microRNA-279 acts through the JAK/STAT pathway to regulate circadian behavioral output in Drosophila (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 17.
Summary
Although molecular components of the circadian clock are known, mechanisms that transmit signals from the clock and produce rhythmic behavior are poorly understood. We found through a genetic screen that over-expression of microRNA miR-279 disrupts rest:activity rhythms in Drosophila. Deletion of miR-279 also attenuates rhythms, which are rescued by a miR-279 transgene. Oscillations of the clock protein PERIOD are normal in pacemaker neurons of miR-279 nulls, suggesting that miR-279 acts downstream of the clock. Through an RNAi screen of putative miR-279 targets, we identified circadian effects of the JAK/STAT ligand, upd, and show that knockdown of upd rescues the behavioral phenotype of miR-279 mutants. Manipulations of the JAK/STAT pathway also disrupt behavioral rhythms. Furthermore, JAK/STAT signaling is regulated by the clock, and central clock neurons appear to project to _upd_-expressing cells. These findings identify a circadian output pathway in which JAK/STAT signaling is regulated by miR-279 to drive rest:activity rhythms.
Introduction
The endogenous system that generate circadian rhythms has historically been depicted in the form of a heuristic model in which an endogenous clock receives environmental signals through an input pathway and transmits signals through an output pathway (Eskin, 1979). The overt rhythm is the product of this system, which is, of course, more complicated than the simplest model would predict. The past two decades have seen major advances in our understanding of how a clock is assembled, and how the clock perceives the primary synchronizing/entraining signal, light (Zheng and Sehgal, 2008). However, the pathway that carries time-of-day signals away from the clock to produce overt circadian rhythms is less well understood.
Output pathways have been characterized in some organisms and also, to some extent, for cellular metabolic pathways in mammals (Asher and Schibler, 2011; Harmer, 2009; Vitalini et al., 2006). However, outputs are difficult to tackle when the overt rhythm measured is that of behavioral rest:activity, as it is usually in rodents and in Drosophila. Although Drosophila has a much simpler nervous system, the circadian system still involves many different cell types in a neural network (Nitabach and Taghert, 2008), making it quite difficult to identify molecular components. In addition, the most successful method used to identify clock molecules in _Drosophila_-forward genetic screens- may have limited use for identification of output molecules. This is because many clock components are largely dedicated to timekeeping, and so their loss is not lethal, while most output components are likely to be essential genes, involved in signal transduction, synaptic transmission etc., whose levels/activity are regulated by the clock. Thus, only a limited number of output molecules have been described for the Drosophila rest:activity rhythm and most of these have not been placed in specific signaling pathways (Allada and Chung, 2010).
microRNAs (miRNAs) are a class of small, non-coding RNAs which usually repress messenger RNA (mRNA) translation or cause mRNA decay by directly binding to the 3′ Untranslated Region (UTR) of target mRNAs (Carthew and Sontheimer, 2009). It is becoming increasingly clear that most biological processes involve regulation by microRNAs. One would expect this to be particularly true for circadian rhythms, where maintenance of appropriate levels can be critical for timekeeping. For instance, in Drosophila, the loss of or over-expression of the rhythmically expressed clock genes, period and timeless, results in arrhythmia (Yang and Sehgal, 2001; Zheng and Sehgal, 2008). A few microRNAs that regulate circadian rhythms have, in fact, been described in mammals and in flies (Cheng et al., 2007; Kadener et al., 2009; Kojima et al., 2010; Shi et al., 2009; Yang et al., 2008). However, only a few have been linked to rest:activity rhythms (Cheng et al., 2007; Kadener et al., 2009) and these are implicated in clock function or input to the clock, but not in output. Since mutations in miRNA genes are difficult to come by, loss of function circadian phenotypes in flies have not been reported for any Drosophila miRNA.
We report here the identification of a microRNA, miR-279, whose loss does not affect the central clock, but abolishes rest:activity rhythms, indicating that it is part of the output pathway. We identified the circadian-relevant target of this miRNA as unpaired (upd), the ligand for the JAK/STAT pathway, and show that manipulations of this pathway also disrupt rest:activity rhythms. These findings are the first to demonstrate a role for JAK/STAT signaling in circadian rhythms, and they also identify a novel miRNA that regulates this pathway in a circadian output circuit.
Results
Identification of miR-279 as a Circadian Rhythm-affecting microRNA
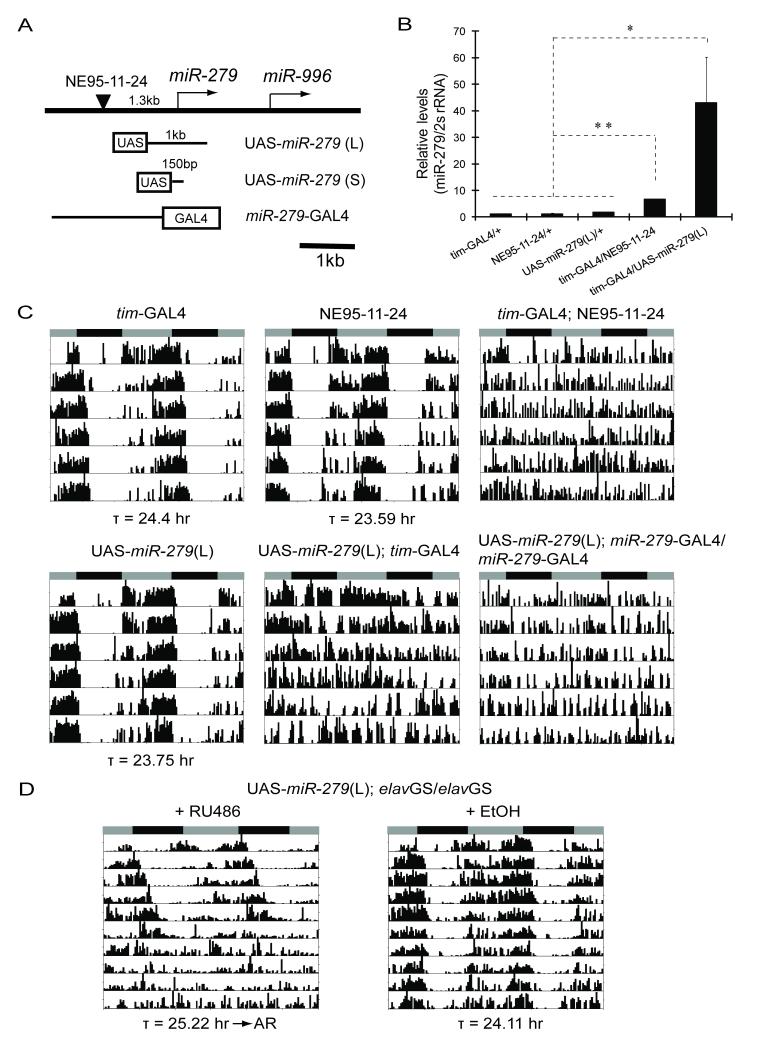
To identify genes affecting locomotor activity rhythms, we previously performed a forward genetic screen of 3662 EP (enhancer and promoter) lines in which expression of each randomly inserted EP element was driven by a _timeless_-GAL4 driver (_tim_-GAL4) (Zheng et al., 2007). One strain (NE95-11-24) that displayed a disrupted locomotor activity rhythm in this screen contains an EP-element insertion 1.3kb upstream of a microRNA gene, miR-279 (Figure 1A, 1C, Table 1).
Figure 1.
Over-expression of miR-279 disrupts locomotor activity rhythms. (A) Map of the miR-279 locus and transgenic constructs. An EP element (NE95-11-24, black triangle) is mapped 1.3kb upstream of miR-279. Two UAS-miR-279 transgenic lines were generated with a 1kb (L) or a 150bp (S) genomic region of miR-279 fused to UAS. A _miR-279_-GAL4 reporter (Cayirlioglu et al., 2008) comprises the entire promoter of miR-279 fused to GAL4. (B) Over-expression of miR-279 by driving expression of NE95-11-24 or a UAS-miR-279 (L) transgene. q-PCR analysis of total RNA prepared from adult heads. The ratio of mature miR-279/2s rRNA was plotted as mean ± SD (** P<0.001, * P<0.01, by Student’s t-test). Levels were normalized relative to the levels seen in _tim_G4/+ control flies (set as 1). (C) Over-expression of miR-279 leads to behavioral arrhythmia in DD. The genotypes are indicated on top of the panels. The grey and black bars indicate subjective day and night respectively. Average periods (τ) of rhythmic flies are shown at the bottom of the panels. Representative activity records are shown. (D) Panneuronal induction of miR-279 in adulthood leads to a long period, which eventually degenerates into arrhythmia. Flies were reared and then aged for 3 days following eclosion on regular food. They were fed either 500μM RU486 or ethanol (EtOH, vehicle control) from the time of entrainment. Average periods of the first 5 days in DD (τ) are shown beneath the panels. See also Figure S1.
