Metagenomic Analysis of Viruses from Bat Fecal Samples Reveals Many Novel Viruses in Insectivorous Bats in China (original) (raw)
Abstract
Increasing data indicate that bats harbor diverse viruses, some of which cause severe human diseases. In this study, sequence-independent amplification and high-throughput sequencing (Solexa) were applied to the metagenomic analysis of viruses in bat fecal samples collected from 6 locations in China. A total of 8,746,417 reads with a length of 306,124,595 bp were obtained. Among these reads, 13,541 (0.15%) had similarity to phage sequences and 9,170 (0.1%) had similarity to eukaryotic virus sequences. A total of 129 assembled contigs (>100 nucleotides) were constructed and compared with GenBank: 32 contigs were related to phages, and 97 were related to eukaryotic viruses. The most frequent reads and contigs related to eukaryotic viruses were homologous to densoviruses, dicistroviruses, coronaviruses, parvoviruses, and tobamoviruses, a range that includes viruses from invertebrates, vertebrates, and plants. Most of the contigs had low identities to known viral genomic or protein sequences, suggesting that a large number of novel and genetically diverse insect viruses as well as putative mammalian viruses are transmitted by bats in China. This study provides the first preliminary understanding of the virome of some bat populations in China, which may guide the discovery and isolation of novel viruses in the future.
INTRODUCTION
Bats, the only flying mammal and the second most diverse group of mammals, are natural reservoirs of many emerging viruses (7, 38). Some bat viruses cause severe human diseases, such as lyssavirus, Hendra virus, Nipah virus, Ebola virus, and Marburg virus (16, 23, 29, 41, 47). Most of the known bat viruses have been discovered in apparently healthy bats (10, 14, 19, 20, 26, 34, 49). When bats are experimentally infected with henipavirus or the rabies virus, the bats shed the virus but do not produce any clinical syndrome like those observed in other animals and humans (1, 17, 30, 40). This phenomenon may be due to the adaption of viruses to their host species, preinfection with related nonpathogenic viruses, or some unique characteristics of the bat immune system to viral infection. Considering the species richness and wide geographic distribution of bats, bats may be a source of many additional unknown viruses.
The techniques for isolating viruses are straightforward but are not always successful because the majority of viruses are unable to grow in cell culture due to a lack of susceptible cell lines, a low titer of virus, or toxicity of environmental samples. PCR assays are sequence-dependent and extremely sensitive and rapid but have been complicated by the lack of a universally conserved gene or gene marker for all types of viruses. Recently, the sequence-independent amplification of viral nucleic acids followed by shotgun sequencing or pyrosequencing has been used to discover an enormous diversity of virus species and genotypes in marine and fresh water (2, 6, 12, 37, 39, 45), human feces (3, 5, 43), animal tissues, and bat feces (11, 13, 24, 32, 42).
In this study, six bat fecal samples were collected from six locations in China and used for metagenomic analysis by sequence-independent amplification followed by high-throughput sequencing (Solexa, Illumina). Our results revealed that the bat fecal sample contains a large number of novel viruses, which are dominated by densoviruses, dicistroviruses, coronaviruses, parvoviruses, and tobamoviruses and cover a range of viruses from invertebrate and vertebrate to plant viruses.
MATERIALS AND METHODS
Sample collection.
Six bat fecal samples were collected from six different locations in China. All of the locations were close to human residences. The sample information, including locations, dominant bat species, and collection dates, is listed in Table 1. To collect fecal samples from grouped bats, clean plastic sheets measuring 2.0 by 2.0 m were placed under known bat roosting sites at approximately 18:00. The fresh fecal samples were collected at approximately 5:30 to 6:00 the following morning and were stored in liquid nitrogen. The samples were transported to the laboratory and stored at −80°C until analysis.
Table 1.
Sample collection locations and bat community
| Location | Abbreviation | Dominant bat species | Collection date |
|---|---|---|---|
| Hubei | HB | Myotis ricketti | May 2007 |
| Tianjin | TJ | Myotis ricketti | July 2007 |
| Hainan | HN | Scotophilus kuhlii | August 2007 |
| Yunnan | YNH | Hipposideros armiger | August 2007 |
| Yunnan | YNM | Myotis spp. | August 2007 |
| Guangdong | GD | Scotophilus kuhlii | October 2007 |
Viral particle purification.
Each fecal sample (15 g, about 200 to 225 fecal droppings) from 6 locations was resuspended (1:10, wt/vol) in Hanks' balanced salt solution (Gibco BRL) and homogenized thoroughly. The suspension was centrifuged at 5,000 × g for 10 min (Sigma 3-18K). The supernatant was transferred to fresh tubes and centrifuged at 13,000 × g for 15 min. A volume of 150 ml of the supernatant was filtered through a 0.45-μm filter and a 0.22-μm filter (Millipore) to remove eukaryotic and bacterial cell-sized particles, followed by fraction ultracentrifugation at 184,000 × g for 3 h at 8°C (Beckman Coulter Optima Ty-70) to pellet the viral particles. The pellets were resuspended in 400 μl Hanks' solution and filtered with a 0.22-μm filter. The filtrate was treated with Turbo DNase (final concentration, 20 U/ml; Ambion) and RNase A (final concentration, 0.1 mg/ml; Fermentas) at 37°C for 30 min to digest non-particle-protected nucleic acids. The total viral nucleic acids (both RNA and DNA) were extracted from a 140-μl sample with the QIAamp viral RNA minikit (Qiagen) according to the manufacturer's protocol and eluted into 60 μl buffer AVE (Qiagen).
Sequence-independent amplification of viral nucleic acids.
