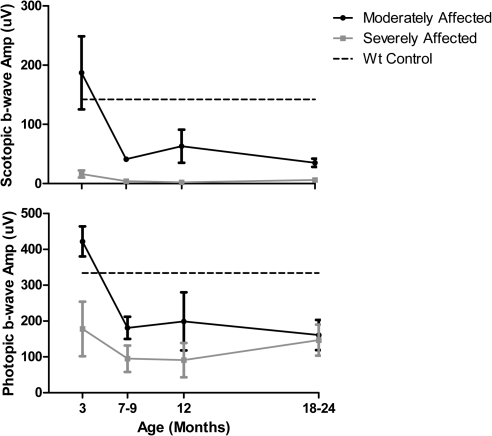
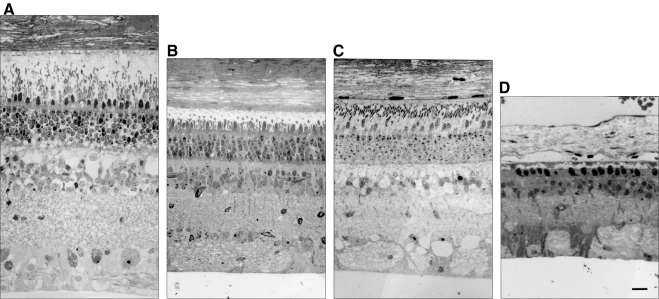
ffERGs were used to assess changes in retinal function resulting from expression of the P23H RHO transgene and to characterize the progression of the RP phenotype in all six founders. ffERGs were recorded at 3 months of age and at regular intervals thereafter for 2 years. WT littermates served as controls, and the amplitudes of their b-wave were stable from 3 months of age and upward. In Figures 4A and 4B, the average scotopic (rod) and photopic (cone) b-wave amplitudes are plotted for two groups of founders. In two founders, 63-1 and 51-1, both scotopic and photopic ffERGs were initially normal, and the onset of photoreceptor dysfunction was delayed until 7 to 9 months of age. In the other four founders (53-1, 62-1, 63-2, and 64-1) both scotopic and photopic ffERGs were significantly reduced as early as 3 months and declined further with age, especially in the scotopic ERG. The retinal morphology of three of the six founders (51-1 at 18 months, 63-1 at 22 months, and 63-2 at 12 months) was evaluated after the last ERG assessment. Figure 5 compares the morphology of an adult domestic swine with the morphology of each of these three founders. The founder 63-2 in the ERG severely affected group showed advanced retinal degeneration (Fig. 5D). A single layer of cone nuclei remains in the outer nuclear layer, and there is a significant reduction in the number of nuclei in the inner nuclear layer. In contrast, the two founders in the moderately affected group (Figs. 5B, 5C) show much less photoreceptor degeneration, although the outer segments are significantly shorter and the ONL thickness is reduced compared to the normal adult (Fig. 5A). Thus, there is a general correlation of changes in retinal structure and function in this model.
Figure 4.
Time course of decrease in retinal function differs in P23H transgenic minipig founders. (A) Scotopic (rod) and (B) photopic (cone) full-field ERGs were recorded during the first 2 years of life under standard ISCEV protocols. The scotopic ERGs were evoked with a flash intensity of 0.01 cd · s · m−2, after 20 minutes of dark-adaptation. The photopic ERGs were measured using flashes of 3 cd · s · m−2 presented on a background of 30 cd · m−2. The founders fell into two groups, based on both scotopic and photopic ffERGs, compared with WT littermates at 3 months of age (n = 3; dashed lines). Average b-wave amplitudes are plotted for the two transgenic founders who were moderately affected (63-1 and 51-1) and the four who were severely affected (53-1, 62-1, 63-2, and 64-1). Moderately affected founders had a later onset of rod and cone dysfunction and retained some degree of both rod and cone response through 12 months of age.
Figure 5.
Retinal histology of three of six P23H founders at death. (A) Retinal morphology of a normal adult domestic sow. (B) 51-1 (18 months) and (C) 63-1 (22 months). The outer segments of the photoreceptors were shortened in the moderately affected ffERG group in the two founders, and the ONL thickness was reduced compared with that in the normal adult. (D) 63-2 (12 months). Only a single layer of cone nuclei remained in the ONL and both the ONL and INL were greatly reduced in thickness in the founder with a severely affected ffERG. Thus, there is a correlation between retinal structure and function in the P23H model. Scale bar, 50 μm.
