miR-758 regulates cholesterol efflux through post-transcriptional repression of ABCA1 (original) (raw)
. Author manuscript; available in PMC: 2012 Nov 1.
Published in final edited form as: Arterioscler Thromb Vasc Biol. 2011 Nov;31(11):2707–2714. doi: 10.1161/ATVBAHA.111.232066
Abstract
Objective
The ATP-binding cassette transporter A1 (ABCA1) is a major regulator of macrophage cholesterol efflux and protects cells from excess intracellular cholesterol accumulation, however the mechanism involved in posttranscriptional regulation of ABCA1 is poorly understood. We previously showed miR-33 was one regulator. Here we investigated the potential contribution of other microRNAs (miRNAs) to post-transcriptionally regulate ABCA1 and macrophage cholesterol efflux.
Methods and Results
We performed a bioinformatic analaysis for identifying miRNA target prediction sites in ABCA1 gene and an unbiased genome-wide screen to identify miRNAs modulated by cholesterol excess in mouse peritoneal macrophages. Quantitative real-time RT-PCR confirmed that miR-758 is repressed in cholesterol-loaded macrophages. Under physiological conditions, high dietary fat excess in mice repressed mir-758 both in peritoneal macrophages and, to a lesser extent in the liver. In mouse and human cells in vitro, miR-758 repressed the expression of ABCA1 and conversely the inhibition of this miRNA by using anti-miR-758 increased ABCA1 expression. In mouse cells, mir-758 reduced cellular cholesterol efflux to apoA1 and anti-miR-758 increased it. miR-758 directly targets the 3′UTR of Abca1 as assessed by 3′UTR luciferase reporter assays. Interestingly, miR-758 is highly expressed in the brain where also target several genes involved in neurological functions including SLC38A1, NTM, EPHA7 and MYT1L.
Conclusion
We identified miR-758 as a novel miRNA that post-transcriptionally controls ABCA1 levels in different cells and regulates macrophage cellular cholesterol efflux to apoA1, opening new avenues to increase apoA1 and raise HDL levels.
Keywords: Lipid homeostasis, microRNAs, atherosclerosis
Text
Cellular cholesterol levels are tightly regulated and represent the balance between cholesterol uptake, endogenous synthesis and efflux. Many diseases result from perturbations in lipid homeostasis, including atherosclerosis, metabolic syndrome, type II diabetes and Alzheimer's disease. In mammals two transcription factors are recognized to the control cellular cholesterol homeostasis: the sterol response element-binding proteins (SREBPs) and the liver × receptors (LXRs). The SREBP transcription factors control both endogenous cholesterol synthesis and uptake by regulating several sterol dependent genes including HMG-CoA reductase1, 2 and the low-density lipoprotein (LDL) receptor (LDLR). The LXRs are members of the nuclear receptor subfamily and are activated in response to cellular cholesterol3. Under cellular cholesterol excess, LXR targets genes such as the ATP-binding cassette transporter A1 (ABCA1) and G1 (ABCG1), which promote cellular cholesterol efflux and maintain cellular sterol homeostasis4, 5. The removal of excess cholesterol from peripheral tissues, such as macrophage foam cells, has been recognized as a key mechanism to prevent atherogenesis.6 ABCA1 promotes macrophage cholesterol efflux, and deficiency or mutations in this transporter leads to defects in cholesterol efflux, cholesterol ester accumulation in macrophages, and increases the risk of developing cardiovascular diseases7-9.
ABCA1 also plays an important role in regulating cholesterol metabolism in the brain. Cholesterol is required for myelination, dendrite differentiation and synaptic activity. Disturbances in central nervous system (CNS) cholesterol homeostasis are implicated in neurodegenerative disorders, including Alzheimer's and Huntington disease10. Several studies have demonstrated that ABCA1 facilitates the cholesterol efflux of CNS cholesterol to apoE, as the absence of ABCA1 compromises apoE secretion from both astrocytes and microglia11, 12. Moreover, the apoE that is present in the cerebrospinal fluid (CSF) of ABCA1-deficient mice is poorly lipidated11. Interestingly, in amyloid mouse models of Alzheimer's disease (AD), ABCA1 deficiency exacerbates amyloidogenesis, whereas ABCA1 overexpression ameliorates amyloid plaque load, suggesting a role for ABCA1 in Aβ metabolism13, 14.
In addition to classical transcription regulators, a class of non-coding RNAs termed microRNAs (miRNAs), has emerged as critical regulators of gene expression acting predominantly at the post-transcriptional level15. This large family of short (22 nt) double-stranded regulatory non-coding RNAs is encoded in the genome and each member is processed from primary transcripts by the sequential actions of Drosha and Dicer enzymes. Upon incorporation into the cytoplasmic RNA-induced silencing complex (RISC), miRNAs bind to partially complementary target sites in messenger RNA (mRNA) 3′untranslated regions (3′UTRs), which results in translational repression, mRNA destabilization, or a combination of both16, 17. We and others have recently shown that miR-33 regulates cholesterol efflux and HDL biogenesis by down-regulating the expression of ABC transporters, ABCA1 and ABCG118-20. Notably, silencing of miR-33 in vivo increases hepatic ABCA1 expression and plasma HDL levels18, 21.
