Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 13.
Abstract
T-cell development comprises a stepwise process of commitment from a multipotent precursor. To define molecular mechanisms controlling this progression, we probed five stages spanning the commitment process using RNA-seq and ChIP-seq to track genome-wide shifts in transcription, cohorts of active transcription factor genes, histone modifications at diverse classes of cis-regulatory elements, and binding repertoire of GATA-3 and PU.1, transcription factors with complementary roles in T-cell development. The results highlight potential promoter-distal cis-regulatory elements in play and reveal both activation sites and diverse mechanisms of repression that silence genes used in alternative lineages. Histone marking is dynamic and reversible, and while permissive marks anticipate, repressive marks often lag behind changes in transcription. In vivo binding of PU.1 and GATA-3 relative to epigenetic marking reveals distinctive, factor-specific rules for recruitment of these crucial transcription factors to different subsets of their potential sites, dependent on dose and developmental context.
INTRODUCTION
T lymphocyte development illuminates the stepwise process of cell fate choice for descendants of multipotent stem cells. Notch pathway signaling in the thymus causes hematopoietic precursors to become committed to the T-cell fate, while mobilizing a T-cell gene expression program that prepares the cells for T-cell antigen receptor (TCR) expression, TCR-based repertoire selection, and long, versatile careers as immune effectors. Sequential events that exclude alternative lineages occur at phenotypically well-defined stages within the thymus, providing a revealing model for the kinds of events needed to channel multipotent stem cells into a single developmental path (Rothenberg, 2011; Yang et al., 2010). However, major questions about the molecular mechanisms involved in this process have remained.
One question is how commitment works. Regulatory genes that promote access to alternative fates are either expressed or inducible in the precursors entering the thymus, but end up not only repressed but irreversibly silenced as a result of commitment. The mechanisms responsible for these regulatory changes have been unknown.
Another question has been how the T-cell program is deployed. Notch signaling initiates and sustains differentiation. T-cell development also depends on additional transcription factors, including E2A and HEB, TCF-1 and LEF-1, GATA-3, Myb, Runx1, Ikaros, and Gfi1 [rev. in (Rothenberg et al., 2008)]. However, it is not clear if this list is complete, and how these factors work remains murky because so few T-cell-specific cis-regulatory elements have been identified. Almost none have been functionally dissected in enough detail to explain fully the expression of the genes they control.
In other hematopoietic cell types, key cis-regulatory sequences of developmental genes have been identified through the collaborative binding of factors known to confer cell-type identity. For example, combined binding sites of E2A, EBF1, and/or Pax5 predict cis-regulatory elements in developing B cells (Lin et al., 2010; Schebesta et al., 2007). In contrast, no formula known a priori has been useful to define T-lineage specific cis-regulatory elements. However, if all the cis-regulatory elements that are “in play” at crucial transitions of T-cell development could be defined, then the motifs enriched in these elements could be matched with the cognate transcription factors that also change at those stages (Novershtern et al., 2011), thus narrowing the search for the key factors in commitment.
Here we identify the dynamic transformations in transcription and epigenetic marking that occur across the genome through five stages of T-cell differentiation that span lineage commitment. The results provide a genome-wide view of a lineage choice process in unusually fine resolution. To test the functional relevance of the histone marking patterns at potential cis-regulatory elements, we also track in vivo binding of GATA-3 and PU.1, two transcription factors with complementary roles in early T-cell development (Rothenberg and Scripture-Adams, 2008). Recruitment rules for these two factors are revealed to be context-dependent but differently affected by dose. The results also reveal how an initial regulatory phase dominated by stem/progenitor-cell regulatory genes first overlaps with Notch signaling, then is dismantled to establish T-cell identity.
RESULTS
Capturing commitment
Our goals were first, to map comprehensively the genes that undergo transcriptional change during T-lineage choice, especially genes encoding transcription factors; and second, to locate likely cis-regulatory sites mediating these gene expression changes by defining regions where histone marks are altered at each step of the process.
Cells in the first major stage of T-cell development, “early T-cell precursors” or Kit++ DN1 cells, pass through the DN2a stage to the DN2b stage, when they undergo T-lineage commitment. Post-commitment, they accumulate in the DN3 stage, during which they rearrange the TCR genes. Only cells that successfully express TCR proteins ever proliferate again, differentiating to the DP stage in a process called “β-selection” [rev. in (Rothenberg et al., 2008)]. Cells are selected after this based on their TCR recognition specificity, and further differentiation refines their mature immunological roles.
To obtain enough of the earliest cells for genomic analysis, we used an in vitro differentiation system that generates copious yields of early T-cell precursors from fetal liver-derived (FL) hematopoietic progenitor cells. These precursors are co-cultured with lymphoid-permissive cytokines and OP9 stromal cells expressing a Delta-like Notch ligand (OP9-DL1). In these conditions a cohort of FLDN1 and FLDN2a cells is generated by day 4.5 of culture, mostly progressing to FLDN2b cells by day 8.5 (Fig. S1A). For an in vivo counterpart, we purified slightly more advanced DN3 stage cells from freshly isolated adult mouse thymus, and to show the effects of β-selection, DP thymocytes were also purified (ThyDN3, ThyDP)(Fig. S1B, C, see Supplemental Methods).
In vitro differentiated FL-derived DN1 and DN2 cells showed gene expression well matched to that of normal in vivo thymocyte counterparts [(Yui et al., 2010; David-Fung et al., 2009)][http://www.immgen.org (Heng et al., 2008)]. Their lineage commitment status was also in good agreement with that of in vivo counterparts (Rothenberg, 2011; Yui et al., 2010), as shown by shifting cells to non-T conditions (Table S1), despite some minor differences (Table S1 legend). As in adult thymus in vivo, cells became committed from DN2a to DN2b.
Global gene expression analysis: selective changes during early T-cell development
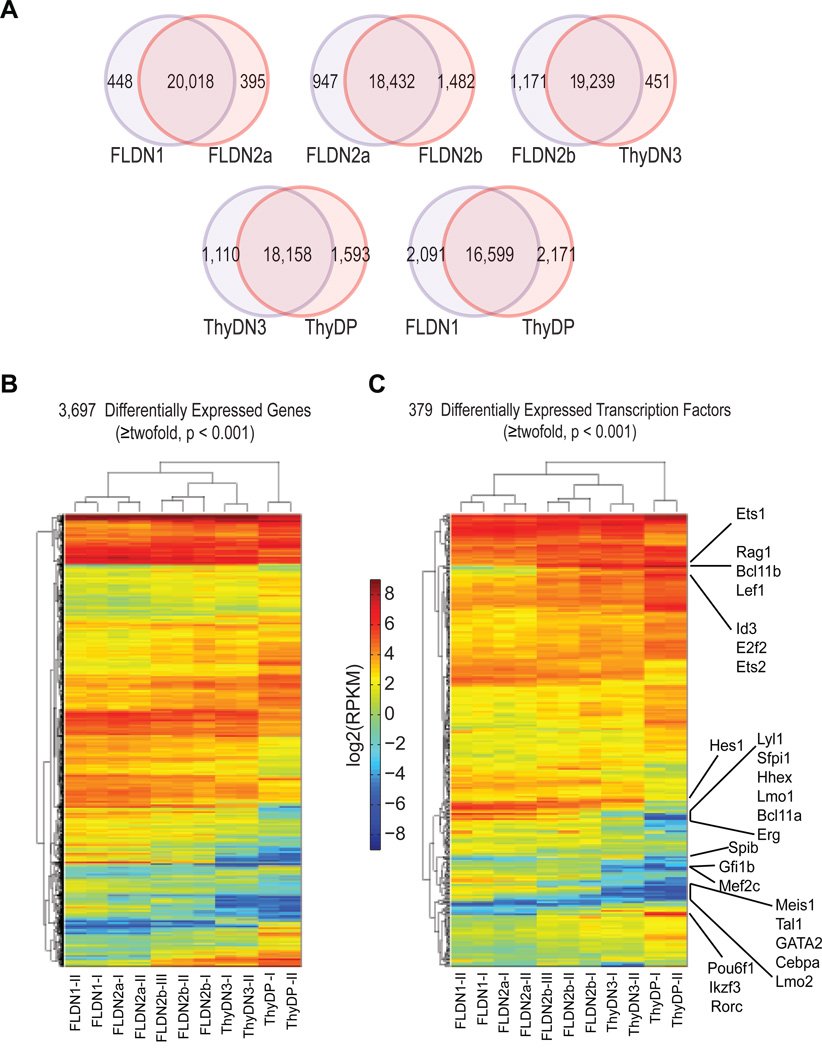
We used RNA-seq (Mortazavi et al., 2008) to identify when major changes in gene expression occurred along the pathway from early T-cell precursor to DP stages, using 2–3 independent biological replicates each of FLDN1, FLDN2a, FLDN2b, ThyDN3, and ThyDP cells (Pearson r>0.97 for independent replicates of the same stages, Fig. S1D). About 10,000 of the 20,861 Refseq genes were detectably expressed (≥ 1RPKM) in each population; of these, ~50% changed significantly in expression (p< 0.001) between at least one pair of stages and ~ 40% changed from FLDN1 to ThyDP (Fig. 1A).
Figure 1. Global comparisons of gene expression among five developmentally related immature T cell populations.
A. Pairwise comparisons in gene expression between successive populations and between initial FLDN1 vs. final ThyDP stages: statistically changed genes defined by DEGseq (p < 0.001).