The variation in both structural and functional phenotype severity is probably the result of different transgene integration sites. In RP there are usually changes in the fundus with disease progression, such as retinal arterial vascular attenuation and intraretinal pigment migration (i.e., bone spicule–like pigmentation). These changes were not observed in any of the transgenic founders through 2 years of age, including posterior subcapsular cataract or optic nerve pallor. Nevertheless, the ffERG phenotype and anatomic degeneration of the neurosensory retina of the founder minipigs is consistent with the presentation of photoreceptor dysfunction in autosomal dominant RP in humans, including variation in phenotype severity.1
Mendelian Inheritance of the Transgene
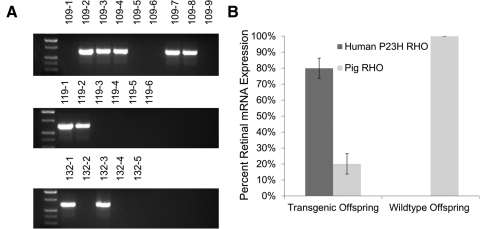
Germline transmission of the transgene was demonstrated in one of the most severely affected founders, 53-1. He was mated using artificial insemination to both a domestic sow and two wild-type NIH miniature sows (SLAC/C haplotype), which resulted in nine (litter 109; sow), six (litter 118; wild-type), and five (litter 132; wild-type) offspring. Of these 20, 9 were positive for the transgene (Fig. 6A), demonstrating the expected ratio of 50% for Mendelian inheritance of the transgenic loci. The matings produced both male and female offspring, indicating that the transgene is located on one of the autosomes of 53-1.
Figure 6.
Characterization of F1 progeny produced by 53-1. (A) Three litters were produced from founder 53-1 after mating to a domestic pig (litter 109) and two miniature pigs (litters 118 and 132). PCR screening of the 20 offspring detected 9 transgenic piglets, suggesting normal Mendelian inheritance. Controls included genomic DNA from a wild-type miniature pig (WT-1), a wild-type domestic pig (WT-2), human genomic DNA, and a no-template control (NTC). Transgenic positive amplicon is 635 bp. (B) Contribution of both human P23H and pig RHO to the total RHO mRNA expression in the neonatal retina of transgenic (109-2, -3, -4, -7, and -8) and wild-type (109-1, -5, -6, and -9) offspring. Retinal RNA extraction, followed by PCR amplification, cloning, and sequencing, identified the mRNA expression ratio of endogenous porcine RHO mRNA versus the transgenic human P23H RHO mRNA.
Level of Expression of the Transgene
Nine offspring from a cross between 53-1 and a domestic sow were euthanized, mRNA isolated from one retina, and the contribution of human P23H RHO mRNA to total RHO mRNA expression estimated from transformations of E. coli that harbored the P23H versus the porcine DNA sequence. From a total of 46 colonies screened in five transgenic piglets, 80% ± 6% had the human P23H transgene sequence. By comparison, of 34 colonies in four wild-type piglets, all had only porcine RHO (Fig. 6B).
Discussion
We created a model of autosomal dominant RP using the most common human mutation, P23H rhodopsin, to best emulate the human disease and facilitate translational research, particularly strategies that target posttranscriptional regulation of the mutated RHO mRNA. We used the NIH mini-pig (SLAC/C haplotype), which is highly inbred, for several important reasons. Transplantation studies of progenitor or retinal stem cells using syngeneic donors avoid the homograft reaction since histocompatibility disparities are avoided. In addition, an inbred host should also result in a more consistent clinical phenotype in heterozygous F1 offspring. We chose the minipig because it weighs approximately 150 pounds at maturity, one-half to one-third the weight of age-matched domestic pigs. This makes the model of more general use in laboratory work and especially in long-term projects. Although in a domestic transgenic pig with a rhodopsin mutation, the proline–leucine substitution at the 347th amino acid residue (P347L) has already been made and is very well characterized, it does not share any of these advantages.15
With the exception of fundus changes, our P23H miniswine model mimics the human disease in many important ways. Our porcine model of RP has a clinical phenotype similar to that observed in human autosomal dominant RP—namely, the early, dramatic reduction in the scotopic b-wave amplitude in the full-field ERG, reflecting rod photoreceptor dysfunction, and a subsequent, albeit delayed, reduction in the photopic b-wave amplitude, reflecting cone photoreceptor dysfunction.16 There is also extensive degeneration of the ONL, with only a single row of cone photoreceptors and reduced thickness of both the ONL and INL at 12 months in a founder with a severely reduced full-field ERG. Interestingly, the six different founder lines have variable time of disease onset and progression, features that may increase their future utility for studying RP.