The synthesis of cDNA from the extracted viral RNA/DNA was performed as described previously (27). A volume of 13 μl of purified viral nucleic acids was mixed with 10 pmol random primer Brs (GCC GGA GCT CTG CAG AAT TCN NNN NN), denatured at 65°C for 5 min, and chilled quickly on ice for 2 min. The 6.5-μl reaction mix contained 4 μl of Moloney murine leukemia virus (M-MLV) reverse transcriptase (RT) 5× buffer (Promega), 1 μl of 10 mM deoxynucleside triphosphate (dNTP) solution, 0.5 μl of RNasin (40 U/μl; Promega), and 1 μl of reverse transcriptase (200 U). The reaction mixture was incubated at 25°C for 10 min, 37°C for 60 min, and 95°C for 5 min and then chilled on ice for 2 min. For the second-strand cDNA synthesis, 0.5 μl of 20 pmol Brs primer, 2.5 μl of 10× Klenow fragment buffer, and 2 μl of Klenow fragment (3.5 U/μl; Takara) were added. The reaction mixture was incubated at 25°C for 10 min and 37°C for 60 min, followed by enzyme inactivation at 75°C for 10 min. Then 2 μl of the reaction product was used as the template in a PCR mix containing 5 μl of 10× KOD-Plus DNA polymerase buffer (Toyobo), 2 μl 25 mM MgSO4, 5 μl of 2 mM dNTP, 2 μl of 10 pmol specific primer Bs (GCC GGA GCT CTG CAG AAT TC), and 1 U of KOD-Plus DNA polymerase (Toyobo). The PCR conditions were as follows: denaturation at 95°C for 5 min, 35 cycles (95°C for 30 s, 54°C for 60 s, 68°C for 2 min 30 s, with a 2-s increase in the extension time with each cycle), and a final extension at 68°C for 10 min.
Solexa sequencing.
The sequencing procedure was conducted according to the Solexa sequencing protocol (8) at the Beijing Institute of Genomics, Chinese Academy of Sciences. Briefly, the PCR products from six samples were equally mixed according to their concentrations. After PAGE purification of the PCR products and ligation of a pair of Solexa adaptors to their 5′ and 3′ ends, the DNA molecules were amplified using the adaptor primers for 17 cycles, and the fragments were isolated on agarose gels. The purified DNA was used directly for cluster generation and sequencing analysis using the Illumina genome analyzer (Illumina, San Diego, CA) according to the manufacturer's instructions. The image files generated by the sequencer were then processed to produce digital-quality data. After masking the adaptor sequences and removing the contaminated reads, the clean reads were processed for computational analysis.
In silico analysis.
Base-calling was performed with the GA Pipeline using standard parameters. Clean reads of 35 bp were obtained after removing the uncalled bases, incorrect primers, and adaptors. The reads and contigs were first aligned against the GenBank nucleotide (nt) database. The reads and contigs with no similarity to the GenBank nt database were then aligned against the GenBank nonredundant (nr) database. The sequences with similarity to eukaryotes, fungi, and bacteria were removed.
Detection of specific viruses in bat fecal samples.
Specific primers were designed based on the sequences of target viruses. The primers and reaction conditions for amplifying densovirus, dicistrovirus, and coronavirus HKU9 are listed in Table S3 in the supplemental material. The cDNA transcribed above from different locations was used as a template. The PCR products were gel-purified using the E.Z.N.A. Gel extraction kit (Omega Bio-Tek) according to the manufacturer's instructions and cloned into the pGEM-T Easy vector (Promega) before sequencing. For each PCR fragment cloned, three independent clones were sequenced to obtain the consensus sequence.
Complete genome sequencing of two insect viruses.
The complete genomes of one dicistrovirus and one densovirus were amplified and sequenced using the nucleic acid extracted from bat feces from Tianjin and Hubei, respectively. The primers were designed based on the sequences obtained in this study and those of target dicistrovirus and densovirus. The 5′ and 3′ ends of the viral genomes were confirmed by rapid amplification of cDNA ends using the 5′/3′ RACE kit (Roche, Germany). The overlapping sequences were confirmed by PCR and sequencing with specific primers. Sequences were assembled and manually edited to produce final viral genomes. The open reading frames (ORFs) of the viral genomes were predicted using the NCBI ORF finder. The amino acid sequences encoded by the ORFs were compared with those of the other dicistroviruses and densoviruses.
Phylogenetic analysis.
Routine sequence management and analysis were performed using DNAStar (DNAStar, Madison, WI). The translated peptide sequences of the contigs and hits with protein sequences available in GenBank were used for the phylogenetic analysis. Sequence alignments were constructed with T-Coffee and corrected manually (33). The phylogenetic trees were constructed using the neighbor-joining (NJ) method with a bootstrap of 1,000 replicates in MEGA version 4.1 (18). Gaps were regarded as a complete deletion unless specifically noted.
Nucleotide sequence accession numbers.
The Solexa sequence data obtained in this study have been deposited in DDBJ Sequence Read Archive (accession no. DRA000500). The trimmed and binned sequence contigs (>200 bp) used for analysis in this study were deposited in GenBank with the accession numbers JN857324 to JN857348.
RESULTS
Solexa sequencing reads.