B. Hierarchical clustering of expression patterns of all differentially expressed genes (DEGseq positive, ≥2× changed).
C. Hierarchical clustering of expression patterns of differentially expressed transcription factors: several key transcription factors indicated.
Fig. 1B shows hierarchical clustering of the expression patterns of the 3,697 genes that change expression ≥2× between any stages. Between DN1 and DN2b key T-cell specific genes involved in pre-TCR expression and function were induced from a low or undetectable level (Table S2), i.e. genes encoding TCR complex components Cd3g, Cd3d, Cd3e, Cd3z (Cd247), T-cell-specific signaling components Itk and Lat, recombinase Rag1, mutagenic DNA polymerase Dntt, and the surrogate α chain (pTα) Ptcra. In addition, a conspicuous group of genes were repressed or silenced during these transitions. They included progenitor-cell specific growth factor receptor genes Kit, Flt3, and Csf2rb, and a set of transcription factor genes described below.
The DN1 to DN2 transition is the first definitive sign of T-lineage entry induced by Notch signaling. However, a much larger difference was seen between pre-commitment FLDN2a and post-commitment FLDN2b cells (2429 genes different, Fig. 1A, B) than between FLDN1 and FLDN2a (<900 genes different), also seen by hierarchical clustering. Conversely, despite their different origins and manipulation, the newly-committed FLDN2b and ThyDN3 populations were more similar to each other as well (Fig. 1A, B). Thus, the major genome-wide transcriptomic changes leading to T-lineage identity do not occur in the DN1 to DN2a transition, but rather in transition to the DN2b or DN3 stages, linked with commitment.
Transcription factor expression dynamics in T-lineage commitment
Genes likely to encode transcriptional regulators (Table 3A, see Supplemental Methods) included 379 that changed expression by ≥2× (Table S3B). Hierarchical clustering of their patterns of expression (Fig. 1C) again showed similarities between FLDN1 and FLDN2a and between FLDN2b and ThyDN3, while the precommitment FLDN2a cells were more different from the newly committed FLDN2b (Fig. 1C); the ThyDP cells were the most different from all. Thus, the two major transitions in regulatory gene expression occur at commitment and at β-selection.
From FLDN1 to FDN2b, the most strongly upregulated “regulatory” loci in the whole genome were found to be Lef1 and Bcl11b (>75× increased). Pou6f1, SpiB, Ikzf3, and Ets1 among others also increased >8×, with weaker increases for Id3, Tcf12, Gfi1, Tcf7, Hes1, and Gata3 (Table S3B). However, many regulatory genes sharply decreased in expression between FLDN1 and FLDN2b, including genes with known, important functions in hematopoietic progenitors, e.g. Gfi1b, Lmo2, Mef2c, Hoxa9, Sfpi1 (PU.1), Gata2, Mycn (N-Myc), Cebpb, Bcl11a, Hhex, Nfe2, Lyl1, and several Irf factors. A major regulatory shift, with broad repression of progenitor-cell transcription factor genes, thus accompanies T-lineage commitment.
Dynamic histone modification changes identify developmentally regulated promoters and distal cis-elements
The specific cis-regulatory elements affected by changing transcription factor action during commitment should be sites of developmentally changing histone modifications [rev. by (Natoli, 2010; Kouzarides, 2007)], of greatest interest where they are linked to differentially expressed genes. We used chromatin immune precipitation and deep sequencing (ChIP-seq) (Johnson et al., 2007)(Barski et al., 2007) to enrich DNA associated with three H3 modifications: H3K(9,14)Ac, H3K4me2, and H3K27me3. Histone H3K(9, 14) acetylation (H3Ac) is functionally linked to activation at transcriptional start sites (TSS), H3K27me3 is used in one mechanism for transcriptional silencing, and H3K4me2 is associated with activation, poising for activation or repression, or repression (Orford et al., 2008; Barski et al., 2007; Johnson et al., 2007; Heintzman et al., 2007). For many enhancers, H3K4me2 provides more precise localization than H3K4me1 (Koche et al., 2011). The results from independent biological replicates again showed excellent correlation (Fig. S1E).
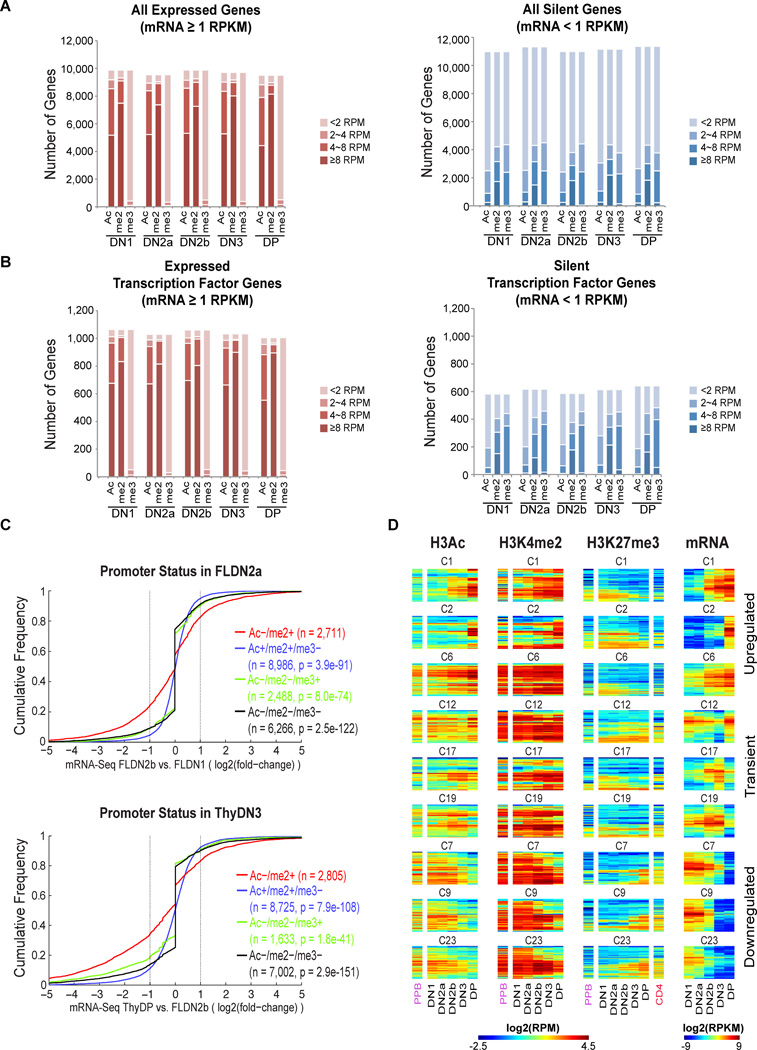
The 42,000 regions with marks were distributed near and distal to the annotated TSS of expressed and silent genes (Fig. 2A, Fig. S2B); tables S2A (TSS) and S2B (non-TSS) report intensities of marking at each stage as correlated with RNA expression of the nearest genes. Fig. S3 presents these comprehensive results for TSS and non-TSS sites for all Refseq loci in hierarchically clustered heat maps.
Figure 2. Distinct gene expression patterns are associated with characteristic histone modifications.
A. Gene expression and histone modifications at the TSS of 20,861 genes. Expressed (≥ 1RPKM) and silent (<1RPKM) genes defined by RNA-seq. Ac: H3Ac, me2: H3K4me2, me3: H3K27me3.
B. Gene expression and histone modifications at the TSS of 1,646 genes encoding DNA-binding proteins or transcription factors.
C. Association of promoter-linked histone modifications with developmental change in expression. Genes are grouped based on histone marks of their TSS in the FLDN2a (top) or ThyDN3 (bottom) stages: H3Ac− H3K4me2+ (H3K27me3+ or −) in red, H3Ac+ H3K4me2+ H3K27me3− in blue, H3Ac− H3K4me2− H3K27me3+ in green, and H3Ac− H3K4me2− H3K27me3− in black. Cumulative distributions of genes in each group are plotted vs. expression changes from FLDN1 to FLDN2b (top), and from FLDN2b to ThyDP (bottom). X axis: gene expression change (log2 ratio of RNA-seq levels), downregulated to the left, upregulated to the right (vertical lines: twofold change). Y axis: fraction of group with expression change ≤x axis value. P values from Kolmogorov-Smirnov (K–S) two-sided tests comparing H3Ac−/H3K4me2+ against each of the other three groups are shown.
D. Heatmaps correlating TSS histone modifications with various patterns of developmentally regulated gene expression for 9 representative expression clusters (from Fig. S5). Hierarchical clustering within individual clusters used Ward linkage and Euclidean distance. Histone modification data from EBF−/− pre-pro B cells (H3Ac, H3K4me2 and H3K27me3; “PPB”) and CD4+ naïve T cells (H3K27me3 only, “CD4”) are also shown for these genes.
Consistent with previous reports, H3Ac and H3K4me2 were overwhelmingly seen at promoters of expressed genes. Silent genes fell into two classes, with only ~35% showing H3K27me3 at their promoters. H3K27me3 was more common at silent regulatory genes than at other silent loci (Fig. 2A,B right panels). However, >25% of the silent genes, including many with H3K27me3, were also marked with H3K4me2 (Figs. S2C,S3), consistent with at least three kinds of repressed states (Filion et al., 2010). The cumulative frequency plots in Fig. 2C show that genes with H3K4me2 but not H3Ac at their promoters in a given stage (red tracks) (±H3K27me3) were most likely to be newly repressed or poised for upregulation in the next stage. Thus, H3K4me2 without H3Ac marks developmentally labile promoters (Koche et al., 2011; Orford et al., 2008).
