Coordinated regulation of neuronal mRNA steady-state levels through developmentally controlled intron retention (original) (raw)
Synaptic transmission is one of the most important activities in the nervous system. This study delineates how Ptbp1-mediated intron retention and subsequent RNA degradation control the neuron-specific expression of critical presynaptic components, including t-SNARE and v-SNARE. The work thus elucidates the underlying mechanism that regulates the coordinated expression of a functionally coherent subset of mammalian presynaptic genes during neuronal differentiation.
Keywords: coordinated gene expression, neuronal differentiation, presynaptic proteins, polypyrimidine tract-binding protein, intron retention, nuclear mRNA surveillance
Abstract
Differentiated cells acquire unique structural and functional traits through coordinated expression of lineage-specific genes. An extensive battery of genes encoding components of the synaptic transmission machinery and specialized cytoskeletal proteins is activated during neurogenesis, but the underlying regulation is not well understood. Here we show that genes encoding critical presynaptic proteins are transcribed at a detectable level in both neurons and nonneuronal cells. However, in nonneuronal cells, the splicing of 3′-terminal introns within these genes is repressed by the polypyrimidine tract-binding protein (Ptbp1). This inhibits the export of incompletely spliced mRNAs to the cytoplasm and triggers their nuclear degradation. Clearance of these intron-containing transcripts occurs independently of the nonsense-mediated decay (NMD) pathway but requires components of the nuclear RNA surveillance machinery, including the nuclear pore-associated protein Tpr and the exosome complex. When Ptbp1 expression decreases during neuronal differentiation, the regulated introns are spliced out, thus allowing the accumulation of translation-competent mRNAs in the cytoplasm. We propose that this mechanism counters ectopic and precocious expression of functionally linked neuron-specific genes and ensures their coherent activation in the appropriate developmental context.
Higher eukaryotes contain multiple cell types that differ in their morphological and physiological properties despite having identical or nearly identical genomes. This phenotypic diversity relies on the expression of distinct repertoires of lineage-specific genes coordinated in a precise spatiotemporal manner. A number of transcriptional and post-transcriptional mechanisms are known to control these elaborate gene networks (Davidson 2006; Moore and Proudfoot 2009; Vaquerizas et al. 2009; Ivey and Srivastava 2010).
Alternative pre-mRNA splicing provides a remarkable example of post-transcriptional regulation operating on a genome-wide scale (Wang and Burge 2008; Nilsen and Graveley 2010). Recent studies suggest that ∼90% of human and mouse genes may give rise to alternatively spliced transcripts, which often encode distinct protein isoforms (Pan et al. 2008; Wang et al. 2008). This effectively increases genome-coding capacity without expanding the number of the genes. Alternative splicing events often occur in a tissue-specific manner, with the largest number of unique splice forms found in the nervous system (Q Li et al. 2007; Pan et al. 2008; Wang et al. 2008).
We and others have previously shown that the polypyrimidine tract-binding protein (PTBP1 in humans and Ptbp1 in mice; other names: PTB and hnRNP I) functions as a global regulator of a neuron-specific alternative splicing program (Boutz et al. 2007; Makeyev et al. 2007; Spellman et al. 2007). Although this protein has traditionally been considered a splicing repressor, recent transcriptome-wide surveys have identified examples of both PTBP1-repressed and PTBP1-activated alternative exons (Xue et al. 2009; Llorian et al. 2010). Ptbp1 tends to be expressed at high levels in nonneuronal and neural stem cells (NSCs), whereas its expression is diminished in differentiating neurons by the nervous system-specific microRNA (miRNA) miR-124 (Makeyev et al. 2007).
PTBP1 also autoregulates its own expression through a negative feedback mechanism, in which an excess of this protein induces skipping of an alternative exon within its own pre-mRNA and leads to the appearance of a premature termination codon (PTC) (Wollerton et al. 2004). The PTC-containing PTBP1 mRNAs are then rapidly degraded in the cytoplasm by the nonsense-mediated decay (NMD) machinery (Isken and Maquat 2008; Nicholson and Muhlemann 2010). Using a similar NMD mechanism, Ptbp1 represses the expression of its neuronal paralog (Ptbp2) and at least two other nervous system-specific genes (Gabbr1 and Dlg4) (Boutz et al. 2007; Makeyev et al. 2007; Spellman et al. 2007; Zheng et al. 2012). Thus, in addition to generating distinct protein isoforms, alternative splicing may cooperate with the cytoplasmic RNA surveillance machinery to control gene expression levels (Lareau et al. 2007; Isken and Maquat 2008; Barash et al. 2010).
Several mechanisms are known to control the quality of RNAs in the nucleus (Sommer and Nehrbass 2005; Egecioglu and Chanfreau 2011). One such mechanism blocks the export of unspliced pre-mRNAs from the nucleus to the cytoplasm, thus preventing their translation into aberrant polypeptides (Chang and Sharp 1989; Legrain and Rosbash 1989). A growing body of evidence suggests that, in addition to correcting splicing errors, nuclear mRNA surveillance may contribute to programmed changes in gene expression levels. For example, human SRSF1/ASF/SF2 protein homeostatically autoregulates its expression at least in part by promoting the accumulation of incompletely spliced SRSF1 mRNA species in the nucleus (Sun et al. 2010). Similarly, the retention of intron 3 in the mouse apolipoprotein E transcripts may interfere with their export from the nucleus to the cytoplasm (Xu et al. 2008).
Regulated intron retention is widely used as a general strategy for coordinated expression of meiotic and ribosomal protein genes in budding and fission yeasts (Averbeck et al. 2005; Moldon et al. 2008; Cremona et al. 2010; Munding et al. 2010; Parenteau et al. 2011). However, it is currently unknown whether higher eukaryotes use similar intron-dependent strategies for orchestrating their cell differentiation programs.
Signal transmission from a presynaptic neuron to a postsynaptic cell is a major nervous system function that depends on coexpression of hundreds of neuronal proteins involved in signal-dependent fusion of synaptic vesicles with synaptic terminals (e.g., vesicle-specific v-SNAREs and calcium sensors and terminal-specific t-SNAREs), trafficking of vesicles along the axon, and their recycling following neurotransmitter release (Sudhof 2004; Wojcik and Brose 2007; DeBoer et al. 2008). The genes encoding these important proteins are tightly regulated during neuronal differentiation; however, relatively little is known about the underlying mechanisms (Waites et al. 2005).
Results
Ptbp1 regulates a set of genes at the level of mRNA abundance
To understand the Ptbp1-controlled post-transcriptional network, we transfected the mouse neuroblastoma CAD cell line with either Ptbp1-specific (siPtbp1) or control (siControl) siRNAs and analyzed the samples by RNA sequencing (RNA-seq). In addition to the expected stimulation of the nervous system-specific alternative splicing program (data not shown), an overall abundance of a number of transcripts either decreased (n = 431; >1.5-fold; P < 0.001) (Supplemental Table S1) or increased (_n_ = 276; >1.5-fold; P < 0.001) (Supplemental Table S2) in the Ptbp1 knockdown sample. As expected, siPtbp1 down-regulated Ptbp1 expression (∼5.4-fold; P = 0) (Supplemental Table S1) and up-regulated the expression of its neuron-enriched paralog, Ptbp2 (∼3.5-fold; P = 2.1 × 10−259) (Supplemental Table S2). Interestingly, these large-scale transcriptome changes were accompanied by an increased propensity of CAD cells to undergo neuron-like differentiation (Supplemental Fig. S1).
