Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing (original) (raw)
Abstract
Homology-dependent RNA silencing occurs in many eukaryotic cells. We reported recently that nodaviral infection triggers an RNA silencing-based antiviral response (RSAR) in Drosophila, which is capable of a rapid virus clearance in the absence of expression of a virus-encoded suppressor. Here, we present further evidence to show that the Drosophila RSAR is mediated by the RNA interference (RNAi) pathway, as the viral suppressor of RSAR inhibits experimental RNAi initiated by exogenous double-stranded RNA and RSAR requires the RNAi machinery. We demonstrate that RNAi also functions as a natural antiviral immunity in mosquito cells. We further show that vaccinia virus and human influenza A, B, and C viruses each encode an essential protein that suppresses RSAR in Drosophila. The vaccinia and influenza viral suppressors, E3L and NS1, are distinct double-stranded RNA-binding proteins and essential for pathogenesis by inhibiting the mammalian IFN-regulated innate antiviral response. We found that the double-stranded RNA-binding domain of NS1, implicated in innate immunity suppression, is both essential and sufficient for RSAR suppression. These findings provide evidence that mammalian virus proteins can inhibit RNA silencing, implicating this mechanism as a nucleic acid-based antiviral immunity in mammalian cells.
RNA silencing is a unique RNA-guided gene regulatory mechanism that operates in a wide range of eukaryotic organisms from plants to mammals (1). A feature common to all RNA silencing processes is the production of 21- to 26-nt small RNAs from structured or double-stranded RNA (dsRNA) by the endoribonuclease Dicer (2–6). These small interfering RNAs (siRNAs) control the specificity of RNA silencing in a homology-dependent manner by means of an RNA-induced silencing complex (RISC), of which Argonaute-2 (AGO2) is an essential protein component (1, 7, 8). RNA silencing in fungi, plants, and worms involves a cellular RNA-dependent RNA polymerase (RdRP); however, the multiple-turnover RISC may mediate RNA silencing in absence of a cellular RdRP in Drosophila and mammalian cells (1, 9–11).
We reported recently that infection of cultured Drosophila cells with the plus-strand RNA Nodavirus flock house virus (FHV), triggers specific silencing of FHV RNAs that is associated with accumulation of 22-nt siRNAs (12). Silencing of the replicating viral RNAs is RISC-dependent and sensitive to inhibition by the FHV B2 protein, as shown by the observation that B2 is essential for FHV infection of WT Drosophila cells but dispensable in cells depleted for AGO2 (12). These findings provided an example indicating an antiviral role for RNA silencing in the animal kingdom (12, 13), as has been established in higher plants (14–18).
In this article, we report that specific RNA silencing was induced in mosquito cells in response to viral RNA replication and show that this mosquito antiviral immunity is RISC-dependent and sensitive to suppression by the B2 protein encoded by either FHV or nodamura virus (NoV). We demonstrate that NoV RNA replication also triggered specific silencing of viral RNAs in Drosophila cells, as has been found after challenge with FHV (12), and we provide further evidence that the RNA silencing-based antiviral response (RSAR) in Drosophila is mediated by the RNA interference (RNAi) pathway. We established a functional screen in cultured Drosophila cells based on the nodaviral RNA silencing system and found that the IFN antagonist proteins encoded by four mammalian viruses are suppressors of the Drosophila RSAR. Our results provide experimental evidence indicating a natural antiviral role for RNA silencing in mammals. A possible role of RNA silencing in the IFN-regulated mammalian innate immunity is discussed.
Materials and Methods
Plasmids. pFR1 and pFR1-ΔB2, encoding the WT and B2-knockout mutant of FHV RNA1, have been described (12). pFR1gfp was obtained by replacing the _Nco_I–_Sac_I fragment (corresponding to nucleotides 2,802–3,001 of RNA1) of pFR1 with the coding sequence for GFP. Full-length cDNA of NoV RNA1 and the mutant 1 (B2–B1+), together with a hepatitis delta virus ribozyme at the 3′ end (19), were cloned immediately downstream of the CuSO4-inducible metallothionein promoter in pMT/V5-HisA vector (Invitrogen) to give pNR1 and pNR1-ΔB2 as described (12). pONR1 and pONR1-ΔB2 contained the same cDNA sequences downstream of the constitutive OplE2 promoter in pIZ vector (Invitrogen). The coding sequence for either fB2 or nB2 was cloned into pIZ vector for expression in mosquito cells. The coding sequence for nB2, p19, E3L (from vaccinia virus Western Reserve strain), A/NS1 (from influenza A/WSN/33 virus), B/NS1 (from influenza B/Yamagata/73 virus), and C/NS1 (from influenza C/JHB/66 virus) was cloned in pMT/V5-HisA for transient expression in S2 cells. pA/NS1 was digested with _Nco_I, end-filled, and religated to create pNS1-A2. pNS1-A1 and pNS1-A3 were generated as described (20, 21). pMTp19 was digested with Asp-718, end-filled, and religated to create pMTp19fs, which led to translational termination after amino acid 24 of the 171-aa residue p19. pgfp and pB2gfp were also constructed in pMT/V5-HisA, and the translational fusion between B2 and GFP in pB2gfp was separated by a _Bam_HI site.