Bioinformatic predictions and experimental approaches indicate that a single miRNA could target more than one hundred mRNAs. Similarly, a single mRNA could be regulated by many miRNAs. This maybe the case of Abca1, which has a very large 3′UTR (3.3 Kb) and over a hundred potential miRNA candidates according the miRNA target prediction algorithms. In the present study, we aim to identify new miRNAs that regulate ABCA1 expression. We show that miR-758 post-transcriptionally regulates the expression of ABCA1 in different cell lines including macrophages, hepatocytes and astrocytes thereby attenuating cholesterol efflux to apoA1. Futhermore, the high expression of miR-758 in the brain suggests an important role for this miRNA in regulating cellular cholesterol metabolism in this organ.
Materials and Methods
See online Data Supplements for an extended description of the methods used in this study, available at http://atvb.ahajournals.org
miRNA mimics, miRNA inhibitors and ABCA1 siRNA transfection
Cells (50-70% confluence) were transfected with 40 nM mi_RIDIAN_ miRNA mimics, mi_RIDIAN_ miRNA inhibitors (Dharmacon), and/or siRNA ABCA1 (smart pool, Dharmacon) using Lipofectamina RNAiMAX (Invitrogen) using manufacturer protocols. In all experiments an equal concentration of a non-targeting control mimics sequence (Con-miR) or inhibitor negative control sequence (Con-Inh) were used as controls for non-sequence-specific effects in miRNA experiments. Cells were transfected for 24 h and treated with or without either T0901317 (3μM) or acetylated LDL (120 μg/mL) for an additional 24h.
3′UTR Luciferase Reporter Assays
cDNA fragments corresponding to the entire 3′UTR of human Abca1 was amplified by RT-PCR from total RNA extracted from HepG2 cells with XhoI and NotI linkers. The PCR products were directionally cloned downstream of the Renilla luciferase open reading frame in the psiCHECK2TM vector (Promega) that also contains a constitutively expressed firefly luciferase gene, which is used to normalize transfections. Site-directed mutations in the seed region of predicted miR-758 sites within the 3′UTR of human Abca1, were generated using Multisite-Quickchange (Stratagene) according to the manufacturer's protocol. All constructs were confirmed by sequencing. COS-7 cells were plated into 12-well plates (Costar) and co-transfected with 1μg of the indicated 3′UTR luciferase reporter vectors and the miR-758 mimic or negative control mimic (Con-miR) (Dharmacon) using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized to the corresponding firefly luciferase activity and plotted as a percentage of the control (Con-miR). Experiments were performed by triplicate and at least three independent experiments were performed.
Cholesterol Efflux Assays
J774 macrophages and human H4 neuroglioma cell line were plated in a 12-well plates (1 × 106 cells/well) and transfected with either a Con-miR, miR-758, a Con-Inh or a anti-miR-758 (Dharmacon) for 24h. Cells were loaded with 0.5μCi/ml 3H-cholesterol (PerkinElmer) for an additional 24 h. Then, cells were washed twice with PBS and incubated with 2% fatty-acid free BSA (FAFA, Sigma) in media in the presence of ACAT inhibitor (2 μM) for 2 h prior to the addition of 50μg/ml human apoAI or 50μg/ml HDL in FAFA-media with or without the indicated treatments. Supernatants were collected after 6 hours, radioactivity counted and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol+cell-associated 3H-cholesterol).
Statistical Analysis
The results are expressed as a mean±SEM. Statistical comparisons between groups were done by Student's t test or analysis of variance (ANOVA) and post hoc multiple comparisons with Student-Newman-Keuls test, by using Statgraphics Plus v5.0 program (Statistical Graphics, USA).
Results
ABCA1 expression is highly regulated by miRNAs
The 3′UTR of Abca1 is particularly long and we hypothesized that it is susceptible to be targeted by microRNAs. To determine the potential miRNA candidates that regulate ABCA1 expression, we used a combination of bioinformatic tools for miRNA target prediction [TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org)]. TargetScan revealed 136 predicted miRNAs that target ABCA1, whereas miRanda predicted 57. To assess whether these predicted miRNAs were modulated by cellular cholesterol content, we undertook an unbiased genome-wide screen. Interestingly, miR-291b-5p, miR-672, miR-673-5p, miR-758 and miR-33 were down-regulated (Supplemental table I), suggesting a physiological role for these miRNAs in regulating cellular cholesterol homeostasis. In our previous work, we studied the role of miR-33 in regulating ABCA1 and cellular cholesterol efflux and were motivated to do so by its intriguing genomic location (intron 16 of Srebf2). To directly examine the effects of the other miRNAs identified in our screening (miR-291b-5p, miR-672, miR-673-5p, and miR-758) on ABCA1 expression, we transfected human macrophages (THP-1) with these miRNAs and treated them with AcLDL or T0901317 to stimulate ABCA1 expression. Transfection of human macrophages with miR-758 (190-fold increase expression), but not a control miRNA (Con-miR) or other miRNAs strongly decreased the stimulation of ABCA1 protein expression (Figure 1A and 1B). miR-33 was used as a positive control for inhibition of ABCA1 expression (Figure 1A). Interestingly, miR-291b-5p transfection instead to inhibit ABCA1 increase its expression. This finding suggests that miR-291b-5p may regulate ABCA1 expression by targeting other genes involved in the regulation of ABCA1 expression. miR-758 also repressed ABCA1 protein in mouse peritoneal macrophages (Figure 1B) and hepatic cell lines (HepG2 and Hepa) (Figure 1C and D), indicating that its effect is not cell type specific. Notably, inhibition of endogenous miR-758 by anti-miR-758 (3-fold decrease expression) increased the expression of ABCA1 in macrophages and hepatic cells (Figure 1E and 1F), suggesting a physiological role of miR-758 in regulating the expression of this transporter.
