T cell-specific FADD-deficient mice: FADD is required for early T cell development (original) (raw)
Abstract
FADD/Mort1, initially identified as a Fas-associated death-domain containing protein, functions as an adapter molecule in apoptosis initiated by Fas, tumor necrosis factor receptor-I, DR3, and TRAIL-receptors. However, FADD likely participates in additional signaling cascades. FADD-null mutations in mice are embryonic-lethal, and analysis of_FADD_−/− T cells from_RAG-1_−/− reconstituted chimeras has suggested a role for FADD in proliferation of mature T cells. Here, we report the generation of T cell-specific FADD-deficient mice via a conditional genomic rescue approach. We find that FADD-deficiency leads to inhibition of T cell development at the CD4−CD8− stage and a reduction in the number of mature T cells. The FADD mutation does not affect apoptosis or the proximal signaling events of the pre-T cell receptor; introduction of a T cell receptor transgene fails to rescue the mutant phenotype. These data suggest that FADD, through either a death-domain containing receptor or a novel receptor-independent mechanism, is required for the proliferative phase of early T cell development.
Fas-associated death domain (FADD)/Mort1 was initially isolated as a protein that interacts with the cytoplasmic tail of the Fas receptor in a yeast two-hybrid system (1–3). FADD consists of a “death effector domain” at its amino terminus and a “death domain” at its carboxyl terminus. The death domain was shown to serve as a protein–protein interaction region with several death-domain containing members of the tumor necrosis factor receptor (TNFR) family. The FADD death effector domain recruits caspase-8 (4, 5). Upon interaction of Fas by FasL, for example, FADD, and subsequently caspase-8, are brought to the “death-inducing signaling complex” at the membrane (3, 6, 7). It was suggested that proximity among caspase-8 molecules within the death-inducing signaling complex leads to intermolecular processing and activation of caspase-8 (8, 9). This eventually leads to cleavage of the downstream targets and apoptosis. In addition to the Fas pathway, FADD also is required for signaling of cell death initiated by TNFR-I, DR3, and the TNF-related apoptosis-inducing ligand (TRAIL) receptors DR4 and DR5. FADD-deficient MEFs or_FADD_−/− Jurkat T cells are completely resistant to apoptosis mediated by anti-Fas agonist antibody, TNFα,or TRAIL (10–15).
Apart from its role in apoptosis, analysis of_FADD_−/− mice showed that FADD is also involved in other biological processes (10, 11). FADD-deficient embryos die of heart defects at day 10 of gestation. This could be caused by either a requirement of a death-domain containing receptor in heart development or an involvement of FADD in other unknown processes during differentiation (10, 11). Additionally, FADD appears to play a role in T cell proliferation and/or lymphocyte development. In_RAG-1_−/− chimeras reconstituted with_FADD_−/− embryonic stem cells, B cell development is blocked. T cell development appears to be normal in newborn chimeras, but no thymocytes can be found in animals of 5 wk or older (11). Mature CD4+ or CD8+ T cells, derived presumably from the early waves of T cell progenitors, are found in the peripheral lymphoid organs but are functionally defective. When stimulated through their antigen receptors,_FADD_−/− T cells fail to proliferate to the same extent as their wild-type counterparts (11).
The impaired T cell development observed in_FADD_−/−→RAG-1_−/− chimeras could be caused by the early effect of the FADD mutation in lymphoid progenitor cells or later during T cell development. To explore this issue in more detail, we developed and characterized T cell-specific FADD knockout mice. We first generated transgenic mice carrying a 12-kilobase (kb) fragment of the_FADD genomic locus flanked by two loxP recombination sites, henceforth termed Tg-FADD mice. We show that this transgene is sufficient to rescue_FADD_−/− mice from embryonic lethality and that FADD transgenic mice are phenotypically normal. To induce deletion of the transgene in T cells,FADD_−/− mice bearing the FADD transgene were further crossed to the_lck-Cre transgenic mice, which express the Cre recombinase in T cells (16) to produce Tg-FADD; _lck-Cre; FADD_−/− mice. We show that early T cell development is inhibited in these T cell-specific FADD knockout mice. The reduction in total thymocyte cellularity coincides with the inhibition of a proliferative stage of T cell development between double negative (DN, CD4−CD8−) and double positive (DP, CD4+CD8+) stage. FADD deficiency does not alter apoptosis at this stage. These data suggest that FADD is essential for proliferation of immature thymocytes.
Materials and Methods
Plasmids.