siRNAs and dsRNAs. siRNAs were synthesized by Qiagen (Valencia, CA), targeting nucleotides 1,695–1,717 (aatgcagtgcgtggaggttgttt) and 3,293–3,315 (aacccaagatttgctgcgtgatt) of Drosophila AGO2, and nucleotides 1,717–1,737 (aagcgcaagatcctcgacctg) and 3,120–2,142 (aatcatcgacacggttaataatt) of Anopheles gambiae AGO2. An siRNA (aattatcgatgagcgtggtggtt) targeting nucleotides 819–841 of the lacZ mRNA was used as a control. Typically, a plasmid DNA (0.3 μg) was mixed with siRNA (0.3 μg) or a dsRNA of ≈500 bp (0.3 μg) in cotransfection experiments. dsRNAs corresponding to the PAZ and PIWI domains of both A. gambiae AGO1 and AGO2 were synthesized in vitro as described for Drosophila (12).
Cell Culture, Transfection, and Analyses. Both Drosophila S2 cells and A. gambiae 4a-2s4 cells were cultured at 27°C in Schneider's insect medium (Sigma) supplemented with FBS. The Effectene transfection reagent (Qiagen) and CellFectin reagent (Invitrogen) were used for transfection of the fruit fly and mosquito cells, respectively, according to the manufacturer's instructions. RNA transcription from pMT-derived plasmids was induced by overnight incubation with CuSO4 at 0.5 mM. No induction was necessary for transfection with pIZ-derived plasmids in the mosquito cells. Two days after induction in Drosophila cells or 3 days after transfection in mosquito cells, total RNA was extracted by using the Trizol reagent, and 10 μg from each sample was fractionated and analyzed by Northern blot hybridization as described (22). DNA fragments corresponding to the B2 coding sequence of either FHV or NoV were labeled as probes by using the Rediprime II random prime labeling system (Amersham Biosciences). RNA quantification from Northern blot hybridizations was carried out with a Molecular Imager FX Phosphor-Imager (Bio-Rad).
Electrophoretic Mobility-Shift Assay. The assay was carried out as described (23). Briefly, 0.4 μM of GST fusion protein, NS1 or NS1-A1, was incubated on ice for 30 min in 20 μl of binding buffer with 32P-radiolabeled (10,000 cpm, 1 nM) 76-nt dsRNA, or 21-nt siRNA that was produced with the Silencer siRNA construction kit (Ambion, Austin, TX) according to the manufacturer's instructions. siRNA was targeted at nucleotides 123–144 of eGFP (aagctgaccctgaagttcatc). The protein–RNA complexes were resolved in an 8% nondenaturing polyacrylamide gel run at 4°C for 4 h at 150 V in 45 mM Tris-borate/1 mM EDTA.
Results
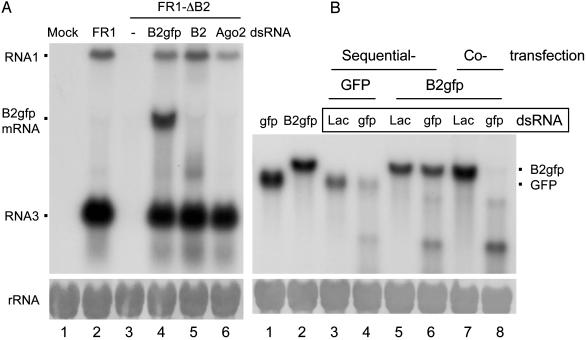
B2 of FHV Is a Suppressor of RNAi. The biochemical framework of the core RNA silencing pathway is based mostly on the experimental induction of RNA silencing by exogenous dsRNA, commonly referred to as RNAi (1, 24). Thus, we first investigated whether B2 of FHV, known to inhibit RNA silencing induced by viral RNA replication, also suppresses RNAi. To this end, Drosophila cells were transfected with a plasmid encoding either GFP or B2 fused with GFP (B2-GFP) under the control of an inducible promoter, and 1 day after induction of expression of GFP or B2-GFP, dsRNA targeting the GFP coding sequence was introduced (Fig. 1_B_). B2-GFP remained active in suppressing virus RNA silencing as cotransfection with either pB2-GFP or pB2 rescued the in vivo accumulation of the self-replicating FHV RNA1 mutant carrying an untranslatable B2, called FR1-ΔB2 (Fig. 1 A, lanes 4 and 5). Suppression of AGO2 expression by the specific dsRNA also rescued the accumulation of FR1-ΔB2 RNAs (Fig. 1 A, lane 6), as expected (12). In the GFP RNAi assay (Fig. 1_B_), targeting of the B2-GFP mRNA by RNAi occurred in cells that were also expressing B2-GFP. Northern blot hybridizations found that accumulation of GFP mRNA was significantly reduced 2 days after GFP dsRNA was introduced as compared with treatment with lacZ dsRNA (compare lanes 3 and 4 in Fig. 1_B_), indicating an effective RNAi against GFP mRNA by this sequential transfection protocol. However, no obvious difference in the accumulation of B2-GFP mRNA was observed between cells treated with either GFP or lacZ dsRNA (compare lanes 5 and 6 in Fig. 1_B_). Thus, the specific degradation of mRNA targeted by a homologous dsRNA did not occur in Drosophila cells expressing the B2 fusion protein, establishing FHV B2 as a viral suppressor of RNAi in an animal system.
