Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions (original) (raw)
Abstract
Dengue is a mosquito-borne flavivirus that is spreading at an unprecedented rate and has developed into a major health and economic burden in over 50 countries. Even though infected individuals develop potent and long-lasting serotype-specific neutralizing antibodies (Abs), the epitopes engaged by human neutralizing Abs have not been identified. Here, we demonstrate that the dengue virus (DENV)-specific serum Ab response in humans consists of a large fraction of cross-reactive, poorly neutralizing Abs and a small fraction of serotype-specific, potently inhibitory Abs. Although many mouse-generated, strongly neutralizing monoclonal antibodies (mAbs) recognize epitopes that are present on recombinant DENV envelope (E) proteins, unexpectedly, the majority of neutralizing Abs in human immune sera bound to intact virions but not to the ectodomain of purified soluble E proteins. These conclusions with polyclonal Abs were confirmed with newly generated human mAbs derived from DENV-immune individuals. Two of three strongly neutralizing human mAbs bound to E protein epitopes that were preserved on the virion but not on recombinant E (rE) protein. We propose that humans produce Abs that neutralize DENV infection by binding a complex, quaternary structure epitope that is expressed only when E proteins are assembled on a virus particle. Mapping studies indicate that this epitope has a footprint that spans adjacent E protein dimers and includes residues at the hinge between domains I and II of E protein. These results have significant implications for the DENV Ab and vaccine field.
Dengue viruses (DENVs) are emerging arboviruses and the causative agents of dengue fever and dengue hemorrhagic fever (DHF). The DENV complex consists of four distinct but related viruses, designated as serotypes (1, 2). A person infected with DENV develops an antibody (Ab) response that, to varying degrees, cross-reacts with all four serotypes. Despite the cross-reactivity, Abs that are produced durably only prevent reinfection by the same homologous serotype. Serotype-specific neutralizing Abs can be detected 60 y after a primary infection, suggesting that Abs provide lifelong protection against the homologous serotype (3). People experiencing a secondary DENV infection with a different (heterologous) serotype face a greater risk for developing DHF. Ab-dependent enhancement by cross-reactive, weakly neutralizing Abs is the most widely suggested theory explaining the higher risk for DHF associated with secondary infection (4). The identity of DENV epitopes recognized by human Abs responsible for potent and long-term neutralization remains unknown. This is a significant knowledge gap impeding the current global effort to develop dengue vaccines that induce protective neutralizing Abs and not cross-reactive Abs with potential to enhance disease.
The DENV envelope contains two integral membrane proteins designated envelope (E) and premembrane/membrane (prM/M) proteins. DENV E protein, which binds to cellular receptors and mediates viral fusion during entry, is thought to be the major target of neutralizing Abs (5). The ectodomain of E proteins has been crystallized, and atomic structures have been determined for several flaviviruses (6–9). Individual subunits of E protein consist of three β-barrel domains designated domains I (EDI), II (EDII), and III (EDIII), with the native protein forming a head-to-tail homodimer on the mature virion. The mature DENV particle consists of 90 dimers that cover the surface of the virion (10). Although several groups have characterized mouse monoclonal antibodies (mAbs) that neutralize DENV infection (4, 5) and mapped them to all three domains on the E protein (5, 11, 12), the strongest neutralizing mouse mAbs were serotype-specific and bound to two overlapping and adjacent epitopes on the lateral ridge and A-strand of EDIII (11–16).
To understand how human Abs neutralize DENV, investigators have begun to characterize human immune sera and human monoclonal Abs (hmAbs) (17–19). Humans also produce EDIII-reactive Abs, including strongly neutralizing mAbs that bind to similar epitopes recognized by murine EDIII Abs (18, 20). However, several recent observations indicate that EDIII-specific Abs alone are unlikely to account for the strong type-specific neutralizing Ab responses observed in people following natural infections. DENV-immune humans have low levels of serum EDIII-specific Abs, and these sera retained potently neutralizing activity even after depletion of EDIII-binding Abs (21–23). Moreover, recombinant DENVs with mutations in EDIII epitopes recognized by neutralizing Abs remained sensitive to neutralization by human DENV-immune sera (24). Collectively, these observations suggest that humans produce neutralizing Abs that bind to epitopes other than those on EDIII. Here, we characterized polyclonal sera and hmAbs generated from DENV-immune individuals to identify DENV epitopes engaged by potently neutralizing human Abs. We demonstrate that human neutralizing Abs recognize a complex epitope that is preserved on the intact virion but is not present on the soluble E protein.
Results
Depletion of Homologous DENV-Specific Abs from Immune Sera.
