Roles of Hydrophobicity and Charge Distribution of Cationic Antimicrobial Peptides in Peptide-Membrane Interactions (original) (raw)
Background: Cationic antimicrobial peptides offer an alternative to conventional antibiotics, as they physically disrupt bacterial membranes, causing cell death.
Results: Peptides designed with high hydrophobicity display strong self-association that is minimized by distribution of positive charges at both peptide termini.
Conclusion: Balancing peptide hydrophobicity and charge distribution promotes efficient antimicrobial activity.
Significance: Routes to optimization of peptide sequences are valuable for devising therapeutic strategies.
Keywords: Antimicrobial Peptides, Atomic Force Microscopy, Circular Dichroism (CD), Fourier Transform Infrared (FTIR), Gel Electrophoresis, Peptide Interactions, Antimicrobial Activity, Bacterial Membranes, Peptide-Membrane Interactions
Abstract
Cationic antimicrobial peptides (CAPs) occur as important innate immunity agents in many organisms, including humans, and offer a viable alternative to conventional antibiotics, as they physically disrupt the bacterial membranes, leading to membrane lysis and eventually cell death. In this work, we studied the biophysical and microbiological characteristics of designed CAPs varying in hydrophobicity levels and charge distributions by a variety of biophysical and biochemical approaches, including in-tandem atomic force microscopy, attenuated total reflection-FTIR, CD spectroscopy, and SDS-PAGE. Peptide structural properties were correlated with their membrane-disruptive abilities and antimicrobial activities. In bacterial lipid model membranes, a time-dependent increase in aggregated β-strand-type structure in CAPs with relatively high hydrophobicity (such as KKKKKKALFALWLAFLA-NH2) was essentially absent in CAPs with lower hydrophobicity (such as KKKKKKAAFAAWAAFAA-NH2). Redistribution of positive charges by placing three Lys residues at both termini while maintaining identical sequences minimized self-aggregation above the dimer level. Peptides containing four Leu residues were destructive to mammalian model membranes, whereas those with corresponding Ala residues were not. This finding was mirrored in hemolysis studies in human erythrocytes, where Ala-only peptides displayed virtually no hemolysis up to 320 μm, but the four-Leu peptides induced 40–80% hemolysis at the same concentration range. All peptides studied displayed strong antimicrobial activity against Pseudomonas aeruginosa (minimum inhibitory concentrations of 4–32 μm). The overall findings suggest optimum routes to balancing peptide hydrophobicity and charge distribution that allow efficient penetration and disruption of the bacterial membranes without damage to mammalian (host) membranes.
Introduction
Antimicrobial peptides occur naturally as important innate immunity agents in a wide range of living organisms ranging from plants to insects to mammals, including humans (1); as such, they have become increasingly recognized in current research as templates for prospective antibiotic agents. Also termed cationic antimicrobial peptides (CAPs),2 they are characterized by their positive net charge, modest length (12–50 residues), and good solubility in water (2–4). CAPs offer a viable alternative to conventional antibiotics, as they bind to and penetrate bacterial cell membranes, physically disrupting them, leading to membrane lysis and eventually cell death (5). The advantages of these CAPs are that (i) they are highly selective against the negatively charged bacterial membrane versus the zwitterionic mammalian membranes of a human host and that (ii) there is no specificity in targeting, as the peptides function by physically disrupting the bacterial membrane and therefore are unlikely to evoke bacterial resistance (6).
Our laboratory previously designed a novel category of synthetic hydrophobic membrane-active CAPs that display antimicrobial activity against multiple bacteria and yeast strains without toxicity to mammalian cells (7, 8), of which a lead candidate is 6K-F17 (sequence KKKKKKAAFAAWAAFAA-NH2). In designing this peptide, Ala was chosen as the background residue due to its midrange hydrophobicity and frequent occurrence in membrane domains. As well, the A_xxx_A (“small-_xxx_-small”) sequence motif promotes peptide dimerization in membranes (9, 10), which has been suggested to enhance antimicrobial activity (11). The Trp residue in the hydrophobic core serves as the fluorescent probe, whereas the positively charged Lys residues were included at the terminus to maintain cationic properties and enhance peptide solubility. On the basis of the highly active CAP 6K-F17, we went on to design a series of CAPs with a range of hydrophobicity values and observed a “threshold hydrophobicity” for selective bacterial membrane insertion (11–13). However, once the core segment hydrophobicity of the CAPs is beyond an upper threshold, as in the case in which the sequence contains two or more Ala-to-Leu substitutions, the CAPs generally have reduced antimicrobial activity and display increased toxicity to mammalian membranes (14). In agreement with these findings, other workers confirmed the importance of peptide hydrophobicity in membrane selectivity and insertion and for antimicrobial activity (15, 16). In addition to hydrophobicity, the net positive charge of a given CAP also plays an important role in peptide-membrane interactions, particularly in attracting the CAP efficiently to the anionic surface of bacterial membranes (17–19). However, the interplay of CAPs with varying hydrophobicity levels versus varying distributions of positive residues along the CAP sequence as a determinant of bioactivity remains to be clarified.
In this work, we report the biological activity (and the biophysical characteristics of peptide-membrane interactions) of selected designed CAPs varying systematically in hydrophobicity levels and charge distributions to answer the following questions. 1) Why does increasing hydrophobicity lead to poorer antimicrobial activity and greater hemolytic toxicity? 2) Will altered charge distribution improve the activity of CAPs with the same hydrophobicity level? 3) Which factor, hydrophobicity or charge distribution, is ultimately the more important contributor to effective CAP design and bioactivity?
EXPERIMENTAL PROCEDURES
Reagents
The reagents for peptide synthesis, cleavage, and purification included (Fmoc-aminomethyl-3,5-dimethoxyphenoxy)valeric acid-polyethylene glycol-polystyrene resin and piperidine (Applied Biosystems); Fmoc-protected amino acids and 2-(7-aza-1_H_-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (GL Biochem (Shanghai) Ltd.); N,_N_-diisopropylethylamine and triisopropylsilane (Aldrich); UltrapureTM buffer-saturated phenol (Invitrogen); TFA (DURAN Group); and peptide-grade N,_N_-dimethylformamide, methanol, diethyl ether, and acetonitrile (Caledon Laboratories Ltd.). Reagent kits for micro-BCA protein assays were purchased from Pierce. Cation-adjusted Mueller-Hinton broth was from Sigma. Lipids, 1-palmitoyl-2-oleoyl-_sn_-glycero-3-phosphoethanolamine (POPE), and 1,2-dioleoyl-_sn_-glycero-3-phospho-(1′-_rac_-glycerol) (DOPG) were from Avanti Polar Lipids. Buffers were prepared in double-distilled water with pH adjustment when necessary.