Fig. 1.
B2 of FHV suppresses RNA silencing induced by a replicating virus RNA (A) or dsRNA (B) in cultured fruit fly cells. (A) Cells were transfected with buffer (Mock), pFR1, pFR1-ΔB2 alone or with pB2gfp, pfB2, or dsRNA of AGO2, as indicated at the top of each lane. Total RNA was extracted 2 days after induction for Northern blot analysis by a probe specific to the B2 coding region of FHV. (B) Cells were either cotransfected or sequentially transfected with a protein-expressing plasmid (pB2gfp or pgfp) and a dsRNA (targets GFP or lacZ mRNA as a control). Total RNA was extracted 2 days after the last transfection and analyzed by Northern blot hybridization with a probe specific to the GFP mRNA. RNA species corresponding to FHV RNAs 1 and 3, mRNA of B2-GFP, and GFP were indicated. Note that the mRNA from pB2 comigrated with RNA3 (A, lane 5). Equal RNA loading was shown by methylene blue staining of rRNA (Lower).
When GFP dsRNA was cotransfected with pB2-GFP, however, B2-GFP mRNA was effectively destroyed by RNAi (compare lanes 7 and 8 in Fig. 1_B_). Thus, RNAi suppression requires B2 expression before dsRNA is introduced into cells to initiate RNAi, suggesting that B2 suppression may occur before the production or RISC loading of siRNAs. Collectively, our data show that RNA silencing triggered by either dsRNA or viral RNA replication not only is AGO2-dependent, but also is sensitive to B2 suppression, providing compelling evidence that the RNA silencing antiviral immunity detected in Drosophila cells is mediated by the RNAi pathway.
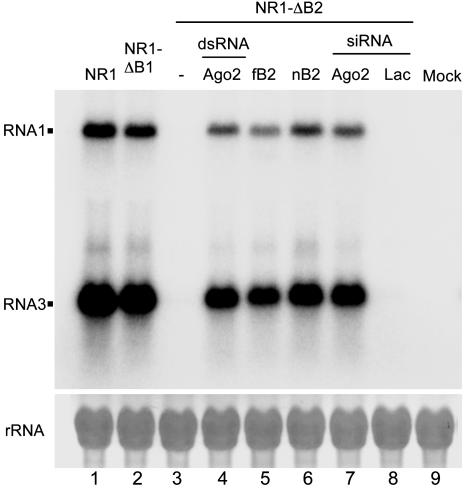
NoV Induces and Inhibits RNA Silencing in Drosophila. We next investigated whether the RNA silencing antiviral response is elicited when Drosophila cells are challenged with another virus. NoV is the only member of the Alphanodavirus genus that can lethally infect both insects and mammals. However, the sequence identity between the encoded proteins of NoV and FHV is either low (44% for the viral RdRP) or minimal (<19% for B2) (25). We constructed a full-length NoV RNA1 cDNA clone (pNR1) encoding a self-replicating RNA, essentially as described (12) for FHV. After transfection into the Drosophila S2 cells and transcriptional induction of the viral cDNA, pNR1 directed RNA1 self-replication and transcription of RNA3, the subgenomic mRNA for B2 (Fig. 2, lane 1). Also as expected (19), a point mutation, as in NR1-ΔB1, that abolished translation of the overlapping B1 ORF from RNA3, corresponding to the C-terminal portion of the viral RdRP, had no effect on the accumulation of either RNA1 or RNA3 (Fig. 2, lane 2). However, a B2-knockout mutant, referred to as NR1-ΔB2, was hardly detectable in transfected Drosophila cells by Northern blot hybridizations (Fig. 2, lane 3). NR1-ΔB2 contained point mutations that prevented B2 translation but did not change any amino acid in the –1 reading frame that codes for RdRP, as described (19). The defect of NR1-ΔB2 accumulation in WT Drosophila cells was complemented in trans by cotransfection of a plasmid expressing the NoV B2 protein (nB2), indicating that nB2 is required for NoV RNA accumulation (Fig. 2, lane 6).
Fig. 2.
Induction and suppression of RNA silencing by NoV. Fruit fly cells were transfected with pNR1, pNR1-ΔB1 or pNR1-ΔB2 alone or with a B2-expressing plasmid (fB2 or nB2), and either dsRNA or siRNA specific for AGO2. An siRNA targeting lacZ mRNA was used as a control. Total RNA was extracted 2 days after induction for Northern blot analysis by a probe specific to the B2 coding region of NoV.