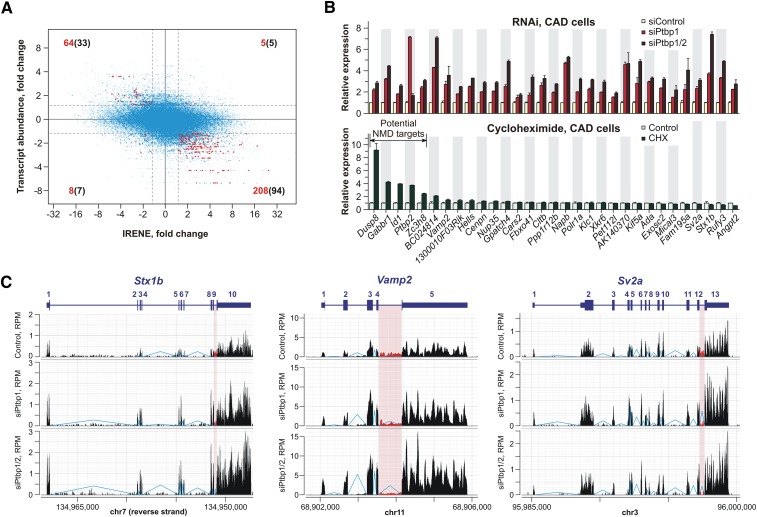
The RNA-seq data contained a substantial number of intronic RNA-seq reads likely derived from the nuclear (pre-)mRNA fraction. We reasoned that the ratio between intronic reads and reads originating from the adjacent exons—the statistic that we refer to as IRENE (intronic reads normalized to exons)—should be a faithful indicator of post-transcriptionally regulated genes. Strikingly, the statistically significant IRENE changes induced by siPtbp1 correlated inversely with statistically significant changes in the corresponding mRNA expression levels (P = 3.2 × 10−47, Fisher's exact test) (Fig. 1A, red dots).
Figure 1.
Ptbp1 regulates the expression levels of an extensive set of genes. (A) Relationship between changes in the IRENE values and in the corresponding mRNA expression levels following the Ptbp1 knockdown. (Red) Introns associated with a ≥1.5-fold change in IRENE (P < 0.001) and a simultaneous ≥1.5-fold change in the mRNA abundance (P < 0.001); (blue) the rest of the genes. Numbers of significantly regulated introns within specific quadrants are shown in red, and the corresponding gene numbers are shown in black. (B, top) CAD cells were transfected with Ptbp1-specific siRNA (siPtbp1), an siRNA mixture against both Ptbp1 and Ptbp2 (siPtbp1/2), or a control siRNA (siControl), and the expression levels of 31 genes from the top left quadrant in A were analyzed 72 h post-transfection by RT-qPCR. (Bottom) CAD cells were treated with CHX or DMSO (control), and the expression levels of the same 31 genes were measured using Agilent gene expression microarrays. In both analyses, data were averaged from two to three independent experiments ±SD, and the control expression levels were set to 1. (C) RNA-seq read density plots for the Stx1b, Vamp2, and Sv2a genes. Reads corresponding to the 3′-terminal introns are shown in red.
We focused on the subset of genes showing increased expression levels and reduced IRENE scores (33 genes) (Fig. 1A, top left quadrant; Supplemental Table S3). Since Ptbp1 and Ptbp2 may regulate overlapping pre-mRNA sets and the loss of Ptbp1 leads to the Ptbp2 up-regulation (Makeyev et al. 2007), we treated CAD cells with a mixture of Ptbp1- and Ptbp2-specific siRNAs (siPtbp1/2) and repeated the RNA-seq analysis. In this case, increased mRNA expression and reduced IRENE scores were detected for 102 genes (>1.5-fold changes; P < 0.001) (Supplemental Table S4), which included 19 of the above 33 genes.
To confirm that these effects were due to increased transcript abundance rather than altered splicing patterns, we analyzed the mRNA expression levels for the 31 Mouse Genome Informatics (MGI)-annotated genes by RT-qPCR using primers specific to constitutively spliced regions. All of these genes were significantly up-regulated upon Ptbp1 or Ptbp1/2 knockdown (Fig. 1B, top graph; Supplemental Fig. S2A). We concluded that Ptbp1 and, possibly, Ptbp2 regulate the expression levels of extensive sets of genes.
Ptbp1 represses the expression of a number of genes in an NMD-independent manner
Ptbp1 protein has previously been shown to reduce the expression of Ptbp2 and Gabbr1 mRNAs through the NMD pathway (Makeyev et al. 2007). Since NMD is thought to function in the cytoplasm without affecting nuclear (pre-)mRNA levels, Ptbp1 knockdown was expected to increase the abundance of Ptbp2 and Gabbr1 mRNAs and simultaneously decrease the corresponding IRENE statistics. Both genes were indeed present among the up-regulated genes with reduced IRENE values (Fig. 1B, top graph; Supplemental Table S3).
To examine whether the remaining 29 genes were also regulated by NMD, we treated CAD cells with cycloheximide (CHX), a protein synthesis inhibitor that also blocks NMD-mediated mRNA degradation, and analyzed the effect of this treatment on the gene expression using an Agilent gene expression microarray. To our surprise, only three additional genes (Dusp8, Id1, and Zc3h8) significantly increased their expression levels in response to CHX treatment (Fig. 1B, bottom graph; Supplemental Fig. S2B). Thus, the 26 remaining genes that failed to respond to the CHX treatment were likely regulated by mechanisms other than NMD.
Expression levels of several NMD-independent genes positively correlate with the 3′-terminal intron splicing efficiency
Gene ontology (GO) analysis of the 26 NMD-independent genes returned “neurotransmitter transport” (GO:0006836) as the highest-scoring term (44.3-fold enrichment over the background frequency; Bonferroni-corrected P = 3.54 × 10−4) (Supplemental Table S5). This corresponded to a subset of four neuron-specific genes encoding critical presynaptic proteins: Stx1b (a t-SNARE), Vamp2 (a v-SNARE), Sv2a (a synaptic vesicle-associated regulator of Ca2+ levels), and Napb/βSNAP (a SNARE recycling protein) (Sudhof 2004; Wojcik and Brose 2007). Similar to the siPtbp1 effect, down-regulation of the Ptbp1 expression by the miRNA miR-124 led to a significantly elevated expression of these genes in CAD cells (Supplemental Fig. S2C; data not shown).
Interestingly, for the Stx1b, Vamp2, and Sv2a genes, the most dramatic decrease in the IRENE scores following Ptbp1 knockdown was observed for the 3′-terminal introns (Supplemental Table S3). This effect was due to a simultaneous decrease in the density of intronic reads and an increase in the density of exonic reads (Fig. 1C). At least three other protein-coding genes from the NMD-independent list followed a similar trend: the Kif5a gene encoding a nervous system-specific kinesin heavy chain involved in anterograde axonal transport of synaptic vesicles and other membrane-bound organelles (DeBoer et al. 2008), the Exosc2 gene encoding the Rrp4 subunit of the RNA exosome complex, and a nucleoporin gene, Nup35 (Supplemental Table S3; Supplemental Fig. S2D; data not shown).