Table 1.
Alterations in levels of miR-279 disrupt locomotor rhythms in flies
| Genotype | R % (n)a | Period ± SEM (hr) | FFT ± SEM |
|---|---|---|---|
| _tim_-GAL4/+ | 100 (18/18) | 24.4 ± 0.07 | 0.122 ± 0.009 |
| NE95-11-24/+ | 100 (26/26) | 23.59 ± 0.03 | 0.135 ± 0.011 |
| _tim_-GAL4/+; NE95-11-24/+ | 0 (0/26) | - | - |
| UAS-miR-279(S)/Y | 100 (51/51) | 23.8 ± 0.03 | 0.111 ± 0.006 |
| UAS-miR-279(L)/Y | 100 (28/28) | 23.75 ± 0.04 | 0.099 ± 0.007 |
| UAS-miR-279(S)/Y; _tim_-GAL4/+ | 2.1 (1/48) | 24.67 | 0.05 |
| UAS-miR-279(L)/Y; _tim_-GAL4/+ | 30 (6/20) | 25.28 ± 0.17 | 0.037 ± 0.007 |
| _miR-279_-GAL4/_miR-279_-GAL4 | 90 (18/20) | 23.65 ± 0.06 | 0.035 ± 0.005 |
| _miR-279_-GAL4/_miR-279_-GAL4;NE95-11-24/+ | 0 (0/12) | - | - |
| UAS-miR-279(S)/Y; _miR-279_-GAL4/_miR-279_-GAL4 | 11.1 (2/18) | 24.08 ± 0.75 | 0.015 ± 0.002 |
| UAS-miR-279(L)/Y; _miR-279_-GAL4/_miR-279_-GAL4 | 0 (0/7) | - | - |
| _elav_GS/_elav_GS (+EtOH) | 100 (8/8) | 23.96 ± 0.12 | 0.095 ± 0.018 |
| _elav_GS/_elav_GS (+RU486) | 100 (7/7) | 24.63 ± 0.13 | 0.069 ± 0.015 |
| UAS-miR-279(S)/Y; _elav_GS/_elav_GS(+EtOH) | 87.5 (7/8) | 24.13 ± 0.09 | 0.048 ± 0.009 |
| UAS-miR-279(S)/Y; _elav_GS/_elav_GS(+RU486) | 0 (0/8)b | 25.42 ± 0.07b | 0.038 ± 0.009b |
| UAS-miR-279(L)/Y; _elav_GS/_elav_GS(+EtOH) | 100 (8/8) | 24.11 ± 0.09 | 0.081 ± 0.014 |
| UAS-miR-279(L)/Y; _elav_GS/_elav_GS(+RU486) | 0 (0/8) b | 25.22 ± 0.12b | 0.056 ± 0.01b |
| _w_1118 (sibling) | 100 (21/21) | 23.72 ± 0.04 | 0.095 ± 0.011 |
| miR-279 ex117-1/ex117-1 | 33.3 (7/21)c | 23.79 ± 0.28c | 0.031 ± 0.008c |
| genomic miR-279/genomic miR-279;miR-279 ex117-1/ex117-1 | 95.5 (21/22) | 23.82 ± 0.08 | 0.103 ± 0.008 |
The miR-279 gene mediates development of Drosophila CO2 sensory neurons in the antenna through down-regulation of the transcription factor Nerfin-1 (Cayirlioglu et al., 2008); however, it was recently also identified as one of several microRNAs that are expressed highly in tim neurons (Kadener et al., 2009). The miR-279 locus produces a ~100nt-long precursor microRNA, pre-miR-279, which is processed to mature 22nt miR-279. To examine whether mature miR-279 levels were increased by expression of NE95-11-24, we conducted quantitative real-time PCR (q-PCR) analysis of total RNA from heads of flies expressing NE95-11-24 under the control of _tim_-GAL4. As shown in Figure 1B, mature miR-279 RNA was more abundant in these flies than in wild-type controls. Since a second microRNA gene, miR-996, located 1.6kb downstream of miR-279 (Figure 1A), could also be induced by the _tim_-GAL4 driver, we sought to determine whether over-expression of miR-279 alone was responsible for the circadian rhythm phenotype. Thus, we generated transgenic miR-279 expressing lines in which either a 1kb (L) or a 150bp (S) genomic sequence that fully covers the miR-279 coding region was fused to an Upstream Activating Sequence (UAS) (UAS-miR-279, Figure 1A). In combination with _tim_-GAL4, UAS-miR-279 also increased expression of mature miR-279 and led to arrhythmic locomotor activity in most flies (Figure 1B and 1C, Table 1). Those few flies that still maintained weak rhythms showed a slightly longer period than controls (Table 1).
To exclude the possibility that the behavioral phenotype was caused by ectopic induction of miR-279 in neurons where it is normally not expressed, we utilized a GAL4 driver under the control of the endogenous miR-279 promoter (Cayirlioglu et al., 2008) (Figure 1A and S1A). Because _miR-279_-GAL4 is a much weaker driver than _tim_-GAL4, we used two copies of this driver to induce miR-279 over-expression. As shown in Figure 1C and Table 1, over-expression of miR-279 by _miR-279_-GAL4 phenocopies the effect of _tim_-GAL4, suggesting that miR-279 regulates circadian rhythms in regions of its normal expression. Since microRNAs frequently play a crucial role in brain development (Coolen and Bally-Cuif, 2009), we sought to distinguish between developmental and adult effects by restricting the expression of miR-279 to adult neurons. We first used a drug (RU486) inducible panneuronal driver, _elav_GeneSwitch (_elav_GS) (McGuire et al., 2004), which lengthened the period by ~0.6 hr during the first 5 days in constant darkness (DD) and then eventually caused arrhythmia (Figure 1D, Table 1). To determine the effects of adult expression in specific neurons, we used a temperature sensitive _tubulin_-GAL80ts coupled with a _tim_-GAL4 driver (_tim_-UAS-GAL4, TUG) (Blau and Young, 1999) or with _miR-279_-GAL4. The TUG driver is weaker than the _tim_-GAL4 used above (data not shown) and thus can be effectively repressed by _tubulin_-GAL80ts at 18oC. Adult-specific over-expression of miR-279 in tim or miR-279 cells, by shifting flies carrying these constructs to 29oC, also resulted in long periods and arrhythmicity (Figure S1B). This indicates that adult over-expression of miR-279 in relevant cells disrupts circadian behavioral rhythms.
A Loss-of-Function Mutation in miR-279 Disrupts Behavioral Rhythms
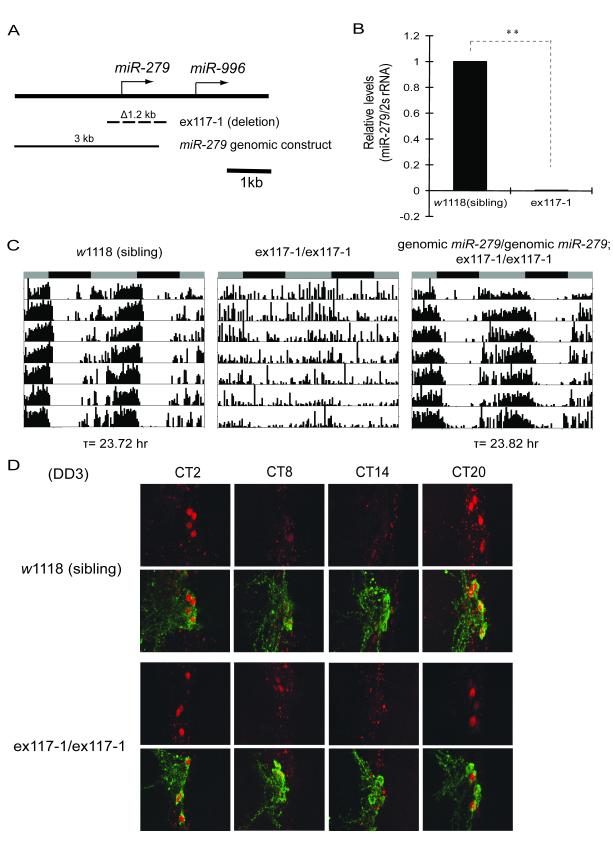
Deletions of the miR-279 gene were generated by imprecise excisions of two different P element insertions, both of which are downstream of miR-279 (Cayirlioglu et al., 2008). We outcrossed the 1.9kb (ex36-2) and 1.2kb (ex117-1) deletions seven times into an isogenic _w_1118 stock, which we use as our standard wild type stock for circadian rhythms. Loss of miR-279 expression causes lethality at the late pupal stage in its original background (Cayirlioglu et al., 2008). We observed the same lethality in the ex36-2 flies after outcrossing; however, a small percentage of outcrossed ex117-1 flies (Figure 2A) survive up to 2 weeks as adults, thus allowing us to perform behavioral analysis.