The 12.2-kb _Eco_RV fragment from the FADD gene, which includes both exons, was cloned into the_Eco_RI/_Sma_I site of pSP72 (Promega) plasmid. For the 5′-loxP site, a 44-bp double-stranded oligonucleotide (5′-GTACCCGGGATAACTTCGTATAGCATACATTA TACGAAGTTATG) was inserted into the 5′-_Kpn_I (_Asp_718) site. Another subclone, MortE4.7, which is the 4.7-kb Eco_RI fragment of the_FADD gene cloned into the _Eco_RI site of pSP72 (3), was used to introduce the 3′-loxP site. A 44-bp double-stranded oligonucleotide (5′-CCTAAGCTTATAACTTCGTATAGCATACATTATACGAAGTTATAGG) was inserted into the _Stu_I site of the MortE4.7 subclone. Upon insertion of the loxP oligonucleotides, each of the positive clones was sequenced to ensure that only one oligonucleotide was inserted into the site in the appropriate orientation. The 4.7-kb _Eco_RI fragment of the clone with a 3′-loxP site was then used to replace the_Eco_RI fragment of the _Eco_RV 12.2-kb fragment that had the 5′-loxP site. The final construct was digested with_Sal_I/_Cla_I, and the 12.2-kb fragment was injected into pronucleus of (C57BL/6 × CBA)F1 fertilized eggs to generate transgenic mice.
Mouse Typing.
Tail DNA was analyzed for multiple alleles. All of the PCR reactions were carried out for 1 min at 95°C, 1 min at 59°C, and 1 min at 72°C for 29 cycles. For the FADD transgenic mice, the PCR primers used were CATTGAGGCCTAAGCTTATAAC and GGCTTGAGGCCAACCTAGGTTAC (195-bp product). For lck-Cre, the primers used were CCAGCTAAACATGCTTCATCGTC and CCTGATCCTGGCAATTTCGG (265-bp product). For typing FADD+/− and_FADD_−/− mice, the oligonucleotides used were ACTGTAGTGCCCAGCAGAGACCAGC and CGCTCGGTGTTCGAGGCCACACGC (380-bp product).
Quantitative PCR.
The ABI 5700 with the geneamp 5700 sds software, version 1.1, was used to distinguish_FADD_+/− from_FADD_−/− DNA. γ-Actin primers were used as loading controls (GCACCTAGCACGATGAAGATTAAG and GCCACCGATCCAGACTGAGT). The primers in the neomycin gene were TGCTCCTGCCGAGAAAGTATC and GCCGGATCAAGCGTATGC. The tail DNA from a FADD+/− mouse was used to derive a standard curve whereby one vs. two copies of neomycin (FADD+/− vs._FADD_−/−) could be deduced on an unknown tail sample.
Flow Cytometry.
Cell suspensions were prepared from lymphoid organs of mice 3–8 wk of age. In brief, thymocytes, lymph node cells, and splenocytes were harvested and depleted of erythrocytes by treatment for 5 min at room temperature with red blood cell lysis buffer (Sigma). Cells were filtered, washed, and resuspended in PBS, 4% FCS, and 1 mM NaN3 for analysis. One million peripheral lymphoid cells were incubated with anti-FcγII/III receptor antibody (2.4G2, PharMingen) and normal rabbit serum for 10 min 4°C before cell surface staining. CD4, CD8, B220, and streptavidin-tricolor antibodies were purchased from Caltag. CD25 and CD44 were purchased from PharMingen. Anti-IgM antibody was purchased from Southern Biotechnology Associates. Cell surface staining was carried out for an additional 20 min in staining buffer with the appropriate antibodies. Cells were washed twice, and analysis was performed on the Coulter EPICS XL-MCL machine.
Western Blot Analysis.
For anti-FADD Western blot, 1.0–2.5 × 107 cells were lysed in 0.5% Nonidet P-40, 150 mM NaCl, 1 mM EDTA, 50 mM Tris (pH 7.5), 1 mM DTT, 0.1 M NaF, 1 mM Na3VO4, and 1 mM PMSF. Lysates from 2.5 or 5 million cells were loaded per lane and resolved in 12% SDS/PAGE. Gels were transferred to Optitran membrane (Schleicher & Schuell), blocked for 2 h at room temperature, and incubated with primary antibody (polyclonal anti-FADD) at 4°C overnight. The blot was washed and probed with horseradish peroxidase-conjugated anti-rabbit antibodies (Amersham Pharmacia). Chemiluminescence was carried out by using Renaissance Western blot chemiluminescence reagent (New England Nuclear).
Apoptotic Assays.
Thymocytes were stained with 20 μg/ml 7-aminoactinomycin D (Sigma) in PBS, 4% FCS, and 1 mM NaN3 for 15 min at 4°C. Cells were washed twice, and annexin V staining was performed as per manufacturer's protocols except that 2 μl of annexin V biotin was used instead of the recommended 5 μl. Annexin V biotin conjugate was purchased from PharMingen. The secondary reagent was streptavidin PE purchased from Dako. This reagent was added directly during the annexin V-binding reaction at 3 μl/sample. Analysis was carried out within 1 h of post-staining on the Coulter EPICS XL-MCL.
Results
Genomic Rescue of FADD−/− Mice by a Tg-FADD Construct.