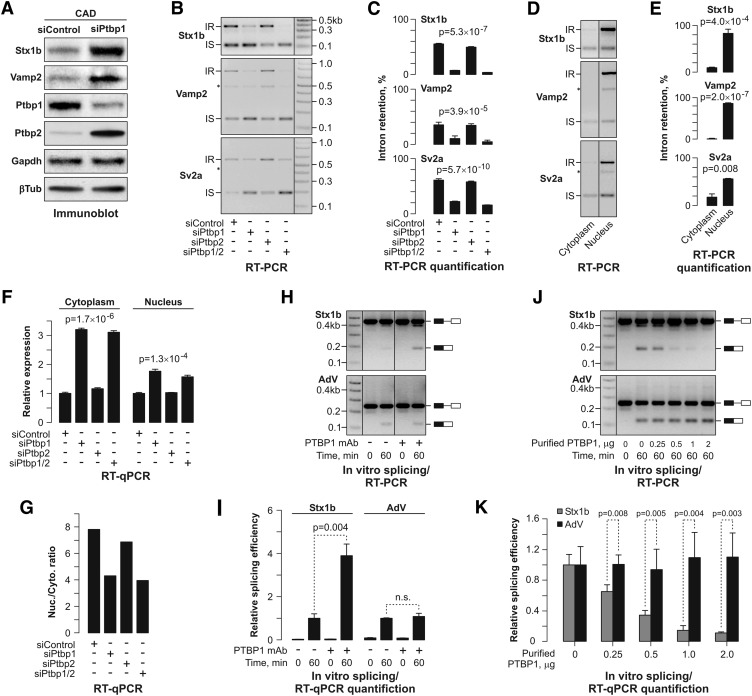
To examine whether the increase in mRNA abundance of the NMD-independent genes led to the accumulation of the corresponding proteins, we knocked down the Ptbp1 protein in CAD cells by RNAi and analyzed the expression of Stx1b and Vamp2 proteins by immunoblotting (Fig. 2A). The levels of both SNARE proteins increased noticeably in the Ptbp1 knockdown sample as compared with the siControl-treated cells (Fig. 2A). Taken together, these data indicated that Ptbp1 might control the expression levels of a subset of genes by modulating the 3′-terminal intron splicing efficiency.
Figure 2.
Ptbp1 controls the expression of neuronal genes by repressing splicing of the 3′-terminal introns. (A) Down-regulation of the Ptbp1 protein levels in CAD cells by RNAi elevates the expression of the Stx1b and Vamp2 proteins. The Ptbp1 knockdown efficiency is controlled by Ptbp1- and Ptbp2-specific antibodies. Gapdh and β-tubulin were used as lane loading controls. (B) CAD cells treated with siControl, siPtbp1, siPtbp2, or siPtbp1/2 were analyzed by RT–PCR using primer pairs designed to estimate the splicing efficiency of the 3′-terminal introns. Note that both siPtbp1 and siPtbp1/2 increase the ratio between the intron-spliced (IS) and intron-retained (IR) forms. (C) Quantification of the results in B. Data are averaged from three independent experiments, ±SD. (D) Transcripts retaining the last intron are readily detectable in the nucleus but not in the cytoplasm of the CAD cell. (E) Quantification of the results in D. Data are averaged from three independent experiments, ±SD. (F) Cytoplasmic and nuclear fractions were prepared form CAD cells treated with siRNAs as in B, and the Stx1b expression levels were analyzed by RT-qPCR. (G) Quantification of the RT-qPCR data in F shows that the ratios between the nuclear and the cytoplasmic expression levels of Stx1b, Vamp2, and Sv2a transcripts reduce following Ptbp1 or Ptbp1/2 knockdown. (H,I) Splicing efficiency of a synthetic RNA substrate comprising the wild-type Stx1b exon 9–intron 9–exon 10 segment was assayed in vitro using either control-treated or PTBP1 monoclonal antibody-treated HeLa S3 nuclear extracts, and the reaction products were analyzed by either RT–PCR with a primer pair specific to exons 9 and 10 (H) or RT-qPCR with an exon junction-specific primer pair (I). As expected, no spliced products were detected at the time point 0. Note that the PTBP1 immunodepletion activates Stx1b splicing while having no effect on the splicing efficiency of the control AdV substrate. Data in I are averaged from three amplification experiments (±SD) and compared using the _t_-test. Splicing efficiencies in the presence of endogenous PTBP1 levels were set to 1. (J) In vitro splicing of the Stx1b and AdV substrates was assayed as in H using PTBP1-immunodepleted nuclear extracts supplemented with the indicated amounts of recombinant PTBP1. Note that the addition of PTBP1 back to the immunodepleted extracts inhibits Stx1b but not AdV splicing. (K) Data from J were averaged from three independent experiments (±SD) and compared using the _t_-test. Splicing efficiencies in the absence of recombinant PTBP1 were set to 1.
Ptbp1 represses the 3′-terminal intron splicing in the Stx1b, Vamp2, Sv2a, and Kif5 pre-mRNAs
Since the 3′-terminal introns of Stx1b, Vamp2, Sv2a, and Kif5 contained multiple consensus Ptbp1-binding sites (Supplemental Fig. S3), we reasoned that Ptbp1 may directly inhibit splicing of these introns. To begin addressing this hypothesis, we first determined the steady-state splicing efficiency of the four short 3′-proximal introns within the Stx1b pre-mRNA using RT–PCR analysis of the CAD cell nuclear fraction with corresponding exon-specific primers (Supplemental Fig. S4A,B). Interestingly, the last intron (intron 9) was retained in >75% of the Stx1b transcripts, in contrast to the three upstream introns (5, 6, and 8) that were retained in <25% of the Stx1b transcripts (P = 4.6 × 10−8, single-factor ANOVA test) (Supplemental Fig. S4A,B). This difference could not be attributed simply to the 5′-to-3′ directionality of the pre-mRNA synthesis, since the steady-state splicing efficiency of the 3′-terminal intron of the β-actin (Actb) pre-mRNA was not significantly different from that of an upstream intron (Supplemental Fig. S4C,D).
We then treated CAD cells with siPtbp1, siPtbp2, siPtbp1/2, or siControl siRNAs and analyzed the Stx1b, Vamp2, Sv2a, and Kif5a 3′-terminal intron splicing status in the whole-cell RNA samples by RT–PCR (Fig. 2B,C; Supplemental Fig. S4E,F, first four samples). The 3′ intron-retained forms of all four transcripts were readily detectable in the siControl transfected samples. Down-regulation of Ptbp1 or both Ptbp1 and Ptbp2 significantly decreased the relative abundance of the intron-retained species (P < 4 × 10−5, single-factor ANOVA test) (Fig. 2B,C; Supplemental Fig. S4E,F, first four Kif5a samples). Treating the cells with siPtbp2 only had no detectable effect, consistent with the low background Ptbp2 expression in the presence of Ptbp1 (Makeyev et al. 2007). We concluded that Ptbp1 represses splicing of the Stx1b, Vamp2, Sv2a, and Kif5a 3′-terminal introns.
Retention of the 3′-terminal introns inhibits mRNA accumulation in the cytoplasm
Incompletely spliced mRNA species often fail to be efficiently exported from the nucleus to the cytoplasm. In line with this trend, the intron-retained forms of the Stx1b, Vamp2, and Sv2a transcripts were up to 10 times more abundant in the nucleus than in the cytoplasm of untreated CAD cells (P < 0.01, _t_-test) (Fig. 2D,E). Nucleocytoplasmic fractionation of the siRNA-treated CAD cells further revealed that knocking down Ptbp1 alone or both Ptbp1 and Ptbp2 dramatically reduced the percentage of the intron-retained species in the nucleus (P = 5 × 10−9, single-factor ANOVA) (Supplemental Fig. S4E,F; data not shown). These treatments also appeared to reduce the fraction of the intron-retained forms in the cytoplasm (P = 0.04, single-factor ANOVA) (Supplemental Fig. S4E,F; data not shown), although the effect was difficult to quantify accurately due to the overall low abundance of the intron-retained form in this cellular compartment. Notably, we failed to detect Actb transcripts retaining the 3′-terminal intron in the whole-cell and cytoplasmic samples (Supplemental Fig. S4E,F, Actb panels). These species were present in the corresponding nuclear fractions, but their abundance was not affected by changes in the Ptbp1 and Ptbp2 expression (Supplemental Fig. S4E,F, Actb panels).