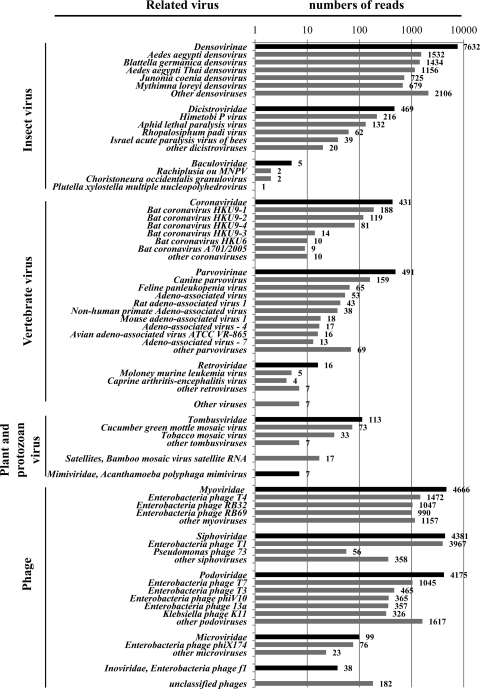
A total of 8,746,417 reads (35 bp each) with a total length of 306,124,595 bp were obtained. Among them, 13,541 (0.15%) reads had similarity to phages and 9,190 (0.1%) reads had similarity to eukaryotic viruses (E value, <0.01 for BLASTn and BLASTx), including densoviruses, dicistroviruses, coronaviruses, parvoviruses, dependoviruses, and tobamoviruses, covering viruses from invertebrates, vertebrates, and plants. Most of the reads were related to insect viruses, particularly members of the subfamily Densovirinae, family Parvoviridae (Fig. 1; see also Table S1 in the supplemental material).
Fig 1.
Schematic summary of the number of reads related to viruses classified by viral genus, subfamily, and family. The reads were compared with GenBank using BLASTn and BLASTx searches (E value, <0.01).
Assembled contigs and BLAST analysis.
A total of 129 assembled contigs (>100 nt) were constructed and compared with GenBank. Among them, 32 contigs were homologous to phages and 97 were homologous to eukaryotic viruses. The longest contig was 1,927 bp, and 6 contigs were longer than 1,000 bp, 16 contigs were between 400 and 1,000 bp, 20 contigs were between 200 and 400 bp, and 87 contigs were shorter than 200 bp. The nucleotide BLASTn and BLASTx searches against the GenBank database are listed in Table S2 in the supplemental material.
Among the 97 contigs that were homologous to eukaryotic viruses, 23 showed nucleotide identities of 68 to 99% with known virus genomic sequences (E value, <1E-16) and 74 showed no nucleotide identities with known sequences, but their translated amino acid sequences showed identities of 20 to 95% with viral protein sequences (E value, <0.001). Most of the contigs had low similarities with the protein sequences of known viruses, suggesting that these sequences represent novel viruses. Like the reads, most of the contigs were related to members of the subfamily Densovirinae of the family Parvoviridae.
Contigs related to novel insect viruses of the subfamily Densovirinae, Parvoviridae.
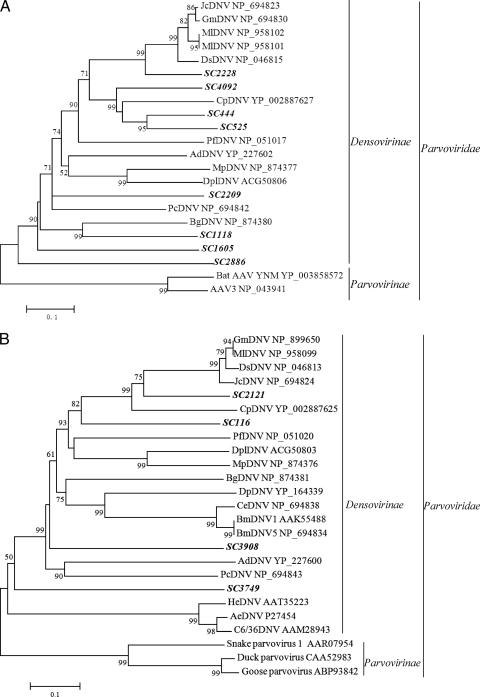
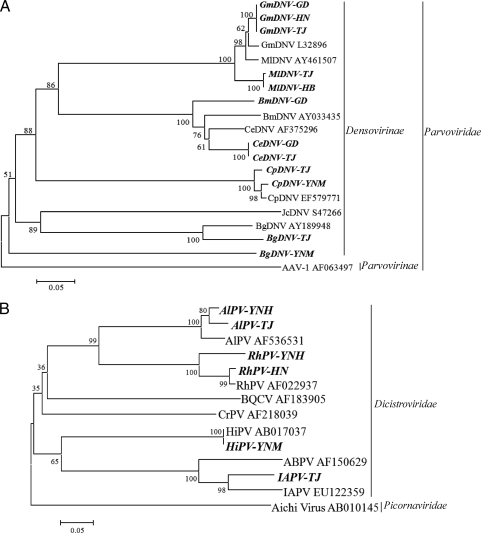
Members of Densovirinae, which are commonly referred to as densoviruses, infect invertebrates and belong to one of four genera: Brevidensovirus, Densovirus, Iteravirus, and Pefudensovirus. The densovirus genome consists of a 3.8- to 6-kb single-stranded linear DNA that has two major ORFs: the left ORF encodes the nonstructural protein NS1, while the right ORF encodes the structural protein VP1. Among the 66 contigs related to densoviruses, 22 had significant hits to 7 species of the genus Densovirus, 3 were related to 2 members of the genus Brevidensovirus, 6 were related to 4 members of the genus Iteravirus, 5 were related to 2 unclassified members of the genus Iteravirus (see Table S2 in the supplemental material), and 30 were related to unclassified members of the subfamily Densovirinae. Most of the contigs had low amino acid identities (<50%) with the known viruses (see Table S2B). Contigs SC444, SC525, SC1118, SC1605, SC2209, SC2228, SC2886, and SC4092 cover the same region of the VP1 gene, and contigs SC116, SC2121, SC3749, and SC3908 cover the same region of NS1. Except for the SC2228 and SC2121 that had respective identities of 70 and 71% with known densoviruses at the nucleotide level, all of the other contigs had low amino acid identities of less than 50% with known densoviruses. The translated sequences of these contigs were used for phylogenetic analysis with reference sequences of the members of Densovirinae. As shown in Fig. 2A and B, all of the selected contigs were distantly related to the known densoviruses, suggesting that these sequences represent novel insect viruses belonging to the Densovirinae subfamily.