Histone modification at promoters was relatively stable across development, more than levels of corresponding RNAs (Fig. S3A). However, distal elements were more dynamically marked: >1/3 of all regions marked with H3K4me2 or H3K27me3 in one stage lacked those marks in at least one other stage (Fig. S3B). Thus regulatory shifts occurring in development most sensitively affect histone marking at non-promoter elements (Heinz et al., 2010; Lin et al., 2010). H3K4me2-marked distal elements in fact included a number of previously noted regulatory elements (Fig. S4A,B): the DP-specific Rag1–Rag2 gene antisilencer 71–75kb 5’ of the Rag2 gene (Yannoutsos et al., 2004), and the DN2b/3-specific E1a promoter for the Notch1 gene (Gomez-del Arco et al., 2010). Both of these discrete cis-elements acquired H3K4me2 specifically at stages when they contribute to gene regulation (Fig. S4A,B). Other non-TSS regions with developmentally dynamic marking may thus locate stage-specific cis-regulatory elements as well.
Timing of TSS epigenetic changes relative to transcriptional changes
To relate the timing of changes in TSS marks with changes in RNA expression during T-cell commitment, we focused on 3,697 differentially regulated genes. First, these were subdivided by K-means clustering into 25 clusters based on expression pattern (Fig. S5; genes listed in Table S4A). Fig. 2D tracks histone marks from stage to stage at the TSS of genes undergoing upregulation (clusters 1, 2, 6), downregulation (clusters 7, 9, 23), and transient decreases (cluster 12) or increases in expression (clusters 17, 19). H3Ac modification (Fig. 2D, first group of columns) was tightly coordinated with presence of RNA (last columns), but H3K4me2 was often present before and after expression (second columns, e.g. clusters 1 & 6). Though H3K27me3 was inversely correlated with expression, only a fraction of repressed genes ever acquired this mark (Fig. 2D, third columns), as also seen at promoters genome wide (Fig. S3A).
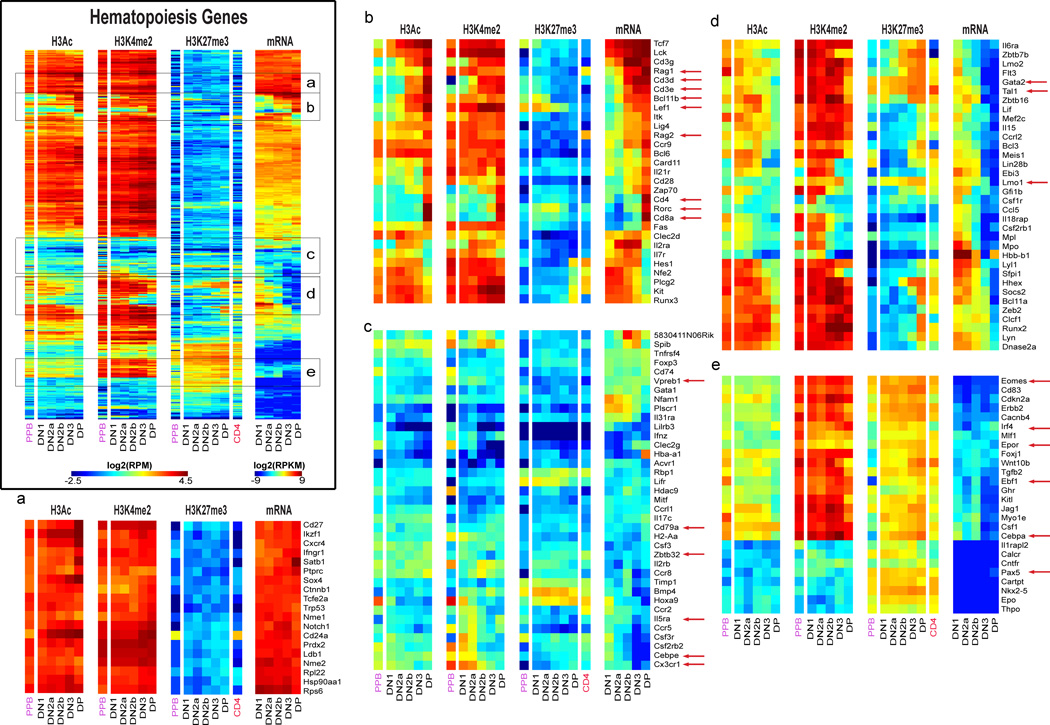
Most relevant to the regulatory decisions in T-lineage commitment (Rothenberg, 2011; Yang et al., 2010) are effects on genes needed for other hematopoietic cell fates, options which are shed in an ordered process. We identified 389 key hematopoietic genes by Gene Ontology (Table S4B), including “signature” regulators of erythroid cells (Gata1, Nfe2, Epor), myeloid cells (Sfpi1, Cebpa, Cebpe, Csf1r), B cells (Pax5, Ebf1), NK cells (Eomes, Il2rb), and stem cells (Gata2, Tal1, Lmo2), and tracked their expression from DN1 to DP in parallel with the status of histone marks at their promoters (Fig. 3). Full results are shown in Table S4B and the Fig. 3 master panel, while zoom-in panels allow individual genes to be identified. Again, H3Ac modification at promoters was tightly correlated with transcription, while H3K4me2 marking also preceded and persisted after transcription.
Figure 3. Histone modifications and gene expression profiles of genes characterizing hematopoiesis.
Results for 379 “hematopoietic” genes are processed and displayed as in Fig. 2D. Master panel: results for all 379 genes. Panels (a)–(e): zoomed in to indicated cluster regions of master panel to allow individual genes to be seen.
To explain alternative-lineage exclusion in T-cell commitment, either one or diverse mechanisms of silencing of non-T regulatory genes might be used. In fact, H3K27me3 use at these functionally relevant loci was both variable and dynamic. Some genes were silent throughout T-cell specification, and many had strong H3K27me3 marks at the promoter, either apparently with H3K4me2 (e.g. Group e, Epor, Irf4, Ebf1, and Eomes) or without (e.g. Group e, Pax5). Other regulatory genes were turned off during development, often gaining H3K27me3 while they lost H3Ac (Group d). Some genes poised for early silencing already had some H3K27me3 at the TSS from FLDN1 stage (e.g. Cebpa in Group e_, Gata2, Lmo1, Tal1_ in Group d), suggesting repression already underway in at least part of the population. However, H3K27me3 did not mandate future silencing, for some T-cell genes like Lef1 were strongly activated during commitment despite initially strong H3K27me3 marking (Group b). Furthermore, other genes stayed silent from FLDN1 to ThyDP without any H3K27me3 at the TSS (e.g. Cebpe, Cx3cr1, Zbtb32, Cd79a, and VpreB1; Group c). Unexpectedly, these variations in H3K27me3 marking cut across myeloid, erythroid, NK cell, and B-cell program boundaries.
Most epigenetic change in T-cell development occurs from DN1 to DP
Both the foreshadowing of future expression by H3K4me2 marking of promoters and the ability of some genes to be repressed without appearance of H3K27me3 (Figs. 2D, 3) raised the question of whether we might be missing changes in promoter status either before DN1, or after DP stage. We therefore compared our results with H3Ac, H3K4me2, and H3K27me3 ChIP-seq data for a prethymic lymphoid precursor population, “PPB” [EBF−/− pre-pro B cells (Lin et al., 2010; Heinz et al., 2010)] and H3K27me3 data for post-thymic naïve CD4 T cells, “CD4” (Wei et al., 2009), shown in flanking columns in Figs. 2D and 3. The prethymic lymphoid precursor data in general concurred with the FLDN1 patterns for all three histone marks. Furthermore, repressed genes that lacked H3K27me3 marks by the DP stage in our samples also remained silent without H3K27me3 marks in the mature T cells (the uniquely regulated Rag genes were an exception). The FLDN1 to ThyDP interval thus encompasses the crucial epigenetic changes for the great majority of genes affected by T-cell specification.