We further assayed the effect of Ptbp1 and Ptbp2 on mRNA expression levels by RT-qPCR using Gapdh mRNA as a normalization control (Fig. 2F). This analysis showed that the overall abundance of the cytoplasmic Stx1b mRNA rose more than threefold in the siPtbp1 and siPtbp1/2 samples as compared with the siControl-treated cells (P = 1.7 × 10−6, single-factor ANOVA) (Fig. 2F). The Stx1b transcript expression also increased in the corresponding nuclear fractions, albeit to a lesser extent (less than two times; P = 1.3 × 10−4, single-factor ANOVA) (Fig. 2F). Interestingly, the apparent abundance of the Stx1b transcripts was sevenfold to eightfold higher in the nucleus than in the cytoplasm of the siControl- and siPtbp2-treated CAD cells (Fig. 2G). This difference decreased to approximately fourfold following the Ptbp1 or the double Ptbp1/2 knockdown (Fig. 2G). The regulation of the Stx1b, Vamp2, Sv2a, and Kif5a mRNA abundance by Ptbp1 was not specific to the CAD cells, since similar results were obtained using a clone of the L929 mouse fibrosarcoma cell line (Supplemental Fig. S4G).
The above results are consistent with the model that the Ptbp1-stimulated retention of 3′-terminal introns inhibits the export of the incompletely spliced Stx1b, Vamp2, Sv2a, and Kif5a mRNAs from the nucleus to the cytoplasm and eventually leads to their nuclear degradation.
Ptbp1 directly inhibits splicing of the Stx1b 3′-terminal intron
To test whether Ptbp1 repressed the 3′-terminal intron splicing directly, we synthesized a transcript comprising the Stx1b exon 9, intron 9, and exon 10 sequences and assayed splicing of this substrate in vitro followed by RT–PCR and RT-qPCR analyses (Fig. 2H–K). Splicing of this RNA was fairly inefficient in a control-treated HeLa S3 nuclear extract but was dramatically activated by immunodepleting the endogenous PTBP1 protein (Fig. 2H,I; Supplemental Fig. S5A). In contrast, PTBP1 depletion had no effect on splicing of a control adenovirus-specific RNA substrate (AdV) (Fig. 2H,I). To ensure that the stimulation of the Stx1b intron 9 splicing following PTBP1 immunodepletion was a specific effect, we added various amounts of purified recombinant PTBP1 to the immunodepleted splicing reactions and repeated the analysis (Fig. 2J,K). Satisfyingly, increasing the PTBP1 concentrations efficiently repressed splicing of the Stx1b but not AdV RNA substrates. Similar results were obtained using in vitro splicing assays with recombinant radioactive RNA substrates (Supplemental Material; Supplemental Fig. S5B–F). Overall, these data indicated that Ptbp1 directly represses splicing of the Stx1b intron 9.
Stx1b intron 9 in its natural context is sufficient for the regulation
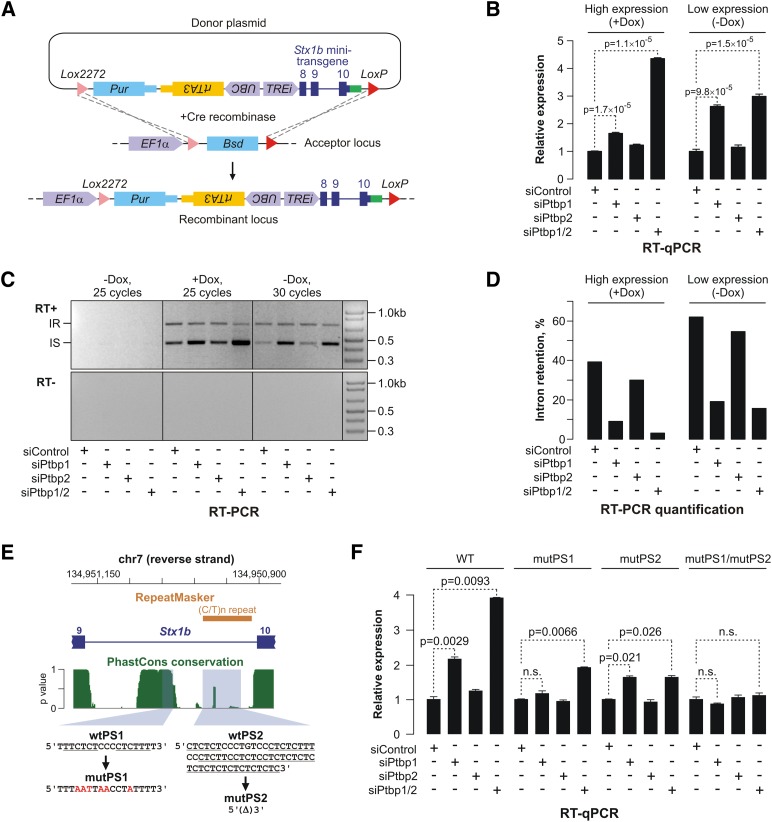
To identify _cis_-elements involved in the regulation of the intron 9 splicing and the Stx1b mRNA abundance, we turned to the minigene approach. Since transiently transfected Stx1b constructs failed to recapitulate the regulation, possibly as a result of titrating out a saturable nuclear factor (data not shown), we took advantage of the single-copy transgene knock-in technology based on high-efficiency and low-background recombination-mediated cassette exchange (HILO-RMCE) (Khandelia et al. 2011). Using this approach, we generated a population of CAD cells expressing a “minitransgene” cassette in which a short intron 9-containing fragment of the Stx1b gene was placed under the control of a doxycycline (Dox)-inducible promoter element containing a recombinant constitutive intron (TREi) (Fig. 3A).
Figure 3.
Stx1b intron 9 is sufficient and necessary for the Ptbp1-dependent regulation. (A) Integration of the Stx1b minitransgene into the CAD-A13 cell genome using RMCE. (B_–_D) Recombinant CAD-A13 cells containing a single copy of the Stx1b minitransgene were treated with the indicated siRNAs for 48 h, followed by 24-h incubation with or without Dox. (B) RT-qPCR analysis showing that the minitransgene expression levels increase following the Ptbp1 or the double Ptbp1/2 knockdown. (C, top) The splicing status of the transgenic intron 9 was assayed by RT–PCR. (Bottom) No PCR signal was detected when reverse transcriptase was omitted from the RT reactions. (D) Quantification of the data in C indicates that a large fraction of transgenic transcripts retain intron 9 in siControl- and siPtbp2-treated samples, whereas siPtbp1 and siPtbp1/2 dramatically stimulate the intron splicing. (E) Diagram of the exon 9–intron 9–exon 10 fragment of the mouse Stx1b gene. The phastCons plot (green) shows the probability of sequence conservation across placental mammalian species (Siepel et al. 2005). Note that a fragment of intron 9 is conserved across species. The orange rectangle marks the position of a poorly conserved (C/T)n repeat predicted by RepeatMasker (http://www.repeatmasker.org). The wild-type PS1 and PS2 sequences containing consensus Ptbp1-binding sites (underlined) and the corresponding mutations are indicated at the bottom. (F) Recombinant CAD-A13 cells containing a single copy of either the wild-type minitransgene or minitransgenes containing the PS1 and/or PS2 mutations were treated with the indicated siRNAs for 72 h, and the expression levels of the transgenic transcripts were analyzed by RT-qPCR. Note that the individual mutations reduce the minitransgene response to the Ptbp1 or the Ptbp1/2 knockdown, whereas the PS1/PS2 double mutation completely abolishes the regulation. Data in B and F are averaged from three experiments, ±SD.