Figure 2.
Deletion of miR-279 reduces behavioral rhythmicity without changing PER cycling in small LNvs. (A) The miR-279 excision allele and genomic rescue construct (Cayirlioglu et al., 2008). The null allele ex117-1 (dashed line, Δ1.2kb) completely deletes the miR-279 gene. The miR-279 genomic construct (filled line, 3kb) comprises the entire promoter and coding region of miR-279. (B) The ex117-1 allele completely abolishes production of mature miR-279. q-PCR analysis of total RNA prepared from adult brains. The ratio of mature miR-279/2s rRNA was plotted as mean ± SD (** P<0.001, by Student’s t-test). Levels were normalized relative to those seen in w1118 control (set as 1). (C) Deletion of miR-279 disrupts activity rhythms, which can be rescued by introducing a miR-279 genomic transgene. After being outcrossed 7 times into a w1118 background, flies homozygous for ex117-1 live up to two weeks as adults. w1118 flies derived from siblings during the last outcross were used as wild-type control (indicated as “sibling”). (D) The null allele of miR-279 shows normal PER oscillations and PDF expression in central clock cells. Brains were dissected and stained with PER (red) and PDF (green) antibodies on the third day of DD at the indicated circadian times (CT). See also Figure S2.
Brains of homozygous ex117-1 flies do not express any detectable levels of mature miR-279 by q-PCR (Figure 2B). Behavioral analysis of these flies revealed that ~67% of the flies have very weak to no rhythms in DD (Figure 2C, Table 1). A previously identified phenotype of these flies-ectopic CO2 neuron formation- was rescued by a 3kb genomic miR-279 transgene (Cayirlioglu et al., 2008). We found that the same genomic construct also rescued the locomotor activity rhythm phenotype of ex117-1 (Figure 2A, 2C, and Table 1), indicating that the miR-279 gene is responsible for the behavioral phenotype of ex117-1 and is necessary for circadian rhythms.
The miR-279 Mutation Does Not Affect the Central Clock
A central pacemaker in the small ventral lateral neurons (sLNvs) of the Drosophila brain drives free-running locomotor activity rhythms in DD (Nitabach and Taghert, 2008). To determine whether miR-279 affects the central pacemaker itself or downstream output pathways that ultimately drive locomotor rhythms, we assayed oscillations of the central clock protein, PERIOD (PER), in sLNvs of ex117-1 adult flies. Entrained ex117-1 and sibling control flies were placed in DD and subject to immunocytochemistry of brain whole mounts on the first and third days in DD. As shown in Figures 2D and S2A, mutant flies and wild-type controls exhibit similar daily PER cycling in sLNvs. No obvious differences in protein levels or protein localization were found at any time point assayed. Furthermore, PER oscillations in other major clock neuron groups, including dorsal lateral neurons (LNds) and group 1 dorsal neurons (DN1s), were also robust and synchronized between the two hemispheres in mutant fly brains (data not shown). In addition, loss of miR-279 does not alter the viability or morphology of any known clock neuron groups, nor the expression of Pigment-Dispersing Factor (PDF), a neuropeptide expressed in LNvs (Figure 2D and S2B). The absence of a GFP signal in sLNvs of _miR-279_-GAL4:UAS-nuclear GFP (nGFP) flies (Figure S1A) further supports the idea that miR-279 does not function in the central pacemaker, but rather in output pathways.
To test whether the biogenesis of miR-279 itself is regulated by the circadian clock, we examined the expression profile of mature miR-279 over the course of a light:dark cycle (LD) and in constant darkness (DD). Under neither condition was significant cycling of mature miR-279 detected (Figure S2C and data not shown). Also, the level of mature miR-279 was not altered in clock gene mutants, _tim_01 and ClkJrk (Figure S2D). The results suggest that levels of miR-279 are not controlled by the circadian clock.
miR-279 Targets Unpaired to Regulate Circadian Behavioral Rhythms
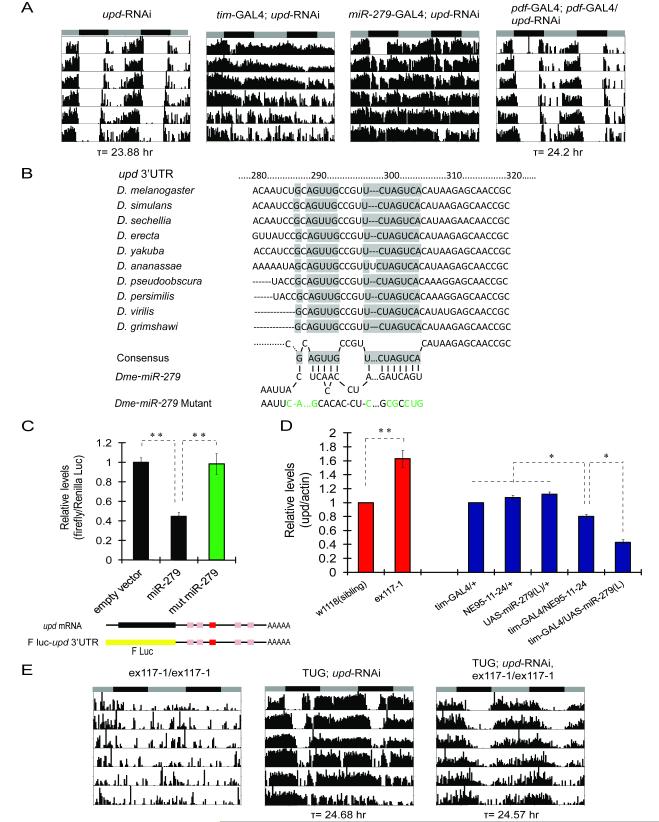
With few exceptions, animal miRNAs recognize miRNA-binding sites in the 3′UTR of target mRNAs and cause mRNA degradation or translational repression (Carthew and Sontheimer, 2009). A previously identified target of miR-279, relevant for its effects on the development of CO2 neurons, is the mRNA encoded by the Nerfin-1 gene (Cayirlioglu et al., 2008). We tested flies expressing a Nerfin-1 RNAi construct for circadian behavior and found that their behavioral rhythm was normal (data not shown), suggesting that Nerfin-1 does not mediate effects of miR-279 on circadian rhythms. We therefore sought to identify other possible targets of miR-279. Using three computational algorithms (i.e. PicTar, TargetScanFly5.1, and MiRanda) (Krek et al., 2005; Ruby et al., 2007), we discovered that ~35 mRNAs contain putative binding sites for miR-279 in their 3′ UTRs (Figure S3A, Supplemental Table). We obtained fly stocks carrying RNAi knockdown constructs for each of these mRNAs from the NIG-FLY stock center (~75 lines), introduced a _tim_-GAL4 or a _miR-279_-GAL4 transgene into each stock and screened the resulting progeny for locomotor rhythms. Rhythms were impaired when expression of the unpaired (upd) gene was knocked down (Figure 3A and S3B).
Figure 3.
Unpaired (upd) is a direct target of miR-279. (A) RNAi knock-down of the JAK/STAT ligand upd abolishes behavioral rhythms. Expression of _upd_-RNAi in either miR-279 or _tim_-expressing neurons, but not in Pdf neurons, causes arrhythmia. (B) A highly conserved 8 nucleotide miR-279 target site in the 3′UTR of upd mRNA. A predicted miR-279 binding site (shaded boxes) is conserved within the 3′UTR sequences of upd mRNAs from different Drosophila species (TargetScanFly 5.1). Base-pairing between the consensus target site and the mature miR-279 is shown. A miR-279 mutant containing mismatched nucleotides (green) is shown at the bottom of the panel. (C) miR-279 inhibits expression of an upd 3′UTR-luciferase reporter in cultured Drosophila S2 cells. The entire 750bp sequence of upd 3′UTR, containing one conserved 8mer (red box, by TargetScanFly 5.1) and 4 other putative miR-279 target sites (pink boxes, by RNAhybrid), was fused to a firefly luciferase (F luc, yellow bar) reporter construct (shown beneath the panel). An empty vector, or a wild-type or a mutant miR-279 (as shown in panel B) expression plasmid was cotransfected with the firefly luciferase reporter plasmid. Renilla luciferase was used as transfection control. The ratio of firefly/Renilla luciferase is plotted as mean ± SD of triplicate data points (** P<0.001, by Student’s t-test). The control with the empty vector is set as 1. (D) miR-279 is a negative regulator of upd mRNA in vivo. q-PCR analysis of total RNA prepared from fly brains (red bars) or fly heads (blue bars). The level of upd in ex117-1 was normalized to that in w1118 control (set as 1, red bars). The levels of upd in flies over-expressing miR-279 were normalized to those in _tim_G4/+ control (set as 1, blue bars). The ratios of upd/actin mRNA are plotted as mean ± SD (** P<0.001, *P < 0.01, by Student’s t-test). (E) RNAi knock-down of upd rescues the arrhythmia of miR-279 nulls. TUG is a comparatively weak _tim_-GAL4 driver (see text). The ex117-1 mutant and flies expressing _upd_-RNAi with TUG alone served as controls. See Also Figure S3 and Supplemental Table.