Distinct mechanisms control key developmental genes
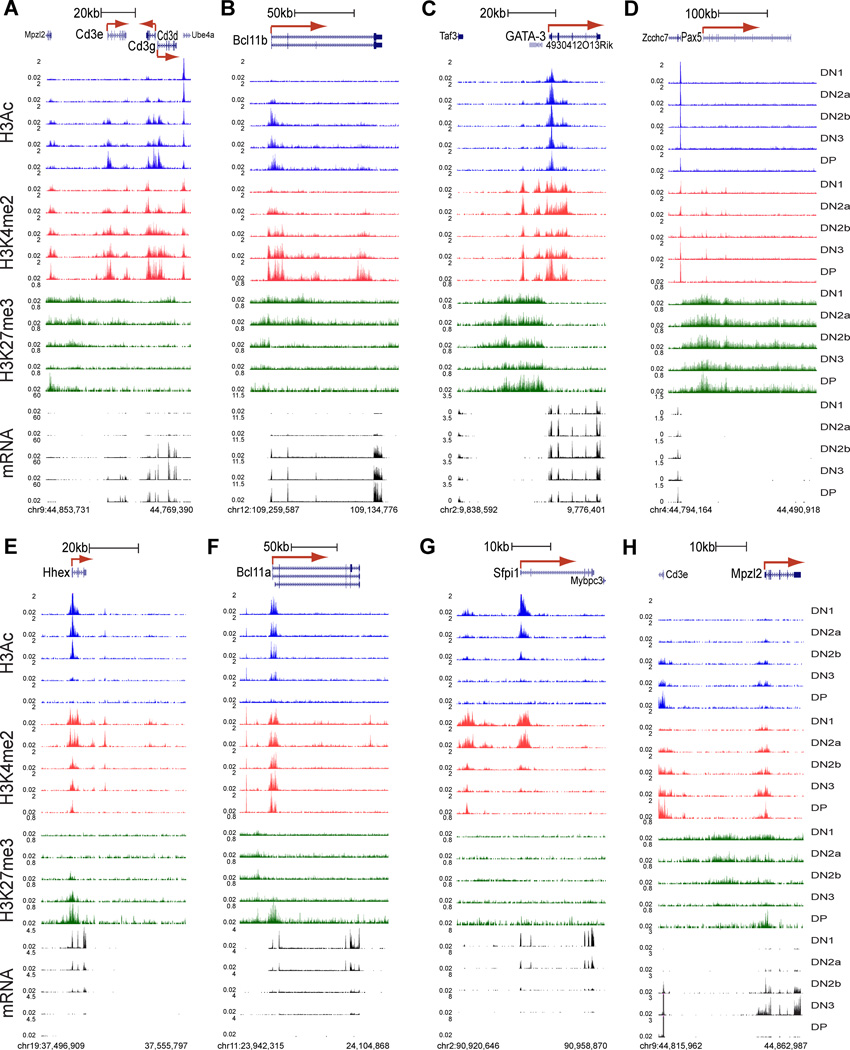
Changes in modification at distal sites (compiled in Table S2B) as well as TSS sites (Table S2A) often appeared implicated in gene regulation, as shown for key genes in Fig. 4. Fig. 4A, B profile two highly T-cell-specific loci activated in parallel from DN2a to DN2b, the Cd3gde gene cluster (A) and Bcl11b (B). These genes initially lack RNA transcripts (black tracks) and H3Ac marks (blue tracks) in FLDN1 cells, but then are strongly upregulated and kept on thereafter.
Figure 4. Portraits of key T-lineage and alternative-lineage genes.
A – H. Distinct epigenetic marking and gene expression patterns at eight different loci: Bcl11b (A), Cd3e/d/g cluster (B), Gata3 (C), Pax5 (D), Hhex (E), Bcl11a (F), Sfpi1 (G) and Mpzl2 (H), in all five immature T-populations (top to bottom, DN1, DN2a, DN2b, DN3 and DP)(coordinates below each panel). Red arrow: TSS and direction of transcription. H3Ac: blue, H3K4me2: red, H3K27me3: green, RNA-seq: black. Uniform scales are used for histone marks in all panels, and mRNA scales are uniform within each panel (y axis units in RPM).
For the Cd3 genes, there was no H3K4me2 (red tracks) at the promoters and light H3K27me3 marking across the locus (Fig. 4A, green tracks) during the initial silence, but the classic enhancer elements at the 3’ ends of Cd3e and Cd3d (Georgopoulos et al., 1988; van de Wetering et al., 1991) were already marked by focal H3K4me2. These enhancers were already accessible to transcription factor binding even in the FLDN1 stage, as shown by binding of the factor GATA-3 (Fig. S4C; see below). Marking of these H3K4me2 sites intensified while H3Ac and H3K4me2 were recruited to the promoters of the genes during the DN2a/2b stages, when transcription began. A similar pattern for activation without initial promoter marking was seen for Il2ra (Fig. S4D).
In contrast to the Cd3gde cluster, the Bcl11b gene (Fig. 4B) began with substantial H3K27me3 (green tracks) over its promoter and the whole gene body at FLDN1 stage. However, its TSS had a cryptic positive cis-regulatory element marked by H3K4me2. Bcl11b then was activated from FLDN2a to FLDN2b stage through a process that swept back the H3K27me3 repressive marks off the promoter, while expanding the H3K4me2 marks into the first intron and creating a new H3K4me2 marked region in the third intron.
The changes in histone marks at these loci contrast with the precisely positioned but virtually unchanging H3K27me3, H3Ac, and H3K4me2 marks that characterized the Gata3 gene (Fig. 4C). Despite a block of H3K27me3 close to the major promoter, this gene was already activated by the time of the FLDN1 stage, and underwent only a fewfold increase in expression after that.
Repression of essential B-cell regulatory factors, myeloid-cell regulatory factors, and stem or progenitor-cell regulatory factors is central to T-lineage commitment. This clearly entailed a variety of distinct mechanisms (Fig. 4D–H). The Pax5 gene, crucial for the B-cell program, had no H3Ac modified regions at any stages (Fig. 4D; cf. neighboring Zcchc7 TSS mark). Small peaks of H3K4me2 marking were seen in intronic regions, one of them corresponding to a known hematopoietic enhancer (Decker et al., 2009). However, the gene was buried in H3K27me3 at all stages. Ebf1 (Fig. S4E) was also repressed from FLDN1 on despite H3K4me2 at several sites.
Hhex and Bcl11a (Fig. 4E, F), in contrast, were expressed strongly in FLDN1 cells but then downregulated sharply by the FLDN2b/ThyDN3 stages, showing evidence of distinct modulating roles for distal and TSS elements. For Hhex (Fig. 4E), the TSS and two H3K4me2-marked distal regions lost activation marks as expression decreased, while H3K27me3 appeared focally at the TSS and then spread. A similar pattern was seen for Flt3 and the Zbtb7b (Thpok) gene, both active in FLDN1 and then repressed (Fig. S4F,I). For Bcl11a (Fig. 4F), H3Ac persisted at the promoter while RNA expression declined during commitment, reflecting a tail of low-level expression through DN3. H3K27me3 marks only appeared at the last stage of silencing in the DP stage. However, the H3K4me2 modification just downstream of the last exon decreased sharply between FLDN2a and FLDN2b, in parallel with RNA expression, suggesting a potential regulatory role for a distal element here.
The myeloid and progenitor-cell transcription factor gene Sfpi1 (encoding PU.1), silenced in parallel with Hhex and never re-expressed in most T-cell lineages, used a different mechanism of repression (Fig. 4G). H3Ac disappeared from the promoter while H3K4me2 marks in the upstream cis-regulatory elements of the gene (Rosenbauer et al., 2006; Zarnegar et al., 2010) narrowed as transcription declined (Fig. 4G). Yet minimal H3K27me3 was ever seen.
Not only were H3K27me3 marks dispensable for repression; they were also labile. Fig. 4H shows that dense H3K27me3 marks on Mpzl2 (same as Eva1) diminished during a spike of RNA expression in the DN2b and DN3 stages, then returned during re-silencing in DP stage. Conversely, despite silencing during commitment, Zbtb7b (Fig. S4I) is later activated for CD4+ cell positive selection.
Early T-cell specific sites for PU.1: a positive role
The significance of epigenetic marks depends on their impact on transcription factor access and their own emplacement via transcription factor binding. We therefore correlated chromatin marks with binding of GATA-3 and PU.1 (encoded by Sfpi1), two factors needed for early T-cell development, which play contrasting roles in the context of Notch signals [rev. by (Rothenberg and Scripture-Adams, 2008; Hosoya et al., 2010)]. PU.1 is one of the progenitor-associated transcription factors in early pro-T cells, but is even more critical for B, dendritic and myeloid cell development. A key question is whether it has distinct T-lineage target genes or simply carries over a multipotent state.
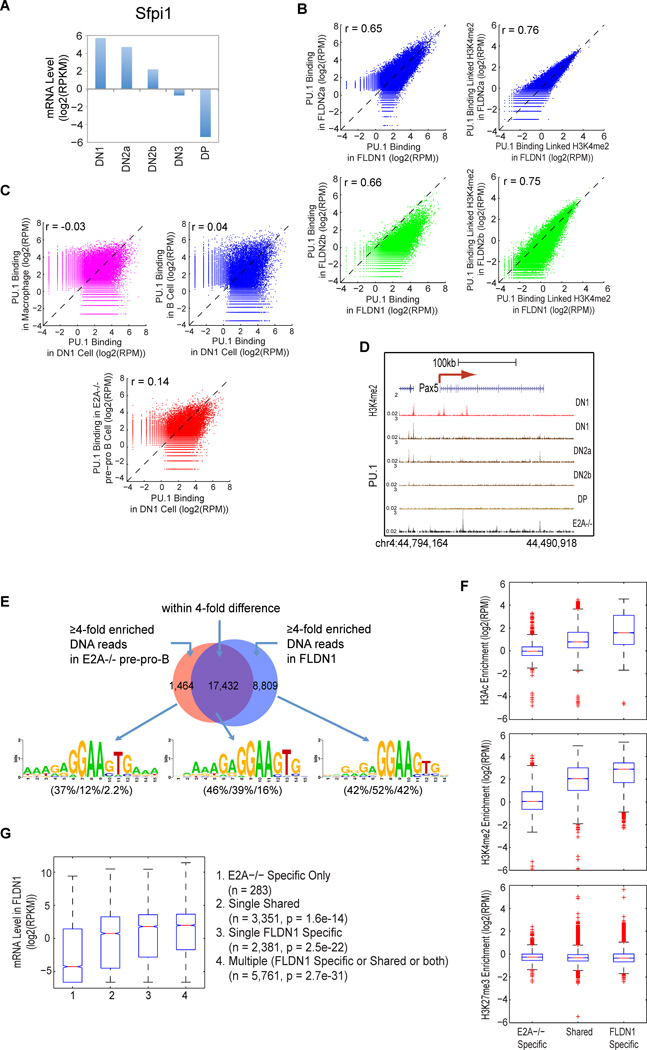
PU.1 bound to ~34,000 sites in DN T cells, comparable to B and myeloid cells (Heinz et al., 2010). Although PU.1 RNA and protein levels decline sharply during T lineage commitment (Fig. 5A)(Yui et al., 2010), PU.1 site binding preferences remained consistent from stage to stage. We compared FLDN1 and FLDN2a cells; FLDN2b cells, where PU.1 is 4–5× downregulated; and DP cells, where PU.1 is absent. Although PU.1 binding intensity per site was ~4–5× lower in the FLDN2b cells, its site choices remained correlated with those in the earlier stages (r=0.65–0.66)(Fig. 5B).