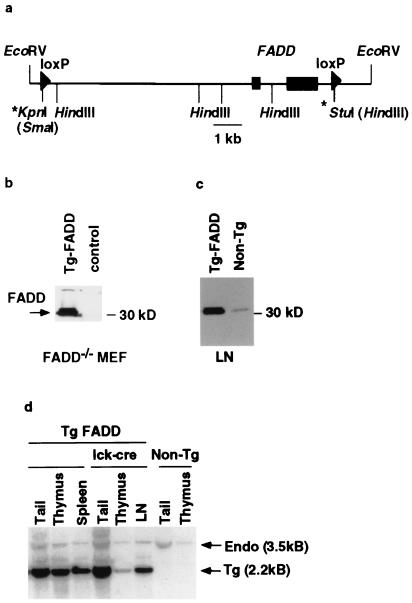
We used a 12-kb Eco_RV fragment of the FADD gene as the basis for a transgenic rescue construct. Two loxP recombination sites were introduced to flank both FADD exons (Fig.1a) such that recombination between these sites would lead to deletion of the FADD gene. To test whether this construct (Tg-FADD) in its unrecombined configuration can drive functional FADD expression, the plasmid was transiently transfected into_FADD_−/− MEF cells. The transfected cells were subsequently analyzed by immunoblotting using FADD-specific polyclonal antibodies (3). As shown in Fig.1b, the Tg-FADD construct reconstituted FADD expression in_FADD_−/− MEF cells. Expression of this 12-kb construct is indistinguishable from that of a larger FADD genomic cosmid clone (data not shown). We conclude that the 12-kb FADD genomic fragment contains all of the regulatory elements needed for controlling FADD transcription, at least in fibroblasts. Four transgenic founders with varying copy numbers (16, 8, 5, and 4 copies) were generated by using this Tg-FADD construct. In the lowest copy mice, FADD expression was slightly elevated in comparison to the nontransgenic_FADD+/− control (Fig. 1c). Three of these founders were bred to_FADD_+/− mice, and the resulting Tg-FADD/FADD+/− were bred again to_FADD_+/− mice to generate Tg-FADD/_FADD_−/− mice. Tg-FADD/_FADD_−/− mice from all of the lines are viable and exhibit no gross abnormalities in T cell development or in T cell proliferation (data not shown), indicating that the FADD transgene functions equivalently to the endogenous gene.
Figure 1.
Generation of T cell-specific_FADD_−/− mice. (a) A schematic diagram of the 12-kb_Eco_RV genomic fragment from the mouse_FADD_ locus used to reconstitute_FADD_−/− mice. The two rectangular boxes represent the two exons that encode the FADD protein. Arrowheads represent the loxP sites. The * at the 5′-end denotes the loxP and a _Sma_I site introduced at the_Kpn_I site, and the * at the 3′-end denotes the loxP and a Hin_dIII site introduced at the_Stu_I site. (b) Western blot analysis of FADD expression in FADD_−/− MEF cells. FADD-deficient MEF cells were transiently transfected with the 12-kb FADD-loxP construct or an irrelevant DNA control. Whole cell extracts were made 2 days post-transfection. FADD expression was assessed by a Western blot analysis with anti-mouse FADD rabbit antibodies. Arrow indicates the phosphorylated and unphosphorylated FADD proteins. (c) Whole cell extracts from lymph nodes (LN) of line 33 FADD transgenic mice or a nontransgenic_FADD+/− littermate control were subjected to Western blot analysis by using anti-FADD rabbit polyclonal antibodies. (d) Southern blot analysis of genomic DNA from tail, thymus, and peripheral immune organs (either spleen or purified lymph node T cells = LN) of t_FADD_−/− mice and their transgenic FADD wild-type littermate or nontransgenic_FADD heterozygous mice. The genomic DNA was digested with _Eco_RV and Hin_dIII. The resulting Southern blot was probed with a fragment from the second exon of_FADD. Arrows indicate the endogenous 3.5-kb band and the 2.2-kb transgenic band.
Generation of Tissue-Specific_FADD−/−_ Mice.
To generate T cell-specific_FADD_−/− mice, we used the previously described lck-Cre transgenic mice (16). In these mice, Cre recombinase expression, controlled by the_lck_ proximal promoter, begins at the DN stage and continues in DP and SP (CD4+CD8− and CD4−CD8+) thymocytes. The Tg-FADD line that carried the lowest copy number (line 33, 4 copies) was bred to lck-Cre/FADD+/− animals to generate Tg-FADD/_FADD_−/−/lck-Cre mice (henceforth called tFADD−/−) and their FADD+ littermate controls (Tg-FADD/_FADD_−/− or Tg-FADD/FADD+/−). Southern blot analysis confirms that the transgenic FADD band is decreased but not completely eliminated in thymus DNA of lck-Cre transgenic mice compared to tail DNA (Fig. 1d). In contrast, no change of the FADD transgene band was evident in the thymus DNA of littermates without the lck-Cre transgene (Fig.1d). The incomplete deletion of Tg-FADD could be caused by variegated expression of the Cre transgene. Alternatively, this could be caused by preferential selection of FADD+ thymocytes during proliferation at the DN/DP transition (see below). In fact, the ratio of transgenic and endogenous bands in peripheral T cells of tFADD−/− mice is similar to that of their Cre-deficient controls (Fig. 1d), suggesting that FADD+ T cells do indeed have a survival advantage over FADD-deficient T cells.
Analysis of tFADD−/− Mice.