Figure 1.
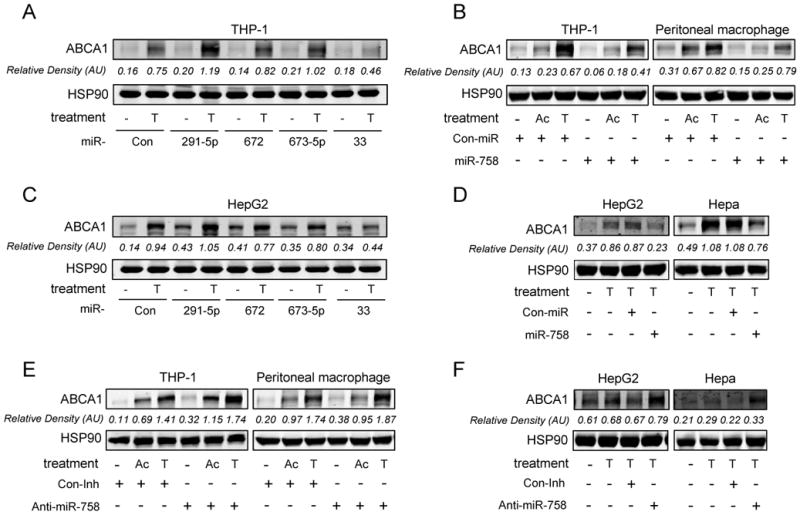
ABCA1 is post-transcriptionally regulated by miR-758. A to C, Protein analysis by Western Blotting of ABCA1 and HSP90 in THP-1-derived macrophages (A and B), mouse peritoneal macrophages (B), HepG2 cells (C and D) and Hepa cells (D) transfected with either Con-miR or miR-758 mimic and induced with acetylated LDL (Ac) or T0901317 (T). E to F, ABCA1 expression in THP1 derived macrophages (E), mouse peritoneal macrophages (E), HepG2 (F) and Hepa cells (F) transfected with Con-Inh or anti-miR-758, in the presence or absence of AcLDL (Ac) or T0901317 (T).
We further assess whether the co-transfection of miR-33 and miR-758 has an additive inhibitory effect on the ABCA1 protein expression. As seen in Supplemental Figure I, Huh7 cells transfected with miR-33 and miR-758 expressed reduced ABCA1 levels compared with cells transfected with the individual miRNAs, suggesting an additive inhibitory effect of both miRNAs on ABCA1 expression.
miR-758 directly targets the 3′UTR of Abca1
The human Abca1 3′UTR has two computationally predicted miR-758 binding sites (Figure 2A). Site 2 is highly conserved between species whereas site 1 is only conserved in human and non-human primates (Figure 2B). To assess the effects of miR-758 on the 3′UTR of human Abca1, we used luciferase reporter constructs. miR-758 markedly repressed the activity of the Abca1 3′UTR reporter construct (Figure 2C). Specific site-directed mutations in site 2, which is widely conserved amongst several species, abolishes the miR-758 repression of Abca1 3′-UTR activity, suggesting that this site is the most important in the post-transcriptional repression of Abca1 by miR-758 (Figure. 2D).
Figure 2.
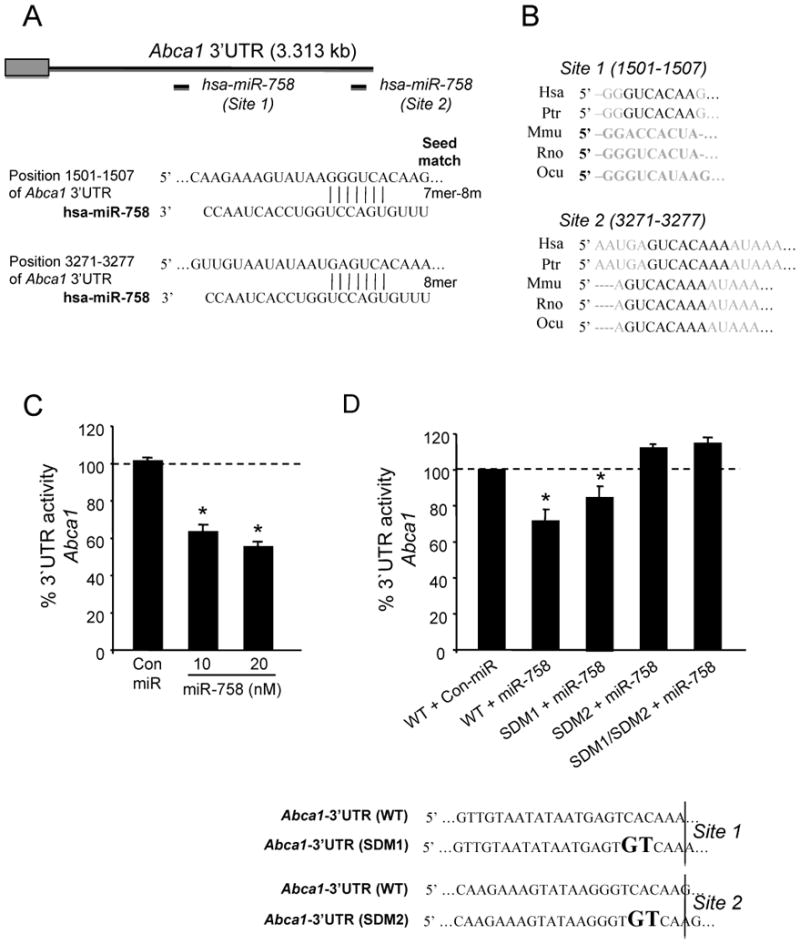
miR-758 directly targets the 3′UTR of Abca1. A, Sequence alignment of the human hsa-miR-758 mature sequence with the binding sites of the human Abca1 3′UTR. Relative position of the binding sites are indicated (top). B, Sequence alignment region of the two human (Hsa) seed sites and the indicated species [Pan troglodites (Ptr), Mus musculus (Mmu), Rat norvegicus (Rno) and Oryctolagus cuniculus (Ocu)]. C, Activity of the luciferase reporter construct fused to the 3′UTR of human Abca1 in COS-7 cells. Cells were transfected with Con-miR or miR-758. D, Abca1 3′UTR containing the indicated site-directed mutations (SDM) in the miR-758 target sites (SDM1: site-directed mutation site 1 and SDM2: site-directed mutation site 2). Luciferase activity was normalized with Renila. Relative luciferase activity is presented and data are the mean ± SEM of three independent experiments in triplicate. Statistical comparisons between groups by ANOVA and Student-Newman-Keuls text; * denote statistically significant differences (p<0.05).