We then knocked down the expression of Ptbp1 or/and Ptbp2 in the transgenic cells (Supplemental Fig. S6) and assayed the minitransgene expression under the conditions in which it was either fully activated (Dox+) or transcribed at a low background level (Dox−) (Fig. 3B–D). RT-qPCR analysis using transgene-specific primers showed that, both with and without Dox, the expression of the Stx1b minitransgene was significantly up-regulated following Ptbp1 or double Ptbp1/2 knockdown as compared with the siControl-treated cells (Fig. 3B). RT–PCR analysis of the same samples indicated that splicing of the transgenic intron 9 was noticeably more efficient in the Ptbp1 or double Ptbp1/2 knockdown samples (Fig. 3C,D). Thus, intron 9 within its natural context is sufficient for Ptbp1-dependent regulation of Stx1b mRNA expression.
Polypyrimidine sequences within the Stx1b intron 9 are necessary for the regulation
To test whether intron 9 was necessary for Stx1b gene regulation, we integrated an ∼170-kb transgenic fragment containing the entire mouse Stx1b gene into the CAD cell genome using an RMCE protocol modified for bacterial artificial chromosome (BAC)-encoded transgenes (Supplemental Fig. S7A,B; K Yap, ZQ Lim, and EV Makeyev, in prep.). To distinguish between the recombinant and endogenous copies, we marked the 3′ untranslated region (UTR) of the transgenic Stx1b with a 68-nucleotide (nt) sequence encoding an FRT site (Supplemental Fig. S7A). _Stx1b_-FRT transgene expression was up-regulated following Ptbp1 or Ptbp1/2 knockdown, similar to the endogenous Stx1b gene (Supplemental Fig. S7C).
Importantly, deletion of either the entire intron 9 (Δintron 9, 226-nt deletion) or its fragment containing Ptbp1 consensus binding sites (ΔPS, 125-nt deletion) from the Stx1b transgene (Supplemental Fig. S7A,B) completely abolished the regulation of mRNA abundance in response to siPtbp1 or siPtbp1/2 (Supplemental Fig. S7D). Notably, splicing of the wild-type transgenic intron 9 was regulated by Ptbp1 similarly to the endogenous intron 9, whereas the ΔPS intron was spliced in a constitutive manner (Supplemental Fig. S7E,F). As expected, no retention products were detected for the Δintron 9 transgene (Supplemental Fig. S7E).
A more detailed bioinformatic analysis of the mouse Stx1b intron 9 sequence revealed two putative Ptbp1-binding surfaces: one highly conserved across placental mammals (PS1), and the other one poorly conserved (PS2) (Fig. 3E). To examine the significance of these elements, we mutated them in the minigene context. Interestingly, mutating PS1 and PS2 individually reduced the response of minitransgene expression to Ptbp1 or Ptbp1/2 knockdown, whereas the mutPS1/mutPS2 double mutation abolished the regulation completely (Fig. 3F). We concluded that intron 9 is necessary for mRNA abundance control by Ptbp1 and Ptbp2 and that both PS1 the PS2 elements are likely required for optimal regulation.
Intron 9 recognition by the splicing machinery is required for nuclear retention and degradation of the incompletely spliced Stx1b mRNA
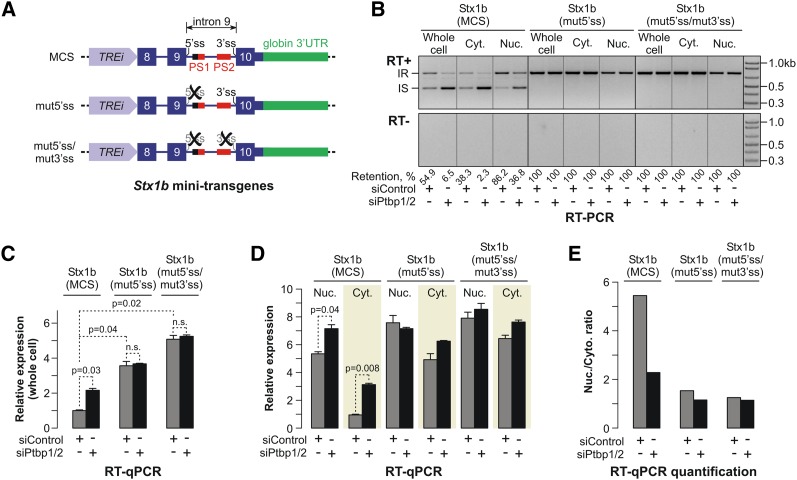
Two models could account for the repressive effect of intron 9 retention on mRNA export: (1) Intron-bound Ptbp1 completely inhibits the intron interaction with the splicing machinery and simultaneously hinders the export of the incompletely spliced mRNA to the cytoplasm. (2) Alternatively, the intron is recognized by the early components of the splicing machinery (e.g., by U1 snRNP, U2AF, and/or U2 snRNP), but the subsequent splicing and mRNA export steps are arrested in the presence of intron-bound Ptbp1.
To distinguish between these possibilities, we inactivated the 5′ splice site (5′ss) within the Stx1b(MCS) minitransgene intron 9 by changing the consensus GU sequence to CC and analyzed the expression of the mutated mRNA [Stx1b(mut5′ss)] in CAD cells treated with siControl or siPtbp1/2 siRNAs (Fig. 4A). RT–PCR analysis confirmed that the mutation completely inactivated intron splicing (Fig. 4B). Notably, in siControl transfected cells, overall Stx1b(mut5′ss) expression increased 3.6-fold as compared with the Stx1b(MCS) minitransgene (P = 0.04, _t_-test) (Fig. 4C). Moreover, Ptbp1/2 knockdown had no effect on Stx1b(mut5′ss) transcript levels, unlike Stx1b(MCS), whose expression increased 2.2-fold following this treatment (P = 0.03, _t_-test) (Fig. 4C).
Figure 4.
Intron 9 recognition by the splicing machinery is necessary for the Ptbp1-dependent regulation of Stx1b mRNA abundance. (A) Diagram of the Stx1b minitransgenes. (B, top) CAD-A13 cells containing the transgenes depicted in A were treated with either siControl or siPtbp1/2 for 72 h, and the total RNAs from the whole cells or the two subcellular fractions were analyzed by RT–PCR to examine the transgenic intron 9 splicing. (Bottom) No PCR products were detected in the absence of reverse transcriptase. (C) RT-qPCR analysis showing that the mutation of either 5′ss or both 5′ss and 3′ss significantly increases the transgene expression levels in the siControl-treated cells. Moreover, unlike Stx1b(MCS), the expression of Stx1b(mut5′ss) and Stx1b(mut5′ss/mut3′ss) minitransgenes is not activated by siPtbp1/2. (D) RT-qPCR analysis showing that siPtbp1/2 stimulates the expression levels of Stx1b(MCS) but not Stx1b(mut5′ss) or Stx1b(mut5′ss/mut3′ss) in the nucleus and the cytoplasm. (E) Quantification of the RT-qPCR data in D suggesting that the siPtbp1/2 treatment also reduces the originally high ratio between the nuclear and cytoplasmic levels of Stx1b(MCS) but not Stx1b(mut5′ss) and Stx1b(mut5′ss/mut3′ss) transcripts.
siPtbp1/2 also had little effect on the apparent abundance of the Stx1b(mut5′ss) transcript in the cytoplasmic and nuclear fractions. In contrast, Stx1b(MCS) transcript levels were significantly up-regulated in both the cytoplasm (3.3-fold; P = 0.008, _t_-test) and the nucleus (1.3-fold; P = 0.04, _t_-test) (Fig. 4D). The apparent abundance of the Stx1b(mut5′ss) transcripts was comparable between the nuclear and cytoplasmic fractions in both siControl- and siPtbp1/2-treated cells (Fig. 4E). On the other hand, Stx1b(MCS) transcripts were enriched 5.6-fold in the nuclei of the siControl-treated cells and 2.3-fold in the nuclei of siPtbp1/2-treated cells (Fig. 4E). Similar to the Stx1b(mut5′ss) behavior, Stx1b(mut5′ss/mut3′ss) transcripts with mutations in both the 5′ss and the 3′ss were efficiently expressed in the nuclear and the cytoplasmic compartments regardless of the Ptbp1 and Ptbp2 concentrations (Fig. 4A–E). These results suggested that the nuclear retention and nuclear degradation of incompletely spliced Stx1b mRNAs require a functional interaction between intron 9 and the splicing machinery.