Studies were undertaken to characterize Abs in human immune sera responsible for potent and long-term neutralization of the homologous virus serotype. We assembled a panel of eight immune sera from healthy volunteers who had been exposed to primary DENV2 or DENV3 infections ∼2–9 y before blood collection (Table S1). Human serum from individuals lacking a past history of DENV infections (confirmed by ELISA and neutralization assays) was used as a negative control.
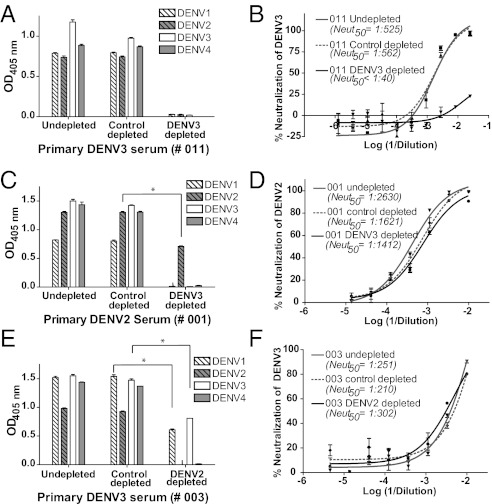
To define the Ab subpopulation in immune sera responsible for DENV neutralization, we developed a bead-based technique to fractionate DENV-specific Abs in immune sera. Polystyrene beads coated with virions of the homologous serotype were incubated with immune sera at 37 °C to deplete DENV-binding Abs. Untreated and control-depleted serum samples bound to whole virus from each of the four DENV serotypes by ELISA and efficiently neutralized DENV (Fig. 1 A and B). Serum samples depleted using beads coated with the homologous DENV displayed greatly reduced binding and neutralization of DENV (Fig. 1 A and B), indicating that beads coated with the homologous serotype successfully removed most DENV-specific Abs from immune sera.
Fig. 1.
Binding and neutralization properties of primary DENV-immune sera depleted of total or cross-reactive DENV-binding Abs. Total DENV-specific Abs were removed from DENV3 primary immune serum (e.g., subject 011) using polystyrene beads coated with purified DENV3 and tested for DENV binding (A) and neutralization (B). The sera depleted with the homologous serotype did not bind to any of the four DENVs and failed to neutralize DENV3. Similar results were observed for four other primary immune sera (2 DENV2 and 2 DENV3 sera) depleted with the homologous serotype responsible for infection. Primary DENV2 (C and D) and DENV3 (E and F) immune sera were depleted of cross-reactive Abs using beads coated with virus of a heterologous serotype and tested for DENV binding (C and E) and neutralization of the homologous serotype (D and F). Immune sera depleted of cross-reactive Abs contained type-specific Abs that bound to virus from the homologous serotype only. Immune sera depleted of cross-reactive Abs were as potently neutralizing as undepleted or control-depleted sera. Results presented here for cross-reactive Ab depletions are representative of data obtained with four primary DENV2 and three primary DENV3 human immune sera (Table 1). *P < 0.001 by an unpaired Student t test of mean binding values.
Depletion of Heterologous DENV-Specific Abs from Immune Sera.
Next, we assessed the contribution of DENV cross-reactive Abs in immune sera to virus binding and neutralization. We used polystyrene beads coated with virus of a heterologous serotype (a serotype that has not infected the DENV-immune subject) to deplete cross-reactive Abs from primary immune sera (Fig. 1 and Table 1). Depletion of primary DENV2-immune sera with DENV3-coated beads led to the removal of all cross-reactive Abs, with the remaining Abs binding to DENV2 in a type-specific manner (Fig. 1_C_). Reciprocal depletion of primary DENV3-immune sera with DENV2-coated beads removed all binding to DENV2 and DENV4 but not to DENV3 and, to a lesser extent, DENV1 (Fig. 1_E_). This residual DENV1-binding signal may be attributable to Abs targeting subcomplex epitopes that are preferentially shared between DENV1 and DENV3 (12, 13, 25). Removal of cross-reactive Abs from primary immune sera did not change the capacity of the sera to neutralize the virus responsible for infection (Fig. 1 D and F, Fig. S1, and Table 1). These results demonstrate that the DENV-specific human Ab response consists of both cross-reactive and type-specific Abs. Although the serotype cross-reactive Abs were abundant, in the samples we analyzed, their contribution to neutralization was negligible. Thus, type-specific Abs appear to be primarily responsible for neutralizing the homologous serotype.
Table 1.