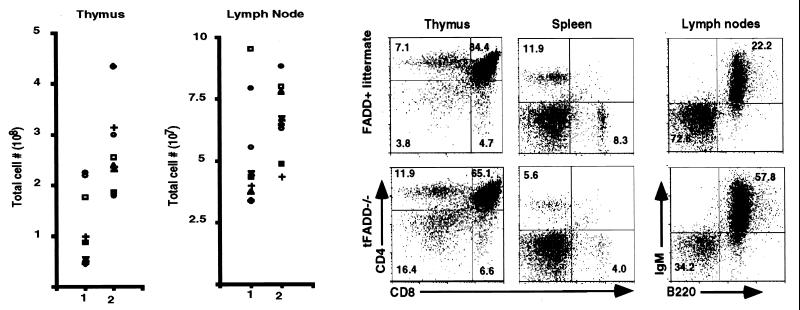
The T cell compartments of mice with tFADD−/− genotype were examined. The total cell numbers of thymocytes, splenocytes, and lymph nodes were determined, and flow cytometric analysis was carried out. As shown in Fig. 2a, the overall thymocyte cellularity is reduced in tFADD−/− mice. As compared to their non-Cre transgenic littermates, there is an average of a 2.2-fold reduction in their thymocyte cellularity. The cellularity of the lymph nodes or spleen does not change dramatically (Fig.2a and data not shown). Nevertheless, flow cytometric analysis with CD4- and CD8-specific antibodies or with IgM and B220-specific antibodies shows that there are lower percentages of CD4 and CD8 T cells and a higher percentage of B cells in the periphery (Fig. 2b).
Figure 2.
tFADD−/− mice exhibit abnormal T cell development. (a) Thymic cellularity and lymph node cell number of t_FADD_−/− mice. The scatter plots depict the thymus (Left) and lymph node (Right) total cellularity. Column 1 represents the tFADD−/− (Tg-FADD/_FADD_−/−/lck-Cre) mice, and column 2 represents the age-matched littermate control (either Tg-FADD/_FADD_−/− or Tg-FADD/FADD+/−). Each symbol represents the particular tFADD−/− and its littermate control from the same experiment. (b) Representative flow cytometric analysis of thymus, spleen, and lymph nodes of tFADD−/− mice and their littermate control (Tg-FADD/FADD+/−). Thymocytes or splenocytes were stained by using antibodies to CD4 and CD8. Lymph node cells were stained with antibodies specific for IgM and B220. The percentage for each quadrant is displayed in the appropriate quadrant. Each plot represents at least 20,000 events.
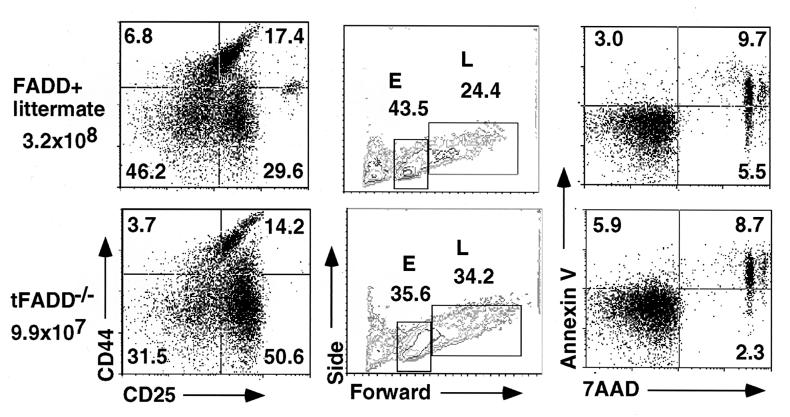
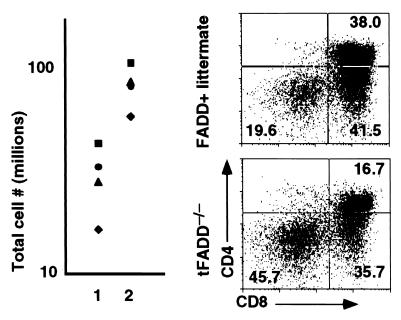
Further analysis of tFADD−/− thymocytes shows an increase in the percentage of DN and a decrease in DP T cells as compared to their FADD+ littermate controls (Fig.2b). In terms of absolute cell numbers, this translates into fewer DP thymocytes in tFADD−/− mice. These data suggest that FADD deficiency results in an inhibition of the DN to DP transition in T cell development. During early T cell development, DN thymocytes start as CD44+CD25− (DN1) population that progresses into CD44+CD25+ (DN2), which then differentiates into CD44−CD25+ (DN3) and finally progresses into CD44−CD25− (DN4) population (17). The DN3 population can be further subdivided into E and L cells based on their cell size (E and L correspond to small and large cells, respectively) (18, 19). Pre-T cell receptor (pre-TCR) signaling initiates between the E and L DN3 stage. Development of DP thymocytes begins with the proliferation at the L cell stage and continues through the DN4 stage (18, 19). Mutations of many genes, including T cell receptor β gene, RAG-1, RAG-2,SLP-76, lck/fyn kinases, and_BRCA-1_, result in a block at the DN3 stage (20–25). To see how the FADD mutation affects early T cell development, tFADD−/− thymocytes were stained for CD4, CD8, CD44, and CD25. The expression of CD44 and CD25 on DN (CD4−CD8−) T cells was then analyzed. As shown in Fig.3a, we found an accumulation of DN3 thymocytes and a corresponding decrease of DN4 T cells in tFADD−/− thymus as compared to their littermate controls (Fig. 3a). The initial pre-TCR signaling appears to be intact as the progression of E to L cells in the DN3 population is normal (Fig. 3b). A slight increase in the L cell population of the tFADD−/− thymus is consistent with a proliferation defect between DN3 (L)/DN4 to DP stages. No significant changes in apoptotic cells are found in the T cell-specific FADD knockout mice as detected by a combination staining of annexin V and 7-aminoactinomycin D in DN thymocytes (Fig.3c). To confirm that the tFADD−/− phenotype is not due to a problem with the initial pre-TCR signaling and/or pre-TCR assembly, we crossed tFADD−/− mice to the 2C TCR transgenic