Stx1b expression is regulated by the nuclear mRNA surveillance machinery
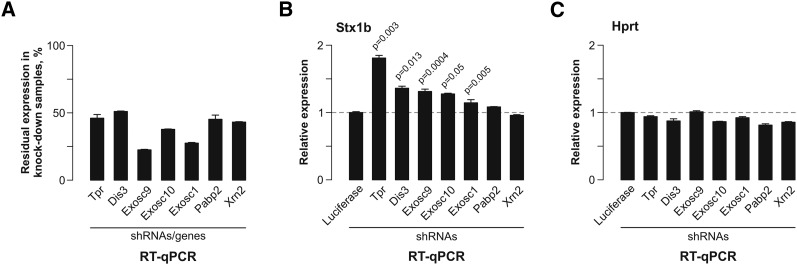
The data presented above are consistent with the model that incompletely spliced mRNAs encoding presynaptic proteins are retained in the nucleus and degraded by the nuclear mRNA quality control machinery (e.g., see Fig. 2F). To identify factors involved in this process, we knocked down several known nuclear RNA surveillance components, including catalytic (Exosc10/Rrp6 and Dis3/Rrp44) and structural (Exosc1/Csl4 and Exosc9/Rrp45) subunits of the exosome complex; a nuclear 5′-to-3′ exonuclease, Xrn2; a nuclear pore protein, Tpr (Mlp1/2 homolog); and a nuclear poly(A)-binding protein, Pabp2 (Pab2 homolog) (Fig. 5A; Galy et al. 2004; Houseley and Tollervey 2009; Tomecki and Dziembowski 2010; Coyle et al. 2011; Kiss and Andrulis 2011; Lemieux et al. 2011). Strikingly, the most prominent increase in the Stx1b mRNA expression was detected in the Tpr knockdown samples (Fig. 5B). This was not an off-target effect, since an shRNA against a distinct Tpr-specific sequence stimulated Stx1b expression in a similar manner (data not shown). Stx1b expression was also stimulated by the Dis3 and Exosc9 knockdowns and, to a lesser extent, the Exosc10 and Exosc1 knockdowns (Fig. 5B). No significant changes in the Stx1b levels were detected in the samples expressing Xrn2- and Pabp2-specific shRNAs (Fig. 5B). Largely similar effects were observed for the Vamp2, Sv2a, and Kif5a mRNAs, except for the lack of significant effect of the Exosc9 knockdown in the case of Vamp2 and the Exosc1 knockdown in all three cases (Supplemental Fig. S8). Importantly, the levels of the Hprt and Actb “housekeeping” mRNA controls were not significantly up-regulated in the knockdown samples (Fig. 5C; Supplemental Fig. S8). We concluded that intron-retained RNAs are specifically destabilized in the nucleus in a Tpr- and exosome-dependent manner.
Figure 5.
Nuclear RNA surveillance machinery is involved in the Stx1b regulation. (A) CAD-A13 cells encoding stably integrated shRNAs under the control of a Dox-inducible promoter (Khandelia et al. 2011) were treated with 2 μg/mL Dox for 72 h, and the knockdown efficiencies were analyzed by RT-qPCR. Shown are residual mRNA expression levels normalized to the expression of the corresponding mRNAs in the presence of a firefly luciferase-specific shRNA control. Values are averaged from three amplification experiments, ±SD. Note that in all five cases, mRNA expression is noticeably diminished by the gene-specific shRNAs. (B) Stx1b mRNA expression levels were assayed by RT-qPCR following the knockdown of the indicated components of the RNA surveillance machinery. Note that shRNA specific to the nuclear pore-associated protein Tpr and the exosome subunit Dis3 leads to statistically significant accumulation of the Stx1b mRNA. (C) None of the shRNAs tested in this experiment led to significant up-regulation of the control Hprt mRNA. In B and C, the _P_-values were calculated using a two-tailed _t_-test assuming unequal variances and shown only for the samples showing significant (P ≤ 0.05) up-regulation of mRNA expression in response to shRNA treatment as compared with the luciferase shRNA control. Data are normalized to the expression levels of Gapdh mRNA and are averaged from at least three amplification experiments, ±SD.
Ptbp1 expression correlates with 3′-terminal intron splicing and mRNA abundance of neuronal genes in vivo
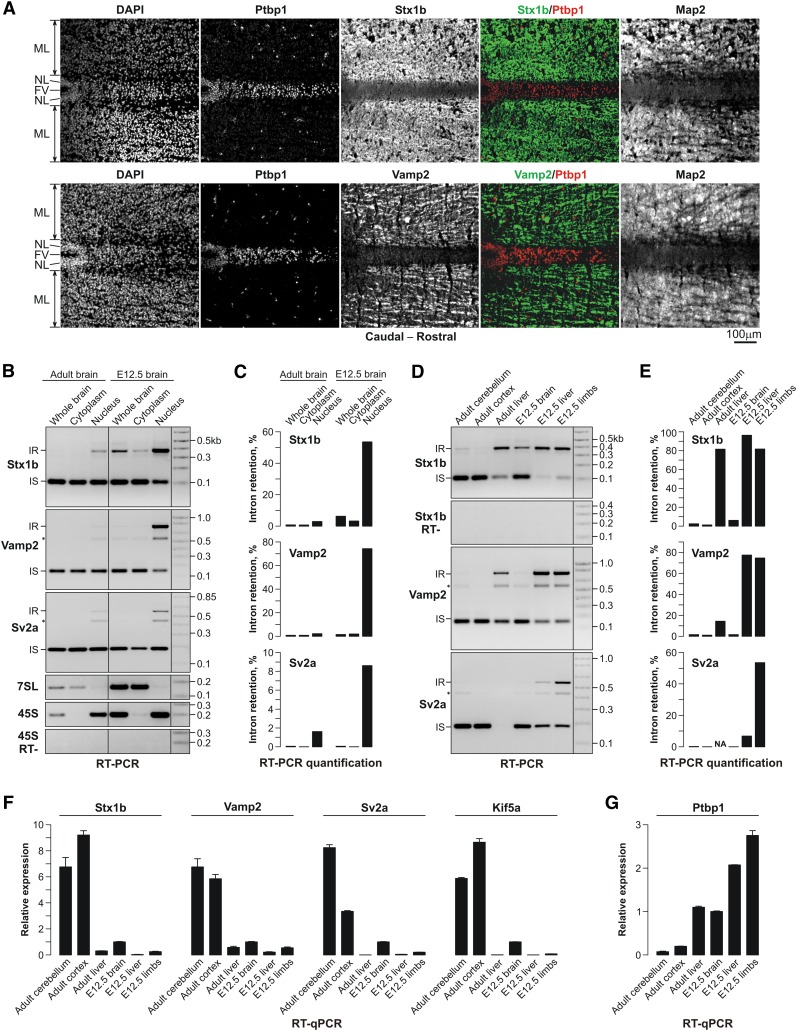
To test whether the above mechanisms functioned in the context of development, we analyzed the expression of the Ptbp1 protein and its SNARE targets, Stx1b and Vamp2, in embryonic day 13.5 (E13.5) mouse brains (Fig. 6A). Ptbp1 was expressed predominantly in the neuroepithelial layer containing proliferating NSCs, whereas Stx1b and Vamp2 showed reciprocal staining patterns, with the maximal expression in the mantle layer containing neurons at different stages of differentiation (Fig. 6A). The expression of mature miR-124 miRNA in the developing neural tube was reciprocal to the Ptbp1 expression pattern, as reported previously (Makeyev et al. 2007), and overlapping with the Stx1b expression pattern (Supplemental Fig. S9A,B).