miR-758 regulates macrophage cellular cholesterol efflux to apolipoprotein A1
The ability of ABCA1 to stimulate the efflux of cholesterol from cells in the periphery, particularly cholesterol-laden macrophages in atherosclerotic plaques, is an important anti-atherosclerotic mechanism6, 22. Transfection of J774 macrophages with miR-758 reduced ABCA1 expression (Figure 3A, upper panel) and attenuated cholesterol efflux to apolipoprotein A1 (apoA1) (Figure 3A, left panel). miR-758 did not impair cholesterol efflux to HDL in J774 macrophages, consistent with the lack of miR-758 binding sites in the mouse/human Abcg1 3′UTR and other compensating mechanisms (Figure 3A, right panel). Notably, antagonism of endogenous miR-758 increased ABCA1 protein (Figure 3B, upper panel) and cholesterol efflux to apoA1, but not to HDL (Figure 3B). Similar results were obtained using human THP-1 macrophages (data not shown). Furthermore, to directly assess the role of ABCA1 in the miR-758 repression of cholesterol efflux, we co-transfected J774 cells with miR-758 and ABCA1 siRNA. As seen in Figure 4, the effect of miR-758 on cholesterol efflux to ApoA1 was significantly attenuated after ABCA1 silencing, suggesting that miR-758 regulates cholesterol efflux via the ABCA1 transporter (Figure 4). Thus, manipulation of cellular miR-758 levels alters macrophage cholesterol efflux, a critical step in the reverse cholesterol transport pathway for the delivery of cholesterol excess to the liver.
Figure 3.
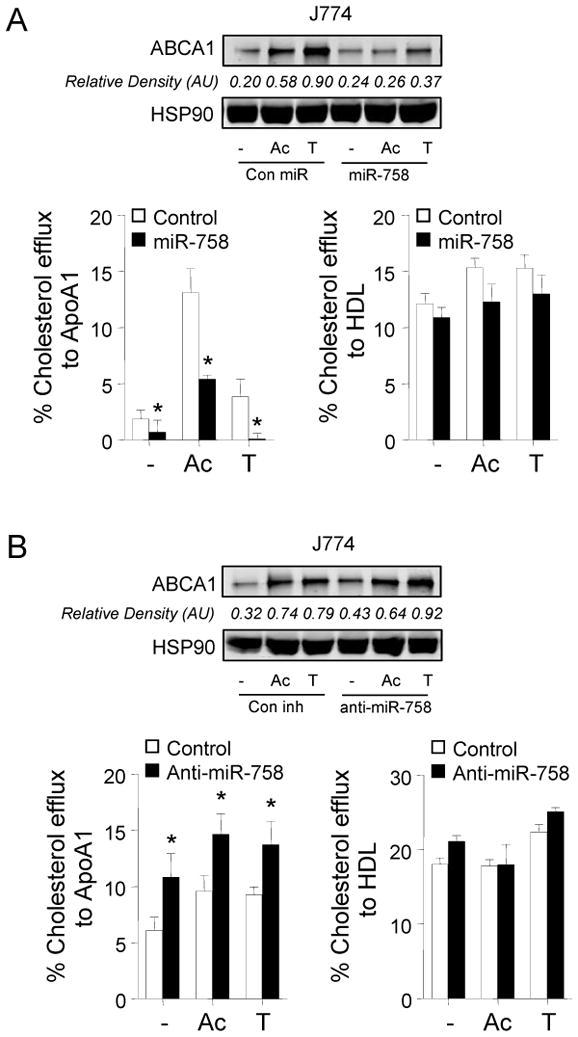
Modulation of miR-758 regulates macrophage cellular cholesterol efflux. Transfection of mouse J774 macrophages with miR-758 (A) or anti-miR-758 (B) modifies ABCA1 expression (top inserts). Total cholesterol efflux from AcLDL (Ac) or T0901317 (T)-treated J774 macrophages was evaluated after 6h incubation of apoA1 or HDL in the presence of miR-758 mimic (A) and anti-miR-758 mimic (B). Data are the mean ± SEM of three independent experiments in triplicate. *, P<0.05 from control.
Figure 4.