Unpaired (upd, also called outstreched) mRNA was predicted as a potential target of miR-279 by all three computational algorithms we employed (Supplemental Table). In fact, the upd 3′ UTR contains multiple putative miR-279 binding sites, one of which is highly conserved across different Drosophila species (Figure 3B and 3C). To determine whether miR-279 directly binds to upd 3′ UTR and inhibits its expression, we performed a luciferase reporter assay in Drosophila S2 cells. A chimeric mRNA was made by fusing the firefly luciferase open reading frame to the full-length upd 3′ UTR that contains all potential miR-279 binding sites (Figure 3C). Compared to the control, luciferase activity of the chimeric mRNA was significantly suppressed when miR-279 was cotransfected into the cells (Figure 3C). However, a mutant form of miR-279, in which sites complementary to those in the upd 3′ UTR were altered (Figure 3B), failed to suppress luciferase activity (Figure 3C). We further examined whether miR-279 affects upd mRNA levels in vivo. As shown in Figure 3D, upd mRNA was up-regulated in adult brains of miR-279 null mutants. On the other hand, miR-279 over-expression by a _tim_-GAL4 driver reduced mRNA levels of upd, with a higher dose of miR-279 producing a stronger effect (Figure 1B and 3D). We conclude that upd is a direct target of miR-279 in vitro and in vivo.
To test whether high levels of upd are responsible for the activity rhythm phenotype of miR-279 mutants, we reduced expression of upd with the _upd_-RNAi construct (Figure S3B) in an ex117-1 background. As shown in Figure 3E and Table 2, knocking down upd with a relatively weak tim promoter (TUG), prevented the abolition of activity rhythms in ex117-1 flies. These data provide strong in vivo evidence that miR-279 regulates circadian rhythms by modulating levels of upd.
Table 2.
miR-279 acts through the JAK/STAT pathway to regulate locomotor activity rhythms
| Genotype | R % (n) | Period ± SEM (hr) | FFT ± SEM |
|---|---|---|---|
| UAS-_upd_RNAi/+ | 100 (42/42) | 23.88 ± 0.03 | 0.102 ± 0.005 |
| _tim_-GAL4/+; UAS-_upd_RNAi/+ | 21.4 (9/42) | 24.83 ± 0.16 | 0.038 ± 0.005 |
| _miR-279_-GAL4/+ | 100 (17/17) | 23.53 ± 0.07 | 0.096 ± 0.008 |
| _miR-279_-GAL4/+; UAS-_upd_RNAi/+ | 42.9 (15/35) | 24.04 ± 0.1 | 0.07 ± 0.008 |
| _Pdf_-GAL4/+; _Pdf_-GAL4/+ | 100 (10/10) | 24 ± 0.08 | 0.114 ± 0.012 |
| _Pdf_-GAL4/+; _Pdf_-GAL4/UAS-_upd_RNAi | 100 (17/17) | 24.2 ± 0.05 | 0.07 ± 0.008 |
| TUG/+ | 100 (18/18) | 24.06 ± 0.08 | 0.124 ± 0.012 |
| TUG/+; UAS-_upd_RNAi/+ | 86.7 (13/15) | 24.68 ± 0.08 | 0.057 ± 0.007 |
| TUG/TUG; UAS-_upd_RNAi/+ | 42.1 (8/19) | 26.12 ± 0.25 | 0.044 ± 0.006 |
| miR-279 ex117-1/ex117-1 | 33.3 (7/21)a | 23.79 ± 0.28a | 0.031 ± 0.008a |
| TUG/+; UAS-_upd_RNAi, _miR-279_ex117-1/ex117-1 | 91.7 (11/12) | 24.57 ± 0.17 | 0.051 ± 0.012 |
| UAS-upd/+ | 100 (18/18) | 23.57 ± 0.05 | 0.145 ± 0.009 |
| _tim_-GAL4/UAS-upd | 0 (0/24) | - | - |
| UAS-upd/+; _elav_GS/_elav_GS (+EtOH) | 87.5 (14/16) | 23.63 ± 0.09 | 0.056 ± 0.009 |
| UAS-upd/+; _elav_GS/_elav_GS(+RU486) | 33.3 (5/15) | 23.45 ± 0.11 | 0.03 ± 0.011 |
| _dome_-GAL4/+ | 75 (12/16) | 23.8 ± 0.12 | 0.031 ±0.008 |
| UAS-_dome_ΔCYT/+ | 100 (17/17) | 23.28 ± 0.07 | 0.142 ± 0.015 |
| _dome_-GAL4/+; UAS-_dome_ΔCYT/+ | 15.4 (2/13) | 24.09 ± 0.09 | 0.03 ± 0.009 |
| _tim_-GAL4/UAS-_dome_ΔCYT | 100 (16/16) | 23.83 ± 0.05 | 0.192 ± 0.016 |
| _w_1118 (sibling) | 97.1 (34/35) | 23.57 ± 0.05 | 0.088 ± 0.007 |
| _w_1118, _hop_25/Y | 32 (8/25) | 23.62 ± 0.18 | 0.038 ± 0.005 |
| UAS-_hop_Tuml/+; _elav_GS/+ (+EtOH) | 100 (10/10) | 23.6 ± 0.09 | 0.085 ± 0.012 |
| UAS-_hop_Tuml/+; _elav_GS/+ (+RU486) | 16.7 (2/12) | 23 ± 0.25 | 0.018 ± 0.002 |
| _upd_-GAL4/Y | 100 (19/19) | 23.7 ± 0.03 | 0.095 ± 0.006 |
| UAS-NachBac/+ | 100 (15/15) | 23.6 ± 0.06 | 0.08 ± 0.009 |
| _upd_-GAL4/Y; UAS-NachBac/+ | 26.7 (4/15) | 23.77 ± 0.22 | 0.034 ± 0.009 |
| UAS-dTrpA1/+ (21°C) | 100 (16/16) | 23.75 ± 0.07 | 0.05 ± 0.005 |
| UAS-dTrpA1/+ (28°C) | 100 (16/16) | 23.69 ± 0.06 | 0.075 ± 0.009 |
| _upd_-GAL4/Y; UAS-dTrpA1/+ (21°C) | 86.7 (13/15) | 23.61 ± 0.07 | 0.034 ± 0.004 |
| _upd_-GAL4/Y; UAS-dTrpA1/+ (28°C) | 35.7 (5/14) | 23.28 ± 0.15 | 0.021 ± 0.003 |
| UAS-_shi_TS/+ ; UAS-_shi_TS/+ (21°C) | 93.8 (15/16) | 24.01 ± 0.12 | 0.033 ± 0.004 |
| UAS-_shi_TS/+ ; UAS-_shi_TS/+ (29°C) | 100 (15/15) | 23.31 ± 0.13 | 0.062 ± 0.008 |
| _upd_-GAL4/Y; UAS-_shi_TS/+; UAS-_shi_TS/+ (21°C) | 93.3 (14/15) | 23.96 ± 0.09 | 0.047 ± 0.007 |
| _upd_-GAL4/Y; UAS-_shi_TS/+; UAS-_shi_TS/+ (29°C) | 100 (15/15) | 24.63 ± 0.09 | 0.066 ± 0.01 |
Manipulations of the JAK/STAT Pathway Affect Behavioral Rhythms
The miR-279 target we identified, upd, is one of the three cytokine ligands (i.e. upd, upd2, and upd3) that activate the JAK/STAT signaling cascade in Drosophila (Arbouzova and Zeidler, 2006; Harrison et al., 1998). Compared to its mammalian counterpart, the canonical JAK/STAT pathway in Drosophila is relatively simple with only a few key components. Ligand binding activates a transmembrane receptor, Domeless (DOME) that recruits the Janus kinase protein, Hopscotch (HOP), to its intracellular domain. This allows HOP to phosphorylate itself, and the receptor DOME, and a cytoplasmic protein, signal transducers and activators of transcription (STAT92E). Upon phosphorylation by HOP, STAT92E dimerizes and translocates to the nucleus to regulate transcription (Arbouzova and Zeidler, 2006).