Several lines of evidence indicate that NoV RNAs were targeted for silencing by the AGO2-dependent RNAi pathway in Drosophila cells and that nB2 suppressed the RNAi antiviral response to ensure successful NoV RNA replication and transcription. First, the FHV B2 protein (fB2), shown to suppress RNAi (12), rescued accumulation of NR1-ΔB2 in Drosophila cells (Fig. 2, lane 5). Second, depleting RISC by either dsRNA or siRNA targeting AGO2 efficiently rescued accumulation of NR1-ΔB2 (Fig. 2, lanes 4 and 7). Such a rescue was not observed by cotransfection with an unrelated lacZ siRNA (Fig. 2, lane 8) or dsRNA (data not shown). These results indicate that rescue of NR1-ΔB2 by cotransfection of dsRNA and siRNA targeting AGO2 is caused by a specific AGO2 depletion, rather than a nonspecific effect of dsRNA. Similar specific rescue of FR1-ΔB2 in Drosophila cells by either the dsRNA or siRNA targeting AGO2, but not by the dsRNA and siRNA targeting lacZ, was also observed (see also Fig. 5, lanes 14–17). In addition, nB2 suppressed RNA silencing targeted against (i) FR1-ΔB2 in transfected Drosophila cells (see Fig. 5, lane 4) and (ii) a GFP transgene in transgenic plants (data not shown), as has been found for fB2 (12).
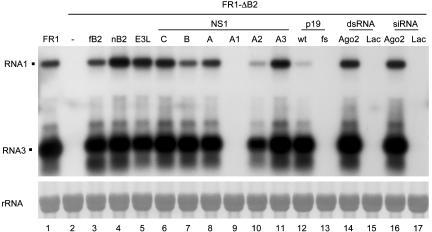
Fig. 5.
Suppression of the RNAi antiviral response by E3L and NS1 proteins. Fruit fly cells were transfected with a pFR1-derived plasmid plus either another plasmid expressing a viral RNAi suppressor (fB2, nB2, E3L, C/NS1, B/NS1, A/NS1, or p19) or a dsRNA/siRNA (to mRNA of AGO2 or lacZ). NS1-A1 contained the R38A/K41A mutation, NS1-A2 corresponded to the N-terminal 82 aa of WT A/NS1, and NS1-A3 contained the D92E mutation. p19fs was a frameshift mutant of p19 that terminates after the first 24 aa. RNA was extracted and analyzed as described for Fig. 2.
Remarkably, we found an effective suppression of RSAR targeting FR1-ΔB2 in Drosophila by the tombusvirus 19-kDa protein (p19) (26, 27), but not after it was truncated (see Fig. 5, lanes 12 and 13). This finding demonstrates cross-kingdom suppression of RNA silencing in an animal system by a plant viral suppressor. Together with the observations that the B2 proteins encoded by two animal nodaviruses suppress RNA silencing in both Drosophila and tobacco plants (12), our results show that the RSAR mechanism is conserved between the plant and animal kingdoms.
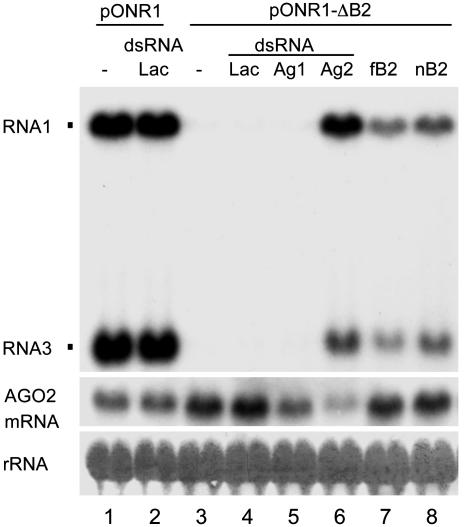
RNA Silencing Is a Natural Antiviral Response in Mosquito Cells. Because the genome of the mosquito A. gambiae, which transmits both malaria and viruses (28), encodes a functional RNAi pathway similar to Drosophila (29–31), we investigated whether RNAi also protects A. gambiae against NoV. Self-replication of NoV RNA1 and transcription of RNA3 were detected (Fig. 3, lane 1) in cells after transfection with pONR1, which contained the NoV RNA1 cDNA under the control of the OplE2 promoter. pNR1 failed to initiate RNA replication in A. gambiae cells (data not shown), possibly because of a lack of transcriptional induction. Neither RNA1 replication nor RNA3 transcription was detected in A. gambiae cells transfected with pONR1-ΔB2 (Fig. 3, lane 3), which encoded the B2-knockout mutant of NoV RNA1. However, the defect was rescued by cotransfection with either a plasmid expressing a B2 (Fig. 3, lanes 7 and 8) or a dsRNA corresponding to mRNA of the A. gambiae AGO2 (Fig. 3, lane 6). The rescue of ONR1-ΔB2 was also observed by cotransfection with an siRNA targeting AGO2, although with an efficiency ≈50% lower than the long dsRNA (data not shown). ONR1-ΔB2 rescue by AGO2 depletion was specific as cotransfection with dsRNA to neither lacZ (Fig. 3, lane 4) nor AGO1 mRNA (Fig. 3, lane 5) was effective. Thus, as found in Drosophila, a self-replicative NoV RNA also triggered the RNAi antiviral response in A. gambiae cells, which is both AGO2 dependent and sensitive to B2 suppression, establishing this immunity pathway in two different insect cell lines. This finding opens up the possibility of targeting this pathway to prevent transmission of mosquito-borne human viral diseases such as dengue.
Fig. 3.