Figure 5. Lineage specific PU.1 DNA binding is associated with lineage specific histone modifications and gene expression.
A. Mean RNA-seq level of PU.1 (Sfpi1) at each stage of early T-cell development.
B. (Left) Comparisons of PU.1 DNA binding site distributions between FLDN1 and FLDN2a or FLDN2b, with Pearson’s correlation coefficients (r). (Right) Comparisons of PU.1 DNA binding-associated H3K4me2 enrichment between FLDN1 and FLDN2a or FLDN2b. H3K4me2 signal densities were from ±1kb of the summit of a PU.1 bound region.
C. Comparisons of PU.1 DNA binding between FLDN1 and E2A−/− pre-pro B, macrophage or mature B cells.
D. Lineage-specific PU.1 binding at the Pax5 locus. B-cell specific Pax5 intronic enhancer is bound by PU.1 (black arrow) in E2A−/− pre-pro pre-pro B cells (Heinz et al., 2010) (black track), but not in DN cells (brown tracks). PU.1 ChIP-seq in ThyDP is used as a negative control. For orientation, H3K4me2 pattern in DN1 stage is included (red track).
E. Lineage specific and shared PU.1 binding sites between FLDN1 and E2A−/− pre-pro B cells. Lineage specific: ≥4× difference in PU.1 occupancy between populations. Sequence logos show the most highly enriched sequence motif for each occupancy subgroup. Percentages of regions from the three subgroups with ≥1 instance of each motif are given in parentheses beneath each sequence logo (E2A−/− specific/Shared/FLDN1 specific).
F. Distribution of the enrichment of specified histone modification over genomic regions within ± 1kb of lineage specific and shared PU.1 binding sites in FLDN1 cells.
G. Correlation of mRNA expression levels in FLDN1 with presence of lineage-specific or shared PU.1 sites. Distribution of mRNA value in FLDN1 for subgroups of genes that are linked to either E2A−/− pre-pro B specific, FLDN1 specific, or shared PU.1 binding sites, and genes linked to more than one PU.1 site occupied in FLDN1 cells (Multiple). K-S test compares E2A−/− Specific Only with each of the other three subgroups (n and p values in parentheses).
Even so, the PU.1 binding sites in FLDN1 and FLDN2a cells were distinct from those reported in B cells, macrophages or even E2A−/− pre-pro B cells [representing prethymic lymphoid progenitors (Heinz et al., 2010)](Fig. 5C). Although the sites bound in the pre-pro B cells were most related, key PU.1 target sites occupied in pre-pro B cells were not bound by PU.1 in FLDN1 cells, e.g. the intronic enhancer of Pax5 (Fig. 5D). De novo motif analysis showed that PU.1 target sites in FLDN1 cells had a different hierarchy of preferred sequences than in pre-pro B cells (Fig. 5E). Thus, the consistent site choices of PU.1 from DN1 stage through commitment include a distinct T lineage-specific component.
PU.1 is needed to generate T-cell precursors, but at high levels it inhibits expression of many T-cell specific genes, particularly if Notch signaling is interrupted (Franco et al., 2006). To test whether the T-lineage specific sites of PU.1 may be repressive, delaying T-cell gene activation in early stages, we asked whether PU.1 binding specific to early T cells was linked to genes that are active or silent, as compared with sites bound by PU.1 in pre-pro B cells but empty in T cells. In fact, the sites occupied by PU.1 in FLDN1 cells, including FLDN1-specific sites, were mostly associated with “positive” marks (H3Ac and/or H3K4me2) and completely uncorrelated with H3K27me3 marking, more than sites bound by PU.1 only in E2A−/− pre-pro B cells (Fig. 5F). In the aggregate, genes with sites of PU.1 binding in FLDN1 cells were also more likely to show strong expression, with higher expression the more sites bound, including T-lineage specific sites (Fig. 5G). Thus, PU.1 binding globally correlates with target gene expression in FLDN1 cells.
PU.1 binding dynamics and temporal control of target gene expression
Experimental perturbation analyses have shown many specific genes in pro-T cells that are activated or repressed by manipulations of PU.1 level (Franco et al., 2006)(A. Champhekar, M. M. Del Real, and E. V. R., unpublished data). However, with so many binding sites for PU.1, binding alone clearly could not define genes that depend on PU.1 for positive or negative regulation. Most PU.1 binding sites were linked to genes expressed stably in all stages whether PU.1 is present or not, like the majority of genes expressed in T-cell development overall. PU.1 could thus be opportunistically recruited to many active genes where it has no required role. To define properties of likely functional sites, we used the dynamics of PU.1 expression itself to filter the binding sites identified genome-wide.
Because PU.1 expression declines, functionally important PU.1 sites should be enriched near genes which themselves change in RNA expression, up or down, during development as a function of PU.1 binding. We therefore compared changes in local PU.1 occupancy from FLDN1 to FLDN2b (Fig. 6A) with the direction and magnitude of changes in RNA expression of the linked genes, in two complementary ways. First, focusing only on the PU.1 site-linked genes that change expression from DN1 to DN2b (Fig. 6B), we grouped them according to whether their linked sites all lost PU.1 occupancy faster (Fig. 6B–C, blue) or slower (red) than the global ~4× average decrease (green=genes with both kinds of sites). Cumulative frequency plots were used to test if changes in PU.1 occupancy predicted the direction of changes in gene expression (Fig. 6C), i.e., whether genes losing PU.1 first all turn off like PU.1 itself, or become activated as PU.1 repression might be relieved. Such analyses could detect both activated and repressed subgroups within a group as well as general trends. However, genes which lost PU.1 binding most rapidly (blue curve) were uniformly more downregulated and less upregulated than those with mixed sites. Conversely, almost 80% of genes with sites that retained PU.1 best (red curve) increased their expression from FLDN1 to FLDN2b.
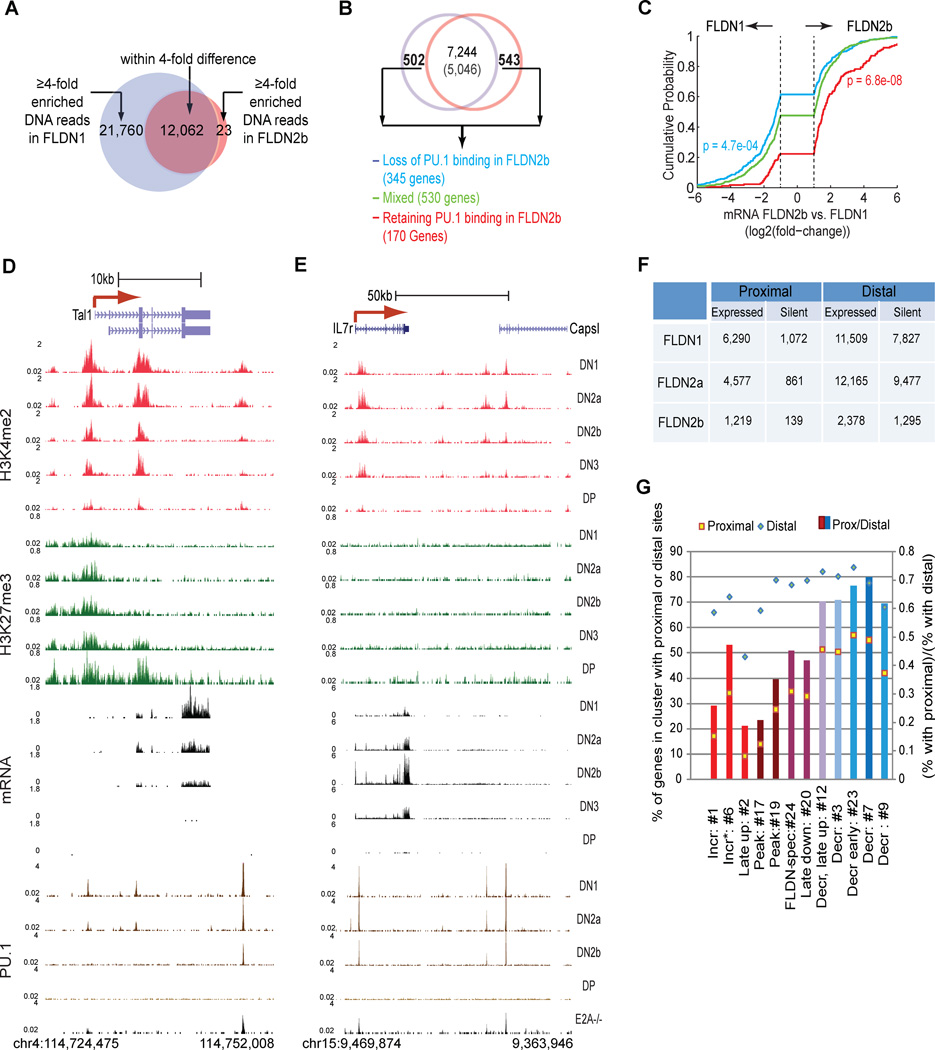
Figure 6. Functional and stage-dependent PU.1 binding in early T-cell development.