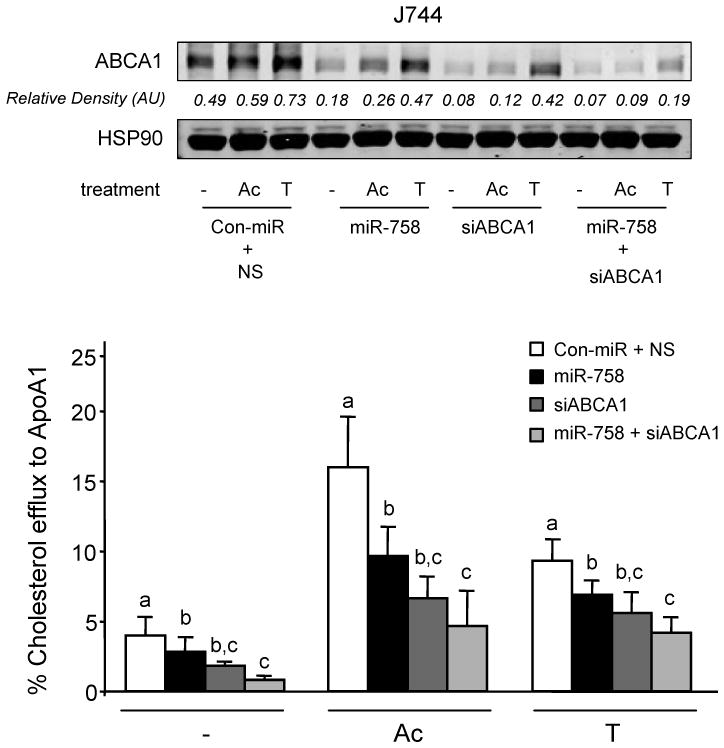
ABCA1 knockdown suppress the effect of miR-758 effect on apoA1-mediated cellular cholesterol efflux. Protein analysis by Western Blotting of ABCA1 and HSP90 and cellular cholesterol efflux in J774 cells transfected with ABCA1 siRNA, miR-758 mimic or both and induced with acetylated LDL (Ac) or T0901317 (T). Statistical comparisons between groups by ANOVA and Student-Newman-Keuls analysis; different letters denote statistically significant differences (p<0.05).
miR-758 tissue and cellular expression and its regulation by plasma lipid levels in vivo
Next, we examined the _in viv_o expression of miR-758 in mice. miR-758 was widely expressed in mouse tissues, and particularly abundant in the brain, heart and aorta (Figure 5A). Interestingly, whole brain tissue expressed very high levels of miR-758 compared to other tissues. We also evaluated the miR-758 expression in different cell lines from mouse (Figure 5B) and human origin (Figure 5C). In accordance with the high expression of miR-758 in the brain, neurogliomas, particularly the astrocyte cell line CCF-STTG1, expressed very high levels of miR-758 (≈75-fold) compared to the other human cell lines analyzed, suggesting an important role for this miRNA in the brain physiology.
Figure 5.
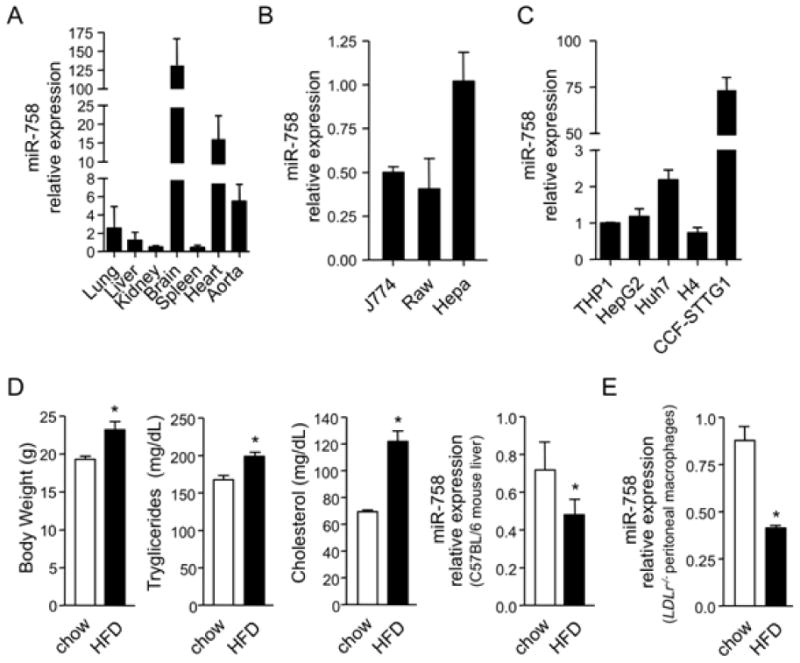
Distribution and dietary regulation of miR-758. (A-C), qRT-PCR Expression profile of miR-758 in selected mouse tissue (A) and selected mouse (B) and human (C) cell lines. D, Real time RT-PCR analysis of miR-758 in the liver of C57BL6 mice (n=5 per group) fed a chow diet (control) or high fat diet (HFD) for five weeks. Left panel represent the body weight, the second to the left panel represent the triglycerides plasma level, the third panel the cholesterol plasma levels and the right panel the miR-758 expression. E, miR-758 expression in peritoneal macrophages from hypercholesterlemic LDLr-/- mice fed a chow or HFD for 12 weeks (n=6 per group).
To determine whether miR-758 is regulated under different physiological conditions, we measured its expression in mice fed either a chow or high fat diet for 5 weeks. As expected, treatment of C57BL6 mice with a high fat diet increased body weight and plasma cholesterol and triglyceride levels (Figure 5D). Consistent with our in vitro observations, hepatic miR-758 levels are regulated by dietary cholesterol in vivo (Figure 5D right panel). Levels of miR-758 were inversely correlated with the expression of ABCA1 (11-fold increase). Moreover, miR-758 levels were reduced in peritoneal macrophages from hypercholesterolemic _LDLr_-/- mice that were fed a high-fat diet (Figure 5E).