Fig 2.
Phylogenetic analysis of the selected contigs related to densoviruses based on an alignment with 150 aa of the partial VP1 (A) and 170 aa of the partial NS1 region (B). AeDNV, Aedes densonucleosis virus; AdDNV, Acheta domestica densovirus; BgDNV, Blattella germanica densovirus; BmDNV, Bombyx mori densovirus; C6/36DNV, Aedes albopictus C6/36 cell densovirus; CeDNV, Casphalia extranea densovirus; CpDNV, Culex pipiens densovirus; DpDNV, Dendrolimus punctatus densovirus; DplDNV, Dysaphis plantaginea densovirus; DsDNV, Diatraea saccharalis densovirus; GmDNV, Galleria mellonella densovirus; HeDNV, Haemagogus equinus densovirus; JcDNV, Junonia coenia densovirus; MlDNV, Mythimna loreyi densovirus; MpDNV, Myzus persicae densovirus; PcDNV, Planococcus citri densovirus; PfDNV, Periplaneta fuliginosa densovirus.
Contigs related to Dependovirus of Parvovirinae, Parvoviridae.
Adeno-associated virus (AAV) belongs to the genus Dependovirus in the subfamily Parvovirinae. The genome of AAV is approximately 4.7 kb and contains two major ORFs, which encode the replication-related protein (Rep) and the capsid protein (Cap). AAV usually requires coinfection with a helper adenovirus for replication. AAV has been found in many vertebrate species, including the bat (28). In this study, 7 contigs (ranging from 103 to 369 bp) showed high similarities at the nucleotide level (ranging from 83% to 100%) with the Cap and Rep proteins of bat AAVs (BtAAV) (28) (see Table S2A in the supplemental material). Contig SC4399 (213 bp) had no identity at the nucleotide level with any known AAV but 58% identity at the amino acid level with the Cap protein of human AAV-3, consistent with our previous report that bat AAVs display great genetic diversity (28).
In addition to the genus Dependovirus, the subfamily Parvovirinae includes four other genera: Amdovirus, Bocavirus, Erythrovirus, and Parvovirus. There were 5 contigs that had similarities to 4 members of Parvovirinae (see Table S2B in the supplemental material). Contig SC5510 had an amino acid identity of 51% with VP1 of H-1 parvovirus, which belongs to the genus Parvovirus, and SC2718 had an amino acid identity of 67% with NS1 of human parvovirus B19, a member of the genus Erythrovirus. Contigs SC462 and SC2596 both had an amino acid identity of 51% to NS1 of parvovirus YX-2010/CHN, which was recently isolated in swine serum (44). Contig SC630 had 41% amino acid identity with VP1 of porcine bocavirus 2 pig/ZJD/China/2006, genus Bocavirus, which was recently identified from swine stool samples (9).
Contigs related to novel insect viruses of the family Dicistroviridae or unclassified single-stranded positive-sense RNA viruses.
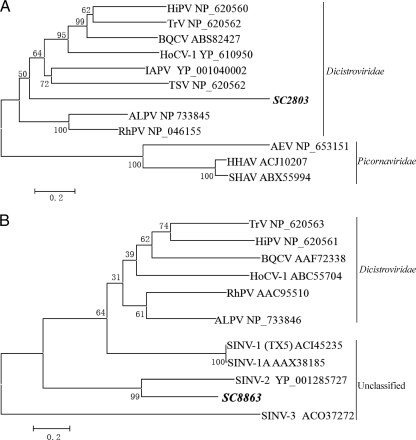
The members of Dicistroviridae have single-stranded positive-sense RNA genomes and infect arthropods. The genomes of dicistroviruses are approximately 8 to 10 kb and contain two nonoverlapping ORFs: the 5′-proximal ORF encodes a nonstructural protein, and the 3′-proximal ORF encodes a structural protein (31). There were 7 contigs that were related to 6 dicistroviruses and 5 contigs to unclassified single-stranded positive-sense RNA virus, Solenopsis invicta virus (SINV) (infecting fire ant Solenopsis invicta) (see Table S2 in the supplemental material). Except for two that had high nucleotide identities to known dicistroviruses (see Table S2A), all contigs had amino acid identities of 27 to 67% with the protein sequences of known dicistroviruses or SINV (see Table S2B). These contigs do not share any common protein-encoding region, and SC2803 and SC8863 were selected for phylogenetic analysis based on the translated amino acid sequence. The analysis showed that these sequences were distantly related to other members of the Dicistroviridae family or SINV (Fig. 3A and B). These results suggest that these contigs represent new members of the family Dicistroviridae or unclassified single-stranded positive-sense RNA viruses.
Fig 3.
Phylogenetic analysis of dicistrovirus-related sequences. (A) Cripavirus-related contig SC2803 based on the 382 aa of partial NS region. (B) SINV-related contig SC8863 based on the 113 aa of partial ORF2 (coat protein). AEV, avian encephalomyelitis virus; ALPV, aphid lethal paralysis virus; BQCV, black queen cell virus; HHAV, human hepatitis A virus; HiPV, Himetobi P virus; HoCV-1, Homalodisca coagulata virus-1; IAPV, Israel acute paralysis virus of bees; RhPV, Rhopalosiphum padi virus; TrV, Triatoma virus; TSV, Taura syndrome virus; SHAV, simian hepatitis A virus; SINV-1, Solenopsis invicta virus 1; SINV-2, Solenopsis invicta virus 2; SINV-3, Solenopsis invicta virus 3.
Circovirus-like virus detection.