Peptide Synthesis and Purification
Peptides were synthesized by the continuous flow Fmoc solid-phase method on a Protein Technologies PS3 peptide synthesizer using the standard cycle (20). (Fmoc-aminomethyl-3,5-dimethoxyphenoxy)valeric acid-polyethylene glycol-polystyrene resin was used to produce an amidated C terminus. 2-(7-Aza-1_H_-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate and N,_N_-diisopropylethylamine were used as the activation pair. Deprotection and cleavage of peptides were performed in a mixture of 88% TFA, 5% phenol, 5% water, and 2% triisopropylsilane for 2 h in the dark at room temperature. The crude peptides were purified on a reverse-phase C4 preparative HPLC column using a linear gradient of acetonitrile in 0.1% TFA. The purities of the peptides were confirmed by their molecular weights using MALDI-MS. The concentrations of the peptides were determined using the micro-BCA protein assay.
Circular Dichroism
CD spectra were recorded on a Jasco J-810 spectropolarimeter using a 1-mm path length quartz cell at 25 °C. Peptide (20 μm each) in 20 mm Tris buffer and 10 mm NaCl with and without 10 mm SDS at pH 7.0 was measured in an average of three scans with the buffer background subtracted.
SDS-PAGE
50-ng portions of each peptide were prepared in Novex sample buffer and incubated at room temperature for >30 min prior to being loaded onto NuPAGE Novex 12% BisTris precast gels (1.0-mm thickness, 10 wells) in MES buffer according to the manufacturer's protocols. Silver staining was performed using the SilverXpress staining kit (Invitrogen) to visualize peptides on gels. Apparent molecular weights were estimated from the migration of Novex sharp unstained protein standard (Invitrogen). The gel was analyzed using the NIH ImageJ program, and MWexp/MWtheor values were calculated from the ratios of the experimental to theoretical molecular weights of the CAPs.
Liposome Preparation
To mimic bacterial and mammalian membranes, desired ratios of lipid solutions in chloroform were mixed and dried by rotary evaporation for a minimum of 1 h. HEPES buffer (10 mm HEPES, 150 mm NaCl (pH 7.4), and 1 m CaCl2) was added to rehydrate the lipid mixture to a final concentration of 1 μm, followed by sonication at a temperature higher than the melting temperatures of all of the lipids for 40 min prior to atomic force microscopy (AFM) measurements. For the FTIR experiments, the lipids were rehydrated in CaCl2-free deuterated HEPES buffer to a final concentration of 2 μm and used immediately.
AFM
AFM images were acquired in fluid tapping mode using a Digital Instruments MultiMode scanning probe microscope comprising a Nanoscope IIIA controller equipped with either a J scanner (maximum scan area of 116 × 16 μm) or an E scanner (maximum lateral scan area of 14.6 × 14.6 μm). All images were acquired using an SNL-10 short thin tip (Veeco Probes, Camarillo, CA) fitted to a combined tapping mode/contact mode glass fluid cell fitted with inlet and outlet ports. Images were collected at a resolution of 512 × 512 pixels using a scan angle of 0º and a scan rate of 1 Hz. The tip drive frequency was generally set between 7 and 10 kHz with a drive amplitude set point of 0.2–0.6 V. The flow-through glass fluid cell was sealed against freshly cleaved mica with a silicone O-ring to create a 200-μl sample compartment. To facilitate liposome fusion and bilayer formation, the freshly cleaved mica sealed in the fluid cell was incubated for ∼10 min with HEPES buffer prior to the introduction of ∼500 μl of hydrated liposomes heated to ∼70 °C. After ∼30 min of incubation at room temperature to allow for bilayer formation, the fluid cell was flushed with liposome-free buffer. Reference AFM images of the bilayers were then acquired prior to the addition of an ∼300-μl aliquot of the CAP of interest at 8 μm in HEPES buffer to the fluid cell. All AFM images were acquired at room temperature. Image analysis was performed using the Nanoscope software (version 5.30r1, Digital Instruments) on images that had been subjected to zero-order flatten and second-order plane fit filters. Relative height differences were determined using a horizontal section line along the slow scan axis.
Attenuated Total Reflection (ATR)-FTIR
The FTIR spectra were collected using a Nexus 670 FTIR spectrometer equipped with an EverGlo mid-IR source, a liquid N2-cooled MCT/A detector, a KBr beam splitter, a SmartOrbit single-bounce ATR accessory fitted with a diamond internal reflectance element, and a custom flow-through sealed fluid cell at a resolution of 2 cm−1 using an average of 128 scans over a scan range of 4000–700 cm−1, referenced against the spectra of a clean diamond internal reflectance element. 500 μl of freshly sonicated liposomes in 10 mm deuterated HEPES and 150 mm NaCl (pH 7.4) were injected into the custom fluid flow cell (as described above) covering the diamond internal reflectance element and flushed with buffer after the lipids fused onto the surface, as confirmed with IR scans. Each tested peptide (8 μm) in HEPES buffer was added to the fused lipids, and scans were taken over time. All spectra were analyzed using Omnic software (version 5.2a, Nicolet Instrument Inc.) with lipid spectra subtracted and H2O- and base line-corrected. Fourier self-deconvolution was performed on the amide peak from 1700 to 1600 cm−1 using a full-width at half-height of 13 cm−1 (typically 12–20 cm−1 (21)) and an enhancement factor (K) of 2.4 (typically 2–3 (22)).