mice. Transgenic TCR has been shown to mimic pre-TCR signaling, leading to an acceleration of T cell development. Thymocyte cell number and CD4 vs. CD8 profiles of 2C TCR transgenic mice between tFADD−/− and FADD positive littermates were compared. Consistent with the notion that the initial pre-TCR signaling is not affected by the FADD mutation, introduction of the 2C prerearranged TCR transgene does not rescue the _FADD_−/− phenotype (Fig. 4). Again, a decrease in thymus cellularity and an increase in percentage of DN thymocytes are evident in the _FADD_-mutant 2C transgenic mice. We conclude from these results that a FADD-null mutation leads to an impairment of early T cell development between the DN3 (L)/DN4 to DP stages.
Figure 3.
Inhibition of early T cell development in tFADD−/− mice. (a) Thymocytes were stained with FITC-conjugated anti-CD4 and anti-CD8 antibodies, PE-conjugated anti-CD44, and biotin-conjugated anti-CD25 antibodies with a streptavidin tri-color secondary. The plots represent CD25 vs. CD44 staining of the gated CD4−CD8− thymocyte population. Each dot plot represents at least 10,000 events in the DN-gated population. The genotype for tFADD−/− is Tg-FADD/_FADD_−/−/lck-Cre whereas its littermate control is Tg-FADD/_FADD_−/−. (b) The CD44lowCD25+ cells (DN3) in the CD4−CD8− population were gated to show their forward vs. side scatter for 15,000 events. E represents the small DN3 thymocytes, and L represents the large DN3 thymocytes. The genotype for the FADD+ control is Tg-FADD/_FADD_−/−. (c) Thymocytes were stained with FITC-conjugated anti-CD4 and anti-CD8 antibodies, followed by staining with 7-aminoactinomycin D (7-AAD) and annexin V. The profiles represent thymocytes gated for the CD4−CD8− cells. The percentage for each quadrant is displayed in the appropriate quadrant. The genotype for the FADD+ control is Tg-FADD/FADD−/−.
Figure 4.
Introduction of a TCR transgene does not rescue the tFADD−/− phenotype. (a) Thymic cellularity of 2C T cell receptor transgenic mice (2C) between t_FADD_−/− and their littermate controls. The scatter plots depict the total thymus cellularity. Column 1 represents the 2C/tFADD−/− mice, and column 2 represents the age-matched littermate control (the genotypes of these mice are either 2C/Tg-FADD/FADD+/−/lck-Cre, 2C/Tg-FADD/FADD+/+, 2C/FADD+/−/lck-Cre, or Tg-FADD/FADD+/−/lck-Cre). Each symbol represents the corresponding littermate. (b) A representative flow cytometric analysis from the thymus of a 2C TCR tFADD−/− transgenic mouse is displayed along with its appropriate 2C transgenic FADD+ littermate control. Thymocytes were stained by using antibodies to the CD8 and CD4. The percentage for each quadrant is displayed. Each plot represents at least 20,000 events.
Discussion
Members of the TNFR superfamily have been shown to participate in a variety of cellular processes, including apoptosis, differentiation, and proliferation (26–30). Those involved in apoptosis contain cytoplasmic tails with a death domain. This domain serves as a protein–protein interacting region to recruit death-domain containing signaling molecules that include FADD and TRADD, an adapter protein that binds to the TNFR-1 death domain (1, 2,31). TRADD also recruits FADD to the signaling complex (32). Genetic and biochemical studies have shown that FADD is involved in signal transduction of all death-domain containing TNFR family members, including Fas, TNF receptor type I, DR3, and TRAIL receptors DR4 and DR5. FADD-deficient MEF and Jurkat cells are resistant to anti-Fas apoptosis as well as TNFα- and TRAIL-mediated death (10–15). Upon ligand-receptor engagement, FADD and its downstream effector caspase, caspase-8, are found in the receptor-signaling complex of Fas, TNF, DR4, and DR5 (4, 5, 7, 33, 34). Thus, FADD is a universal adapter protein that mediates signaling of all known death-domain containing members of the TNF receptor superfamily.