Homologous DENV serotype neutralization titers of immune sera depleted of cross-reactive Abs from subjects following primary infection
| Infection serotype | Sample ID | Reciprocal of Neut50 titer against the homologous virus (SEM)*† | ||
|---|---|---|---|---|
| Undepleted | Control-depleted | Cross-reactive Ab-depleted | ||
| Primary DENV2 | 001 | 2600 | 1650 | 1412 |
| (2,040–2,700) | (1,100–1,650) | (1,060–1,600) | ||
| 013 | 350 | 320 | 420 | |
| (260–470) | (260–380) | (370–550) | ||
| 019 | 1202 | 1047 | 1000 | |
| (1,000–1,550) | (930–1,580) | (800–1,420) | ||
| 031 | 1150 | 790 | 640 | |
| (1,000–1,310) | (650–950) | (540–740) | ||
| Primary DENV3 | 003 | 250 | 210 | 300 |
| (230–350) | (160–260) | (250–360) | ||
| 011 | 320 | 300 | 252 | |
| (265–390) | (260–380) | (211–300) | ||
| 118 | 628 | 720 | 618 | |
| (510–770) | (610–860) | (500–750) |
Depletion of DENV Recombinant E Protein-Binding Abs from Immune Sera.
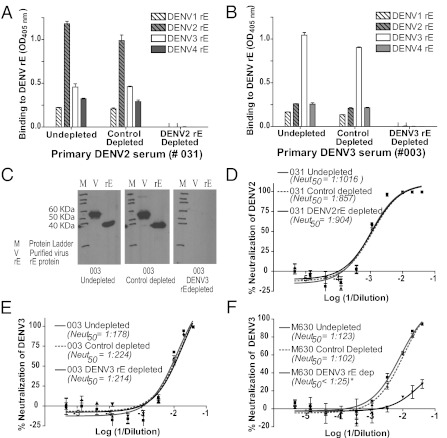
The organization of DENV E protein dimers on the surface of the infectious virus has been modeled using crystal structures of DENV recombinant E (rE) and cryo-EM reconstructions of the virion (8, 10, 26, 27). Furthermore, neutralizing mouse mAbs have been mapped extensively to the rE protein, and DENV subunit vaccines using the rE protein are currently being developed (11–15, 28–30). We next assessed whether epitopes targeted by neutralizing Abs in human immune sera were preserved on the rE protein. DENV rE protein that was covalently coupled to agarose beads was used to deplete Abs in immune sera. Sera were incubated with either control beads or homologous rE-conjugated beads at 37 °C. The structure of DENV rE on the beads was confirmed to be conformationally preserved, and rE dimers were confirmed to be intact by successfully depleting mouse mAbs previously mapped to the fusion loop (mAb 4G2), EDIII (mAb 9F16) (12), and E dimer interface (mAbs DV2-10, DV2-46, and DV2-58) (11) (Fig. S2). We also titrated the amount of rE protein on the beads required to deplete rE-binding Abs efficiently from immune sera (Fig. S3). Both untreated and control-depleted immune sera bound to rE from all four serotypes, but the binding was greatest for the homologous serotype (Fig. 2 A and B). Depletion of primary immune sera using homologous rE ablated binding to rE from each of the four serotypes (Fig. 2 A and B). Successful depletion of rE-binding Abs was also confirmed by Western blot, where rE and solubilized virions were used as the antigen on the blot (Fig. 2_C_). By Western blot, we could not detect binding to rE protein (which is missing 20% of the native protein at the C terminus) or to full-length E protein from the virus (Fig. 2_C_). These results established that beads coated with the rE from the homologous serotype efficiently removed all Abs recognizing purified rE protein. We also measured the relative proportion of virion-binding Abs in human immune sera that bound to rE by comparing the binding of untreated, control-depleted, and rE-depleted sera with the homologous virus by ELISA. Results demonstrated an approximate 45 ± 7% reduction in DENV binding following the removal of rE-binding Abs (Fig. S4 and Table S2), indicating that approximately half of the DENV-specific Abs in primary immune sera recognized the intact virus but not rE protein.
Fig. 2.
Binding and neutralization properties of primary DENV-immune sera depleted of rE-binding Abs. DENV rE from the homotypic strain was coupled covalently to agarose beads and incubated with the relevant DENV-immune sera to deplete DENV rE-specific Abs. (A and B) Binding of immune sera to rE protein. Primary DENV2 (A) and DENV3-immune (B) sera were depleted with DENV2 and DENV3 rE proteins, respectively, and binding to rE protein from each of the four serotypes was measured by ELISA. Depletion with the rE from the homologous serotype led to a loss of binding to rE protein from each of the four serotypes. (C) Successful removal of all rE-reactive Abs from sera (e.g., primary DENV3-immune subject 003) also was confirmed by Western blot analysis. Purified homotypic DENV (700 ng per well) and rE protein (500 ng per well) were electrophoresed, transferred to nitrocellulose membrane, and probed with undepleted, control-depleted, or rE-depleted sera (at a 1:1,000 dilution). (D and E) Neutralization of the homologous DENV by rE-depleted sera was measured using a U937 + DC-SIGN flow cytometry-based assay. Homologous DENV neutralization by primary DENV2 (D; subject 031) and primary DENV3 (E; subject 003) human immune sera depleted of rE-binding Abs was tested. No reduction in neutralization potency was observed following removal of rE-binding Abs from either of these two serum samples. A total of six primary immune sera were depleted of rE-binding Abs and tested (Table 2). (F) Nonhuman primates vaccinated with rE develop neutralizing Abs that can be depleted with rE antigen. Rhesus macaques (M. mulatta) were vaccinated and boosted with an α-virus vector expressing DENV3 E ectodomain, and sera were collected 10 wk postvaccination. Depletion of rE-binding Abs from sera of vaccinated animals (e.g., M630) removed greater than 98% (value estimated by comparing Neut50 values between control-depleted and rE-depleted sera) of the neutralizing Abs. Data are representative of two vaccinated rhesus macaque controls.