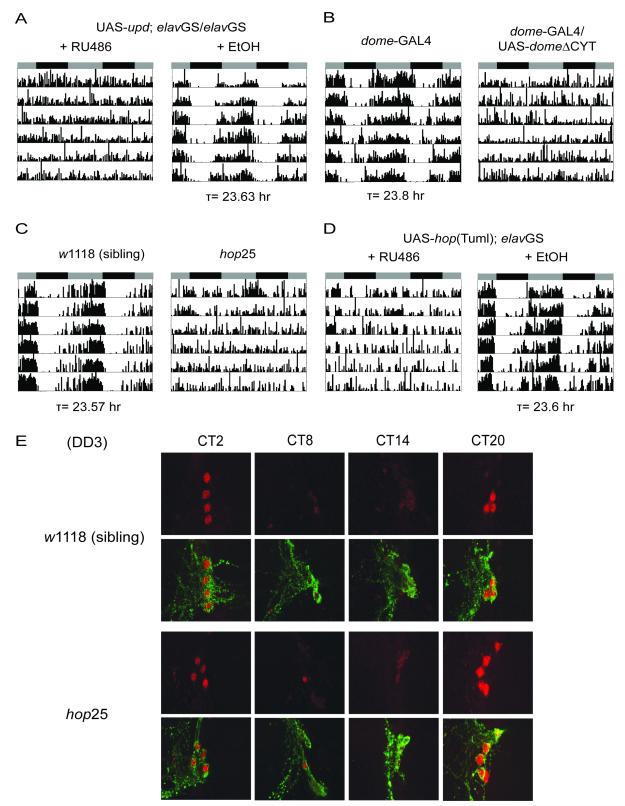
Given that upd is part of the JAK/STAT pathway, we asked if manipulating components of the JAK/STAT would alter locomotor activity rhythms in flies. As noted above, knockdown of upd in clock neurons with the strong _tim_-GAL4 driver disrupted rhythms (Figure 3A, Table 2). A slow breakdown of rhythms was also observed with the miR-279 driver (Figure 3A, Table 2). Since _tim_-GAL4 is expressed at higher levels but may show some ectopic expression (data not shown), we also tested two copies of the TUG driver, which likely better represents the circadian clock network. This manipulation also disrupted behavioral rhythms and produced overall high activity in wild-type flies (Table 2; notice higher activity in Figure 3E even with one copy of TUG). The _upd_-RNAi transgene yielded no phenotype with the _Pdf_-GAL4 driver, even when two copies were used (Figure 3A, Table 2), indicating that upd function is not restricted to Pdf neurons. Together, these data suggest that appropriate expression of upd is required for normal rhythms; it needs to be down-regulated, but not completely abolished by miR-279, which is also consistent with the results of the in vitro luciferase reporter assay (Figure 3C). This conclusion is further supported by our findings that either constant high levels of upd in tim neurons or induction of upd in adults disrupts activity rhythms (Table 2, Figure 4A). Over-expression of upd in miR-279 neurons throughout development resulted in lethality (data not shown).
Figure 4.
The JAK/STAT signaling pathway is required for locomotor activity rhythms. (A) Panneuronal induction of upd in adulthood leads to arrhythmia. Flies were reared on regular food. 500μM RU486 or ethanol (control) was administered in the food from the time of entrainment. (B) Blocking JAK/STAT signaling with a dominant-negative form of the receptor domeless (_dome_ΔCYT) leads to arrhythmia. The _dome_-GAL4 driver (Ghiglione et al., 2002) causes lethality in males, and thus only females were tested. (C) A hypomorphic mutation of JAK kinase hopscotch (_hop_25) causes loss of activity rhythms. The _hop_25 allele is a Q246K point mutation of the hop gene (Luo et al., 1999). (D) Over-expression of a constitutively active form of hop (hopTuml) in the adult nervous system disrupts activity rhythms. The gain-of-function mutation, hopTuml, is a G341E single amino acid substitution in the hop gene (Harrison et al., 1995; Luo et al., 1995). (E) The _hop_25 mutants show normal PER cycling and PDF expression in central pacemaker neurons. Brains were dissected and stained with PER (red) and PDF (green) antibodies on the third day of DD at the indicated circadian times (CT). See Also Figure S4.
We next examined whether blocking signal transduction downstream of upd affects behavioral rhythms. To investigate the role of DOME, we used a dominant-negative form of this receptor, DOMEΔCYT, which lacks the intracellular domain and the sites for HOP binding. The mutant receptor contains the extracellular and transmembrane portion of the protein, and thus titrates the ligand and acts as a signaling antagonist (Brown et al., 2001). Expression of UAS-_dome_ΔCYT under the control of a dome promoter enhancer trap GAL4 (Ghiglione et al., 2002), but not _tim_-GAL4, dramatically reduced the percentage of rhythmic flies compared to controls (Table 2, Figure 4B), suggesting that normal activity rhythms require a ligand-responsive DOME receptor in non-tim neurons.
We further determined whether the Janus kinase HOP modulates circadian behaviors. Although complete amorphs of hop are lethal, a small percentage of flies carrying a severe hypomorphic allele, _hop_25, emerge as hemizygous males (Perrimon and Mahowald, 1986) and survive up to 2 weeks in an isogenic _w_1118 background. We found that ~68% of the _hop_25 males lost activity rhythms in constant darkness, whereas only 3% of wild-type sibling controls were arrhythmic (Figure 4C, Table 2). We also tested a dominant gain-of-function allele, _hopscotch_Tumorous-lethal (_hop_Tuml), which produces a constitutively active form of HOP and causes hyperactivity of the JAK/STAT pathway (Harrison et al., 1995; Luo et al., 1995). Expression of UAS-_hop_Tuml by _elav_GS resulted in an acute disruption of locomotor activity rhythms in adults (Figure 4D, Table 2). To determine whether hop also affects circadian output pathways downstream of the central clock, we assayed PDF expression and protein oscillations of PER in small LNvs of _hop_25 adult flies. Entrained _hop_25 and sibling control flies were subject to immunocytochemistry of brain whole mounts on the third day in DD. As shown in Figure 4E, mutant flies exhibit PDF expression and daily PER cycling in sLNvs that are similar to those of wild-type controls. All these data are consistent with a major role for JAK/STAT signaling in circadian outputs that drive rhythmic behaviors.
Modulation of _upd_-Expressing Neurons Alters Behavioral Rhythms
To determine whether UPD secreting neurons are important for circadian behaviors, we utilized an upd enhancer trap GAL4 driver, upd (E132)-GAL4 (Halder et al., 1995), and tested whether silencing or activating _upd_-expressing neurons alters activity rhythms. Due to a critical role of upd and JAK/STAT signaling in fly development (Harrison et al., 1998; Hou et al., 1996; Perrimon and Mahowald, 1986), inactivation of UPD-secreting cells by _upd_-GAL4-controlled expression of an inward rectifying potassium channel, Kir2.1 (Baines et al., 2001), caused lethality (data not shown). To conditionally suppress upd neurons at the adult stage, we expressed a temperature-sensitive synaptic blocker, _shibire_TS (_shi_TS), under the control of _upd_-GAL4. The _shi_TS allele is defective in synaptic vesicle recycling at restrictive temperatures (>29°C), and leads to a rapid and reversible inhibition of synaptic transmission (Koenig et al., 1983). Upon a shift to 29°C, the period of the activity rhythm was lengthened ~0.7 hrs compared to that of control flies, and this phenotype reverted to normal after flies were transferred back to permissive temperature (21°C) (Figure S4A, Table 2). The long period phenotype produced by blocking _upd_-expressing neurons is consistent with the effects of miR-279 over-expression, which lowers the level of upd, in the small percentage of rhythmic flies (Figure 1D, Table 1). To confirm the importance of _upd_-expressing neurons in regulating circadian rhythms, we then activated these neurons. Under the control of _upd_-GAL4, the expression of a sodium channel, NaChBac, which depolarizes neurons and increases their excitability (Ren et al., 2001), abolished free-running rhythms (Table 2). In addition, acute excitation of upd neurons in adult flies by activating a temperature-gated cation channel dTrpA1 at 28°C (Hamada et al., 2008) also disrupted activity rhythms (Figure S4B, Table 2). The effect of dTrpA1 was reversible, with normal rhythms restored at the permissive temperature of 21°C. We conclude that _upd_-expressing cells are necessary for normal circadian locomotor rhythms.