The role of several TNFR family members in the immune system has been elucidated through phenotypic analysis of the corresponding mutant mice. Mice with mutations at the Fas or FasL locus develop severe autoimmunity with symptoms that include splenomegaly, lymphadenopathy, increased serum immunoglobulins, development of anti-double-stranded DNA antibodies, and glomerulonephritis (35, 36). Mice deficient in both TNFR-I and Fas develop autoimmunity at an accelerated pace (37). These data suggest that Fas and TNFR play a crucial role in the maintenance of peripheral tolerance and apoptosis of mature T and B lymphocytes. In contrast to Fas- and TNFR-I-deficient mice, however, FADD-null mice die during gestation and_FADD_−/−→_RAG-1_−/− chimeras do not develop any autoimmunity (10, 11). Because the characterization of DR3- or DR4/5-null mice has not been reported, the requirement of FADD during embryogenesis could be caused by the need of one of these TNFR family members during development. The phenotype observed in_FADD_−/− embryonic stem cell reconstituted_RAG-1_−/− chimeras has suggested novel roles for FADD. In addition to problems in B cell development,_FADD_−/− mature T cells fail to proliferate normally when challenged with anti-CD3 and anti-CD28 antibodies. However, the lack of proliferation could formally be due to secondary defects of abnormal T cell development. FADD−/− thymocyte populations appear to be normal in newborn chimera but 5-wk-old animals do not have any thymocytes (11). The absence of DP in these animals could be due to a block in DN to DP transition. Alternatively, the observed FADD−/− T cell phenotype also could be caused by a defect in the common lymphoid progenitors or an accelerated process of apoptosis in the absence of FADD.
Several groups have attempted to examine the role of FADD in T cell development by over-expressing a FADD dominant negative mutant using the lck proximal promoter (38–41). However, the reported phenotypes of these FADD dominant negative transgenic mice vary to some extent. Despite a very high level of transgene expression, no changes in T cell development was observed in mice expressing the mouse FADD dominant negative protein (41). By using the human version of FADD mutant, Walsh et al. (39) found an inhibition of T cell development at the DN stage. Others reported that the FADD dominant negative mutant might inhibit DN to DP transition but can also induce differentiation of DN thymocytes into DP thymocytes in RAG-1−/− mice (38, 42). To get a clearer picture of whether FADD functions in T cell development, we have generated T cell-specific FADD-deficient mice. Instead of introducing loxP sites into the FADD gene, we chose to use a conditional genomic rescue construct containing the_FADD_ gene flanked by two loxP sites. We showed that expression of the transgenic FADD in different copy numbers can rescue the embryonic lethality of_FADD_−/− mice. We then used Cre transgenic mice (lck-Cre) to delete the loxP flanked genomic FADD construct in a T cell-specific manner. Analysis of these mice showed consistently that FADD deficiency in T cells results in several-fold decrease of DP thymocytes and an inhibition of pre-T cell development at the DN3 stage. Cre-mediated deletion of the Tg-FADD is not complete, possibly because of many factors. The copy number of the transgenic FADD (four copies) could contribute to incomplete deletion. As the lck proximal promoter does not contain locus control region activity, variegated expression of the Cre transgene could be another contributing factor. Finally, as FADD deficiency impairs the DN to DP thymocyte transition, FADD-null DN cells could be at a selective disadvantage; those that retain the FADD transgene would preferentially proliferate to populate the thymic compartment. Consistent with this latter idea, peripheral T cells in tFADD−/− mice express FADD at a level indistinguishable from that of the wild-type littermate T cells (data not shown). Southern blot analysis of lymph node DNA also shows quantitatively the same ratio of transgenic band to endogenous band as that of the tail DNA. In addition, no proliferative defects have been detected in the peripheral T cells of tFADD−/− mice (data not shown). It is therefore possible that the emergence of mature FADD-null T cells in the young_RAG-1_−/− chimeras is caused by a lack of competition. Despite a lack of complete Cre-mediated deletion in tFADD−/− mice, the data obtained argue that the abnormal T cell development in 2 wk or older_FADD_−/−_→RAG-1_−/− chimeras is caused by a complete block of DN to DP transition.
One of the crucial checkpoints during early T cell development is proper assembly of the T cell receptor β chain gene segments (18,19). Association of a productively rearranged TCR β product with the pre-T cell α chain is an obligatory event for DN thymocytes to become DP T cells (43). Signals from the pre-TCR lead to proliferation and differentiation of DN T cells (18, 19). We showed that the differentiation stage that requires pre-TCR signals, E to L DN3 stage, is normal in tFADD−/− mice. Furthermore, introduction of the rearranged 2C TCR transgene does not rescue the FADD-mutant phenotype. These data indicate that TCR β gene rearrangement and the initial pre-TCR signaling proceed normally in the absence of FADD. Although cell cycle analysis failed to reveal any differences between tFADD−/− and FADD+ DN thymocytes (N.H.K., unpublished data), the increased percentage of DN3 population and the decrease of the absolute cell number of DP thymocytes in the FADD-deficient thymus would indicate that proliferation between DN3 (L)/DN4 to DP is compromised. Interestingly, RAG-2-deficient DN3 developing in normal thymic environment of a chimeric animal showed normal cell cycle profile (44), suggesting that pre-TCR signaling is not required for progression of G1 to S transition in pre-T cells. We also showed that in tFADD−/− mice, apoptosis of DN thymocytes does not change substantially. Consistent with this idea, DN thymocytes from tFADD−/− mice did not exhibit any changes in p53 tumor suppressor (data not shown), a protein that has been shown to mediate apoptosis of DN T cells in mice with mutation in the pre-TCR complex or in the RAG-1 or RAG-2 gene (45,46).