Next, we assessed the neutralizing activity of six immune sera depleted of rE-binding Abs. Unexpectedly, four of the six immune sera displayed no loss of neutralization potency after removal of rE-binding Abs (Fig. 2 D and E and Table 2). One of the three primary DENV2-immune sera and all three of the primary DENV3-immune sera tested displayed no significant loss of neutralization against the homotypic virus after removal of rE-specific Abs. In contrast, two of the three primary DENV2-immune sera displayed a statistically significant two- to threefold drop (P < 0.05) in the 50% neutralization (Neut50) titer when rE-specific Abs were removed (Table 2). Sera from rhesus macaques (_Macaca mulatta_) immunized with Venezuelan equine encephalitis virus (VEEV) replicons expressing DENV3 E85 protein were used as a positive control in these experiments. These animals should develop neutralizing Abs that bind to rE protein; accordingly, rE-coated beads removed >98% of the neutralizing Abs from these vaccine sera (Fig. 2_F_). We conclude that although there was some variation among human immune sera in the contribution of rE-reactive Abs to homotypic DENV neutralization, a large fraction of DENV neutralizing Abs in humans consists of neutralizing Abs that bind to intact virions but not the rE protein.
Table 2.
Homologous DENV serotype neutralization titers of primary immune sera depleted of rE-binding Abs
| Infection serotype | Sample ID | Reciprocal of Neut50 titer against the homologous virus (SEM)* | ||
|---|---|---|---|---|
| Undepleted | Control-depleted | rE-depleted | ||
| Primary DENV2 | 001† | 660 | 850 | 260 |
| (510–840) | (690–950) | (210–310) | ||
| 019‡ | 1,250 | 1,120 | 500 | |
| (1,010–1,500) | (950–1,350) | (400–600) | ||
| 031 | 1,020 | 860 | 900 | |
| (790–1,300) | (670–1,150) | (690–1,190) | ||
| Primary DENV3 | 003 | 180 | 225 | 215 |
| (155–250) | (210–310) | (150–220) | ||
| 105 | 180 | 200 | 160 | |
| (140–235) | (160–275) | (130–185) | ||
| 118 | 1,580 | 1,480 | 1,120 | |
| (1,180–2,020) | (1,200–1,790) | (900–1,400) |
Characterization of hmAbs That Strongly Neutralize DENV.
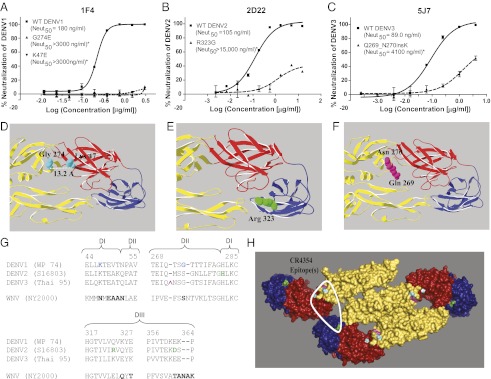
As an alternate approach to identify neutralizing viral epitopes targeted by DENV-immune individuals, we generated a panel of hmAbs that strongly neutralized DENV. These Abs were generated by transforming memory B cells from DENV-immune subjects with EBV and generating hmAbs by electrofusion as previously described (17). Because strongly neutralizing hmAbs comprise a minor fraction of the total hmAbs isolated from immune subjects (18–20), we used a two-step screen to isolate strongly inhibitory Abs: We first identified Abs that bound to DENV virions and then tested them for neutralizing activity. We isolated three strongly neutralizing type-specific hmAbs (Neut50 value <0.2 μg/mL), designated 1F4, 2D22, and 5J7, that inhibited infection of DENV1, DENV2, and DENV3, respectively. Two of these hmAbs bound to the intact virus but not to rE (Table 3).
Table 3.