Germline transmission of the P23H rhodopsin mutation in this inbred NIH mini-pig provides the scientific community with a large-animal model of the most common form of autosomal dominant human RP. This line (53-1) is available from the National Swine Resource and Research Center (http://nsrrc.missouri.edu; University of Missouri, Columbia). Several important questions should be addressed with this novel model, including the electrophysiologic function of piglets shortly after birth, the anatomic changes in the neural retina over time, and the integrity of the retinal circuitry, since the P347L model possess a PSD95 abnormality at birth, a finding not obvious in humans.17
Although there are several models of RP in mice and rats, the small eye of the rodent, with very limited access to the subretinal space, hinders surgical therapeutic intervention. In addition, the limited number of cells that can be injected into the subretinal space or vitreous cavity because of the small volume of the inoculum, as well as the difficulty in injecting the inoculum into a localized space beneath the retina, make evaluation of a treatment effect problematic in these species. In contrast, the pig eye is large, is similar in size to the human eye, and is a more appropriate biologic model, since approximately 14% of the photoreceptors in the pig are cones, compared to only 1% in mice, and most of these cones in the pig are reported to be located in the visual streak.4 Although several canine and feline models of inherited RP have been identified and in some cases colonies of affected animals have been established,18–20 there are several major limitations in these species, such as limited genetic information, the presence of major and minor histocompatibility differences using cell-based therapies, the societal concern over experimentation in dogs and cats, and the limited access to retinal progenitor and stem cells in these species.
The eye of the minipig will allow the study of various medical and surgical therapeutic approaches, as well as the development and use of novel intraocular surgical techniques applicable to humans. The technique of SCNT in the minipig to produce ocular models of human disease has exciting potential application in translational research besides regenerative medicine, including the ability to restore vision with slow-release intravitreal devices using therapeutic drugs, the retinal prosthesis, and gene therapy.
Supplementary Material
Supplementary Data
Acknowledgments
The authors are thankful to Kevin Wells, Clifton N. Murphy, Lee Spate, David Wax, Rong Feng Li, Yanhong Hao, Jeffrey Whyte, Kyle Dobbs, Kristen Whitworth, S. Clay Isom, Michael Linville, Kristina Narfstrom, Man Bok Jeong, Scott Korte, Bethany Bauer, Elane C. Wright, and Robin Scanlon for their excellent assistance in completion of this project.
Footnotes
Supported by National Institutes of Health (NIH) Grants U42-RR18877 (RSP), EY018608 (MAM), EY-020647 (HJK), and HL076138-08 (JPF); Discovery Eye Foundation; Research to Prevent Blindness, New York, NY; Department of Ophthalmology & Visual Sciences, UofL, Moran Eye Center, UofU and Career Development Award (BWJ); NIFA (National Institute of Food and Agriculture) Grant 2008-35205-18712 (JWR); Kentucky Research Challenge Trust Fund (HJK); Kentucky Science and Engineering Foundation (HJK); EY015128 (RM), NIH EY02576 (RM), EYO14800 Vision Core, an unrestricted grant from Research to Prevent Blindness to the Moran Eye Center; Edward N. and Della L. Thome Memorial Foundation grant for Age-Related Macular Degeneration Research (BWJ), a Research to Prevent Blindness Career Development Award (BWJ), Moran Eye Center Tiger Team Translational Medicine Award (BWJ); and a University of Louisville Clinical and Translational Science Grant (HJK).