Antibacterial Activity
The antimicrobial activity of each peptide was tested in sterile 96-well plates by standard microtiter dilution protocols in cation-adjusted Mueller-Hinton broth (23). Pseudomonas aeruginosa strain PAO1 (a kind gift from Dr. Lynne Howell, Hospital for Sick Children) was grown in cation-adjusted Mueller-Hinton broth at 37 °C overnight and diluted to a final concentration of 5 × 104 to 1 × 105 colony-forming units/ml. 11 μl of peptides at 2-fold serial dilutions were added to 100 μl of diluted bacterial suspension. Plates were incubated at 37 °C overnight for 20 h, and the minimum inhibitory concentration (MIC) was taken as the concentration at which the bacterial growth was fully inhibited, as detected at _A_600 using a Genesys 5 microplate autoreader spectrophotometer.
Hemolytic Activity
The hemolytic toxicity levels of the designed CAPs were measured in human RBCs as described previously (14). Freshly collected venous human blood with heparin was centrifuged at 1000 × g for 5 min at 11 °C to remove the buffy coat, and the RBCs obtained were washed three times with PBS. Peptides were diluted in PBS to 100 μl and mixed with 100 μl of 4% (v/v) RBC suspension to final concentrations ranging from 320 to 5 μm in 96-well polystyrene microtiter plates (Nunc). PBS and 0.1% Triton X-100 were used as negative and positive lysis controls, respectively. The plates were incubated at 37 °C for 1 h, and unlysed RBCs were removed by centrifugation at 1000 × g on a Beckman Model TJ-6 microplate centrifuge for 5 min. 100 μl of the supernatant aliquots were transferred to new microplates, and the release of hemoglobin was monitored at _A_540 on a Genesys 5 microplate autoreader spectrophotometer. The percent hemolysis at each peptide concentration was measured in duplicate according to the following equation: hemolysis = ((_A_exp − _A_PBS)/(_A_100% − _A_PBS)) × 100%.
Computational Modeling of CAPs
Energy-minimized models of the interaction between two identical α-helices of CAP sequence were generated using the global conformation search program CHI as described (24). The resulting CAP homodimer models with energetically favorable packing interfaces were analyzed and visualized using PyMOL (DeLano Scientific).
RESULTS
Helical Induction of CAPs in SDS Micelles
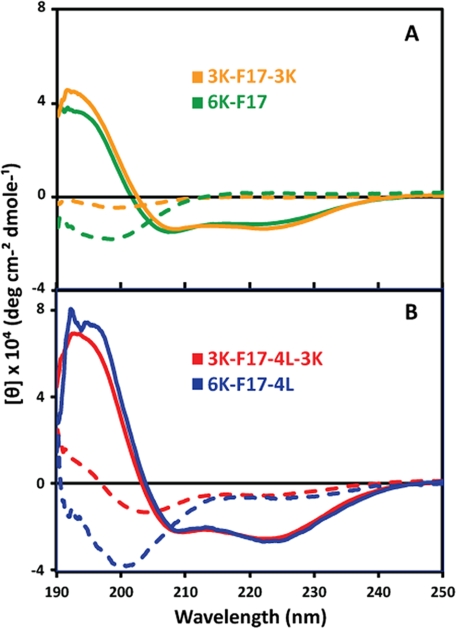
The TM-Finder program (25) predicts that the hydrophobic core segments of the peptides should adopt an α-helical secondary structure in membrane-mimetic environments. Both the 6K peptides and their derived 3K-3K partners (Table 1) displayed “random coil” structures in aqueous buffer (Fig. 1, dashed lines). In the membrane-mimetic environment of SDS micelles, the CAPs all underwent an α-helical conformational transition, signified by the minima of their CD spectra at 208 and 222 nm (Fig. 1, solid lines). There was no significant difference between the induced helicity of the 6K peptides versus their 3K-3K analogs (Fig. 1, A and B).
TABLE 1.
Sequences, molecular weights, and hydrophobicity values of designed CAPs
| Peptide | Amino acid sequence | Molecular weight | Hydrophobicitya |
|---|---|---|---|
| 6K-F17 | KKKKKKAAFAAWAAFAA-NH2 | 1836 | 1.48 |
| 6K-F17-4L | KKKKKKALFALWLAFLA-NH2 | 2005 | 3.14 |
| 3K-F17-3K | KKKAAFAAWAAFAAKKK-NH2 | 1836 | 1.48 |
| 3K-F17-4L-3K | KKKALFALWLAFLAKKK-NH2 | 2005 | 3.14 |
FIGURE 1.
CD spectra of CAPs in aqueous solution and SDS micelles. The spectra were measured at 20 μm each CAP in 10 mm Tris-HCl and 10 mm NaCl (pH 7.0) in the presence (solid lines) and absence (dashed lines) of 10 mm SDS. The peptide sequences are given in Table 1. The curves are based on triplicate measurements with the buffer background subtracted. deg, degrees.
Designed CAPs Are Dimeric on SDS-PAGE
The sequences of the designed peptides contain at least one pair of “small” residues separated by three residues (A_xxx_A motifs) that are known to promote helix-helix dimerization in transmembrane segments (9, 10). The F17 sequence contains three A_xxx_A motifs (two AAFAA sequences and a central AAWAA sequence); the F17-4L sequence retains the central motif (ALWLA). Consistent with our previous findings (11, 14), the MWexp/MWtheor values for both 6K-F17 and 6K-F17-4L are 2.4 (Fig. 2), indicating that these two CAPs form SDS-resistant dimers on SDS-polyacrylamide gels, even at the low loading levels used in silver stain analysis. Peptides 3K-F17-3K and 3K-F17-4L-3K migrate with MWexp/MWtheor ratios are 2.0 and 2.1, respectively, also indicative of dimer formation, but the slower movement of the 6K versus 3K-3K peptides may signal some qualitative difference in the strength of dimer interfaces (see “Discussion”).
FIGURE 2.