An RNA-based antiviral immunity in mosquito cells. A. gambiae 4a-2s4 cells were transfected with pONR1, pONR1-ΔB2 alone or with a pIZ-based plasmid coding for fB2 or nB2, plus dsRNA targeting the PAZ domain of A. gambiae AGO1 or AGO2. A lacZ dsRNA was used as a control. An identical blot was probed for the accumulation of AGO2 mRNA with DNA corresponding to the C-terminal PIWI domain of A. gambiae AGO2. RNA was extracted and analyzed as for Fig. 2.
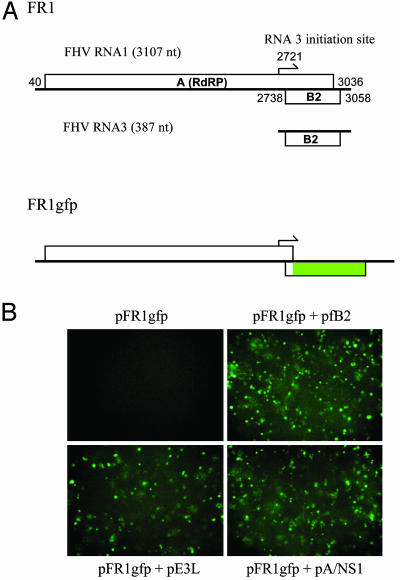
A Cell-Based Screen for Identifying Animal RNA Silencing Suppressors. To facilitate screening for new animal RNAi suppressors, we fused the coding sequence of GFP in-frame with the start of the B2 ORF of FHV RNA1 to yield pFR1gfp (Fig. 4_A_). pFR1gfp was essentially a B2-knockout mutant carrying a visual marker because only the first 23 aa of the 106-aa residue fB2 were translated in the fusion protein. Indeed, GFP cells were not visible after pFR1gfp was transfected alone (Fig. 4_B_ Upper Left) but became abundant when it was cotransfected with a B2-expressing plasmid (Fig. 4_B_ Upper Right). GFP expression was detected also when pFR1gfp was cotransfected with the AGO2 dsRNA (data not shown). Replication and accumulation of FR1gfp in S2 cells cotransfected with either the AGO2 dsRNA or the fB2-expressing plasmid were confirmed by Northern blot detection of both positive- and negative-strand viral RNAs (data not shown). Thus, FR1gfp was defective in suppressing RSAR in Drosophila cells as was FR1-ΔB2. Importantly, B2 suppression of the Drosophila RNA silencing response targeting either FR1gfp or FR1-ΔB2 did not require prior B2 expression, which was found to be necessary for RNA silencing induced by dsRNA (Fig. 1_B_). This finding is consistent with the natural situation in which the expression of B2 after viral RNA replication but early in infection is sufficient to allow productive FHV infection (12) and establishes an easy cell-based assay for the identification of animal RNAi suppressors, simply by detection of GFP after cotransfection of pFR1gfp with a plasmid expressing a candidate protein.
Fig. 4.
Suppression of the RNAi antiviral response visualized by GFP expression. (A) Genome organization and expression of FHV RNA1 and R1gfp. RNA1 encodes protein A, the catalytic subunit of the viral RdRP, and directs transcription of RNA3, mRNA for protein B2. Nucleotides 2,802–3,001 of RNA1 as encoded in pFR1 were replaced with the coding sequence for enhanced GFP (eGFP) to give pFR1gfp so that eGFP was translated from a chimeric RNA3 as a fusion protein with the N terminus of B2. (B) Detection of GFP expression in fruit fly cells from a FHV RNA1 mutant defective in silencing suppression. Cells were transfected with pFR1gfp alone or with a plasmid expressing fB2, E3L, or NS1A and photographed 2 days after induction.
Identification of Mammalian Viral Proteins That Suppress the Drosophila RSAR. The nodaviral silencing system established in Drosophila cells was used in a search for mammalian viral suppressors of RNA silencing because the RNAi machinery is highly conserved between Drosophila and humans and the available mammalian RNA silencing system induced by siRNAs (32) will not be able to identify suppressors that act upstream of siRNA production. NS1 encoded by influenza A virus was among those first selected because it is encoded by an overlapping gene and influences virulence and virus accumulation but is not essential for viral replication (33), which are common features of many known viral silencing suppressors such as the nodaviral B2 and plant viral 2b proteins (12, 16, 34). We found that NS1 protein expressed from a cotransfected plasmid indeed rescued GFP expression from pFR1gfp in Drosophila cells (Fig. 4_B_ Lower Right). GFP expression was also observed in cells cotransfected with a plasmid expressing E3L of vaccinia virus (Fig. 4_B_ Lower Left), which shares key functional features with NS1 (35, 36). The RSAR suppressor activity of both NS1 and E3L was confirmed further by the rescue of FR1-ΔB2 in Drosophila cells, leading to detection of both the genomic (RNA1) and subgenomic (RNA3) RNAs of FHV, as illustrated by Northern blot hybridization (Fig. 5, lanes 5 and 8). The cotransfection experiments also revealed that the NS1 proteins encoded by the related influenza B and C viruses were also suppressors of RSAR (Fig. 5, lanes 6 and 7). Efficient rescue of NR1-ΔB2 by these vertebrate viral suppressors was also observed in cotransfected Drosophila cells (W.-X.L. and S.-W.D., unpublished observations). These experiments identify mammalian viral proteins capable of suppressing RNA silencing in an animal system and indicate that this activity is an evolutionarily conserved function of NS1 encoded by the three influenza virus genera in the Orthomyxoviridae family.