Binding and neutralization properties of strongly neutralizing hmAbs
| mAbs | Binding, 2 μg/mL* | Neut50 titer, μg/mL‡ | Escape mutant | Escape mutation | Mutation location | Analogous residue in WNV (comparison to CR4354 epitope)§ | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Virus | rE | EDIII | prM† | DENV1 | DENV2 | DENV3 | DENV4 | |||||
| 1F4 | Type-specific (DENV1) | − | − | − | 0.11 | >10 | >10 | >10 | 1 | G274E | DI-DII hinge | 276 (contact site) |
| 2 | K47E | DI | 49 (contact site) | |||||||||
| 2D22¶ | Type-specific (DENV2) | − | − | − | >10 | 0.08 | >10 | >10 | 3 | R323G | DIII | 326 (near contact site 328) |
| 5J7¶ | Type-specific (DENV3) | + | − | − | >10 | >10 | 0.10 | >10 | 4 | Q269_N271insK | DI-DII hinge | Before 274 (near contact site 276) |
Generation of DENV Mutants That Escape Neutralization by hmAbs.
To map the epitopes engaged by neutralizing hmAbs, we subjected the appropriate DENV serotype to Ab pressure and selected for neutralization escape mutant viruses in vitro. DENV1, DENV2, or DENV3 was passaged several times under varying concentrations (0.2–10 μg/mL) of the neutralizing hmAb 1F4, 2D22, or 5J7, respectively. The original WT virus was passaged in parallel in the absence of hmAb treatment. Structural genes of the mutant and WT viruses were sequenced and compared to identify the mutation(s) responsible for neutralization escape. We successfully isolated two escape mutants against DENV1 type-specific mAb 1F4, with two independent single-nucleotide mutations resulting in amino acid changes at position 274 (G→E) in the DI-DII hinge and 47 (K→E) in DI of the E protein (Fig. 3 A and D) that conferred loss of neutralization. K47 and G274 are located 13.2 Å apart and likely comprise part of the same 1F4 epitope. For the DENV2-specific neutralizing hmAb 2D22, we isolated one mutant with an EDIII mutation at residue 323 (R→G) that resulted in neutralization escape (Fig. 3 B and E). Selection with the neutralizing DENV3-specific hmAb 5J7 resulted in an escape mutant with a lysine insertion in the E DI-DII hinge region between the amino acid residues Q269 and N270 (Fig. 3 C and F). All the mutated residues are surface-exposed on the structure of the E protein dimer and within the footprint of a complex epitope described for an hmAb (CR4354) that strongly neutralized West Nile virus (WNV) (31, 32) (Fig. 3 G and H and Table 3).
Fig. 3.
Epitope mapping of escape mutants generated from type-specific neutralizing hmAbs. Neutralization profiles of respective WT and escape mutants against 1F4 (A), 2D22 (B), and 5J7 (C). Neutralization escape by the mutant viruses was confirmed using U937 + DC-SIGN cells in a flow cytometry-based neutralization assay for 1F4 and 2D22, and by focus reduction neutralization assay (FRNT) for 5J7. Display enlarged views indicate the positions of the original amino acids of the escape mutations on EDIII and the EDI-EDII hinge region for 1F4 (D), 2D22 (E), and 5J7 (F). Images were generated with DENV1, DENV2, and DENV3 E dimer structures, respectively. The DENV2 and DENV3 E dimer structures [Research Collaboratory for Structural Bioinformatics (RCSB) accession nos. 1OAN and 1UZG, respectively] (8, 9) were modeled using the UniProt protein database viewer and PyMOL (Schrödinger) to generate structures for DENV1 and DENV3 (Thai 95) E dimers. (G) Alignment of E protein segments from DENV and WNV identified in the neutralizing hmAb-binding epitope of CR4354. Mutations leading to escape from 1F4 (blue), 2D22 (green), or 5J7 (pink) are highlighted on relevant regions of the aligned DENV E protein sequences. A portion of the CR4354 epitope that overlaps with the corresponding DENV escape mutations described here is highlighted in bold on the aligned WNV (New York 2000) sequence. (H) Escape mutations were mapped onto the E polymeric structure generated for TBEV (RCSB accession no. 1K4R) (10). The positions of escape mutations generated from 1F4, 2D22, and 5J7 are highlighted on the structure in blue (Gly274, K47), green (Arg323, His282, Asp362), and pink (Gln271, Asn272) (i.e., residues surrounding the lysine insertion), respectively. The footprint of the anti-WNV CR4354 hmAb that spans E protein dimers is circled with a white line. Note that all escape mutationsfor 1F4, 2D22, and 5J7 fall within the CR4354 footprint. *Neut50 values for each escape mutant differed significantly from the respective WT virus (P < 0.0001).