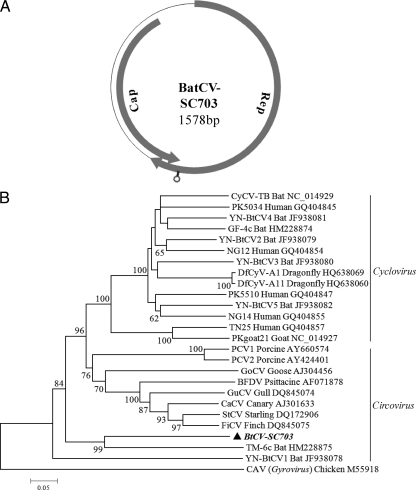
Recently, multiple diverse circovirus-related sequences were found in stool samples from humans, wild chimpanzees, and bats (15, 24, 25), muscle tissue of hoofed farm animals and chickens from Pakistan and Nigeria (25), and water samples (36). In our Solexa sequencing data, we found a small, circular DNA virus with a full genome size of 1,578 bp (SC703) that was related to the bat circovirus-like virus and temporarily named BtCV-SC703. The virus has two major ORFs arranged in opposite directions: a Rep protein of 309 amino acids (aa) and a Cap protein of 209 aa (Fig. 4A). The stem-loop structure also has the conserved nonanucleotide motif (5′-TAATACTAT-3′) and is located within the 3′ region of the Rep ORF. The phylogenetic analysis constructed based on the alignment of the complete Rep protein of BtCV-SC703 and other circoviruses or circovirus-related viruses showed that BtCV-SC703 formed a distinct group with the bat circovirus-like virus TM-6C (24) (Fig. 4B). BtCV-SC703 Rep had an amino acid identity of 48% with TM-6C. In addition, contig SC3668 (143 bp) had 41% identity with the Rep protein of circovirus NG11, which was detected in stool samples of children from Nigeria (25).
Fig 4.
Genomic organization (A) and phylogenetic analysis (B) of a bat circovirus-like sequence (BtCV-SC703). The phylogenetic tree was constructed based on the alignment of the complete amino acid sequence of the Rep protein (267 aa). CaCV, canary circovirus; CAV, chicken anemia virus; CyCV-TB, cyclovirus bat/USA/2009; BFDV, beak and feather disease virus; DuCV, duck circovirus; DfCyV, dragonfly cyclovirus DfCyV; GF-4c, bat cyclovirus GF-4c; GoCV, goose circovirus; GuCV, gull circovirus; FiCV, finch circovirus; NG12, cyclovirus NG12; NG14, cyclovirus NG14; PKgoat21, cyclovirus PKgoat21/PAK/2009; PCV1, porcine circovirus 1; PCV2, porcine circovirus 2; PK5034, cyclovirus PK5034; RaCV, raven circovirus; StCV, starling circovirus; TM-6c, bat circovirus-like virus; TN25, cyclovirus TN25; YN-BtCV-1-5, circovirus-like virus Yunnan strain 1-5.
Low-abundance contigs related to other viruses.
Contigs SC2797 and SC3066 had amino acid identities of 45% and 66%, respectively, with the picorna-like virus Eptesicus fuscus/P5/InLV/IT/USA/2009, which was identified from Eptesicus fuscus samples (big brown bat) by metagenomic analysis (13). Considering the diversity of picornaviruses, these viruses found in bat may be come from other dietary eukaryotic hosts or bat origin as found by the Hong Kong group (19). Contig SC2842 was homologous to E1 of human papillomavirus, which belongs to the Papillomaviridae family of double-stranded DNA (dsDNA) viruses, with an amino acid identity of 62%.
Identification of bacteriophage sequences.
There were 32 contigs that had high similarities (>90%) with the sequences of known phages, including members of Myoviridae (18 contigs), Podoviridae (12 contigs), and Siphoviridae (2 contigs). Most of the sequences were closely related to the T4-like and T7-like enterobacterial phages (Fig. 1).
Amplification of insect viruses in bat fecal samples.
We conducted a retrospective investigation of the virus distribution in the six bat communities. We chose 10 insect viruses and designed 10 specific primers based on the sequences of target viruses. The primer sequences targeted the NS1 sequence of densoviruses, including Blattella germanica densovirus (BgDNV), Bombyx mori densovirus (BmDNV), Casphalia extranea densovirus (CeDNV), Culex pipiens densovirus (CpDNV), Galleria mellonella densovirus (GmDNV), and Mythimna loreyi densovirus (MlDNV), or the RdRp sequence of dicistroviruses, including aphid lethal paralysis virus (ALPV), Himetobi P virus (HiPV), Israel acute paralysis virus of bees (IAPV), and Rhopalosiphum padi virus (RhPV) (see Table S3 in the supplemental material). Viral genomic fragments were amplified and confirmed by sequencing. The PCR result is summarized in Table 2. A total of 12 different sequences related to densoviruses and 6 different sequences related to dicistroviruses were obtained (GenBank accession numbers JN857349 to JN857358, JN857319 to JN857323). All target viruses were detected in at least two samples, and all of the amplicons except one had high nucleotide identities of 86 to 100% with the respective target viruses (Table 2). However, amplicons targeting the same viruses and amplified from different samples showed great genetic diversity (see Tables S4 and S5). The phylogenetic analysis showed that the detected viral sequences clustered together with the reference viruses except YNM-BgDNV, which was amplified from a Myotis community in Yunnan Province (Fig. 5A and B). These results indicate that a large number of genetically diverse insect viruses are present in bat feces.
Table 2.
PCR screening of samples with 10 sets of primers for insect viruses
| Target virus_a_ | PCR detection (nucleotide identity with target viral sequence)b | |||||
|---|---|---|---|---|---|---|
| TJ | HB | GD | HN | YNH | YNM | |
| BgDNV | +(92%) | − | − | − | − | +(54%) |
| BmDNV | − | − | +(86%) | − | − | − |
| CeDNV | +(91%) | − | +(91%) | − | − | − |
| CpDNV | +(97%) | +(100%) | − | − | − | +(98%) |
| GmDNV | +(97%) | − | +(98%) | +(95%) | − | − |
| MlDNV | +(91%) | +(92%) | − | − | − | − |
| AlPV | +(94%) | − | − | − | +(95%) | − |
| HiPV | − | − | − | − | − | +(100%) |
| IAPV | +(86%) | − | − | − | − | − |
| RhPV | − | − | − | +(90%) | +(98%) | − |
Fig 5.