NS1 Suppression of RNAi Requires the N-Terminal dsRNA-Binding Domain. Both NS1 and E3L bind dsRNA in vitro, although each involves a distinct protein structure (37). E3L contains the dsRNA-binding motif found in many cellular proteins and the reovirus σ3 protein, which was recently found to suppress transgene RNA silencing in plants with a coinfiltration protocol (38). Further, the NS1 proteins encoded by influenza A, B, and C viruses, designated A/NS1, B/NS1, and C/NS1, share few sequence similarities beyond the N-terminal dsRNA-binding domain (39). To analyze the role of the dsRNA-binding domain of NS1 in RSAR suppression, two mutants of A/NS1 were created. The first contained alanine substitution of Arg-38 and Lys-41 (designated NS1-A1), which are essential for dsRNA binding, whereas the second contained only the N-terminal fragment (82 aa) of A/NS1 (designated NS1-A2), which possesses all of the in vitro RNA-binding properties of the full-length 230-aa protein (39).
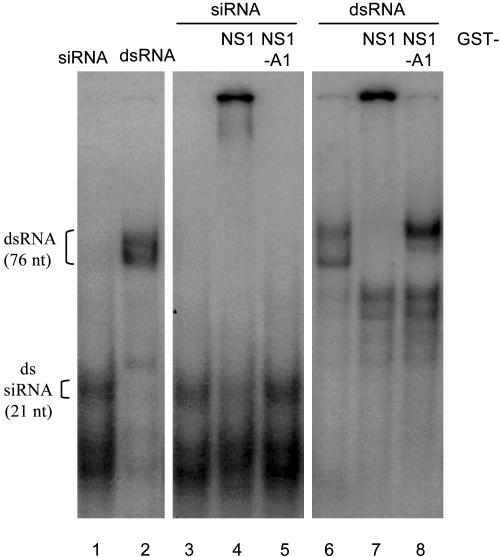
We found that the R38A/K41A mutant (NS1-A1) was unable to rescue FR1-ΔB2 in transfected Drosophila cells (Fig. 5, lane 9). In contrast, the N-terminal fragment (NS1-A2) retained the activity of A/NS1 to rescue FR1-ΔB2 in transfected cells (Fig. 5, lane 10). These results indicate that the N-terminal dsRNA-binding domain of A/NS1 is both essential and sufficient in the suppression of RSAR in the invertebrate animal cells. Interestingly, A/NS1, but not NS1-A1, was found to bind synthetic siRNA (Fig. 6). This finding indicates that NS1 suppression of RSAR may require siRNA binding, as proposed for p19 (27), and is unlikely caused by a simple dsRNA sequestration, the latter of which is also supported by the failure to rescue FR1-ΔB2 in Drosophila cells by cotransfection with plasmids that code for any of the five cellular dsRNA-binding motif-containing Drosophila proteins (X. Wang, F.L., and S.-W.D., unpublished data).
Fig. 6.
Binding of GST-NS1 fusion protein to long and short dsRNAs in vitro. GST (0.4 μM, lanes 3 and 6), GST-NS1 (lanes 4 and 7), or GST-NS1-A1 (lanes 5 and 8) was incubated with 32P-radiolabeled dsRNA (Right) or siRNA (Center) before the RNA–protein complexes were resolved in a nondenaturing polyacrylamide gel electrophoresis. The positions of the free 76-nt dsRNA and the 21-nt siRNA are indicated. The lower band in lanes 6–8 migrates in the same position as T7-generated single-stranded RNA.
By comparison, NS1-A2 was weaker than WT A/NS1 in RSAR suppression (Fig. 5, compare lanes 8 and 10). Intriguingly, NS1-A2 exhibited a significantly reduced activity in preventing RNA silencing that targeted the genomic RNA1, similar to that of the plant virus protein p19 (Fig. 5, compare lane 8 with lanes 10 and 12). Furthermore, we found that a single Asp-to-Glu mutation at position 92 of A/NS1 enhanced the rescue efficiency as compared to WT A/NS1, indicated by an ≈100% increase in the accumulation of RNA1 observed in two independent experiments (Fig. 5, compare lanes 8 and 11). Position 92 is located near the known RNA-binding domain, and the D92E mutation was associated with the virulence of the lethal H5N1 influenza A virus isolate transmitted to humans in 1997 (21). These findings indicate that the amino acid sequence of NS1 that is C terminal to the core RNA-binding domain is able to modulate the activity of NS1 in RNAi suppression.
Discussion
Members of Orthomyxoviridae have segmented negative-strand RNA genomes, whereas vaccinia virus, which is used as the vaccine for smallpox, contains a double-stranded DNA genome. Our work shows that these diverse mammalian viruses encode proteins that are potent suppressors of the Drosophila RSAR. It is known that mammals encode a functional RNA silencing pathway (1) and that NS1 and E3L are both essential for viral pathogenesis in mammalian hosts (40). Thus, we postulate that RNA silencing is a major antiviral response in mammals and that encoding a suppressor of RNAi is an essential feature of mammalian viruses, as has been established for viruses that infect plants and invertebrates (refs. 12, 16, 17, and 41 and this work).