The signal transduction pathways of pre-TCR and mature TCR share many similarities. Many proteins implicated in signaling of mature T cells are also involved in pre-TCR signaling. For example, T cell development of the lck/fyn double-mutant mice is arrested at the DN3 stage (24). A similar block was found for mice deficient in SLP76, another protein important for transmitting TCR signals (23). Recently, the mutation of BRCA-1, a gene implicated in breast cancer and DNA repair/damage, was shown to affect not only proliferation of mature T cells but also development of DN thymocytes (25). We have previously shown that FADD-deficient mature T cells, which develop in_FADD_−/−_→RAG-1_−/− chimeras are defective in their capacity to proliferate in response to anti-TCR stimulation (11). TCR- and IL-2-signaling pathways seem to be intact in these cells, suggesting that FADD is required for downstream signals that lead to proliferation. Although this could be formally caused by an indirect effect of abnormal T cell development, we show here that early T cell development in tFADD−/− mice is inhibited at the DN3 (L)/DN4 to DP transition, a critical proliferative stage in early T cell development. Thus, FADD may regulate not only proliferation of mature T cells but also immature T cells during development. Whether FADD is required for transmitting signals of a death-domain containing protein in this context is not known. FADD is phosphorylated at the G2/M state of the cell cycle (47), perhaps influencing its activity and cell-cycle progression. Future biochemical and genetic experiments are necessary to resolve this issue.
Acknowledgments
We thank Sue Sohn and Nancy Hong for critical reading of this manuscript, Dr. Jamey Marth for lck-Cre transgenic mice, and members of the Winoto lab for helpful discussion. This work is supported by National Institutes of Health Grant CA75162 and National Science Foundation Presidential Faculty Fellow Award (to A.W.).
Abbreviations
TNFR
tumor necrosis factor receptor
DN
double negative
DP
double positive
MEF
mouse embryonic fibroblast
pre-TCR
pre-T cell receptor
Tg
transgenic
TRAIL
TNF-related apoptosis-inducing ligand
References
- 1.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 2.Chinnaiyan A M, O'Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 3.Zhang J, Winoto A. Mol Cell Biol. 1996;16:2756–2763. doi: 10.1128/mcb.16.6.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muzio M, Chinnaiyan A M, Kischkel F C, O'Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, et al. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 5.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 6.Kischkel F C, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer P H, Peter M E. EMBO J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chinnaiyan A M, Tepper C G, Seldin M F, O'Rourke K, Kischkel F C, Hellbardt S, Krammer P H, Peter M E, Dixit V M. J Biol Chem. 1996;271:4961–4965. doi: 10.1074/jbc.271.9.4961. [DOI] [PubMed] [Google Scholar]
- 8.Muzio M, Stockwell B R, Stennicke H R, Salvesen G S, Dixit V M. J Biol Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 9.Yang X L, Chang H Y, Baltimore D. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 10.Yeh W-C, Pompa J L, McCurrach M E, Shu H-B, Elia A J, Shahinian A, Ng M, Wakeham A, Khoo W, Mitchell K, et al. Science. 1998;279:1954–1958. doi: 10.1126/science.279.5358.1954. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J, Cado D, Chen A, Kabra N H, Winoto A. Nature (London) 1998;392:296–300. doi: 10.1038/32681. [DOI] [PubMed] [Google Scholar]
- 12.Bodmer J-L, Holler N, Reynard S, Vinciguerra P, Schneider P, Juo P, Blenis J, Tschopp J. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 13.Kischkel F C, Lawrence D A, Chuntharapai A, Schow P, Kim K J, Ashkenazi A. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 14.Kuang A A, Diehl G E, Zhang J, Winoto A. J Biol Chem. 2000;275:25065–25068. doi: 10.1074/jbc.C000284200. [DOI] [PubMed] [Google Scholar]
- 15.Sprick M R, Weigand M A, Rieser E, Rauch C T, Juo P, Blenis J, Krammer P H, Walczak H. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 16.Orban P C, Chui D, Marth J D. Proc Natl Acad Sci USA. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Godfrey D I, Zlotnik A. Immunol Today. 1993;14:547–553. doi: 10.1016/0167-5699(93)90186-O. [DOI] [PubMed] [Google Scholar]