A. Stage specific and non stage-specific (shared) PU.1 binding sites: stage specific binding defined by ≥4× difference in signal densities between FLDN1 and FLDN2b.
B. Differential expression of PU.1 binding linked genes. Top: of 13,335 PU.1 binding linked genes in DN cells, the numbers expressed in FLDN1 (blue circle) and FLDN2b cells (red circle) are shown (7,244 stably expressed, 1,045 differentially expressed: ≥2× change). To test whether PU.1 occupancy correlated with positive or negative regulation, all differentially expressed genes were split among 3 subgroups based on changes in PU.1 binding to linked sites (see panel A): genes with FLDN1 specific sites only (Loss of PU.1 binding in FLDN2b, blue), those retaining PU.1 binding at all sites in FLDN2b (red), and genes that rapidly lose PU.1 binding from some sites but not others (Mixed, green).
C. Relationship between PU.1 occupancy changes and mRNA expression changes between FLDN2b and FLDN1: cumulative distributions of expression changes for three groups of genes depicted in B. The number of genes in each group and p values (K-S tests for comparisons with “Mixed”) are indicated next to the plots.
D, E. Developmentally distinct PU.1 binding patterns at the Tal1 (D) and Il7ra (E) loci in FLDN1, FLDN2a, FLDN2b and E2A−/− pre-pro B cells, compared with H3K4me2, H3K27me3 and mRNA in all five immature T-populations.
F. Distribution of PU.1 occupancy relative to TSS sites in expressed and silent genes at individual stages.
G. Location of PU.1 sites in potential target genes according to expression pattern. Clusters of genes with different developmental trajectories (Fig. S5) were scored by the number of genes they include with PU.1 binding sites ± 1kb from the TSS (proximal) or further from the TSS (distal). □,◊ (left axis): % of genes in a cluster with proximal (□) or distal (◊) PU.1 binding. Bar graphs (right axis): (number of genes with TSS sites)/(number of genes with distal sites). Colors of bars relate expression pattern of each cluster to endogenous PU.1 expression (most similar: blue, inverse: red). See Figs. 2D, S5, & Table S6.
Second, reciprocally, we classified individual PU.1 sites according to whether their linked genes were upregulated, downregulated, stably expressed, or silent across the DN1 to DN2b interval, and then assessed whether these sites near developmentally regulated genes tended to lose PU.1 faster or slower than open but nonregulated sites, i.e. those linked to stably expressed genes (Fig. S6A). Downregulated, upregulated, and stably expressed genes relinquished their PU.1 binding very differently, and sites linked to upregulated genes retained their PU.1 even better than fully “accessible”, stably expressed ones, again arguing against a repressive role.
At candidate target genes identified both by co-regulation with PU.1 and by PU.1 perturbation effects, PU.1 typically occupied multiple regions, implying that full PU.1 regulatory function is commonly mediated through combinations of binding complexes. At Tal1, both PU.1 binding and local H3K4me2 were lost jointly from three regions, as transcription also declined (Fig. 6D). At the TSS and intragenic regions, PU.1 loss appeared to open the way for H3K27me3 deposition. Similar patterns were seen at the TSS of the known PU.1 target Flt3, and at a downstream element and a known intronic enhancer of Hhex (Donaldson et al., 2005)(Fig. S4F, G). Other genes with binding sites that lose PU.1 early include Lmo1 and Bcl11a as well as Itgam, which decrease naturally from FLDN1 to FLDN2b; all are sharply downregulated in FLDN2 cells if Sfpi1 is deleted (not shown; A. Champhekar and E. V. R., unpublished). A PU.1 target with a different pattern of expression but a similar relation to PU.1 binding was Il7r, which is upregulated from FLDN1 to FLDN2b. Despite the decreasing level of PU.1 protein, the Il7r gene retained PU.1 through the FLDN2b stage, both at a known TSS positive regulatory site (DeKoter et al., 2002; Xue et al., 2004), and at another putative cis-element within a silent neighboring gene, Capsl (Fig. 6E). Here too PU.1 regions have a positive link to expression, even for this gene integral for the T-cell program.
Histone marking and modes of action
PU.1 binding can recruit histone methyltransferases and create locally “open” chromatin states (Ghisletti et al., 2010; Heinz et al., 2010), and in early T-lineage cells as in non-T cells, PU.1 occupancy was dynamically linked with local H3K4me2 modification. Due to the developmental stability of H3K4me2 modification at TSS’s, the association was clearest at distal PU.1 binding regions, where H3K4me2 modification usually melted away as PU.1 binding decreased (Figs. S6B, bottom & S6D). In contrast to H3K4me2, PU.1 binding had little overlap with H3K27me3, even at silent genes (Fig. S6C1, 2; S6D). PU.1 binding-linked H3K4me2 was not simply an effect of general “accessibility” or expression level of the linked gene (Fig. S6D, “silent” vs. “E2A−/−” sites). Thus, PU.1 occupancy-linked changes in H3K4me2 could be used to screen candidate distal cis-regulatory regions with PU.1-dependent activity.
Globally, with or without distal binding sites, PU.1 binding near the TSS appeared most tightly correlated with a positive role (Fig. 6F). Genes from diverse expression pattern clusters (see Figs. 2D, S5) could all harbor PU.1 binding either within the body of the gene or in flanking regions, but differed sharply in frequencies of genes with PU.1 binding at the TSS (Fig. 6G; Table S6). Genes coregulated with PU.1 itself (cl. 7; also 3, 9 & 23; blue bars) were more likely to have PU.1 binding at the TSS than genes regulated divergently from it (e.g. cl. 1, 2, 6; red bars)(Table S6B). In contrast, many T-cell genes that can be downregulated by high-level PU.1 (Franco et al., 2006) either had no PU.1 binding in early T cells or had binding only in the body or flanking regions of the genes. Genes with particularly low expression in DN1 stage were most impoverished for PU.1 binding at the TSS (Table S6A, χ2 test p <0.0001). Thus, PU.1 binding at the promoter may provide, or indicate, specific antisilencing functions that maintain key stem- and progenitor-cell genes in early FLDN1 and FLDN2a stages.
Developmentally plastic deployment of GATA-3 binding
GATA-3 is needed repeatedly in T-cell stages from ETP/DN1 onward and is crucial for T lineage commitment, but capable of paradoxical effects at high doses (Taghon et al., 2007). Unlike PU.1, it is expressed almost stably across all the stages analyzed (Fig. 4C; and unpublished results). We therefore asked whether it controls the same targets in the distinct regulatory states of FLDN1, FLDN2b, and ThyDP cells.
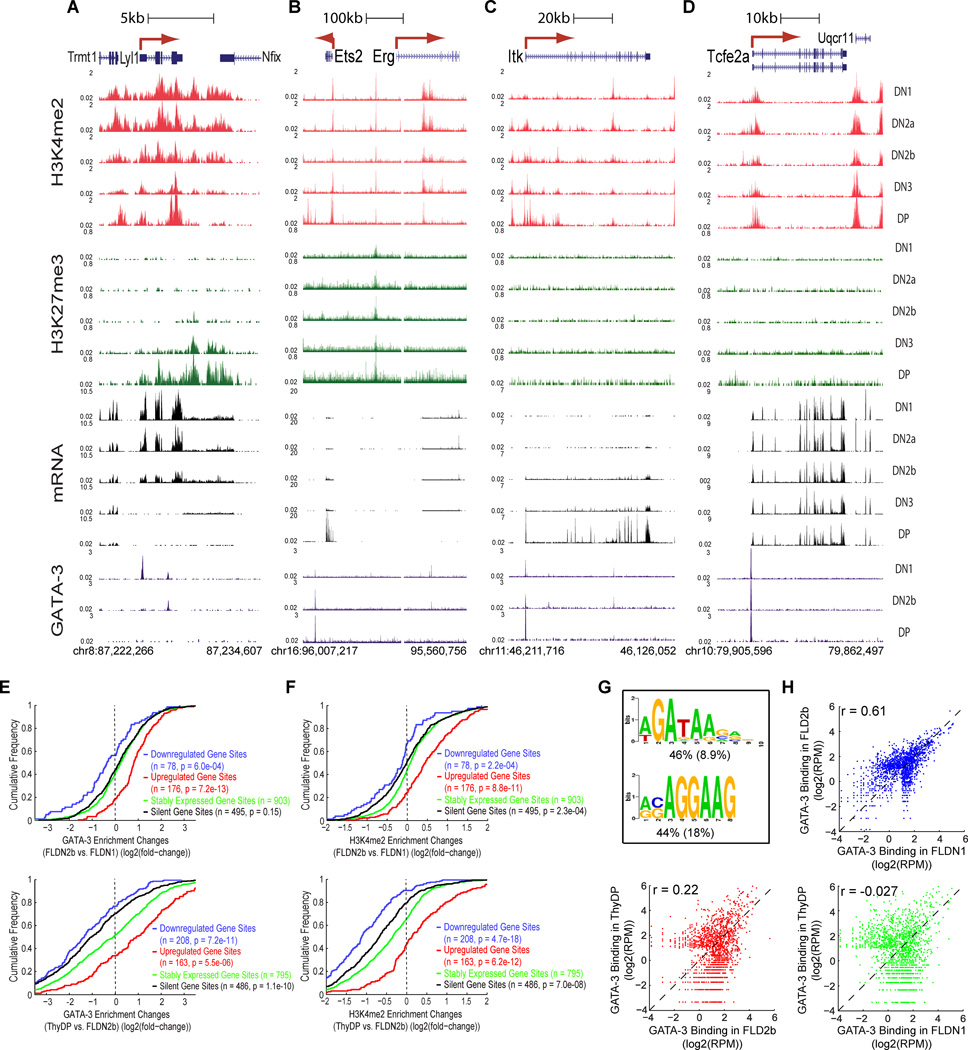
GATA-3 detectably bound only ~1500 regions (Table S7). In accord with its recurrent T-cell roles, these GATA-3 sites were enriched for cis-elements of T-lineage genes including Cd3d, Tcf7, Zbtb7b, and the DP-specific Rag1-Rag2 distal enhancer (Fig. S4C,H,I,B). Occupancy patterns in our ThyDP samples were broadly consistent with those in DP CD3lo samples published elsewhere (Wei et al., 2011)(r=0.60; Fig. S7A). Yet progenitor-specific genes like Lyl1 and Erg (Fig. 7A,B) as well as later-expressed T-cell genes like Ets2 and Itk (Fig. 7C,D) also harbored GATA-3 sites.