Disclosure: J.W. Ross, None; J.P. Fernandez de Castro, None; J. Zhao, None; M. Samuel, None; E. Walters, None; C. Rios, None; P. Bray-Ward, None; B.W. Jones, None; R.E. Marc, None; W. Wang, None; L. Zhou, None; J.M. Noel, None; M.A. McCall, None; P.J. DeMarco, None; R.S. Prather, None; H.J. Kaplan, None
References
- 1.Inglehearn CF. Molecular genetics of human retinal dystrophies. Eye (Lond). 1998;12:571–579 [DOI] [PubMed] [Google Scholar]
- 2.Simonelli F, Maguire AM, Testa F, et al. Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol Ther. 18:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacLaren RE, Pearson RA, MacNeil A, et al. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444:203–207 [DOI] [PubMed] [Google Scholar]
- 4.Hendrickson A, Hicks D. Distribution and density of medium- and short-wavelength selective cones in the domestic pig retina. Exp Eye Res. 2002;74:435–444 [DOI] [PubMed] [Google Scholar]
- 5.Inglehearn CF, Tarttelin EE, Plant C, et al. A linkage survey of 20 dominant retinitis pigmentosa families: frequencies of the nine known loci and evidence for further heterogeneity. J Med Genet. 1998;35:1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartong DT, McGee TL, Sandberg MA, et al. Search for a correlation between telomere length and severity of retinitis pigmentosa due to the dominant rhodopsin Pro23His mutation. Mol Vis. 2009;15:592–597 [PMC free article] [PubMed] [Google Scholar]
- 7.Lai L, Prather RS. Production of cloned pigs by using somatic cells as donors. Cloning Stem Cells. 2003;5:233–241 [DOI] [PubMed] [Google Scholar]
- 8.Olsson JE, Gordon JW, Pawlyk BS, et al. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830 [DOI] [PubMed] [Google Scholar]
- 9.Whitworth KM, Li R, Spate LD, et al. Method of oocyte activation affects cloning efficiency in pigs. Mol Reprod Dev. 2009;76:490–500 [DOI] [PubMed] [Google Scholar]
- 10.Ross JW, Whyte JJ, Zhao J, Samuel M, Wells KD, Prather RS. Optimization of square-wave electroporation for transfection of porcine fetal fibroblasts. Transgenic Res. 2010;19:611–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hoff MJ, Moorman AF, Lamers WH. Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res. 1992;20:2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, Ross JW, Hao Y, et al. Significant improvement in cloning efficiency of an inbred miniature pig by histone deacetylase inhibitor treatment after somatic cell nuclear transfer. Biol Reprod. 2009;81:525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Machaty Z, Wang WH, Day BN, Prather RS. Complete activation of porcine oocytes induced by the sulfhydryl reagent, thimerosal. Biol Reprod. 1997;57:1123–1127 [DOI] [PubMed] [Google Scholar]
- 14.Im GS, Lai L, Liu Z, et al. In vitro development of preimplantation porcine nuclear transfer embryos cultured in different media and gas atmospheres. Theriogenology. 2004;61:1125–1135 [DOI] [PubMed] [Google Scholar]
- 15.Petters RM, Alexander CA, Wells KD, et al. Genetically engineered large-animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nature Biotechnol. 1997;15:965–970 [DOI] [PubMed] [Google Scholar]
- 16.Berson EL. Retinitis pigmentosa. The Friedenwald Lecture. Invest Ophthalmol Vis Sci. 1993;34:1659–1676 [PubMed] [Google Scholar]
- 17.Blackmon SM, Peng YW, Hao Y, et al. Early loss of synaptic protein PSD-95 from rod terminals of rhodopsin P347L transgenic porcine retina. Brain Res. 2000;885:53–61 [DOI] [PubMed] [Google Scholar]
- 18.Aguirre G. Retinal degenerations in the dog. I. Rod dysplasia. Exp Eye Res. 1978;26:233–253 [DOI] [PubMed] [Google Scholar]
- 19.Suber ML, Pittler SJ, Qin N, et al. Irish setter dogs affected with rod/cone dysplasia contain a nonsense mutation in the rod cGMP phosphodiesterase beta-subunit gene. Proc Natl Acad Sci U S A. 1993;90:3968–3972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.May CA, Lütjen-Drecoll E, Narfstrom K. Morphological changes in the anterior segment of the Abyssinian cat eye with hereditary rod-cone degeneration. Curr Eye Res. 2005;30:855–862 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data