Phylogenetic analysis of densoviruses and dicistroviruses amplified with specific primers based on an alignment with 180 aa (A) and 172 aa (B) of the partial NS gene and RdRp gene, respectively. The abbreviations in panel A are the same as those in Fig. 2 and 3.
Genomic sequence analysis of two insect viruses, BgDNV-HB and ALPV-TJ.
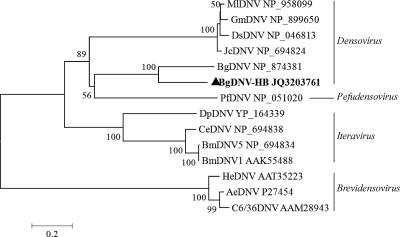
The nearly full-length genomic sequence of a densovirus (BgDNV-HB) was determined from the sample collected in Hubei (5,166 bp, GenBank accession no. JQ320376). Five ORFs were identified in the genome and their arrangement was similar to the known BgDNV which can infect Blattella germanica (German cockroach). ORF1 and 2 (593 and 303 aa, respectively) were on one DNA strand and encoded capsid proteins, and ORF3, 4 and 5 (533, 265 and 170 aa, respectively) were on the complementary strand and encoded nonstructural proteins (NS). All ORFs had low amino acid identities to BgDNV, suggesting that this virus is a new virus strain and was temporarily named BgDNV-like virus. The phylogenetic analysis showed this virus was clustered with other members of Densovirinae and close to BgDNV (Fig. 6). Several trials for obtaining the 5′ and 3′ ends were unsuccessful.
Fig 6.
Phylogenetic analysis of bat stool BgDNV-like virus. The predicted amino acid sequences (534 aa) corresponding to the fragments of nonstructural protein (NS-1) of insect-infecting parvoviruses were aligned to generate a phylogenetic tree. Four genera of Densovirinae were marked. The abbreviations of the densoviruses are the same as those in Fig. 2.
The full-length genomic sequence of a dicistrovirus was determined from the sample from Tianjin [9,819 bp excluding the poly(A) tail, GenBank accession no. JQ320375]. The genomic sequence showed high similarity of 89% to an aphid-infecting virus, ALPV, and was temporarily designated ALPV-TJ. The genome contained two ORFs that were separated by a 176-bp intergenic region. The 5′ ORF (538 to 6,642 nt) encoded the nonstructural proteins and had an amino acid identity of 95% with that of APLV. The 3′ ORF (6,818 to 9,241 nt) encoded the structural protein and had an amino acid identity of 94% with that of ALPV (see Table S6 in the supplemental material). The 5′ and 3′ untranslated regions were 537 and 678 bp, respectively.
Amplification of bat coronaviruses.
Bat coronaviruses have been studied extensively since the global disease outbreak caused by severe acute respiratory syndrome coronavirus (SARS-CoV) (14, 20–22, 35, 46, 48). In this study, a set of primers, BCNF and BCNR (see Table S3 in the supplemental material), which target the nucleocapsid gene of BtCoV HKU9, were designed and used to detect BtCoV HKU9 in the six fecal samples. There were 16 amplicons obtained, all from sample YNH, a community dominated by Hipposideros bats. The alignment of the 384-bp consensus sequences showed that there were 8 variants (BtCoV YNH-1-8) within the 16 amplicons (GenBank accession numbers JN857311 to JN857318). These 8 variants shared nucleotide identities of 75 to 99% and amino acid identities of 79 to 99% and had nucleotide identities of 76 to 96% and amino acid identities of 81 to 96% with BtCoV HUK9. The phylogenetic analysis showed that all of the variants belonged to the bat coronavirus HKU9 clade, which belonged to the novel Betacoronavirus subgroup 2d (Fig. 7) (22). These results further demonstrate the genetic diversity of BtCoV HKU9 in bat populations (22).
Fig 7.
Phylogenetic analysis based on an alignment of the partial BtCoV HKU9 N gene sequences (384 nt) amplified from Yunnan fecal sample and other representative coronaviruses. BtCoV HKU9-1, bat coronavirus HKU9-1; BtCoV HKU9-2, bat coronavirus HKU9-2; BtCoV HKU9-3, bat coronavirus HKU9-3; BtCoV HKU9-4, bat coronavirus HKU9-4; BtCoV HKU9-5-1, bat coronavirus HKU9-5-1; BtCoV HKU9-5-2, bat coronavirus HKU9-5-2; BtCoV HKU9-10-1, bat coronavirus HKU9-10-1; BtCoV HKU9-10-2, bat coronavirus HKU9-10-2; HCoV OC43, human coronavirus OC43; HCoV HKU1, human coronavirus HKU1; SARS CoV SZ16, civet SARS coronavirus SZ16; SARS CoV, SARS coronavirus; SARS CoV HKU3-1, bat SARS coronavirus HKU3-1; BtCoV HKU4-1, bat coronavirus HKU4-1; BtCoV HKU4-2, bat coronavirus HKU4-2; BtCoV HKU4-3, bat coronavirus HKU4-3; BtCoV HKU4-4, bat coronavirus HKU4-4; BtCoV HKU5-1, bat coronavirus HKU5-1; BtCoV HKU5-2, bat coronavirus HKU5-2; BtCoV HKU5-3, bat coronavirus HKU5-3; BtCoV HKU5-4, bat coronavirus HKU5-4; BtCoV HKU7, bat coronavirus HKU7; BtCoV/512/2005, bat coronavirus (BtCoV/512/2005); BtCoV HKU8, bat coronavirus HKU8; BtCoV HKU2, bat coronavirus HKU2; HCoV 229E, human coronavirus 229E; HCoV NL63, human coronavirus NL63.