We further characterize the regulation of miR-758 in macrophages by cholesterol. Since AcLDL binds to scavenger receptors, we assess whether or not receptor-independent cholesterol loading of macrophage also represses miR-758 expression. As shown in Supplemental Figure IIA, expression levels of pri-miR-758 and _miR-75_8 were significantly reduced in mouse peritoneal macrophages treated with AcLDL or Chol:MCD (cholesterol:methyl-beta-cyclodextrin). Similarly, THP-1 human macrophages treated with AcLDL also have reduced levels of miR-758 (Supplemental Figure IIB).
miR-758 controls cellular cholesterol efflux and other targets in neuroglioma cells
To determine the potential role of miR-758 in regulating cellular cholesterol efflux in neural cells, we transfected glioblastoma H4 cells with miR-758 mimics. As seen in Figure 6A, miR-758 over-expression significantly decreased ABCA1 protein expression. Conversely, endogenous inhibition of miR-758 resulted in an increase in ABCA1 protein levels (Figure 6B), confirming that ABCA1 is regulated by miR-758 in neural cell lines. Next, we examined the effects of miR-758 manipulation on cholesterol efflux in H4 cells. As expected, miR-758 over-expression leads to a reduction in cholesterol efflux to ApoA1 (Figure 6A) whereas the endogenous inhibition of miR-758 increased cellular cholesterol efflux (Figure 6B). Thus, these findings demonstrate that miR-758 effects on ABCA1 result in the retention of cellular cholesterol and that manipulating cellular miR-758 levels can influence neuronal cholesterol homeostasis. Interestingly, several genes involved in amino acid synthesis [sodium-coupled neutral amino acid transporter 1 (SLC38A1)], neurite outgrowth [neurotrimin (NTM)] and the development of nervous system [ephrin type-A receptor 7 (EPHA7) and myelin transcription factor 1-like (MYT1L)] are predicted targets for miR-758.
Figure 6.
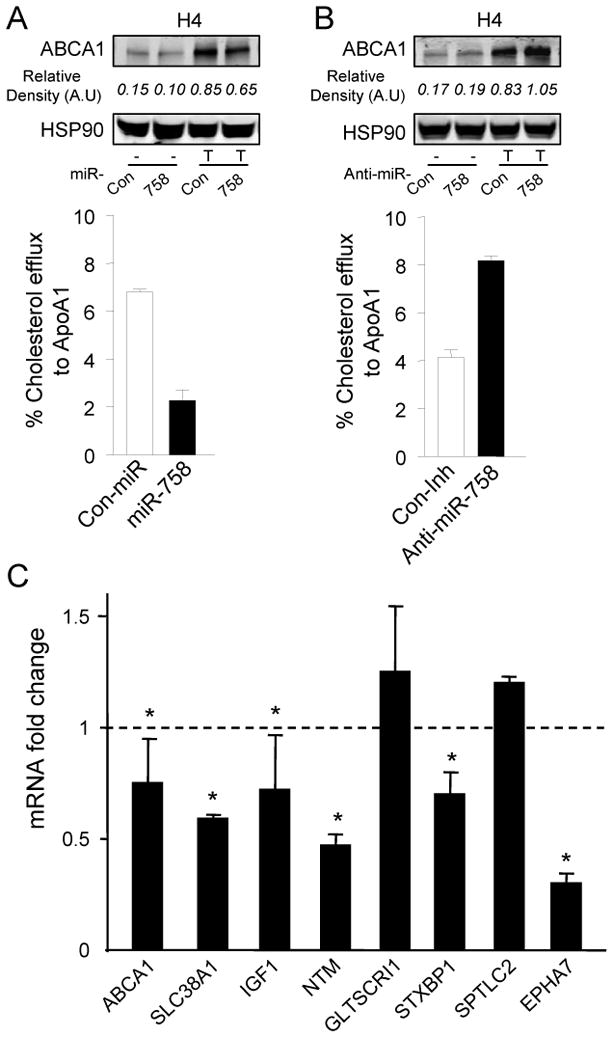
miR-758 targets ABCA1 in neuroglioma cells and regulates cholesterol efflux. A, Western blot analysis of ABCA1 expression and cellular cholesterol efflux in H4 cells transfected with either Con-miR or miR-758 mimic and induced with T0901317 (T). B, Western blot analysis of ABCA1 expression and cellular cholesterol efflux in H4 cells transfected with either Con-miR or miR-758 mimic and induced with T0901317 (T). Data are the mean ± SEM of three independent experiments in triplicate. qRT-PCR analysis of neurological targets from H4 cells transfected with miR-758. Data represent the mRNA fold change versus H4 cells transfected with Con-miR. Mean ± SEM of three independent experiments in triplicate. Statistical comparisons of cells transfected with miR-758 versus Con-miR by Student's t test. *, P<0.05 from control.
To assess whether miR-758 regulates the expression of these targets, we transfected H4 cells with miR-758. As seen in Figure 6C, miR-758 over-expression reduced significantly the mRNA expression of SLC38A1, IGF1, NTM, STXBP1 and EPHA7 suggesting an important role of miR-758 in regulating neurological functions. Whether other miR-758 targets can contribute to this effect and which physiological or pathological conditions underlie the regulation of miR-758 gene expression remains to be determined.