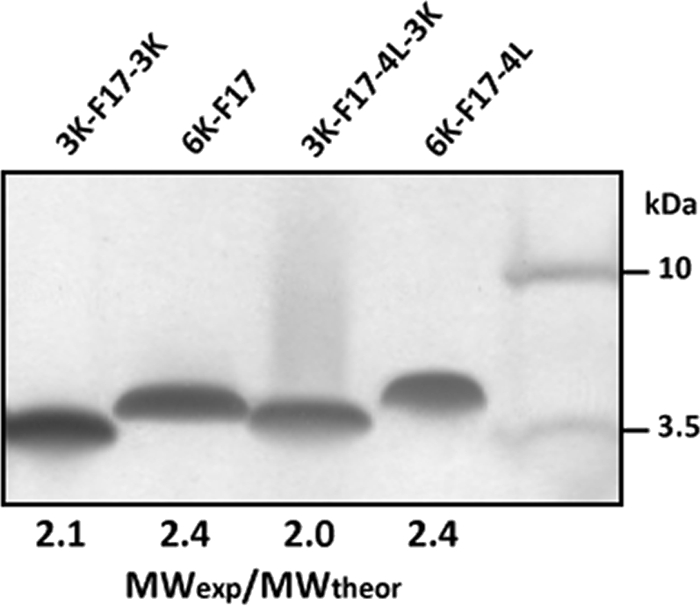
SDS-PAGE (silver stain) analysis of CAPs. Peptides (50 ng) were diluted in 1× SDS-containing sample buffer and incubated for 30 min at room temperature prior to electrophoresis using NuPAGE 12% BisTris gel at 200 V. MWexp/MWtheor is the ratio of experimentally determined to actual molecular weight of each CAP, where a value of ∼2 corresponds to the dimer. Molecular mass standards are given to the right of the gel. The molecular weights of CAPs are given in Table 1.
CAP Interactions with Model Bacterial Membranes
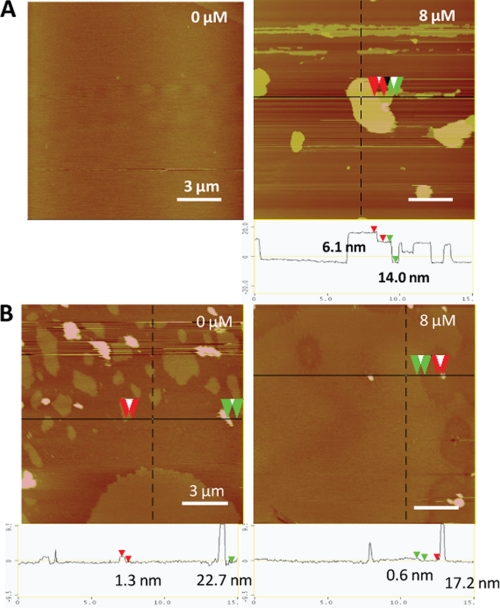
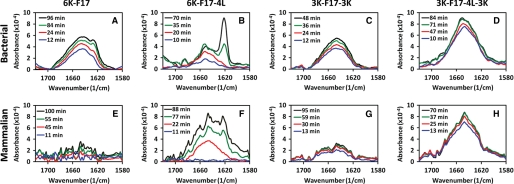
Because the core segment hydrophobicity of CAPs is a key factor that affects bacterial membrane insertion directly and antimicrobial activity indirectly (11, 14), it becomes crucial to understand how peptide hydrophobicity influences their membrane-disruptive ability, particularly where host membranes are at stake. To address this, we compared the mechanistic details of the interactions of two CAPs with contrasting core hydrophobicities (6K-F17 with a “low” core segment hydrophobicity (1.48) and 6K-F17-4L with a “high” core segment hydrophobicity (3.14)) (Table 1) in a bacterial membrane lipid bilayer model using a combination of “simultaneous” AFM and ATR-FTIR techniques. In a typical run, CAPs (8 μm) were added to a lipid bilayer mixture composed of 3:1 POPE/DOPG, which resembles the phosphoethanolamine/phosphoglycerol ratio found in the inner membrane of Gram-positive and Gram-negative bacteria (26, 27). The in situ AFM images revealed that direct fusion of the lipid vesicles onto mica resulted in a molecularly smooth intact surface with no or few defects (Fig. 3A, left panel). The addition of the CAPs to the membrane bilayers led to an immediate remodeling or restructuring of the surface (28). Over time, CAP-induced membrane defects (darker region) were observed in the entire bilayer (Fig. 3A, right panel). Notably, we did not observe detachment of the membrane from the supporting mica; rather, the membrane itself appeared structurally roughened, indicating the destabilization and disruption of the membrane bilayer. In tandem to the AFM measurements that revealed the structural nature of the peptide-bilayer interaction, FTIR spectra were monitored continuously to pinpoint any accompanying structural changes in the CAPs that may underlie their membrane-disruptive properties (29, 30). As shown in Fig. 4A, 6K-F17 adopted a largely α-helical structure (indicated by the amide I band at ∼1650 cm−1) over the corresponding time course, and the spectral intensity increased over time as additional amounts of CAPs penetrated the lipids. In contrast, 6K-F17-4L initially induced an α-helical conformation in the bacterial membrane lipids and eventually exhibited a time-dependent increase in aggregated β-strand species (indicated by the amide I band at ∼1625 cm−1) with a concomitant relatively decreased population of α-helical species (70-min curve in Fig. 4B). Membrane disruption by 6K-F17-4L under comparable conditions was also seen in the topographical defects resolved by in situ AFM (supplemental Fig. S1_A_).
FIGURE 3.
AFM topography images of synthetic CAP 6K-F17 in model bilayers. Images were acquired in the fluid of 6K-F17 at 0 μM (left panel) and 8 μM (right panel) peptide in a bacterial model lipid bilayer (3:1 POPE/DOPG) (A) and in a mammalian model lipid bilayer (1:1:1 DOPC/DSPC/cholesterol) (B). Scale bars = 3 μm. Color indicates the local height of the lipid sample, ranging from 0 nm (lightest brown) to 20 nm (darkest brown). The corresponding section analysis for each CAP is shown below the images. The height differences are indicated between the red or green pairs of arrowheads. The spikes in these mammalian bilayer images (e.g. ∼22.7 nm in B, left panel; and ∼17.2 nm in B, right panel) represent unfused liposomes.
FIGURE 4.