DISCUSSION
High-throughput sequencing is a powerful tool for the microbial metagenomic analysis of a specific environmental sample (11). This technique has been used to discover a large diversity of novel viruses that could not have been identified by traditional virus culture methods in samples collected from water, humans, and animals (5, 11–13, 24, 32). In this study, we conducted a viral metagenomic analysis of bat fecal samples using the Solexa sequencing technique (Illumina). The mixed fecal samples were collected from six different locations, and all of the sampled bats were insectivorous. The data analysis indicated that a large number of viral sequences were discovered, and the most abundant sequences were related to phages, insect viruses, and mammalian viruses. To date, this is the first reported virome analysis in animals in China. Among the obtained sequences, most of the phage-related sequences have high identities at the nucleotide level with enterobacterial phages, indicating that the intestinal bacterial population of bats may be similar to that of humans (4, 5, 43). However, most of the insect virus-related or mammalian virus-related sequences have low identities at the nucleotide or amino acid levels with known viruses that have never been reported in China. These results indicate that a large number of novel and genetically diverse insect viruses and mammalian viruses are circulating among the insect and bat populations in China and provide useful sequence information for future virus isolation and identification. The rich source of insect viruses in bat feces reflects the diet of insectivorous bats, implying that insectivorous bats may serve as natural biological insecticides in at least two ways: as predators and as disseminators of viruses among insects.
To trace the viruses in the samples from different locations, we designed specific primers based on the sequences obtained and detected the insect viruses in 6 fecal samples. Unfortunately, only 10 pairs of primers, which target 6 densoviruses and 4 dicistroviruses, successfully amplified the expected fragments in the samples. While CpDNV and BmDNV have been reported before in China, all of the other viruses detected by sequencing were previously unknown in China. All of the amplified fragments, except BgDNV in the Myotis bat samples collected in Yunnan, have high nucleotide identities with the target viruses. With the exception of BmDNV, HiPV and IAPV, all of the viruses were detected in more than two locations. The BgDNV, MlDNV and BmDNV that were detected in different bat species and different locations displayed great genetic diversity and may represent novel virus species. In contrast, the CeDNV, CpDNV, AlPV and RhPV that were detected in bat samples from different locations are closely related to each other and are likely variants of the respective viruses. Interestingly, with the exception of the target virus HiPV and RhPV, all of the viruses were successfully amplified in Myotis bat samples collected from Tianjin, reflecting a large diversity of insect viruses and their hosts in this region.
In contrast to the insect virus-related reads and contigs, which are of insect origin, the hits against the 48 mammalian viruses should represent viruses of possible bat origin. Coronaviruses and parvoviruses are dominant in our collected samples. We performed a retro-investigation by PCR using several pairs of primers that were designed based on the coronavirus-related sequences. Consistent with the Solexa data, a variety of HKU9 viruses were detected, but only in Hipposideros bat samples collected from Yunnan Province. Our results are consistent with a previous investigation in Hong Kong, China, conducted by Lau et al. (22).
Consistent with our previous report, AAVs are highly prevalent in bat populations (28). In addition, a large numbers of reads that were related to animal or human parvoviruses were discovered in the Solexa data. These sequences have very low identities with known parvoviruses at the amino acid level, indicating that a large diversity of parvoviruses are circulating in bat populations. Because some parvoviruses cause severe diseases in animal or humans, more research should be focused on bat parvoviruses to understand the evolution and pathogenesis of bat parvoviruses. Prior to this study, two metagenomic analyses of bat viromes had been conducted (13, 24). The composition and proportion of these viromes are different from ours. For example, Li et al. (24) analyzed the virome in the feces of bats from California and Texas and found a lower proportion of phages and a larger proportion of eukaryotic viruses than in the virome reported in this study and in the viromes of human and equine feces. In addition, the majority of the insect virus-related sequences in their study were related to members of the viral families Dicistroviridae, Iflaviridae, Tetraviridae, and Nodaviridae and the subfamily Densovirinae, while most of the sequences in this study were related to members of Densovirinae and Dicistroviridae. These differences may be due to differences in the sampled bat species and the dietary habitats of the bats. Donaldson et al. have demonstrated that the viromes of bats vary by species, age, and sample type (13).
We compared in detail the virome from our study with the other two published bat viromes. Two contigs (SC2797, SC3066) in our study match most closely with a picorna-like virus (Eptesicus fuscus/P5/InLV/IT/USA/2009, GenBank no. ADR79388) detected from Eptesicus fuscus (big brown bat) in the virome analyzed by Donaldson et al. (13). The contig BtCV-SC703 most closely matches with TM6, a circovirus-like virus detected in a Texas bat stool sample dominated by Tadarida brasiliensis (24). The other insect and mammalian virus-related sequences in our data did not have significant hits with the viruses in the other two bat virome studies.
Due to the short reads, the low quantity of viral load in the sample, or a lack of known reference sequences, we obtained only 129 (>100-bp) contigs, and no full-length viral genomic sequences were obtained by assembling the data within the 22,711 virus-related reads. Similar results were obtained in the other two bat viromes (13, 24). We noticed a high background of sequences that are related to the genome of diverse hosts, including bacteria, insects, bats, and plants. This may be due to incomplete treatment with DNase and RNase during virus purification or high sensitivity of short-read sequencing compared to 454 sequencing (13, 24). A large proportion of unmatched sequences could come from unknown viral sequences or prokaryotic or eukaryotic genomic sequences not yet determined. The 5 reads that were highly similar to Moloney murine leukemia virus (M-MLV) in our sequencing data may be due to the contamination of the reagents, as M-MLV is commonly found in reverse transcriptase. The other limitations of this study include the sample collection and treatment techniques that were unable to distinguish the virome in a specific bat species, which will be improved in the future work for obtaining more virome data from different bat species and locations.