JAK/STAT Signaling Cycles under the Control of the Circadian Clock
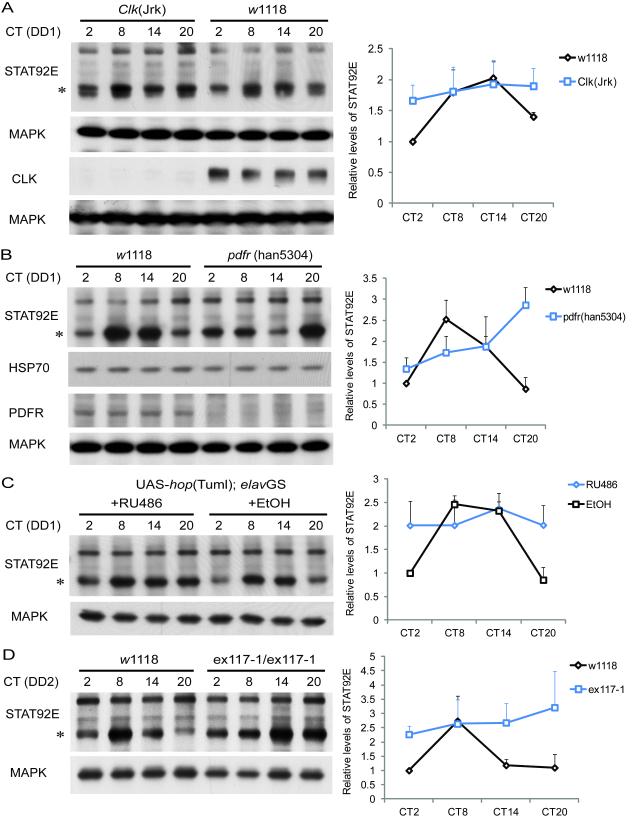
To test the hypothesis that JAK/STAT signaling is an output modulated by the circadian clock, we examined the expression profile of JAK/STAT components over the course of a circadian cycle in DD. Western blot assays of fly brain extracts were performed using antibodies specific for UPD, HOP and STAT92E. Although no oscillations were detectable for the major bands corresponding to UPD and HOP proteins, a higher band, which may correspond to phosphorylated HOP, was expressed cyclically (Figure S5A). We also found that the fastest migrating form of STAT92E, which appears to be a doublet, was expressed cyclically on the first day in DD (Figure 5A-C, S5B), with peak protein levels in the late day/early evening. STAT92E oscillations were dampened in clock gene mutants, more so in _Clk_Jrk flies (Figure 5A) than in per01 (Figure S5B). The cycling of STAT92E persisted on the second day of DD (Figure 5D), further supporting that it is under circadian control. STAT92E cycling was delayed in a null mutant of the PDF receptor, _pdfr_han5304 (Hyun et al., 2005), suggesting that it is regulated by PDF signaling (Figure 5B). To determine if JAK/STAT signaling and miR-279 contribute to oscillations of STAT92E, we assayed STAT92E cycling in flies over-expressing _hop_Tuml and also in the miR-279 mutants. In both genotypes, the cycling of STAT92E was dampened, such that trough levels were increased (Figure 5C and 5D).
Figure 5.
JAK/STAT signaling is downstream of the central clock and the PDF receptor (PDFR). Flies of the indicated genotypes were collected at different circadian times (CT) on the first (DD1) or second (DD2) day of DD. Protein extracts of brains were subjected to western blot analysis using antibodies specific for STAT92E and loading controls (MAPK or HSP70). Multiple bands are detected for STAT92E, of which the fastest mobility forms are expressed cyclically (indicated with asterisks). Each experiment was conducted three times and quantified STAT92E and loading control levels were normalized to those at CT2 of wild-type controls in each gel. The quantification curves in each panel were plotted as the average ± standard error of the mean (SEM) of three independent western blots. (A and B) STAT92E protein levels cycle in fly brains and are regulated by the circadian clock and PDFR. STAT92E levels at the trough (CT2 and CT20) are significantly lower than at the peak (CT8 and CT14) in _w_1118 control flies (P<0.01), but not so in _Clk_Jrk mutants (P=0.39) by Student’s t-test (panel A). Although random fluctuations were observed in _Clk_Jrk mutants, a 24 h oscillation was not detected in any of multiple experiments. One-way ANOVA detects a significant cycle in both _w_1118 and _pdfr_han5304 mutants (panel B), and two-way ANOVA indicates that the oscillation is altered in _pdfr_han5304 (P<0.05). The _Clk_Jrk and _pdfr_han5304 mutations were confirmed by probing with CLK and PDFR antibodies using fly head extracts. (C) The cycling of STAT92E is disrupted when _hop_Tuml is over-expressed in the adult nervous system. 500μM RU486 or ethanol (control) was administered in the food to induce _elav_GS from the time of entrainment. One-way ANOVA detects a cycle in the ethanol control (P<0.05), but not in RU486 treated flies (P=0.93). (D) STAT92E protein levels continue to cycle on DD2 in wild type flies but the cycling is dampened in miR-279 mutants. STAT92E levels at the peak (CT8) are significantly higher than at the trough (CT2, CT14 and CT20) in wild-type flies (P<0.01), but not so in ex117-1 flies (P=0.46) by Student’s t-test. The daily expression of STAT92E in ex117-1 flies is significantly different from that in _w_1118 flies (P<0.05, by two-way ANOVA). See Also Figure S5.
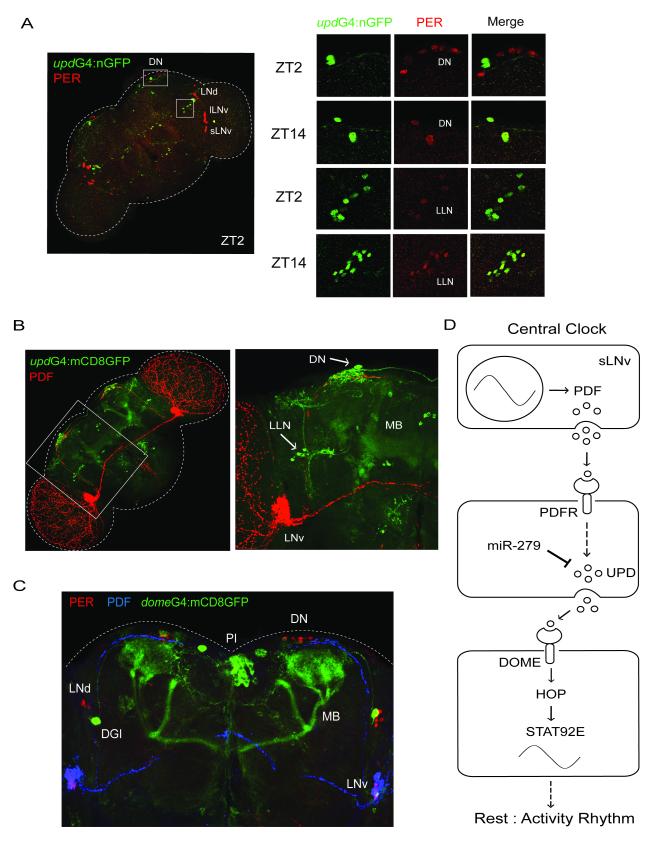
Since JAK/STAT signaling appears to be downstream of the central clock, we sought to determine if _upd_-expressing cells receive input from clock cells. We first labeled _upd_-expressing neurons with nuclear-targeted GFP (nGFP) by crossing UAS-nGFP to _upd_-GAL4, and found that upd is broadly expressed in the adult fly brain. The expression pattern includes cells within two clusters of PER-positive neurons, dorsal neurons (DNs) and some other neurons that we are calling lateral located neurons (LLNs) based upon their position (Figure 6A). Interestingly, the neurons in which upd and PER expression overlap show shifted-phase PER cycling under LD conditions (i.e. protein levels are high in the early night, and low in the early morning) (Figure 6A). We next expressed a cell membrane-targeted GFP (mCD8GFP) under the control of _upd_-GAL4, and found that the PDF-containing dorsal projection from the small LNvs lies within clusters of _upd_-expressing neurites (Figure 6B). Since these projections are the ones typically associated with rest:activity output from small LNvs (Helfrich-Forster et al., 2000; Williams et al., 2001), these data are consistent with a role for _upd_-expressing cells in this output. Using a Denmark reporter (Nicolai et al., 2010) we also confirmed that the upd neurites that contact the PDF projections are dendrites (Figure S6).
Figure 6.
JAK/STAT signaling and miR-279 are in a circadian output circuit. (A) The expression pattern of upd includes dorsal neurons (DNs) and lateral located neurons (LLNs), which show shifted-phase expression of PER. Brains of _upd_-GAL4: nGFP flies were dissected and stained with GFP (green) and PER (red) antibodies at indicated Zeitgeber times (ZT) on the 4th day in LD. PER protein levels are high in the early night in a subset of DNs and LLNs, but high in the early morning in other clock neurons. (B) PDF-containing dorsal projections from the central clock are in the vicinity of _upd_-expressing neurons. A membrane-targeted mCD8GFP was expressed under the control of _upd_-GAL4. Brains were dissected and stained with GFP (green) and PDF (red) antibodies. An enlarged image is shown on the right of the panel. _upd_-expressing neurons are indicated with white arrows. (C) Dome expresses in the mushroom bodies (MBs), pars intercerebralis (PI), and dorsal giant interneurons (DGIs). A mCD8GFP was expressed under the control of _dome_-GAL4. Brains were dissected at ZT2 on the 4th day in LD and stained with GFP (green), PER (red) and PDF (blue) antibodies. (D) Model depicting the role of miR-279 and JAK/STAT signaling in a circadian behavioral output. We propose that the central clock affects cyclic secretion of the UPD protein from cells that act downstream of PDF signaling. The mRNA levels of upd in these neurons are negatively modulated by miR-279. UPD may rhythmically activate the DOME receptor in _dome_-expressing cells, which would lead to daily oscillations of JAK/STAT activity and therefore of STAT92E levels (see Discussion). Rhythmic activity of this pathway is likely required for rest:activity rhythm. See Also Figure S6.