Figure 6.
Ptbp1 expression pattern is consistent with its role as a regulator of presynaptic genes in vivo. (A) Immunofluorescence analyses of E13.5 medulla sections of the hind brain show that the Ptbp1 protein is expressed in the NSC-containing neuroepithelial layer (NL) lining the fourth ventricle (FV) but not in the mantle layer (ML) containing neurons at different stages of differentiation. Stx1b and Vamp2 proteins are expressed in a strictly reciprocal manner. The sections are additionally stained with antibodies against Map2, a marker of mature neurons. (B) Total RNAs were purified from the entire adult and E12.5 embryonic mouse brains or the corresponding cytoplasmic and nuclear fractions, and the Stx1b, Vamp2, and Sv2a 3′-terminal intron splicing was analyzed by RT–PCR. Nuclear (45S pre-rRNA) and cytoplasmic (7SL) RNA markers were analyzed by RT–PCR to confirm the nucleocytoplasmic fractionation quality. (C) Quantification of the results in B. (D,E) The analyses in B and C were repeated for six adult and embryonic tissue samples. (F,G) RT-qPCR analyses of Stx1b, Vamp2, Sv2a, Kif5a, and Ptbp1 expression in vivo.
We further examined 3′-terminal intron splicing for the Stx1b, Vamp2, Sv2a, and Kif5a transcripts in adult and embryonic mouse tissues (Fig. 6B–F; Supplemental Fig. S9C,D). Although the four transcripts were expressed in virtually all samples (except for the lack of the Sv2a expression in the adult liver), the percentage of the transcripts retaining the 3′-terminal introns varied dramatically across the tissues. Virtually no intron-retained products were detected in the total adult brain or its nuclear and cytoplasmic fractions (Fig. 6B,C; Supplemental Fig. S9C,D). We also failed to detect intron-retained Vamp2, Sv2a, and Kif5a RNAs in unfractionated E12.5 brain samples (Fig. 6B,C; Supplemental Fig. S9). However, these species were detected in the corresponding nuclear fractions (Fig. 6B; Supplemental Fig. S9C). Substantial amounts of the intron 9-retained Stx1b transcripts were expressed in both the total E12.5 brain samples and the nuclear fraction, while being noticeably depleted from the cytoplasmic fraction (Fig. 6B,C). Strikingly, all three nonneural tissues contained readily detectable amounts of the intron-retained transcripts (Fig. 6D,E; Supplemental Fig. S9D).
Quantitative analyses showed that the Ptbp1 expression levels correlated positively with the relative abundance of the retained mRNA species in the corresponding tissues (Spearman's correlation coefficient ρ = 0.8, P = 4.0 × 10−6) and negatively with the overall expression levels of the Stx1b, Vamp2, Sv2a, and Kif5a genes (ρ = −0.83, P = 7.0 × 10−7) (Fig. 6G; Supplemental Fig. S9E). These results are consistent with the role of Ptbp1 as a negative regulator of the Stx1b, Vamp2, Sv2a, and Kif5a genes in vivo.
Ptbp1 regulates Stx1b expression in primary mouse cells
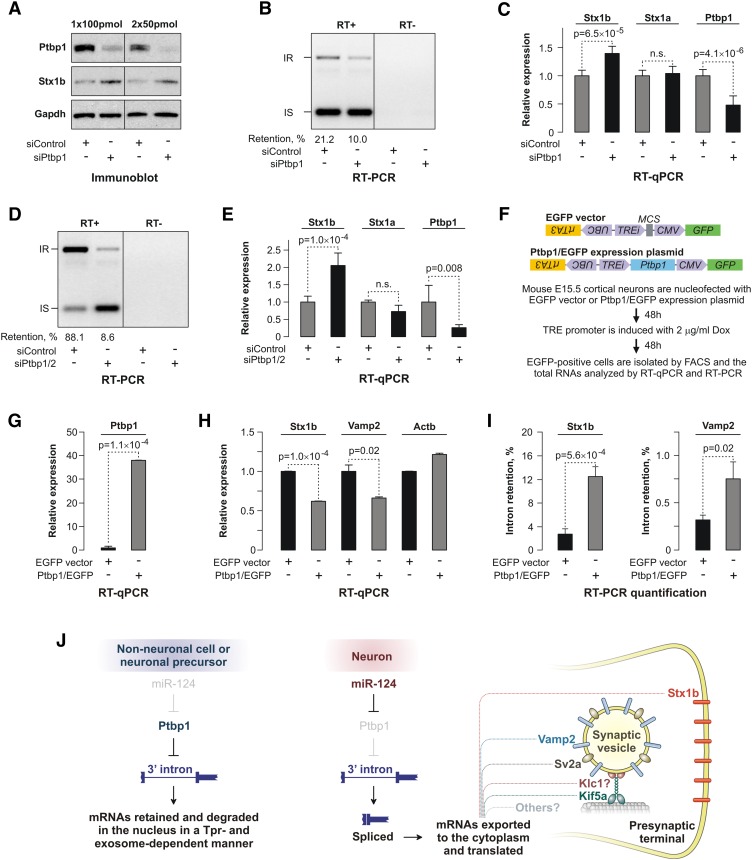
As a direct test of the Ptbp1 repressor activity in primary mouse cells, we knocked down Ptbp1 expression in embryonic cortical NSCs by RNAi. Immunoblot analysis of the siPtbp1- and siControl-treated samples suggested that the reduction in the Ptbp1 protein levels led to a noticeable increase in the Stx1b protein expression (Fig. 7A). This was accompanied by a 2.1-fold decrease in the relative abundance of the intron 9-retained Stx1b transcripts (Fig. 7B) and a modest but significant increase in the overall Stx1b mRNA abundance (P = 6.5 × 10−5) (Fig. 7C). The increased Stx1b expression was not due to siPtbp1-induced differentiation, since virtually all cells in the siControl- and siPtbp1-treated samples expressed the NSC marker nestin (Supplemental Fig. S10A). Ptbp1 knockdown did not change the expression of Stx1a, a Stx1b paralog lacking extensive Ptbp1-binding motifs within its last intron (Fig. 7C). Similarly, transfection of primary mouse embryonic fibroblasts (PMEFs) with siPtbp1/2 decreased the relative abundance of the intron 9-retained species 10-fold (Fig. 7D) and increased the expression of the Stx1b (2.1-fold, P = 1.0 × 10−4, _t_-test) but not the Stx1a mRNA (Fig. 7E).
Figure 7.