Time-dependent ATR-FTIR spectra of synthetic CAPs in model membrane bilayers. Spectra of four CAPs (6K-F17, 6K-F17-4L, 3K-F17-3K, and 3K-F17-4L-3K) are shown in a bacterial-type (anionic) membrane lipid bilayer (3:1 POPE/DOPG) (A–D) and in a mammalian-type (zwitterionic) membrane lipid bilayer (1:1:1 DOPC/DSPC/cholesterol) (E–H). The CAP concentration was 8 μm in each spectrum. FTIR spectra for the amide I region of the CAPs reveal relative content(s) of α-helical conformation (∼1650 cm−1) (31), unordered structure (∼1640 cm−1) (22), and β-sheet-type structure (∼1625 cm−1) (30). The spectral intensity increased over time as additional amounts of CAPs penetrated the lipids, and no further changes were detected beyond the final time point in each diagram (black lines). The 6K-F17 peptide tested in E is the d-isomer; the l-enantiomer gives comparable results (data not shown).
Positive Charges on Both CAP Termini Minimize Aggregation in Bacterial Membranes
Peptide 6K-F17-4L discussed above likely self-oligomerizes and aggregates in an antiparallel manner because the grouping of the six Lys positive charges at the N terminus mitigates against parallel association. Therefore, placement of three positive charges on each of the N and C termini while an identical peptide core sequence is maintained might be expected to produce a mixture of parallel and antiparallel dimers, but likely with reduced higher level aggregation in membranes due to charge repulsion. To test this hypothesis, we designed the CAPs 3K-F17-3K and 3K-F17-4L-3K as charge distribution analogs of the parent CAPs 6K-F17 and 6K-F17-4L, respectively (Table 1). In contrast to 6K-F17-4L, the interactions of 3K-F17-3K and 3K-F17-4L-3K with the bacterial membrane mimic were less well defined, appearing to result in a reduction in the lateral stability of the membrane itself rather than outright restructuring. This was seen in the emergence of sunken regions and the ability of the AFM tip to induce localized distortions (supplemental Fig. S1, B and C). Remarkably, association of these two CAPs with the bacterial membrane did not result in an aggregated β-strand-type structure, as indicated by the predominant α-helical band (∼1650 cm−1) (31) and the absence of the β-strand band (∼1625 cm−1) (30) in the amide I region of their FTIR spectra in bacterial membranes (Fig. 4, C and D).
Antimicrobial Activity
P. aeruginosa is a commonly studied opportunistic pathogen that may cause acute pneumonia in emphysema patients, bacteremia in burn and cancer victims, and lung disease and mortality in cystic fibrosis patients (32). MICs of the novel CAPs were measured against P. aeruginosa strain PAO1 (Table 2), and it was determined that all four CAPs displayed significant antimicrobial activity. 3K-F17-4L-3K showed an improved antimicrobial efficacy (8 μm) versus 6K-F17-4L (16 μm), whereas 3K-F17-3K (32 μm) displayed reduced activity versus 6K-F17 (4 μm).
TABLE 2.
Antimicrobial activities of designed CAPs
| Peptide | MICa |
|---|---|
| μ_m_ | |
| 3K-F17-3K | 32 |
| 6K-F17 | 4 |
| 3K-F17-4L-3K | 8 |
| 6K-F17-4L | 16 |
CAP Interactions with Model Mammalian Membranes
Although interactions with bacterial membranes are of primary interest, it is essential to establish whether a given CAP interacts with a zwitterionic (neutral) mammalian-type membrane, particularly because peptide-mediated hemolysis of host cells cannot be tolerated for a useful therapeutic. To examine the effects of the present CAPs on mammalian membranes, we performed in-tandem AFM and ATR-FTIR experiments on the four designed peptides with a mammalian membrane model (1:1:1 1,2-dioleoyl-_sn_-glycero-3-phosphocholine (DOPC)/1,2-distearoyl-_sn_-glycero-3-phosphocholine (DSPC)/cholesterol) that directly mimics the outer leaflet of eukaryotic cell membranes (27, 33). Before peptide addition, fusion of this lipid mixture to mica resulted in DSPC/cholesterol-rich lipid-ordered domains surrounded by a DOPC-rich liquid-disordered phase, distinguishable by an ∼2-nm height difference (Fig. 3B, left panel). We ascribe the ∼20-nm taller features seen as blobs in the control image to unfused liposomes bound with the bilayer surface. 6K-F17 simply does not interact with mammalian membrane bilayers at 8 μm, as noted by the absence of membrane defects (Fig. 3B, right panel) and the absence of a detectable FTIR signal (Fig. 4E), indicating no membrane penetration. In contrast, 6K-F17-4L is apparently of sufficient core hydrophobicity to use this property (rather than electrostatic binding) as a route to partition into, and produce defects in, mammalian membranes (supplemental Fig. S2_A_), where it gives rise to a mixture of helical and β-strand bands in FTIR spectra (Fig. 4F). 3K-F17-3K and 3K-F17-4L-3K showed comparable effects in mammalian membranes: 3K-F17-3K produced no significant membrane destabilization (supplemental Fig. S2_B_) and very minimum structures in the zwitterionic mammalian membrane model (Fig. 4G), whereas 3K-F17-4L-3K adopted significant helical structure detected by FTIR (Fig. 4H) and induced visible small “holes” in the bilayer (supplemental Fig. S2_C_).
Hemolytic Activity
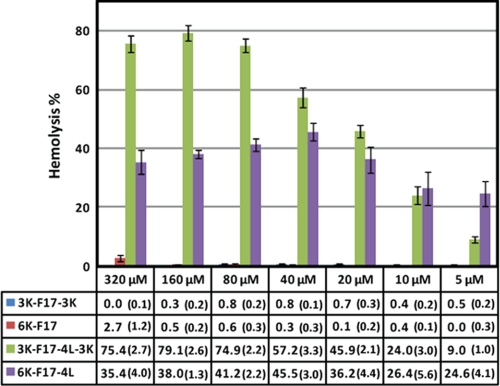
The hemolytic activities of the designed CAPs were measured against human RBCs at concentrations ranging from 320 to 10 μm (Fig. 5). Peptides with low hydrophobicity, namely 3K-F17-3K and 6K-F17, displayed no hemolysis even at high concentrations (320 and 160 μm, respectively). In striking contrast, peptides with high hydrophobicity (3K-F17-4L-3K and 6K-F17-4L) showed hemolytic activities at every concentration tested. In parallel to the mammalian membrane-disruptive effects reported above, these results support the notion that CAPs with greater hydrophobicity are prone to inducing hemolysis against human erythrocytes (14). The 6K-F17 peptide did display a detectable level of hemolysis at the highest concentration tested (2.7 ± 1.2% at 320 μm); the all-d chiral version of this peptide displayed no hemolysis at 325 μm (14).