Discussion
HDL and its principal apolipoprotein, apoA1, play a critical role in the removal of cellular cholesterol excess and its transport to the liver, a process called reverse cholesterol transport (RCT). When the excess cholesterol originates from foam cells in arterial plaques, RCT is though to underlie the atheroprotective effect of HDL6, 22. ABCA1 and ABCG1 transporters promote cellular cholesterol efflux to HDL and its associated apolipoprotein, apo-A1 in peripheral cells including macrophages. ABCA1 is also primarily responsible for initiating HDL formation in the liver, a finding brought to light when mutation in Tangier disease, a condition characterized by low plasma HDL, was mapped to the ABCA1 gene23, 24. Transcriptionally, ABCA1 expression is regulated primarily by the LXR/RXR system and also by other agonists in a LXR-dependant and independent manner in pheripheral cells25. In the liver, evidence suggests that ABCA1 is also regulated by SREBP-226. Post-transcriptional regulation of ABCA1, by calpain-mediated proteolytic degradation, has also been described27, 28.
Very recently four independent groups have shown that microRNAs play an important role in post-transcriptional regulation of ABCA118-21 by targeting the 3′UTR of Abca1. The 3′UTR of Abca1 is particularly long and we hypothesized that it might be targeted by microRNAs besides miR-33. By conducting a whole-genome microRNA microarray in cholesterol-loaded macrophages, here we show that other microRNAs can regulate cholesterol homeostasis by targeting ABCA1, expanding our current knowledge of how cholesterol is modulated and the relevance of cholesterol and lipoprotein metabolism by microRNAs.
Two other microRNAs have been shown to have a direct role in regulating cholesterol homeostasis18, 29. The liver-restricted miR-122 regulates cholesterol and triglyceride metabolism and its silencing in mice and non-human primates reduces cholesterol synthesis, hepatic fatty acids and plasma cholesterol levels, and increases liver fatty acid β-oxidation30-32. In contrast to miR-122, miR-758 is widely expressed in different tissues and cell types and its effect on ABCA1 has a more global effect on cellular cholesterol homeostasis. Similarly, miR-33 has also been reported to be expressed in different tissues and cell lines18. Interestingly, here we have shown that miR-758 is highly expressed in the brain, particularly in astrocytes. The brain accounts for almost 25% of total body cholesterol33. In the developed brain, cholesterol is synthesized by astrocytes and oligodendrocytes, and in contrast to oligodendrocytes, the astrocytes supply cholesterol to other neurons by ABC proteins, ABCA1, ABCG1 and ABCG410, 34. ApoE is an important mediator of cholesterol efflux in the brain, and certain neurodegenerative disorders are associated with disturbances in cholesterol metabolism and the presence of the apoE4 isoform10, 35. Here we show that manipulating miR-758 levels in neuronal cells can modify both ABCA1 expression and cholesterol efflux to ApoA1. However, the in vivo physiological and pathological changes of miR-758 levels in the brain are not known and need to be evaluated.
Compared to miR-33a/b, which are localized within introns of Srebf genes and regulated primarily by host genes18, 19, the highly conserved miR-758 is localized in an intergenic region within chromosome 14 and its direct regulation is not know. Here we confirm that miR-758 is down-regulated upon cholesterol loading in macrophages both in culture and in a mouse model of hypercholesterolemia (_Ldlr_-/-) induced by high fat diet, indicating that in vivo, miR-758 in macrophages is regulated by dietary cholesterol.
So far, only one specific microRNA, miR-33, has been described to target ABCA1. Here we describe and confirm a second microRNA that controls ABCA1 levels and therefore regulates cellular cholesterol homeostasis. Our screening of cholesterol-loaded macrophages revealed other microRNAs that according to different microRNA prediction algorithms also target ABCA1. Why cells expressed several microRNAs (miR-33 and miR-758) regulating this protein and what drives the specificity of these microRNAs in different cell types under physiological and pathological conditions needs to be determined. Through evolution, cholesterol has played a crucial role in the development of both the vertebrate brain33 and special membrane domains36 and its level in our organism is tightly controlled by plasma lipoprotein levels and transcription factors37. Imbalances in cholesterol metabolism are recognized as the major risk factor for developing cardiometabolic disease. Therefore, control of cellular cholesterol homeostasis through microRNAs can be an alternative for cells to regulate cholesterol homeostasis in cholesterol-related pathologies.
Further studies are needed to explore the in vivo effects of miR-758 silencing in HDL biogenesis to decipher the antagonism of endogenous miR-758 as a potentially useful therapeutic strategy to enhance plasma levels of HDL. Understanding the complex network of microRNAs that target ABCA1 may provide new tools to develop therapeutic strategies to combat the burden of cardiovascular disease and atherosclerosis. Our findings increase the knowlwdge of the complex regulation of ABCA1 and therefore the cellular cholesterol homeostasis by post-transcriptional regulation of the 3′UTR of Abca1. They also open new possibilities in therapeutically targeting miR-758 and influencing both HDL levels and macrophage cholesterol efflux in patients with cardiovascular disease.
Supplementary Material
1
Acknowledgments
The authors thank Dr. Edward A. Fisher for helpful discussions.
SOURCE OF FUNDING
This work was supported by grants from the American Heart Association (SDG-0835585D to C.F.-H, grant SDG-0835481N to Y.S.), and the National Institutes of Health (RO1HL107953 and RO1HL106063 to C.F.-H.). A.G.S by Capes Foundation, Ministry of Education of Brazil, Brazil.