Finally, to visualize neurons receiving signals from UPD-secreting neurons, we expressed a mCD8GFP under the control of the _dome_-GAL4 driver. As shown in Figure 6C, the expression pattern of dome does not overlap with the major pacemaker neurons such as the small and large LNvs, LNds, and DN1s (Nitabach and Taghert, 2008), which is consistent with the lack of a phenotype when UAS-_dome_ΔCYT is expressed by _tim_-GAL4 (Table 2). However, the GFP signal is enriched in other structures such as the Kenyon cell bodies and the αand β lobes of the mushroom bodies (MBs), the pars intercerebralis (PI), and a bilateral pair of dorsal giant interneurons (DGIs). Notably, the Kenyon cell bodies of the MBs are adjacent to the dorsal neurons (DNs) and the PDF-containing dorsal projections of small LNvs (Figure 6C).
Discussion
We describe here a miRNA that affects output from the circadian clock. Although it is widely believed that microRNAs play a critical role in most biological processes, few have been specifically implicated in circadian rhythms. In mammals, miR-219 is involved in central clock function and miR-132 in light input to the central clock (Cheng et al., 2007). The Drosophila miRNA, bantam, affects free-running circadian rhythms by targeting the Clock gene (Kadener et al., 2009). Although mutants have not been identified for any of these, the data thus far do not indicate an effect on circadian output. We show that loss or over-expression of miR-279 disrupts rest:activity rhythms, suggesting that it is required to maintain levels of one or more target genes within a specific range. Given that the phenotype produced by inducibly over-expressing miR-279 in adults is weaker than that seen when it is expressed constitutively, developmental effects may also contribute to the circadian phenotype.
Consistent with the effects of manipulating miR-279 levels, up or down regulation of the relevant target gene we identified, upd, also impairs rhythmicity. We know that levels of specific clock molecules are critical for regulating circadian rhythms. Thus, in case of per and tim, not only does loss of these genes cause arrhythmicity, but also over-expression (Yang and Sehgal, 2001), the thinking being that over-expression dampens cycles of protein expression/function that are essential for overt rhythms. However, less is known about over-expression phenotypes of molecules that function solely in output; PDF over-expression causes a phenotype (Helfrich-Forster et al., 2000), but PDF not only transmits output signals, but also synchronizes clock cells (Lin et al., 2004; Stoleru et al., 2005). In Neurofibromatosis 1 (NF1) mutants (further discussed below), elevated MAPK kinase activity contributes to the arrhythmic phenotype, but independent effects of increasing MAPK or NF1 levels were not examined (Williams et al., 2001). Our data here indicate that levels of molecules in the output pathway also have to be appropriately regulated, and this can be accomplished by microRNAs.
UPD signals through the JAK/STAT pathway and we show here that manipulations of other components of this pathway also disrupt behavioral rhythms. In addition, the expression of STAT92E is cyclic. Given the critical need for appropriate expression levels of upd, we speculate that rhythms in STAT92E derive from circadian regulation at the level of upd. While levels of UPD do not appear to cycle in whole brains (Figure S5A; cycling in specific cells cannot be excluded), its release may be cyclic and changes in expression levels could impact the rhythm of release. Cyclic release of UDP would lead to rhythmic activation of DOME and thereby STAT92E (Figure 6D). Since STAT92E regulates its own expression through feedback (Arbouzova and Zeidler, 2006), rhythmic STAT92E activity is expected to result in cyclic expression, as reported here. PDF is also thought to be released cyclically (Park et al., 2000) and it regulates cycling of STAT92E (Figure 5B). This suggests control of the JAK/STAT pathway by central clock cells, which is supported by the proximity of PDF projections and upd neurons (Figure 6B and S6). However, rhythmic regulation of the JAK/STAT pathway could be reinforced by clocks in other cells e.g. those in which PER and _upd_-GAL4 are co-expressed. In addition, miR-279 may regulate more than just upd in the JAK/STAT pathway (Yoon et al., 2011).
Our data are also the first to demonstrate a role of the JAK/STAT pathway in circadian rhythms. To date, most studies of signaling in the circadian system have focused on cAMP or mitogen activated protein kinase (MAPK) pathways, which may function in all aspects of circadian timekeeping (input to the clock, clock function and output) (Allada and Chung, 2010). However, the entire process involved in generating a rhythm is quite complex, especially when the output is rhythmic behavior of the animal. Thus, a network of molecules and cells is likely required, although little is known about these. Indeed, the output pathway of the circadian system remains the most poorly understood aspect of the system in every organism examined. While the past decade or so has seen major advances in our understanding of clock mechanisms and mechanisms that entrain the clock to light, mechanisms that transmit signals from the clock and produce overt rhythms, in particular behavioral rhythms, remain a mystery. Advances in this area, including those reported here, will provide a framework upon which whole circuits can be assembled.
In the Drosophila circadian system, one of the points of MAPK action is downstream of the NF1 gene. NF1 is required downstream of the clock for rhythmic rest:activity and its effects on rhythms are mediated by the RAS/MAPK pathway (Williams et al., 2001). Interestingly, NF1 affects STAT3 activity in mammalian glia and neural stem cells, such that STAT3 signaling is hyperactivated in NF1-deficient cells (Banerjee et al., 2010). While we do not know of a connection between NF1 and JAK/STAT in Drosophila, it is an intriguing possibility that the two are linked in the circadian system. Like STAT92E, MAPK activity cycles in parts of the fly brain (Williams et al., 2001).
Upd and its receptor, Dome, are expressed widely in the fly brain. However, the identity of most positive neurons must await further characterization of fly brain circuitry. It is interesting though that upd is co-expressed with PER in cells where PER cycles with a shifted phase under LD conditions, i.e. with a phase different to that seen in all other neurons, including the central clock cells. Two sites of upd and PER co-expresssion are in some of the DNs and in neurons we have termed LLNs because of their lateral location. The identity of these PER-positive neurons is unclear at this time. The dorsal neurons are unlikely to be the DN2s because although PER expression in DN2s is anti-phase in larvae, it cycles with a normal phase in adult flies (Kaneko et al., 1997). Likewise, the LLNs are probably not lateral posterior neurons (LPNs), located in the same region, because those apparently are three in number in each hemisphere and show normal PER cycling (Shafer et al., 2006). Based upon their position, the dorsal neurons (DNs) that express upd could represent a subset of the dorsal neuron cluster 1 (DN1s). Regardless of the precise cell types in which they are expressed, upd and dome expression provide tools to map the cellular circuitry of the output pathway that drives rhythmic rest:activity.
It is likely that many other microRNAs are involved in circadian rhythms. Since identification of such miRNAs will be difficult through traditional genetic screens, other approaches will have to be devised. Bioinformatics to identify miRNAs that target clock genes is obviously a viable method, but, as noted here, this approach typically identifies many possible targets for each miRNA, only some of which are biologically relevant. Thus, the computational analysis will have to be followed by many additional tests. While this is still doable, another caveat of this approach is that it will restrict analysis to miRNAs that target clock genes. Yet another approach is to identify miRNAs that are expressed cyclically. This was done recently in Drosophila and it identified some miRNAs, which may turn out to be important regulators of circadian rhythms (Yang et al., 2008).
Experimental Procedures
Fly Strains and Behavioral Assays
Transgenic fly lines carrying either the UAS-miR-279 (S) or UAS-miR-279 (L) construct were generated by site-specific PhiC31 Integration System (Rainbow Transgenics) using the attP landing site 2A on the X chromosome (Bischof et al., 2007). Fly lines obtained from other sources were outcrossed 5-7 times into an isogenic _w_1118 (iso31) strain. The EP insertion in NE95-11-24 was mapped by using inverse PCR and a cycle sequencing of P element insertions protocol (E. Jay Rehm, Berkeley Drosophila Genome Project). Locomotor activity rhythms were measured as previously described (Williams et al. 2001). Refer to the Extended Experimental Procedures for details.
Quantitative Real-time PCR
Fly heads or dissected fly brains were collected on dry ice and stored at -80°C until use. Total RNA, including miRNA, was purified with miRNeasy Mini kit (Qiagen). Reverse transcription and real-time PCR of miR-279 and 2s rRNA (or U27) were performed with TaqMan MicroRNA Assays designed for detecting mature miRNAs (Applied Biosystems). RT-PCR was performed on an ABI prism 7100 (Applied Biosystems). Refer to the Extended Experimental Procedures for details of other q-PCR assays.