Discussion
Although it has been known for several decades that humans develop strongly neutralizing Abs following DENV infection, the relevant Abs and epitopes on the virus have not been identified. This is a major gap in knowledge, given the important roles postulated for Abs in clearing DENV or enhancing infection and disease. Here, the results from seven DENV-immune individuals demonstrate that the DENV-specific human Ab response consists of distinct populations of serotype cross-reactive and type-specific Abs. The type-specific Abs were responsible for potent neutralization of DENV. Although cross-reactive Abs were abundant in human immune sera, their contribution to neutralization was relatively negligible.
DENV serotype-specific neutralizing Abs generated in mice bind to epitopes that are present on the soluble form of rE protein (13–19). We report here that a substantial fraction of DENV-reactive Abs in human immune sera, including type-specific neutralizing Abs, bound to the intact virion but not to rE protein. Some of these virion-specific Abs likely bind to prM or M proteins, which also are displayed on the virion surface. However, prM/M is unlikely to be the primary target of neutralizing Abs, because several studies have shown that human anti-prM Abs are highly cross-reactive and weakly neutralizing (18–20). The rE protein used in this study was only 80% of the full-length E protein and lacked the amphipathic helices, conserved stem anchor, and transmembrane regions. Cryo-EM reconstructions of mature virions demonstrate that the amphipathic helices, stem anchor, and transmembrane segments are not exposed on the surface, and therefore are unlikely to be the targets of type-specific neutralizing Abs (8, 10, 26). Based on the results of Ab depletion studies with human immune sera, we suggest that the packing of E proteins on the virion surface creates unique epitopes involving two or more E protein molecules in adjacent symmetry groups, and that these quaternary epitopes are targets of the human neutralizing Ab response. Similar complex quaternary epitopes that are recognized by potently neutralizing hmAbs have been described for several other viruses, including WNV and HIV (31, 33, 34).
Our studies with hmAbs also confirmed that humans can produce strongly neutralizing Abs that bind a quaternary epitope expressed on intact virus particles but not on rE protein. The strongly neutralizing hmAbs 1F4 and 5J7 selected for viruses that escaped neutralization with mutations or insertions either on or near the E DI-DII hinge region. Consistent with this, other mAbs that strongly neutralize flaviviruses also have been reported to recognize the hinge region between EDI and EDII (31, 32, 35–37). Additionally, we identified a mutation at position 323 on EDIII that resulted in neutralization escape from the anti-DENV2 hmAb 2D22. This Ab likely also recognized a structurally complex neutralizing epitope, because it bound to DENV2 particles but not to EDIII or rE from DENV2. Collectively, the location of all four escape mutations identified for hmAbs 1F4, 2D22, and 5J7 (representing Abs that neutralize 3 different serotypes in a type-specific manner) map to a region that overlaps with an epitope recently described for the CR4354 hmAb that strongly neutralized WNV in vitro and in vivo (31, 32), and bound to virions but not to soluble E protein. Cryo-EM studies revealed that the CR4354 footprint spanned the DI-DII hinge from one E protein dimer and DIII of the adjacent E dimer (31). Abs that bind to this epitope were postulated to be strongly neutralizing for two reasons: (i) each mature virion should have 120 of these epitopes available for interaction, of which 90% could be simultaneously occupied by Ab (31), and (ii) Abs binding to this EDI–EDII complex epitope could cross-link adjacent E dimmers and inhibit trimer formation, which is a prerequisite for viral fusion. Because all the escape mutants that we identified localized within the footprint of CR4354, we propose that this region is also a target of human neutralizing Abs against DENV. Structure studies are in progress to define the relationships of 1F4, 2D22, and 5J7 with one another and with CR4354 further.
The EDI/II hinge region is not the only target of neutralizing hmAbs. Strongly neutralizing hmAbs that bind to EDIII (including A strand and lateral ridge epitopes recognized by mouse Abs) have been isolated from memory B cells of DENV-immune subjects (17–20). Further studies are needed to estimate the variety and frequency of DENV-specific memory B cells encoding for strongly neutralizing Abs. Such estimates are not possible from the existing DENV hmAb literature because of the different methods used to screen for DENV-specific memory B cells and the different criteria used to select B-cell clones and hmAbs for in-depth study (17–20). Previous studies have demonstrated that human immune sera depleted of EDIII Abs retained most of their neutralizing activity (21). Similar to WNV, recombinant DENV with mutations in EDIII lateral ridge and A-strand epitopes remained sensitive to neutralization by human immune sera (22–24). In the current study, we report that depletion of Abs that bind homologous rE also had more modest effects on neutralization potency, whereas removal of Abs that bound intact homologous virus resulted in a large decrease in neutralization titer. Collectively, these observations suggest that Abs in human immune sera that recognize the intact virus only account for the majority of the inhibitory activity relative to those binding EDIII and rE protein.