Figure 7. Developmental plasticity of GATA-3 DNA binding and associated epigenetic marking.
A – D. Stage-specific GATA-3 binding (brown) in Lyl1, Ets2-Erg, Itk and Tcfe2a loci of FLDN1, FLDN2b and ThyDP cells, shown with binding associated H3K4me2 (red) and H3K27me3 (green) enrichment and mRNA (black) expression in all five immature T-populations.
E. Cumulative distributions of changes in GATA-3 occupancy between FLDN2b and FLDN1 (top) and between ThyDP and FLDN2b (bottom), for genes differentially regulated across the same intervals. GATA-3 binding sites were divided into 4 subgroups, based on linkage to downregulated genes (blue), upregulated genes (red), stably expressed genes (< 2-fold change in expression, green) and silent gene sites (<1RPKM in both stages, black). P values are from K-S tests between stably expressed gene sites and each of the other three subgroups (n= no. of sites).
F. Cumulative distributions of changes in H3K4me2 marks associated with GATA-3 binding between FLDN2b and FLDN1 (top) and between ThyDP and FLDN2b (bottom) stages. H3K4me2 signal densities were calculated within −/+1kb of the summit of a given GATA-3 bound region (depicted in Figure 7H). P values calculated as in E.
G. Most highly enriched sequence motifs in GATA-3 binding regions (see panel H). The percentages of regions containing ≥1 instance of each motif are indicated beneath each sequence logo, with the expected frequency of the motif in random regions in parentheses.
H. Scatter plots depicting the comparisons in GATA-3 binding between FLDN1, FLDN2b and ThyDP. Pearson correlation coefficients are shown for each comparison.
Changes in GATA-3 binding were strongly positively correlated with expression trajectories of linked genes both from DN1 to DN2b and from DN2b to DP (Fig. 7E), and also correlated with H3K4me2 modification changes (Fig. 7F). GATA-3 binding was even more likely to be a site of H3K4me2 enrichment than PU.1 binding (Fig. S7B). Yet GATA-3 also bound regions linked to silent and active genes alike at early stages, and dramatically differed from PU.1 in its ability to bind regions with H3K27me3 (Fig. S7C). For example, it remained bound to Zbtb7b even as it became silenced with H3K27me3 (Fig. S4I). Intriguingly, GATA-3 occupancy also preceded full cis-element activation for Cd3d and the Rag enhancer (Fig. S4B,C), suggesting a possible “pioneering” role.
Most unlike PU.1, the distribution of GATA-3 occupancies among different regions was strikingly different in FLDN1, FLDN2b, and ThyDP. This was despite nearly constant protein availability, as shown by similar global occupancy levels and peak heights at stably occupied sites such as the Tcf3 (Tcfe2a) promoter (Fig. 7D) and the Tcrb 3’ enhancer (not shown) in all stages. The most common motifs at regions of occupancy were a classic GATA site and an Ets family-like site (Fig. 7G), in FLDN1, FLDN2b, and ThyDP alike. But from FLDN1 to ThyDP, GATA-3 occupancy increased sharply in some regions (e.g. Ets2 in DN2b, Itk promoter in DP; Fig. 7B,C), while disappearing from others entirely (e.g. Lyl1, Erg, Itk introns; Fig. 7A–C). Overall, whereas regions occupied in FLDN1 and FLDN2b stages were moderately well correlated (Pearson r=0.61), sites in FLDN2b and ThyDP were poorly correlated and those in FLDN1 and ThyDP entirely uncorrelated (r=−0.0064, Fig. 7H).
These results locate elements in T and non-T genes where the crucial T-cell factor GATA-3 can be contributing to regulation, from the FLDN1 stage on (Table S7). Nevertheless, they also reveal that a target gene for GATA-3 at one stage of T-cell development may not normally receive input from GATA-3 at another stage, despite similar GATA-3 availability. In contrast to PU.1, GATA-3’s physiological deployment at any given stage depends not only on its own availability but also on a specific developmental regulatory context.
DISCUSSION
Our results provide a resource for T cell development, a new reference case for regulatory epigenomics, and potentially powerful new explanatory elements for a complex developmental process. Our global, base-resolution timecourse of chromatin and transcriptome changes sheds new light on the finely defined stages of T-cell specification. The RNA-seq data not only quantify RNA levels but also provide detailed information about promoter and exon choice that may affect gene regulation as well as function. Note that these data also reveal noncoding transcripts that may be important in various regulatory roles. Here, we focused on the ~400 regulatory gene loci that themselves are developmentally regulated during this process, and also the subset of candidate cis-regulatory genomic sites that undergo developmental changes in histone modifications, revealing changes in local regulatory inputs. These are the most likely trans and cis components of nodes in the gene network that causally drive successive steps of the T-cell program.
Most powerfully, the results reveal the subcomponent processes out of which T-cell specification is built. Relatively few regulatory genes are strongly activated during lineage commitment itself, and the list is now likely to be complete. Furthermore, a major feature of commitment is specific, marked downregulation of progenitor-cell genes, through an unexpectedly complex process. Many important non-T hematopoietic regulatory genes are still expressed in the precursors we examine through 4 days of consistent Notch pathway signaling, and many persist even into the DN2a stage before they are shut off. Importantly, the histone marking status of the promoters and linked cis-elements provides an independent line of evidence about the timing and mechanism of regulatory changes. Loss of H3Ac rules out an artifact of the decay kinetics of old mRNA persisting after transcription has ceased. Our results also show that the repression of progenitor-cell genes is not due to a single switch. The diverse histone mark transformations that are applied to different repression targets imply that a variety of biochemically and temporally distinct silencing mechanisms must be used. Notably, this rules out Notch signaling itself as a common mechanism of repression and implies that the T-lineage program requires multiple distinct repressor functions to establish T-cell identity.
Dynamically regulated transcriptional repression during this process is often separable from “epigenetic silencing”. Repeatedly, deposition of H3K27me3 histone marks follows RNA downregulation, more likely as an effect and stabilizer of repression than an initial cause of repression. De novo H3K27me3 marking appears in two distinct, major patterns. One can be by lateral invasion from a neighboring patch of pre-existing “closed” chromatin, another is by tight focal deposition at a previously active TSS or enhancer, followed by spreading. In other cases repression does not involve H3K27me3 at all, possibly due to the nature of the repressor: e.g. at Sfpi1 and Cd4, two key genes known to be repressed by Runx factors in DN3 cells. Even when H3K27me3 is used, it is readily and precisely reversible. Repression via DNA methylation was not studied here but is also reversible, as shown recently by the cell type-specific demethylation of CpGs in DN2–DN3 cells at loci that include Tcf7 and Bcl11b (Ji et al., 2010)(http://charm.jhmi.edu/hsc/). These examples show that transcriptional repressors must act first to trigger chromatin closing, while transcriptional activators retain power to undo it.
Our results also shed light on the positive regulation of the T-cell program. Despite known essential roles, finding specific cis-regulatory targets for Notch, GATA-3, and TCF-1 has been slow. The identification of a battery of cis-regulatory elements activated de novo from DN1 to DN2b is an important new resource for clarifying these links. GATA-3 effects in early T cells have been especially difficult to dissect, in part due to the profound loss of viability when GATA-3 dose is reduced (Hosoya et al., 2009), and in part due to lineage-inappropriate effects of GATA-3 in gain of function experiments (Taghon et al., 2007). Identification of potential GATA-3 regulatory inputs into Tcf7 as well as Tcfe2a from the earliest stages suggests a new level of regulatory interlinkage, which could explain the acuteness of the GATA-3 requirement. At least in DP cells, data from (Wei et al., 2011) suggest that the GATA-3 sites we see positively regulate Tcf7, Cd3d, and Zfpm1, and may negatively regulate Tcfe2a. Our results may also help to explain GATA-3’s lineage infidelity in gain of function experiments by showing that its recruitment to legitimate target sites, even at a constant level of expression, is intensely stage specific. Altered dosages could thus override the mechanisms that must provide appropriate targeting specificity.