FIGURE 5.
Hemolytic activities of the designed CAPs. The percent hemolysis (±error) was tested at peptide concentrations ranging from 5 to 320 μm with 4% (v/v) RBCs. Values calculated from at least triplicate measurements are listed below the bar graph. See “Experimental Procedures” for details of these experiments.
DISCUSSION
Our laboratory has developed a novel category of synthetic hydrophobic membrane-active peptides that display high antimicrobial activity (7, 8, 12, 14). Relating their biology to their biochemical mechanism(s) of action can aid in optimization of the properties of sequence that underlie their potency. In this study, we examined four such bioactive peptides, selected to highlight the extremes of two such properties: core sequence hydrophobicity (four Ala residues substituted with four Leu residues) and positive charge distribution (six Lys residues on the N terminus versus three Lys residues on each of the N and C termini). The peptides were subjected to microbiological assays and a panel of biophysical experiments, including CD, SDS-PAGE, AFM, and FTIR. Among key results, we found that CAPs can inflict serious membrane damage on model bacterial membranes, yet CAPs with low hydrophobicity (6K-F17 and 3K-F17-3K) are inert to mammalian membranes. It was also observed that a CAP with relatively higher hydrophobicity (i.e. 6K-F17-4L) undergoes a structural transition in contact with bacterial-type membranes from α-helical to β-strand-type structure, whereas the corresponding CAP with lower hydrophobicity (i.e. 6K-F17) largely retains its α-helical conformation upon entering the membrane. This phenomenon is likely attributable to a charge neutralization effect as the peptides bind to the surface of the anionic bacterial membranes, and the resulting dehydrated environment facilitates the formation of β-strand-type aggregates of the CAP with a sequence of higher hydrophobicity (34, 35). This result indicates that (overly) high core segmental hydrophobicity can lead to an increased potential to peptide self-association at the membrane surface (and possibly to precipitation), thus limiting the concentration of peptide actually impacting on the bacterial membrane and consequently reducing antimicrobial activity; this latter notion is supported by the relatively lower MIC for the 6K-F17 peptide (4 μm) versus its Leu-containing counterpart (16 μm).
Because 6K-F17-4L can self-aggregate at the bacterial membrane surface and undergo oligomerization via propagation of antiparallel aggregation (and hence staggered 6K regions), we sought to inquire whether separated charge distribution would prevent such self-association without altering the helix-forming and dimerization ability of the peptide in the membrane environment, which may be the crucial properties for bioactivity in the 6K-F17 series of novel CAPs (11, 14). We found that charge distribution itself does not dominate the impact on activity because it improves the antimicrobial activity of only the CAP with high hydrophobicity (3K-F17-4L-3K) but reduces the activity of the CAP with low hydrophobicity (3K-F17-3K) (Table 2). As described above, this situation can be explained by the fact that CAPs still need to self-dimerize to function upon binding to the bacterial membrane (see below), yet separated charges are seen to mitigate against the parallel or antiparallel aggregation of the 3K-3K peptides, ostensibly via repulsions of the positive changes at the peptide termini.
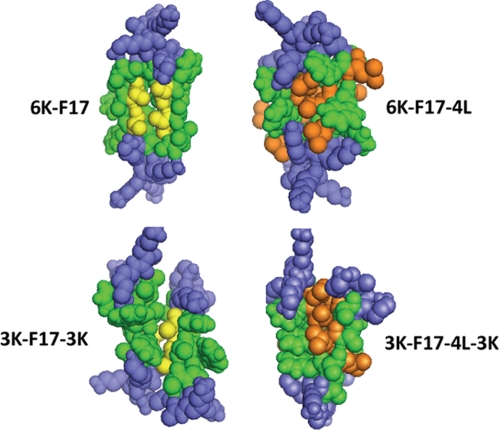
In addition to their ability to induce defects in bacterial membrane mimics, the four peptides examined also display a facile propensity to insert into micelles of the anionic detergent SDS and adopt helical conformations (Fig. 1). As well, all four peptides migrate as dimers on SDS-polyacrylamide gels (Fig. 2). However, when gel migration behavior is examined in detail, several further observations that relate CAP structure to function emerge. First, it was noted that each four-Leu-containing peptide migrated perceptibly slower than its four-Ala counterpart, which likely reflects the fact that the SDS-polyacrylamide gel is detecting the molecular weight difference between the four-Leu versus four-Ala peptides (_M_r 2005 versus 1836). However, hydrophobic peptides and proteins are known to run anomalously on SDS-PAGE due to varying levels of detergent binding as a function of sequence hydrophobicity (36); thus, regardless of the four-Leu positions versus their four-Ala counterparts, the 6K-F17-4L peptide should be migrating significantly more slowly than the 6K-F17 peptide, yet its position is virtually identical. Given that all four peptides are dimeric (Fig. 2), this latter finding may be explicable by the fact that the Leu residues in 6K-F17-4L largely line the peptide-peptide dimer interface, thus exposing mainly Ala rather than Leu residues to detergent, as would be the case for the 6K-F17 peptide as well. The same analysis holds for 3K-F17-3K versus 3K-F17-4L-3K, which also display comparable migration positions. These findings allow us to choose representative CAP dimer models from CHI clusters, as shown in Fig. 6. In CAPs where Ala is not replaced by Leu (e.g. 6K-F17 and 3K-F17-3K), the “A_xxx_A” motif is expected to be involved primarily in the helix-helix packing pocket of the CAP dimer pairs (11). In the four-Leu CAPs, Leu residues become participants in the van der Waals interface that stabilizes the peptide dimer. This effect may be further driven by the hydrophobic effect when the peptides are first solubilized in aqueous media. Dimers of 6K-F17 and 3K-F17-3K show discernibly more “space” between molecules, further suggesting that the Leu residues can effectively “fill” the van der Waals packing interface.
FIGURE 6.