Our findings reporting on a target of neutralizing human Abs after natural infection are relevant to the development of DENV vaccines and the evaluation of vaccines currently under development. The leading dengue vaccine candidates currently being tested in clinical trials consist of tetravalent formulations of live-attenuated dengue or dengue/yellow fever chimeric viruses (38, 39). Studies are needed to determine if these live viral vaccines also induce neutralizing Abs that bind to quaternary epitopes expressed on the intact virion but not on soluble E protein.
Methods
Serum Samples.
Human serum samples were collected from individuals who had experienced a DENV infection during travel to an endemic region. Rhesus macaque (Macaca mulatta) sera were taken from animals vaccinated with a VEEV replicon particle (VRP-rE) expressing 80% of DENV3 E protein. More information is provided in SI Methods.
Virus and rE Proteins.
The DENV1 (West Pac 74), DENV2 (S-16803), DENV3 (CH-53489 and Thailand 95), and DENV4 (TVP-360) strains were used in the present study. All viruses used in the neutralization assays were grown in C6/36 Aedes albopictus mosquito cells at 28 °C and titered on Vero-81 cells as previously described (40). DENV was purified as previously described (21). The rE proteins from each of the four DENV serotypes were purchased from Hawaii Biotech, Inc.
Depletion of DENV-Specific Abs from Human Immune Sera.
Purified DENVs were adsorbed onto 4.0-μm Polybead polystyrene microspheres following the manufacturer's instructions (Polysciences, Inc.). Control beads were adsorbed with BSA instead. Human immune sera were depleted of virus-specific Abs by incubating sera with virus-adsorbed beads at 37 °C. Detailed information is given in SI Methods.
Depletion of DENV rE-Specific Abs from Human and Monkey Immune Sera.
DENV rE proteins were covalently conjugated to cyanogen bromide (CNBr)-activated beads following the manufacturer's protocol (Sigma). Control beads were conjugated with the blocking reagent instead of rE protein. DENV rE-specific Abs were depleted by incubating human and rhesus macaque immune sera with rE-conjugated beads at 37 °C. Detailed information is given in SI Methods.
Detection of DENV or rE-Binding Abs by ELISA.
ELISAs were conducted as previously described (18). Sera were used at dilutions of 1:40 and 1:25 for the depletion confirmation ELISAs in the virus and rE depletion experiments, respectively. More information is provided in SI Methods.
Detection of rE-Binding Abs by Western Blot.
Detailed information is provided in SI Methods.
Supplementary Material
Supporting Information
Acknowledgments
We thank Anne Broadwater and Yang Zhou for their contributions to the initial work in the hmAb screening and characterization process. These studies were supported by National Institutes of Health Grant U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense and a Pediatric Dengue Vaccine Initiative Targeted Research Grant.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: The spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med. 2004;10(Suppl):S98–S109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 2.Beasley DWC, Barrett ADT. The Infectious Agent, Dengue, Tropical Medicine: Science and Practice. In: Halstead SBP, Hoffman G, editors. Vol 5. London: Imperial College Press; 2008. pp. 29–73. [Google Scholar]
- 3.Imrie A, et al. Antibody to dengue 1 detected more than 60 years after infection. Viral Immunol. 2007;20:672–675. doi: 10.1089/vim.2007.0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halstead SB. Neutralization and antibody-dependent enhancement of dengue viruses. Adv Virus Res. 2003;60:421–467. doi: 10.1016/s0065-3527(03)60011-4. [DOI] [PubMed] [Google Scholar]
- 5.Roehrig JT. Antigenic structure of flavivirus proteins. Adv Virus Res. 2003;59:141–175. doi: 10.1016/s0065-3527(03)59005-4. [DOI] [PubMed] [Google Scholar]
- 6.Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J Virol. 2006;80:11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 8.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc Natl Acad Sci USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn RJ, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sukupolvi-Petty S, et al. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J Virol. 2010;84:9227–9239. doi: 10.1128/JVI.01087-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sukupolvi-Petty S, et al. Type- and subcomplex-specific neutralizing antibodies against domain III of dengue virus type 2 envelope protein recognize adjacent epitopes. J Virol. 2007;81:12816–12826. doi: 10.1128/JVI.00432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gromowski GD, et al. Mutations of an antibody binding energy hot spot on domain III of the dengue 2 envelope glycoprotein exploited for neutralization escape. Virology. 2010;407:237–246. doi: 10.1016/j.virol.2010.06.044. [DOI] [PubMed] [Google Scholar]