Whole-mount Brain Immunocytochemistry
Adult fly brains were collected at indicated time points and fixed with 4% para-formaldehyde (PFA) in PBS. After three 15 min washes with 0.3% Triton X-100 in PBS, brains were blocked with 5% normal donkey serum. Samples were then incubated overnight at 4°C with primary antibodies. After three 20 min washes, brains were incubated with secondary donkey antibodies (Jackson ImmunoResearch Laboratories, 1: 1000) for 2 h at room temperature, followed by extensive washes. Samples were imaged using a Leica TCS SP5 confocal microscope. Six to ten fly brains were examined for each time point. Representative images are shown. Refer to the Extended Experimental Procedures for details.
Western Blot Analysis
Ten to fourteen flies were exposed to microwave radiation for 2-3 min at indicated time points in DD. The eyes and fat bodies were carefully removed and dissected fly brains were collected on dry ice. The brain samples were immediately subjected to protein extraction and western blots as previously described (Sathyanarayanan et al. 2004). The STAT92E and loading control bands were quantified using ImageJ software as previously described (Kojima et al., 2010). Western blot assays were repeated three times with similar results. Refer to the Extended Experimental Procedures for details.
Luciferase Reporter Assay in S2 Cells
The full length 3′UTR of upd and a ~600bp coding region of miR-279 were amplified by PCR using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen). The upd 3′ UTR was cloned into a pAc5.1-firefly luciferase-V5-His vector and the miR-279 coding region was cloned into a pAC5.1-V5-His vector (Invitrogen). The miR-279 coding sequence was mutated using the Quickchange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). 10ng pAc-firefly luc-upd 3′ UTR, 90ng pAc-miR-279 (or empty vector control, or pAc-miR-279 mutant) and 50ng pAc-Renilla luc (transfection control) were cotransfected into S2 cells with Effectene Transfection Reagent (Qiagen). Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Refer to the Extended Experimental Procedures for details.
Supplementary Material
01
02
Acknowledgements
We are very grateful to Zhaohai Yang and Zhifeng Yue for generating the NE95-11-24 fly line. We thank L. Zipursky, B. Edgar, D. Bilder, S. Hou, G. Baeg, T. Cline, W. Odenwald, I. Rebay, J. Blau and M. Rosbash for providing miR-279, JAK/STAT and other fly stocks, S. Hou, D. Harrison, P. Taghert, and P. Hardin for STAT92E, UPD, HOP, PDFR, and CLK antibodies, E. Izaurralde and K. Basler for pAc5.1-F. Luc and pUAST_attB_ vectors, X. Zheng and D. Chen for developing PER antibodies, W. Joiner, A. Crocker and S. Kumar for outcrossing some mutants and GAL4 lines and other members of the laboratory for useful discussions. The work was supported by NIH grant 2R01NS048471. A.S. is an Investigator in the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–2616. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Byrd JN, Gianino SM, Harpstrite SE, Rodriguez FJ, Tuskan RG, Reilly KM, Piwnica-Worms DR, Gutmann DH. The neurofibromatosis type 1 tumor suppressor controls cell growth by regulating signal transducer and activator of transcription-3 activity in vitro and in vivo. Cancer Res. 2010;70:1356–1366. doi: 10.1158/0008-5472.CAN-09-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. doi: 10.1016/s0092-8674(00)81554-8. [DOI] [PubMed] [Google Scholar]
- Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–1705. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, Gunning D, Lai EC, Zipursky SL. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science. 2008;319:1256–1260. doi: 10.1126/science.1149483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HY, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, et al. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol. 2009;19:461–470. doi: 10.1016/j.conb.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Eskin A. Identification and physiology of circadian pacemakers. Introduction. Fed Proc. 1979;38:2570–2572. [PubMed] [Google Scholar]
- Ghiglione C, Devergne O, Georgenthum E, Carballes F, Medioni C, Cerezo D, Noselli S. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development. 2002;129:5437–5447. doi: 10.1242/dev.00116. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. The circadian system in higher plants. Annu Rev Plant Biol. 2009;60:357–377. doi: 10.1146/annurev.arplant.043008.092054. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–3263. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Tauber M, Park JH, Muhlig-Versen M, Schneuwly S, Hofbauer A. Ectopic expression of the neuropeptide pigment-dispersing factor alters behavioral rhythms in Drosophila melanogaster. Journal of Neuroscience. 2000;20:3339–3353. doi: 10.1523/JNEUROSCI.20-09-03339.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou XS, Melnick MB, Perrimon N. Marelle acts downstream of the Drosophila HOP/JAK kinase and encodes a protein similar to the mammalian STATs. Cell. 1996;84:411–419. doi: 10.1016/s0092-8674(00)81286-6. [DOI] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR Han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Sugino K, Horwich MD, Weissbein U, Nawathean P, Vagin VV, Zamore PD, Nelson SB, Rosbash M. A role for microRNAs in the Drosophila circadian clock. Genes Dev. 2009;23:2179–2191. doi: 10.1101/gad.1819509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Helfrich-Forster C, Hall JC. Spatial and temporal expression of the period and timeless genes in the developing nervous system of Drosophila: newly identified pacemaker candidates and novel features of clock gene product cycling. Journal of Neuroscience. 1997;17:6745–6760. doi: 10.1523/JNEUROSCI.17-17-06745.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JH, Saito K, Ikeda K. Reversible control of synaptic transmission in a single gene mutant of Drosophila melanogaster. J Cell Biol. 1983;96:1517–1522. doi: 10.1083/jcb.96.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Gatfield D, Esau CC, Green CB. MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One. 2010;5:e11264. doi: 10.1371/journal.pone.0011264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. J Neurosci. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H, Asha H, Kockel L, Parke T, Mlodzik M, Dearolf CR. The Drosophila Jak kinase hopscotch is required for multiple developmental processes in the eye. Dev Biol. 1999;213:432–441. doi: 10.1006/dbio.1999.9390. [DOI] [PubMed] [Google Scholar]
- Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14:1412–1420. doi: 10.1002/j.1460-2075.1995.tb07127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004:pl6. doi: 10.1126/stke.2202004pl6. [DOI] [PubMed] [Google Scholar]
- Nicolai LJ, Ramaekers A, Raemaekers T, Drozdzecki A, Mauss AS, Yan J, Landgraf M, Annaert W, Hassan BA. Genetically encoded dendritic marker sheds light on neuronal connectivity in Drosophila. Proc Natl Acad Sci U S A. 2010;107:20553–20558. doi: 10.1073/pnas.1010198107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH. Organization of the Drosophila circadian control circuit. Curr Biol. 2008;18:R84–93. doi: 10.1016/j.cub.2007.11.061. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Forster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc Natl Acad Sci U S A. 2000;97:3608–3613. doi: 10.1073/pnas.070036197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N, Mahowald AP. l(1)hopscotch, A larval-pupal zygotic lethal with a specific maternal effect on segmentation in Drosophila. Dev Biol. 1986;118:28–41. doi: 10.1016/0012-1606(86)90070-9. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyanarayanan S, Zheng X, Xiao R, Sehgal A. Post-translational modification of Drosophila PERIOD protein by Protein Phosphatase 2A. Cell. 2004;116:603–615. doi: 10.1016/s0092-8674(04)00128-x. [DOI] [PubMed] [Google Scholar]
- Shafer OT, Helfrich-Forster C, Renn SC, Taghert PH. Reevaluation of Drosophila melanogaster’s neuronal circadian pacemakers reveals new neuronal classes. J Comp Neurol. 2006;498:180–193. doi: 10.1002/cne.21021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Ko ML, Ko GY. Rhythmic expression of microRNA-26a regulates the L-type voltage-gated calcium channel alpha1C subunit in chicken cone photoreceptors. J Biol Chem. 2009;284:25791–25803. doi: 10.1074/jbc.M109.033993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, Rosbash M. A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature. 2005;438:238–242. doi: 10.1038/nature04192. [DOI] [PubMed] [Google Scholar]
- Vitalini MW, de Paula RM, Park WD, Bell-Pedersen D. The rhythms of life: circadian output pathways in Neurospora. J Biol Rhythms. 2006;21:432–444. doi: 10.1177/0748730406294396. [DOI] [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Yang M, Lee JE, Padgett RW, Edery I. Circadian regulation of a limited set of conserved microRNAs in Drosophila. BMC Genomics. 2008;9:83. doi: 10.1186/1471-2164-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
- Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat Cell Biol. 2011;13:1062–1069. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Sehgal A. Probing the relative importance of molecular oscillations in the circadian clock. Genetics. 2008;178:1147–1155. doi: 10.1534/genetics.107.088658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Yang Z, Yue Z, Alvarez JD, Sehgal A. FOXO and insulin signaling regulate sensitivity of the circadian clock to oxidative stress. Proc Natl Acad Sci U S A. 2007;104:15899–15904. doi: 10.1073/pnas.0701599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02