The ordered alternative lineage exclusion events in T-lineage commitment are an ideal context to test whether developmental relatedness is preserved in a hierarchy of epigenetic chromatin changes. Clearly, separable events mediate repression of different alternative lineages. The B-cell regulatory genes Pax5 and Ebf1 are silenced by H3K27me3 and rendered inaccessible to PU.1 binding from the start, whereas the myeloid regulatory gene Cebpa is bivalently marked. The myeloid and progenitor regulatory gene Sfpi1 (PU.1), initially fully activated, appears to play a regulatory role even into the DN2b stage, and is silenced only when T-cell gene expression is under way. However, there is no simple mapping of developmental lineage exclusion order with a particular molecular class of repression mechanism. Drivers of the most “distant” fate in developmental terms, the erythroid genes, can be repressed via H3K27me3 (EpoR), or without it (Gata1), as can genes associated with the “closest”, NK-cell fate (Eomes, Il2rb respectively). In an interesting additional case, many multipotent progenitor-cell regulatory genes are expressed throughout the early stages like Sfpi1 and only shut off during commitment itself. Some may be sustained by a common progenitor-cell positive regulator, Lmo2 (McCormack et al., 2010), and our results suggest that many receive input from PU.1 itself. The progenitor-associated genes may thus constitute a discrete early subcircuit within the T-lineage specification network.
Finally, our multistage analysis shows that many mouse hematopoietic genes are each likely controlled by different constellations of cis-regulatory elements at one stage of development versus another, even within the same cell lineage. In this light, the quest for single, minimal sufficient regulatory elements for such genes seems naïve, as it would a priori sacrifice the full range of developmental control. The roles of the candidate cis-elements and their rules for engagement with promoters should be greatly clarified by future extensions of this analysis, to detect specific chromatin looping events, enhancer activation states mapped by association with p300 and H3K4me3, and latent enhancers using H3K4me1 at transcription factor binding sites. Mechanisms of repression could be clarified when effects on a broader range of non-activating cis-elements are mapped based on DNase hypersensitivity and DNA methylation. The mapping of developmentally dynamic histone modification sites provides a new way to locate the sites in cis-regulatory DNA that process distinct inputs for crucial regulatory genes. In this collection of regulatory domains lie the answers to how cells are driven to T-lineage commitment.
METHODS
For full materials and experimental procedures, see Supplement. Briefly, in vitro developing CD4− CD8− TCR− “double negative” populations were generated from fetal liver hematopoietic precursors (Taghon et al., 2007) sorted as ETP/DN1 (Kit++ CD44+ CD25−), “DN2a” (Kit++ CD44+ CD25+), and “DN2b” (Kit+ CD44+ CD25+). “DN3” (Kit− CD44− CD25+) and “DP” (CD25− CD4+ CD8+) cells were sorted from thymus. ChIP-seq and RNA-seq were carried out and analyzed as previously reported (Pepke et al., 2009; Mortazavi et al., 2008; Johnson et al., 2007). The programs ERANGE and DEGSeq were used to compare samples.
Supplementary Material
01
02
03
04
05
06
07
08
Acknowledgments
We thank Lorian Schaeffer and Vijaya Kumar for library preparation and sequencing, Henry Amrhein and Diane Trout for data curation, Igor Antoshechkin for sequencing facility management, Diana Perez, Josh Verceles, and Rochelle Diamond for cell sorting and advice, Tian Ling, Georgi Marinov, and Hao Yuan Kueh for statistical advice and programming, Rothenberg group members for sharing advice and unpublished data, and Robert Butler, Lorena Sandoval and Scott Washburn for care of the mice. Support was from NIH grants R33HL089123, R01CA090233, R01CA90233-08S1, and RC2CA148278, the Beckman Institute, the Millard and Muriel Jacobs Genetics and Genomics Center, the L. A. Garfinkle Memorial Laboratory Fund, the Al Sherman Foundation, the Bren Professorship (B.J.W.), and the A. B. Ruddock Professorship (E.V.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession codes. All data are deposited under GEO:GSE31235.
REFERENCES
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- David-Fung E-S, Butler R, Buzi G, Yui MA, Diamond RA, Anderson MK, Rowen L, Rothenberg EV. Transcription factor expression dynamics of early T-lymphocyte specification and commitment. Dev. Biol. 2009;325:444–467. doi: 10.1016/j.ydbio.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Pasca dM, McManus S, Sun Q, Bonifer C, Tagoh H, Busslinger M. Stepwise activation of enhancer and promoter regions of the B cell commitment gene Pax5 in early lymphopoiesis. Immunity. 2009;30:508–520. doi: 10.1016/j.immuni.2009.01.012. [DOI] [PubMed] [Google Scholar]
- DeKoter RP, Lee H-J, Singh H. PU.1 regulates expression of the Interleukin-7 receptor in lymphoid progenitors. Immunity. 2002;16:297–309. doi: 10.1016/s1074-7613(02)00269-8. [DOI] [PubMed] [Google Scholar]
- Donaldson IJ, Chapman M, Kinston S, Landry JR, Knezevic K, Piltz S, Buckley N, Green AR, Gottgens B. Genome-wide identification of cis-regulatory sequences controlling blood and endothelial development. Hum. Mol. Genet. 2005;14:595–601. doi: 10.1093/hmg/ddi056. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CB, Scripture-Adams DD, Proekt I, Taghon T, Weiss AH, Yui MA, Adams SL, Diamond RA, Rothenberg EV. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc. Natl. Acad. Sci. U. S. A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K, van den EP, Bier E, Maxam A, Terhorst C. A T cell-specific enhancer is located in a DNase I-hypersensitive area at the 3' end of the CD3-δ gene. EMBO J. 1988;7:2401–2407. doi: 10.1002/j.1460-2075.1988.tb03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gomez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, Georgopoulos K. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 2010;33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TSP, Painter MW, Consortium TIGP. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- Hosoya T, Kuroha T, Moriguchi T, Cummings D, Maillard I, Lim KC, Engel JD. GATA-3 is required for early T lineage progenitor development. J. Exp. Med. 2009;206:2987–3000. doi: 10.1084/jem.20090934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T, Maillard I, Engel JD. From the cradle to the grave: activities of GATA-3 throughout T-cell development and differentiation. Immunol. Rev. 2010;238:110–125. doi: 10.1111/j.1600-065X.2010.00954.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–342. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Lin YC, Jhunjhunwala S, Benner C, Heinz S, Welinder E, Mansson R, Sigvardsson M, Hagman J, Espinoza CA, Dutkowski J, Ideker T, Glass CK, Murre C. A global network of transcription factors, involving E2A, EBF1 and Foxo1, that orchestrates B cell fate. Nat. Immunol. 2010;11:635–643. doi: 10.1038/ni.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack MP, Young LF, Vasudevan S, de Graaf CA, Codrington R, Rabbitts TH, Jane SM, Curtis DJ. The Lmo2 oncogene initiates leukemia in mice by inducing thymocyte self-renewal. Science. 2010;327:879–883. doi: 10.1126/science.1182378. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Natoli G. Maintaining cell identity through global control of genomic organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell. 2008;14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F, Owens BM, Yu L, Tumang JR, Steidl U, Kutok JL, Clayton LK, Wagner K, Scheller M, Iwasaki H, Liu C, Hackanson B, Akashi K, Leutz A, Rothstein TL, Plass C, Tenen DG. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- Rothenberg EV. T cell lineage commitment: identity and renunciation. J. Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat. Rev. Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg EV, Scripture-Adams DD. Competition and collaboration: GATA-3, PU.1, and Notch signaling in early T-cell fate determination. Semin. Immunol. 2008;20:236–246. doi: 10.1016/j.smim.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schebesta A, McManus S, Salvagiotto G, Delogu A, Busslinger GA, Busslinger M. Transcription factor Pax5 activates the chromatin of key genes involved in B cell signaling, adhesion, migration, and immune function. Immunity. 2007;27:49–63. doi: 10.1016/j.immuni.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nat. Immunol. 2007;8:845–855. doi: 10.1038/ni1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, Zhu J, Zhao K. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H-H, Bollenbacher J, Rovella V, Tripuraneni R, Du Y-B, Liu C-Y, Williams A, McCoy JP, Leonard WJ. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nat. Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- Yang Q, Bell JJ, Bhandoola A. T-cell lineage determination. Immunol. Rev. 2010;238:12–22. doi: 10.1111/j.1600-065X.2010.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yannoutsos N, Barreto V, Misulovin Z, Gazumyan A, Yu W, Rajewsky N, Peixoto BR, Eisenreich T, Nussenzweig MC. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat. Immunol. 2004;5:443–450. doi: 10.1038/ni1053. [DOI] [PubMed] [Google Scholar]
- Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J. Immunol. 2010;185:284–293. doi: 10.4049/jimmunol.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar MA, Chen J, Rothenberg EV. Cell type-specific activation and repression of PU.1 by a complex of discrete, functionally specialized cis-regulatory elements. Mol. Cell Biol. 2010;30:4922–4939. doi: 10.1128/MCB.00354-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08