Space-filling models of synthetic CAP dimers. Shown are representative structural models of antiparallel dimers of the four CAPs studied in this work. Structures were generated using the global conformation search program CHI. Peptides are as labeled in the diagram. Lys residues are rendered in blue; core hydrophobic residues (for both peptide molecules in the dimer) are shown in green. The packing interfaces of 6K-F17 and 3K-F17-3K are highlighted by the two (underlined) Ala residues (from each peptide molecule) in the motif “_A_AWA_A_” (rendered in gold), whereas 3K-F17-4L-3K and 6K-F17-4L have several Leu residues (from each peptide molecule) embedded in the hydrophobic binding pocket (rendered in orange).
Biophysical characterization further revealed that only CAPs 6K-F17-4L and 3K-F17-4L-3K destabilized mammalian membrane bilayers, which suggests that CAPs with a sufficiently high level of hydrophobicity will likely partition from the bulk water phase for insertion into zwitterionic bilayers via hydrophobic solvation, supporting our previous findings (14). It is interesting to note that the hemolysis percentages induced in RBCs by the four-Leu peptides (Fig. 5) level off near 40% for the 6K-F17-4L peptide and near 80% for the 3K-F17-4L-3K peptide. This indicates that, in each case, the limiting aqueous solubility of the peptide has been reached and that any further addition of peptide results in precipitation such that no further activity will ensue.
CONCLUSION
The data suggest that, in a bacterial membrane model, peptides with higher core hydrophobicity have stronger self-association and aggregation tendencies than those with lower hydrophobicity. On the other hand, when positive charge is distributed equally at both termini, FTIR spectra suggest that peptide aggregation above dimers is eliminated. Our results further indicate that the separated charge distribution of CAPs, which otherwise retain identical hydrophobic cores with A_xxx_A motif(s), does not disrupt their dimerization ability in hydrophobic environments and, accordingly, that primary sequence motifs remain the chief determinant of oligomeric status. Nevertheless, once dimers are established, the inherent charge repulsion extant at both termini versus the six-Lys N terminus mitigates against further oligomerization (Fig. 4, C and D). Our findings also reveal that CAPs with increased hydrophobicity can readily enter zwitterionic membranes and cause hemolytic effects. Separate distribution of positive charges reduces the hemolysis of peptides with the same core hydrophobicity at low concentrations but does not have a favorable effect at higher concentrations.
Given that the maximum antimicrobial activity and minimum hemolytic activity of CAPs do not involve only one factor but rather require a good balance among (i) peptide helicity, (ii) optimum hydrophobicity of the core segment, (iii) positive charges and their distribution, (iv) dimerization and/or oligomerization ability in the membrane, and (v) minimization of aggregation, our overall findings clarify preferable routes to the optimization of sequences most likely to be of value as therapeutic strategies.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Gillian Vanderlee, Michael Wong, and Lauren Lin for assistance with the AFM and FTIR experiments.
*
This work was supported in part by Canadian Institutes of Health Research (CIHR) Grant FRN-79284 (to C. M. D.) and Natural Sciences and Engineering Research Council of Canada (NSERC) Grant RGPIN 194435 (to C. M. Y.).
2
The abbreviations used are:
CAP
cationic antimicrobial peptide
Fmoc
_N_-(9-fluorenyl)methoxycarbonyl
POPE
1-palmitoyl-2-oleoyl-_sn_-glycero-3-phosphoethanolamine
DOPG
1,2-dioleoyl-_sn_-glycero-3-phospho-(1′-_rac_-glycerol)
BisTris
2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
AFM
atomic force microscopy
ATR
attenuated total reflection
MIC
minimum inhibitory concentration
DOPC
1,2-dioleoyl-_sn_-glycero-3-phosphocholine
DSPC
1,2-distearoyl-_sn_-glycero-3-phosphocholine.
REFERENCES
- 1.Zasloff M. (2002) Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 [DOI] [PubMed] [Google Scholar]
- 2.Mookherjee N., Hancock R. E. (2007) Cationic host defense peptides: innate immune regulatory peptides as a novel approach for treating infections. Cell. Mol. Life Sci. 64, 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock R. E. (1997) Peptide antibiotics. Lancet 349, 418–422 [DOI] [PubMed] [Google Scholar]
- 4.Oren Z., Shai Y. (1998) Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 47, 451–463 [DOI] [PubMed] [Google Scholar]
- 5.La Rocca P., Shai Y., Sansom M. S. (1999) Peptide-bilayer interactions: simulations of dermaseptin B, an antimicrobial peptide. Biophys. Chem. 76, 145–159 [DOI] [PubMed] [Google Scholar]
- 6.Hancock R. E. (2001) Cationic peptides: effectors in innate immunity and novel antimicrobials. Lancet Infect. Dis. 1, 156–164 [DOI] [PubMed] [Google Scholar]
- 7.Stark M., Liu L. P., Deber C. M. (2002) Cationic hydrophobic peptides with antimicrobial activity. Antimicrob. Agents Chemother. 46, 3585–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burrows L. L., Stark M., Chan C., Glukhov E., Sinnadurai S., Deber C. M. (2006) Activity of novel non-amphipathic cationic antimicrobial peptides against Candida species. J. Antimicrob. Chemother. 57, 899–907 [DOI] [PubMed] [Google Scholar]
- 9.Lear J. D., Stouffer A. L., Gratkowski H., Nanda V., Degrado W. F. (2004) Association of a model transmembrane peptide containing Gly in a heptad sequence motif. Biophys. J. 87, 3421–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schneider D., Engelman D. M. (2004) Motifs of two small residues can assist but are not sufficient to mediate transmembrane helix interactions. J. Mol. Biol. 343, 799–804 [DOI] [PubMed] [Google Scholar]