Footnotes
References
- 1.Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, Wang X. SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc Natl Acad Sci U S A. 1993;90:11603–11607. doi: 10.1073/pnas.90.24.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yokoyama C, Wang X, Briggs MR, Admon A, Wu J, Hua X, Goldstein JL, Brown MS. SREBP-1, a basic-helix-loop-helix-leucine zipper protein that controls transcription of the low density lipoprotein receptor gene. Cell. 1993;75:187–197. [PubMed] [Google Scholar]
- 3.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Collins HL, Ranalletta M, Fuki IV, Billheimer JT, Rothblat GH, Tall AR, Rader DJ. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J Clin Invest. 2007;117:2216–2224. doi: 10.1172/JCI32057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci U S A. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rader DJ, Alexander ET, Weibel GL, Billheimer J, Rothblat GH. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J Lipid Res. 2009;50(Suppl):S189–194. doi: 10.1194/jlr.R800088-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clee SM, Kastelein JJ, van Dam M, Marcil M, Roomp K, Zwarts KY, Collins JA, Roelants R, Tamasawa N, Stulc T, Suda T, Ceska R, Boucher B, Rondeau C, DeSouich C, Brooks-Wilson A, Molhuizen HO, Frohlich J, Genest J, Jr, Hayden MR. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J Clin Invest. 2000;106:1263–1270. doi: 10.1172/JCI10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mott S, Yu L, Marcil M, Boucher B, Rondeau C, Genest J., Jr Decreased cellular cholesterol efflux is a common cause of familial hypoalphalipoproteinemia: role of the ABCA1 gene mutations. Atherosclerosis. 2000;152:457–468. doi: 10.1016/s0021-9150(99)00498-0. [DOI] [PubMed] [Google Scholar]
- 9.Yvan-Charvet L, Wang N, Tall AR. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler Thromb Vasc Biol. 2010;30:139–143. doi: 10.1161/ATVBAHA.108.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu JP, Tang Y, Zhou S, Toh BH, McLean C, Li H. Cholesterol involvement in the pathogenesis of neurodegenerative diseases. Mol Cell Neurosci. 2010;43:33–42. doi: 10.1016/j.mcn.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J Biol Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- 13.Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- 14.Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 18.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. miR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marquart TJ, Allen RM, Ory DS, Baldan A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc Natl Acad Sci U S A. 2010;107:12228–12232. doi: 10.1073/pnas.1005191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tall AR. Role of ABCA1 in cellular cholesterol efflux and reverse cholesterol transport. Arterioscler Thromb Vasc Biol. 2003;23:710–711. doi: 10.1161/01.ATV.0000068683.51375.59. [DOI] [PubMed] [Google Scholar]
- 23.Brooks-Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO, Loubser O, Ouelette BF, Fichter K, Ashbourne-Excoffon KJ, Sensen CW, Scherer S, Mott S, Denis M, Martindale D, Frohlich J, Morgan K, Koop B, Pimstone S, Kastelein JJ, Genest J, Jr, Hayden MR. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–345. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- 24.Oram JF. Tangier disease and ABCA1. Biochim Biophys Acta. 2000;1529:321–330. doi: 10.1016/s1388-1981(00)00157-8. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama S. Assembly of high-density lipoprotein. Arterioscler Thromb Vasc Biol. 2006;26:20–27. doi: 10.1161/01.ATV.0000195789.39418.e8. [DOI] [PubMed] [Google Scholar]
- 26.Tamehiro N, Shigemoto-Mogami Y, Kakeya T, Okuhira K, Suzuki K, Sato R, Nagao T, Nishimaki-Mogami T. Sterol regulatory element-binding protein-2- and liver × receptor-driven dual promoter regulation of hepatic ABC transporter A1 gene expression: mechanism underlying the unique response to cellular cholesterol status. J Biol Chem. 2007;282:21090–21099. doi: 10.1074/jbc.M701228200. [DOI] [PubMed] [Google Scholar]
- 27.Wang N, Chen W, Linsel-Nitschke P, Martinez LO, Agerholm-Larsen B, Silver DL, Tall AR. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J Clin Invest. 2003;111:99–107. doi: 10.1172/JCI16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwamoto N, Lu R, Tanaka N, Abe-Dohmae S, Yokoyama S. Calmodulin interacts with ATP binding cassette transporter A1 to protect from calpain-mediated degradation and upregulates high-density lipoprotein generation. Arterioscler Thromb Vasc Biol. 2010;30:1446–1452. doi: 10.1161/ATVBAHA.110.203927. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 30.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 31.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 32.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietschy JM, Turley SD. Thematic review series: brain Lipids. Cholesterol metabolism in the central nervous system during early development and in the mature animal. J Lipid Res. 2004;45:1375–1397. doi: 10.1194/jlr.R400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Wang N, Yvan-Charvet L, Lutjohann D, Mulder M, Vanmierlo T, Kim TW, Tall AR. ATP-binding cassette transporters G1 and G4 mediate cholesterol and desmosterol efflux to HDL and regulate sterol accumulation in the brain. FASEB J. 2008;22:1073–1082. doi: 10.1096/fj.07-9944com. [DOI] [PubMed] [Google Scholar]
- 35.Bjorkhem I, Leoni V, Meaney S. Genetic connections between neurological disorders and cholesterol metabolism. J Lipid Res. 2010;51:2489–2503. doi: 10.1194/jlr.R006338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mouritsen OG, Zuckermann MJ. What's so special about cholesterol? Lipids. 2004;39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 37.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci U S A. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1