- 14.Wahala WM, et al. Natural strain variation and antibody neutralization of dengue serotype 3 viruses. PLoS Pathog. 2010;6:e1000821. doi: 10.1371/journal.ppat.1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brien JD, et al. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J Virol. 2010;84:10630–10643. doi: 10.1128/JVI.01190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shrestha B, et al. The development of therapeutic antibodies that neutralize homologous and heterologous genotypes of dengue virus type 1. PLoS Pathog. 2010;6:e1000823. doi: 10.1371/journal.ppat.1000823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith SA, et al. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol. 2012;86(5):2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Alwis R, et al. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis. 2011;5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dejnirattisai W, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science. 2010;328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beltramello M, et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wahala WM, Kraus AA, Haymore LB, Accavitti-Loper MA, de Silva AM. Dengue virus neutralization by human immune sera: Role of envelope protein domain III-reactive antibody. Virology. 2009;392:103–113. doi: 10.1016/j.virol.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliphant T, et al. Induction of epitope-specific neutralizing antibodies against West Nile virus. J Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sánchez MD, et al. The neutralizing antibody response against West Nile virus in naturally infected horses. Virology. 2007;359:336–348. doi: 10.1016/j.virol.2006.08.047. [DOI] [PubMed] [Google Scholar]
- 24.Wahala WM, Huang C, Butrapet S, White LJ, de Silva AM. Recombinant dengue type 2 viruses with altered E protein domain III epitopes are efficiently neutralized by human immune sera. J Virol. 2012;86:4019–4023. doi: 10.1128/JVI.06871-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lok SM, et al. Binding of a neutralizing antibody to dengue virus alters the arrangement of surface glycoproteins. Nat Struct Mol Biol. 2008;15:312–317. doi: 10.1038/nsmb.1382. [DOI] [PubMed] [Google Scholar]
- 26.Zhang W, et al. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat Struct Biol. 2003;10:907–912. doi: 10.1038/nsb990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, et al. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 28.Gromowski GD, Barrett AD. Characterization of an antigenic site that contains a dominant, type-specific neutralization determinant on the envelope protein domain III (ED3) of dengue 2 virus. Virology. 2007;366:349–360. doi: 10.1016/j.virol.2007.05.042. [DOI] [PubMed] [Google Scholar]
- 29.Coller BA, Clements DE, Bett AJ, Sagar SL, Ter Meulen JH. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine. 2011;29:7267–7275. doi: 10.1016/j.vaccine.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valdés I, et al. The chimeric protein domain III-capsid of dengue virus serotype 2 (DEN-2) successfully boosts neutralizing antibodies generated in monkeys upon infection with DEN-2. Clin Vaccine Immunol. 2011;18:455–459. doi: 10.1128/CVI.00382-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaufmann B, et al. Neutralization of West Nile virus by cross-linking of its surface proteins with Fab fragments of the human monoclonal antibody CR4354. Proc Natl Acad Sci USA. 2010;107:18950–18955. doi: 10.1073/pnas.1011036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt MR, et al. Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J Virol. 2009;83:6494–6507. doi: 10.1128/JVI.00286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spurrier B, et al. Structural analysis of human and macaque mAbs 2909 and 2.5B: Implications for the configuration of the quaternary neutralizing epitope of HIV-1 gp120. Structure. 2011;19:691–699. doi: 10.1016/j.str.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dias JM, et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goncalvez AP, et al. Humanized monoclonal antibodies derived from chimpanzee Fabs protect against Japanese encephalitis virus in vitro and in vivo. J Virol. 2008;82:7009–7021. doi: 10.1128/JVI.00291-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliphant T, et al. Antibody recognition and neutralization determinants on domains I and II of West Nile Virus envelope protein. J Virol. 2006;80:12149–12159. doi: 10.1128/JVI.01732-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMinn PC, et al. Murray valley encephalitis virus envelope protein antigenic variants with altered hemagglutination properties and reduced neuroinvasiveness in mice. Virology. 1995;211:10–20. doi: 10.1006/viro.1995.1374. [DOI] [PubMed] [Google Scholar]
- 38.Mantel N, et al. Genetic stability of a dengue vaccine based on chimeric yellow fever/dengue viruses. Vaccine. 2011;29:6629–6635. doi: 10.1016/j.vaccine.2011.06.101. [DOI] [PubMed] [Google Scholar]
- 39.Osorio JE, Huang CY, Kinney RM, Stinchcomb DT. Development of DENVax: A chimeric dengue-2 PDK-53-based tetravalent vaccine for protection against dengue fever. Vaccine. 2011;29:7251–7260. doi: 10.1016/j.vaccine.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kraus AA, Messer W, Haymore LB, de Silva AM. Comparison of plaque- and flow cytometry-based methods for measuring dengue virus neutralization. J Clin Microbiol. 2007;45:3777–3780. doi: 10.1128/JCM.00827-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information