- 11.Glukhov E., Stark M., Burrows L. L., Deber C. M. (2005) Basis for selectivity of cationic antimicrobial peptides for bacterial versus mammalian membranes. J. Biol. Chem. 280, 33960–33967 [DOI] [PubMed] [Google Scholar]
- 12.Liu L. P., Li S. C., Goto N. K., Deber C. M. (1996) Threshold hydrophobicity dictates helical conformations of peptides in membrane environments. Biopolymers 39, 465–470 [DOI] [PubMed] [Google Scholar]
- 13.Liu L. P., Deber C. M. (1998) Guidelines for membrane protein engineering derived from de novo designed model peptides. Biopolymers 47, 41–62 [DOI] [PubMed] [Google Scholar]
- 14.Glukhov E., Burrows L. L., Deber C. M. (2008) Membrane interactions of designed cationic antimicrobial peptides: the two thresholds. Biopolymers 89, 360–371 [DOI] [PubMed] [Google Scholar]
- 15.Chen Y., Guarnieri M. T., Vasil A. I., Vasil M. L., Mant C. T., Hodges R. S. (2007) Role of peptide hydrophobicity in the mechanism of action of α-helical antimicrobial peptides. Antimicrob. Agents Chemother. 51, 1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wieprecht T., Dathe M., Beyermann M., Krause E., Maloy W. L., MacDonald D. L., Bienert M. (1997) Peptide hydrophobicity controls the activity and selectivity of magainin 2 amide in interaction with membranes. Biochemistry 36, 6124–6132 [DOI] [PubMed] [Google Scholar]
- 17.De Kroon A. I., Soekarjo M. W., De Gier J., De Kruijff B. (1990) The role of charge and hydrophobicity in peptide-lipid interaction: a comparative study based on tryptophan fluorescence measurements combined with the use of aqueous and hydrophobic quenchers. Biochemistry 29, 8229–8240 [DOI] [PubMed] [Google Scholar]
- 18.Dathe M., Nikolenko H., Meyer J., Beyermann M., Bienert M. (2001) Optimization of the antimicrobial activity of magainin peptides by modification of charge. FEBS Lett. 501, 146–150 [DOI] [PubMed] [Google Scholar]
- 19.Leptihn S., Har J. Y., Wohland T., Ding J. L. (2010) Correlation of charge, hydrophobicity, and structure with antimicrobial activity of S1 and MIRIAM peptides. Biochemistry 49, 9161–9170 [DOI] [PubMed] [Google Scholar]
- 20.Liu L. P., Deber C. M. (1997) Anionic phospholipids modulate peptide insertion into membranes. Biochemistry 36, 5476–5482 [DOI] [PubMed] [Google Scholar]
- 21.Goormaghtigh E., Raussens V., Ruysschaert J. M. (1999) Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta 1422, 105–185 [DOI] [PubMed] [Google Scholar]
- 22.Tamm L. K., Tatulian S. A. (1997) Infrared spectroscopy of proteins and peptides in lipid bilayers. Q. Rev. Biophys. 30, 365–429 [DOI] [PubMed] [Google Scholar]
- 23.Wu M., Hancock R. E. (1999) Interaction of the cyclic antimicrobial cationic peptide bactenecin with the outer and cytoplasmic membrane. J. Biol. Chem. 274, 29–35 [DOI] [PubMed] [Google Scholar]
- 24.Adams P. D., Engelman D. M., Brünger A. T. (1996) Improved prediction for the structure of the dimeric transmembrane domain of glycophorin A obtained through global searching. Proteins 26, 257–261 [DOI] [PubMed] [Google Scholar]
- 25.Deber C. M., Wang C., Liu L. P., Prior A. S., Agrawal S., Muskat B. L., Cuticchia A. J. (2001) TM Finder: a prediction program for transmembrane protein segments using a combination of hydrophobicity and nonpolar phase helicity scales. Protein Sci. 10, 212–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Does C., Swaving J., van Klompenburg W., Driessen A. J. (2000) Non-bilayer lipids stimulate the activity of the reconstituted bacterial protein translocase. J. Biol. Chem. 275, 2472–2478 [DOI] [PubMed] [Google Scholar]
- 27.Oreopoulos J., Yip C. M. (2009) Combinatorial microscopy for the study of protein-membrane interactions in supported lipid bilayers: order parameter measurements by combined polarized TIRFM/AFM. J. Struct. Biol. 168, 21–36 [DOI] [PubMed] [Google Scholar]
- 28.Yin L. M., Lee S., Edwards M. A., Yip C. M., Deber C. M. (2011) Proceedings of the 22nd American Peptide Symposium, San Diego, CA, June 25–30, 2011, pp. 252–253, American Peptide Society, San Diego, CA [Google Scholar]
- 29.Van Mau N., Vié V., Chaloin L., Lesniewska E., Heitz F., Le Grimellec C. (1999) Lipid-induced organization of a primary amphipathic peptide: a coupled AFM-monolayer study. J. Membr. Biol. 167, 241–249 [DOI] [PubMed] [Google Scholar]
- 30.Verity J. E., Chhabra N., Sinnathamby K., Yip C. M. (2009) Tracking molecular interactions in membranes by simultaneous ATR-FTIR-AFM. Biophys. J. 97, 1225–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vié V., Van Mau N., Chaloin L., Lesniewska E., Le Grimellec C., Heitz F. (2000) Detection of peptide-lipid interactions in mixed monolayers, using isotherms, atomic force microscopy, and Fourier transform infrared analyses. Biophys. J. 78, 846–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govan J. R., Deretic V. (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60, 539–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Florin-Christensen J., Suarez C. E., Florin-Christensen M., Wainszelbaum M., Brown W. C., McElwain T. F., Palmer G. H. (2001) A unique phospholipid organization in bovine erythrocyte membranes. Proc. Natl. Acad. Sci. U.S.A. 98, 7736–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee S., Chowdhury P., Gai F. (2007) Infrared study of the effect of hydration on the amide I band and aggregation properties of helical peptides. J. Phys. Chem. B 111, 4596–4602 [DOI] [PubMed] [Google Scholar]
- 35.Dzwolak W., Ravindra R., Nicolini C., Jansen R., Winter R. (2004) The diastereomeric assembly of polylysine is the low volume pathway for preferential formation of β-sheet aggregates. J. Am. Chem. Soc. 126, 3762–3768 [DOI] [PubMed] [Google Scholar]
- 36.Rath A., Glibowicka M., Nadeau V. G., Chen G., Deber C. M. (2009) Detergent binding explains anomalous SDS-PAGE migration of membrane proteins. Proc. Natl. Acad. Sci. U.S.A. 106, 1760–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data