Our hypothesis is supported by three additional lines of evidence. First, human cells are equipped with an RNAi machinery, including Dicer and RISC, that is most closely related to that found in Drosophila (5, 10, 42–44). Second, recent work found that replication of influenza A virus and of several mammalian RNA and DNA viruses is effectively inhibited by synthetic siRNAs, indicating that viral RNA can be targeted by the RNAi machinery in mammalian cells (45, 46). Third, both NS1 and E3L proteins are known to function in their mammalian hosts as inhibitors of the innate antiviral response regulated by the IFN system (40), a first line of vertebrate defense against virus infections. Notably, the dsRNA-binding domains of NS1 and E3L are also required to inhibit the innate antiviral immunity (20, 35), as found for NS1 suppression of the Drosophila RSAR (Fig. 5). Thus, our observations are more consistent with an antiviral role for RNA silencing in mammals, most likely as part of the mammalian innate antiviral immunity. However, it cannot be ruled out at present that the innate antiviral immunity targeted by NS1 in mammalian cells is unrelated to RNA silencing; for example, NS1 may target an unrelated or shared factor in the two independent mechanisms. In this regard, it is of interest to consider that, similar to the innate antiviral immunity, RSAR is rapid, effective in single cells, and highly specific, and previous studies have indicated the existence of alternative antiviral effector pathways in addition to those mediated by protein kinase PKR, RNase L, and Mx proteins (35, 47).
Important questions that remain to be addressed are whether viral infection indeed triggers specific RNA silencing in vertebrates, whether this antiviral response is regulated by the IFN system, and whether other known viral suppressors of RSAR are also effective IFN antagonists. Such a positive effect on RSAR by IFN could occur before the production of viral siRNAs in the vertebrate RSAR pathway because RNA silencing initiated by synthetic siRNAs is independent of the IFN system (48). In any case, discovery of RNAi suppressor activity will facilitate screening for small molecule inhibitors of these essential mammalian viral virulence factors, which may lead to new treatments of human viral diseases.
Acknowledgments
We thank Hans Mueller for providing the A. gambiae cell line 4a-2s4. This project was supported by National Institutes of Health Grant R01 AI52447 (to S.-W.D.) and U.S. Department of Agriculture Grant 2001-02678 from the Biology of Plant-Microbe Associations panel (to S.-W.D.). Partial support for this work was provided by National Institutes of Health grants (to L.A.B., P.A., A.G.-S., and P.P.). P.P. is an Ellison Foundation Senior Scholar.
Abbreviations: AGO2, Argonaute-2; dsRNA, double-stranded RNA; FHV, flock house virus; NoV, nodamura virus; RdRP, RNA-dependent RNA polymerase; RISC, RNA-induced silencing complex; RNAi, RNA interference; RSAR, RNA silencing-based antiviral response; siRNA, small interfering RNA.
References
- 1.Denli, A. M. & Hannon, G. J. (2003) Trends Biochem. Sci. 28**,** 196–201. [DOI] [PubMed] [Google Scholar]
- 2.Hamilton, A. J. & Baulcombe, D. C. (1999) Science 286**,** 950–952. [DOI] [PubMed] [Google Scholar]
- 3.Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. J. (2000) Nature 404**,** 293–296. [DOI] [PubMed] [Google Scholar]
- 4.Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. (2000) Cell 101**,** 25–33. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409**,** 363–366. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. (2002) EMBO J. 21**,** 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hammond, S. M., Boettcher, S., Caudy, A. A., Kobayashi, R. & Hannon, G. J. (2001) Science 293**,** 1146–1150. [DOI] [PubMed] [Google Scholar]
- 8.Elbashir, S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15**,** 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz, D. S., Hutvagner, G., Haley, B. & Zamore, P. D. (2002) Mol. Cell 10**,** 537–548. [DOI] [PubMed] [Google Scholar]
- 10.Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R. & Tuschl, T. (2002) Cell 110**,** 563–574. [DOI] [PubMed] [Google Scholar]
- 11.Cogoni, C. & Macino, G. (1999) Nature 399**,** 166–169. [DOI] [PubMed] [Google Scholar]
- 12.Li, H. W., Li, W. X. & Ding, S. W. (2002) Science 296**,** 1319–1321. [DOI] [PubMed] [Google Scholar]
- 13.Lindenbach, B. D. & Rice, C. M. (2002) Mol. Cell 9**,** 925–927. [DOI] [PubMed] [Google Scholar]
- 14.Covey, S. N., Al-Kaff, N. S., Langara, A. & Turner, D. S. (1997) Nature 385**,** 781–782. [Google Scholar]
- 15.Ratcliff, F., Harrison, B. D. & Baulcombe, D. C. (1997) Science 276**,** 1558–1560. [DOI] [PubMed] [Google Scholar]