- 18.Haks M C, Oosterwegel M A, Blom B, Spits H, Kruisbeek A M. Semin Immunol. 1999;11:23–37. doi: 10.1006/smim.1998.0153. [DOI] [PubMed] [Google Scholar]
- 19.Hayday A C, Barber D F, Douglas N, Hoffman E S. Semin Immunol. 1999;11:239–249. doi: 10.1006/smim.1999.0180. [DOI] [PubMed] [Google Scholar]
- 20.Mombaerts P, Clarke A R, Rudnicki M A, Iacomini J, Itohara S, Lafaille J J, Wang L, Ichikawa Y, Jaenisch R, Hooper M L. Nature (London) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 21.Mombaerts P, Iacomini J, Johnson R S, Herrup K, Tonegawa S, Papaioannou V E. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 22.Alt F W, Rathbun G, Oltz E, Taccioli G, Shinkai Y. Ann NY Acad Sci. 1992;651:277–294. doi: 10.1111/j.1749-6632.1992.tb24626.x. [DOI] [PubMed] [Google Scholar]
- 23.Pivniouk V, Tsitsikov E, Swinton P, Rathbun G, Alt F W, Geha R S. Cell. 1998;94:229–238. doi: 10.1016/s0092-8674(00)81422-1. [DOI] [PubMed] [Google Scholar]
- 24.Van Oers N S C, Lowinkropf B, Finlay D, Connolly K, Weiss A. Immunity. 1996;5:429–436. doi: 10.1016/s1074-7613(00)80499-9. [DOI] [PubMed] [Google Scholar]
- 25.Mak T W, Hakem A, McPherson J P, Shehabeldin A, Zablocki E, Migon E, Duncan G S, Bouchard D, Wakeham A, Cheung A, et al. Nat Immunol. 2000;1:77–82. doi: 10.1038/76950. [DOI] [PubMed] [Google Scholar]
- 26.Nagata S. Cell. 1997;88:355–365. doi: 10.1016/s0092-8674(00)81874-7. [DOI] [PubMed] [Google Scholar]
- 27.Ashkenazi A, Dixit V M. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 28.Wallach D, Varfolomeev E E, Malinin N L, Goltsev Y V, Kovalenko A V, Boldin M P. Annu Rev Immunol. 1999;17:331–368. doi: 10.1146/annurev.immunol.17.1.331. [DOI] [PubMed] [Google Scholar]
- 29.Lenardo M, Chan K-M, Hornung F, McFarland H, Siegel R, Wang J, Zheng L. Annu Rev Immunol. 1999;17:221–254. doi: 10.1146/annurev.immunol.17.1.221. [DOI] [PubMed] [Google Scholar]
- 30.Krammer P H. Nature (London) 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- 31.Hsu H, Xiong J, Goeddel D V. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 32.Hsu H, Shu H-B, Pan M-G, Goeddel D V. Cell. 1996;84:299–308. doi: 10.1016/s0092-8674(00)80984-8. [DOI] [PubMed] [Google Scholar]
- 33.Varfolomeev E E, Boldin M P, Goncharov T M, Wallach D. J Exp Med. 1996;183:1271–1275. doi: 10.1084/jem.183.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Medema J P, Scaffidi C, Kischkel F C, Shevchenko A, Mann M, Krammer P H, Peter M E. EMBO J. 1997;16:2794–2804. doi: 10.1093/emboj/16.10.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen P L, Eisenberg R A. Annu Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- 36.Nagata S, Golstein P. Science. 1995;267:1449–1455. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 37.Zhou T, Edwards C K, 3rd, Yang P, Wang Z, Bluethmann H, Mountz J D. J Immunol. 1996;156:2661–2665. [PubMed] [Google Scholar]
- 38.Newton K, Harris A W, Bath M L, Smith K G C, Strasser A. EMBO J. 1998;17:706–718. doi: 10.1093/emboj/17.3.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walsh C M, Wen B G, Chinnaiyan A M, O'Rourke K, Dixit V M, Hedrick S M. Immunity. 1998;8:439–449. doi: 10.1016/s1074-7613(00)80549-x. [DOI] [PubMed] [Google Scholar]
- 40.Zornig M, Hueber A-O, Evan G. Curr Biol. 1998;8:467–470. doi: 10.1016/s0960-9822(98)70182-4. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, DeYoung A, Kasler H G, Kabra N H, Kuang A A, Diehl G, Sohn S J, Bishop C, Winoto A. Cold Spring Harbor Symp Quant Biol. 1999;64:363–371. doi: 10.1101/sqb.1999.64.363. [DOI] [PubMed] [Google Scholar]
- 42.Newton K, Harris A W, Strasser A. EMBO J. 2000;19:931–941. doi: 10.1093/emboj/19.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling H J, von Boehmer H. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 44.Petrie H T, Tourigny M, Burtrum D B, Livak F. J Immunol. 2000;165:3094–3098. doi: 10.4049/jimmunol.165.6.3094. [DOI] [PubMed] [Google Scholar]
- 45.Haks M C, Krimpenfort P, van den Brakel J H N, Kruisbeek A M. Immunity. 1999;11:91–101. doi: 10.1016/s1074-7613(00)80084-9. [DOI] [PubMed] [Google Scholar]
- 46.Jiang D, Lenardo M J, Zuniga-Pflucker C. J Exp Med. 1996;183:1923–1928. doi: 10.1084/jem.183.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scaffidi C, Volkland J, Blomberg I, Hoffmann I, Krammer P H, Peter M E. J Immunol. 2000;164:1236–1242. doi: 10.4049/jimmunol.164.3.1236. [DOI] [PubMed] [Google Scholar]