Supplementary Material
Supplemental material
ACKNOWLEDGMENTS
This work was jointly funded by the State Key Program for Basic Research Grant (2011CB504701) and the National Natural Science Foundation of China (30970137).
Footnotes
Published ahead of print 15 February 2012
REFERENCES
- 1.Almeida MF, et al. 2005. Experimental rabies infection in haematophagous bats Desmodus rotundus. Epidemiol. Infect. 133:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bench SR, et al. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blinkova O, et al. 2009. Cardioviruses are genetically diverse and cause common enteric infections in South Asian children. J. Virol. 83:4631–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitbart M, et al. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367–373 [DOI] [PubMed] [Google Scholar]
- 5.Breitbart M, et al. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart M, et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19:531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, et al. 2008. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18:997–1006 [DOI] [PubMed] [Google Scholar]
- 9.Cheng WX, et al. 2010. Identification and nearly full-length genome characterization of novel porcine bocaviruses. PLoS One 5:e13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu DK, Poon LL, Guan Y, Peiris JS. 2008. Novel astroviruses in insectivorous bats. J. Virol. 82:9107–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delwart EL. 2007. Viral metagenomics. Rev. Med. Virol. 17:115–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. 2009. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One 4:e7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donaldson EF, et al. 2010. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84:13004–13018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler JF, et al. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 84:11336–11349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge X, et al. 2011. Genetic diversity of novel circular ssDNA viruses in bats in China. J. Gen. Virol. 92:2646–2653 [DOI] [PubMed] [Google Scholar]
- 16.Halpin K, Young PL, Field HE, Mackenzie JS. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81:1927–1932 [DOI] [PubMed] [Google Scholar]
- 17.Hughes GJ, et al. 2006. Experimental infection of big brown bats (Eptesicus fuscus) with Eurasian bat lyssaviruses Aravan, Khujand, and Irkut virus. Arch. Virol. 151:2021–2035 [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150–163 [DOI] [PubMed] [Google Scholar]
- 19.Lau SK, et al. 2011. Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 85:8819–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau SK, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040–14045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lau SK, et al. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J. Virol. 84:2808–2819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau SK, et al. 2010. Coexistence of different genotypes in the same bat and serological characterization of rousettus bat coronavirus HKU9 belonging to a novel betacoronavirus subgroup. J. Virol. 84:11385–11394 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Leroy EM, et al. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576 [DOI] [PubMed] [Google Scholar]
- 24.Li L, et al. 2010. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84:6955–6965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L, et al. 2010. Multiple diverse circoviruses infect farm animals and are commonly found in human and chimpanzee feces. J. Virol. 84:1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679 [DOI] [PubMed] [Google Scholar]
- 27.Li Y, et al. 2010. Host range, prevalence, and genetic diversity of adenoviruses in bats. J. Virol. 84:3889–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, et al. 2010. Prevalence and genetic diversity of adeno-associated viruses in bats from China. J. Gen. Virol. 91:2601–2609 [DOI] [PubMed] [Google Scholar]
- 29.Luby SP, et al. 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 15:1229–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middleton DJ, et al. 2007. Experimental Nipah virus infection in pteropid bats (Pteropus poliocephalus). J. Comp. Pathol. 136:266–272 [DOI] [PubMed] [Google Scholar]
- 31.Nakashima N, Uchiumi T. 2009. Functional analysis of structural motifs in dicistroviruses. Virus Res. 139:137–147 [DOI] [PubMed] [Google Scholar]
- 32.Ng TF, et al. 2009. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma by using viral metagenomics. J. Virol. 83:2500–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205–217 [DOI] [PubMed] [Google Scholar]
- 34.Poon LL, et al. 2005. Identification of a novel coronavirus in bats. J. Virol. 79:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren W, et al. 2006. Full-length genome sequences of two SARS-like coronaviruses in horseshoe bats and genetic variation analysis. J. Gen. Virol. 87:3355–3359 [DOI] [PubMed] [Google Scholar]
- 36.Rosario K, Duffy S, Breitbart M. 2009. Diverse circovirus-like genome architectures revealed by environmental metagenomics. J. Gen. Virol. 90:2418–2424 [DOI] [PubMed] [Google Scholar]
- 37.Schoenfeld T, et al. 2008. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 74:4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Z. 2010. Bat and virus. Protein Cell 1:109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan MB, et al. 2008. Portal protein diversity and phage ecology. Environ. Microbiol. 10:2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tjørnehøj K, Fooks AR, Agerholm JS, Ronsholt L. 2006. Natural and experimental infection of sheep with European bat lyssavirus type-1 of Danish bat origin. J. Comp. Pathol. 134:190–201 [DOI] [PubMed] [Google Scholar]
- 41.Towner JS, et al. 2009. Isolation of genetically diverse Marburg viruses from Egyptian fruit bats. PLoS Pathog. 5:e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Victoria JG, Kapoor A, Dupuis K, Schnurr DP, Delwart EL. 2008. Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog. 4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Victoria JG, et al. 2009. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 83:4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, et al. 2010. Novel parvovirus sublineage in the family of Parvoviridae. Virus Genes 41:305–308 [DOI] [PubMed] [Google Scholar]
- 45.Williamson SJ, et al. 2008. The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo PCY, et al. 2007. Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J. Virol. 81:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yob JM, et al. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan J, et al. 2010. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J. Gen. Virol. 91:1058–1062 [DOI] [PubMed] [Google Scholar]
- 49.Zhu HC, et al. 2009. Detection of diverse astroviruses from bats in China. J. Gen. Virol. 90:883–887 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material