- 16.Li, W. X. & Ding, S. W. (2001) Curr. Opin. Biotechnol. 12**,** 150–154. [DOI] [PubMed] [Google Scholar]
- 17.Vance, V. & Vaucheret, H. (2001) Science 292**,** 2277–2280. [DOI] [PubMed] [Google Scholar]
- 18.Lindbo, J. A., Silva-Rosales, L., Proebsting, W. M. & Dougherty, W. G. (1993) Plant Cell 5**,** 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, K. L., Price, B. D. & Ball, L. A. (2003) Virology 305**,** 436–451. [DOI] [PubMed] [Google Scholar]
- 20.Wang, X., Li, M., Zheng, H., Muster, T., Palese, P., Beg, A. A. & Garcia-Sastre, A. (2000) J. Virol. 74**,** 11566–11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo, S. H., Hoffmann, E. & Webster, R. G. (2002) Nat. Med. 8**,** 950–954. [DOI] [PubMed] [Google Scholar]
- 22.Li, H. W., Lucy, A. P., Guo, H. S., Li, W. X., Ji, L. H., Wong, S. M. & Ding, S. W. (1999) EMBO J. 18**,** 2683–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donelan, N. R., Basler, C. F. & García-Sastre, A. (2003) J. Virol. 77**,** 13257–13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fire, A., Xu, S. Q., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391**,** 806–811. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, K. N., Johnson, K. L., Dasgupta, R., Gratsch, T. & Ball, L. A. (2001) J. Gen. Virol. 82**,** 1855–1866. [DOI] [PubMed] [Google Scholar]
- 26.Voinnet, O., Pinto, Y. M. & Baulcombe, D. C. (1999) Proc. Natl. Acad. Sci. USA 96**,** 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silhavy, D., Molnar, A., Lucioli, A., Szittya, G., Hornyik, C., Tavazza, M. & Burgyan, J. (2002) EMBO J. 21**,** 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powers, A. M., Brault, A. C., Tesh, R. B. & Weaver, S. C. (2000) J. Gen. Virol. 81**,** 471–479. [DOI] [PubMed] [Google Scholar]
- 29.Levashina, E. A., Moita, L. F., Blandin, S., Vriend, G., Lagueux, M. & Kafatos, F. C. (2001) Cell 104**,** 709–718. [DOI] [PubMed] [Google Scholar]
- 30.Hoa, N. T., Keene, K. M., Olson, K. E. & Zheng, L. (2003) Insect Biochem. Mol. Biol. 33**,** 949–957. [DOI] [PubMed] [Google Scholar]
- 31.Holt, R. A., Subramanian, G. M., Halpern, A., Sutton, G. G., Charlab, R., Nusskern, D. R., Wincker, P., Clark, A. G., Ribeiro, J. M., Wides, R., et al. (2002) Science 298**,** 129–149.12364791 [Google Scholar]
- 32.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4**,** 457–467. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Sastre, A., Egorov, A., Matassov, D., Brandt, S., Levy, D. E., Durbin, J. E., Palese, P. & Muster, T. (1998) Virology 252**,** 324–330. [DOI] [PubMed] [Google Scholar]
- 34.Ding, S. W., Li, W. X. & Symons, R. H. (1995) EMBO J. 14**,** 5762–5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiang, Y., Condit, R. C., Vijaysri, S., Jacobs, B., Williams, B. R. & Silverman, R. H. (2002) J. Virol. 76**,** 5251–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, E. J., Marie, I., Prakash, A., Garcia-Sastre, A. & Levy, D. E. (2001) J. Biol. Chem. 276**,** 8951–8957. [DOI] [PubMed] [Google Scholar]
- 37.Fierro-Monti, I. & Mathews, M. B. (2000) Trends Biochem. Sci. 25**,** 241–246. [DOI] [PubMed] [Google Scholar]
- 38.Lichner, Z., Silhavy, D. & Burgyan, J. (2003) J. Gen. Virol. 84**,** 975–980. [DOI] [PubMed] [Google Scholar]
- 39.Krug, R. M., Yuan, W., Noah, D. L. & Latham, A. G. (2003) Virology 309**,** 181–189. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Sastre, A. (2002) Microbes Infect. 4**,** 647–655. [DOI] [PubMed] [Google Scholar]
- 41.Adelman, Z. N., Sanchez-Vargas, I., Travanty, E. A., Carlson, J. O., Beaty, B. J., Blair, C. D. & Olson, K. E. (2002) J. Virol. 76**,** 12925–12933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hutvagner, G. & Zamore, P. D. (2002) Science 297**,** 2056–2060. [DOI] [PubMed] [Google Scholar]
- 43.Provost, P., Dishart, D., Doucet, J., Frendewey, D., Samuelsson, B. & Radmark, O. (2002) EMBO J. 21**,** 5864–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, H., Kolb, F. A., Brondani, V., Billy, E. & Filipowicz, W. (2002) EMBO J. 21**,** 5875–5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ge, Q., McManus, M. T., Nguyen, T., Shen, C. H., Sharp, P. A., Eisen, H. N. & Chen, J. (2003) Proc. Natl. Acad. Sci. USA 100**,** 2718–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gitlin, L. & Andino, R. (2003) J. Virol. 77**,** 7159–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou, A., Paranjape, J. M., Der, S. D., Williams, B. R. & Silverman, R. H. (1999) Virology 258**,** 435–440. [DOI] [PubMed] [Google Scholar]
- 48.Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H. & Williams, B. R. (2003) Nat. Cell Biol. 5**,** 834–839. [DOI] [PubMed] [Google Scholar]