Ptbp1 regulates the Stx1b expression in NSCs and nonneuronal cells. (A) Neurosphere cultures were nucleofected once with 100 pmol or twice with 50 pmol of either siControl or siPtbp1, and the expression of the Ptbp1 and Stx1b proteins was analyzed by immunoblotting. The Stx1b expression increased noticeably following the Ptbp1 knockdown. (B,C) The neurosphere cultures nucleofected once with 100 pmol of siRNAs were additionally analyzed by RT–PCR to examine the Stx1b intron 9 splicing status (B) and RT-qPCR to assess the Stx1b expression levels (C). (D,E) PMEFs were transfected with siControl or siPtbp1/2 and analyzed by RT–PCR and RT-qPCR as in B and C. (F, top) EGFP and Ptbp1/EGFP expression plasmid used in nucleofection experiments. (Bottom) A flow chart of the experiment carried out to examine the effect of Ptbp1 overexpression in primary cortical neurons. (G) RT-qPCR analysis confirming that Ptbp1/EGFP-nucleofected neurons express larger amounts of Ptbp1 mRNA than their EGFP-only counterparts. (H) RT-qPCR analysis showing that neurons overexpressing Ptbp1 express significantly less Stx1b and Vamp2 mRNA. (I) Quantification of RT–PCR analyses carried out as in B and D, demonstrating significantly elevated retention of the Stx1b- and Vamp2-regulated introns in the presence of Ptbp1. Data in C, E, and G–I are averaged from at least three independent amplification experiments (±SD) and compared using the _t_-test. (J) Model for coordinated regulation of presynaptic genes through Ptbp1-controlled splicing of the 3′-terminal introns.
To test whether rescue of the Ptbp1 levels in post-mitotic neurons—where it is naturally diminished by miR-124 (Makeyev et al. 2007)—would reduce presynaptic gene expression levels, we nucleofected primary cortical neurons from E15.5 mouse embryos (Supplemental Fig. S10B) with a plasmid encoding a constitutively expressed EGFP marker and a Dox-inducible Ptbp1 cDNA lacking its natural 3′ UTR to bypass miR-124 regulation (Fig. 7F). Nucleofected EGFP-positive neurons were then isolated from neuronal cultures by fluorescence-activated cell sorting (FACS) and analyzed by RT-qPCR and RT–PCR. Notably, steady-state levels of Stx1b and Vamp2 but not Actb mRNA decreased significantly in the presence of exogenous Ptbp1 (Fig. 7G,H). Moreover, Ptbp1 overexpression arrested splicing of the regulated Stx1b and Vamp2 introns, as expected (Fig. 7I). We concluded that Ptbp1 represses presynaptic gene expression in vivo.
Discussion
Intron retention is a major form of alternative splicing in metazoan organisms that contributes to proteome diversity, regulates the efficiency of translation, and modulates intracellular mRNA localization (Galante et al. 2004; Denis et al. 2005; Mansilla et al. 2005; Marinescu et al. 2007; Wang and Burge 2008; Bell et al. 2010; Nilsen and Graveley 2010; Buckley et al. 2011). Here we show that the RNA-binding protein Ptbp1 coordinately regulates the expression levels of at least four functionally linked neuron-specific genes (Stx1b, Vamp2, Sv2a, and Kif5a) by inhibiting the 3′-terminal intron splicing and thus promoting nuclear retention and nuclear degradation of the incompletely spliced mRNAs (Fig. 7J).
This regulation mechanism adds a compelling example to the growing list of RNA-binding proteins that function as post-transcriptional master regulators coordinating the expression of functionally linked genes (Keene 2007). There is also an interesting parallel between the Ptbp1-controlled pathway uncovered in our study and the transcriptional repression of neuronal genes in nonneuronal cells by the REST/NRSF complex (Ballas and Mandel 2005). Similar to Ptbp1, components of this transcriptional repressor complex are expressed at high level in nonneuronal cells, and reduced expression of these proteins in mature neurons is essential to activate a number of neuron-specific genes containing the REST/NRSF-binding motifs (Ballas and Mandel 2005).
While further work will be required to address the functional significance of the 3′-terminal location of the regulated introns, at least one additional gene (Klc1) encoding a Kif5a-interacting kinesin light chain appears to be regulated through a related mechanism. Klc1 encodes multiple splice forms, some of which are restricted to the nervous system (DeBoer et al. 2008). Our RNA-seq data indicate that the effect of Ptbp1 and, possibly, Ptbp2 on Klc1 mRNA is threefold: (1) reduced overall abundance, (2) retention of the two 3′-terminal introns flanking the neuron-specific cassette exon, and (3) repression of the neuron-specific penultimate cassette exon and the 3′-terminal alternative exon (Supplemental Fig. S11).
Notably, introns neighboring alternative exons regulated by the TDP-43 and NOVA splicing regulators have recently been shown to change their retention status during brain development (Ameur et al. 2011). This effect might be explained by a largely post-transcriptional splicing of introns adjoining repressed exons demonstrated by Tyagi and colleagues (Vargas et al. 2011). Although not shown experimentally, we predict that regulation of intron splicing in these cases may also be used as a mechanism for regulating the abundance of the corresponding mRNAs and that the mechanistic link between intron retention status, utilization of alternative exons, and mRNA steady-state expression levels in mammalian cells might be substantially more prevalent than currently thought.
Of note, the Stx1b splice form retaining intron 9 has previously been hypothesized to generate a truncated Stx1b protein (Pereira et al. 2008). While we do not exclude the possibility that minute amounts of this protein may indeed be produced under some circumstances, the significant nuclear enrichment of the intron 9-containing Stx1b mRNA species in both neuroblastoma cells and embryonic tissues argues that the main function of this intron is to regulate mRNA abundance.
Several lines of evidence argue that the Stx1b, Vamp2, Sv2a, and Kif5a genes are controlled by a mechanism distinct from the NMD. (1) These genes are not up-regulated by CHX (Fig. 1B). (2) The 3′ position of the regulated introns makes NMD an unlikely scenario, since the 3′-terminal exon–exon junction is located upstream of a translation termination codon regardless of intron splicing status. (3) The Stx1b minitransgene lacking a functional translation initiation codon is regulated at the mRNA abundance level (Fig. 3A–D), and this regulation is abolished by mutation of the intronic 5′ss, which is not expected to result in a PTC (Fig. 4). Moreover, translocation of the intron-retained Stx1b transcripts to the cytoplasm by inactivating the intronic splice sites (Fig. 4) leads to a significant increase in the mRNA levels, which rules out models invoking any form of cytoplasmic instability of intron-containing transcripts.
The requirement for functional splice sites (Fig. 4) suggests that the regulation circuitry identified in our study involves interaction of the retained introns with the pre-mRNA splicing machinery. Interestingly, the U1 snRNP and U2AF complexes interacting with the 5′ss and the 3′ss have been shown to facilitate nuclear retention of incompletely spliced transcripts (Rain and Legrain 1997; Takemura et al. 2011). Nuclear retention of incompletely spliced transcripts has additionally been shown to be modulated by _cis_-regulatory elements. Some intron-containing retroviral and cellular transcripts are known to be efficiently translocated to the cytoplasm due to their interaction with the Tap/NXF1 or Crm1 export factors (Cullen 2003; Li et al. 2006). It is possible that similar strategies are used by other incompletely spliced mRNA species that have been shown to accumulate in the cytoplasm (Denis et al. 2005; Mansilla et al. 2005; Marinescu et al. 2007; Buckley et al. 2011). On the other hand, specialized elements have been described that promote RNA retention in the nucleus (Taniguchi et al. 2007). Further work will be required to understand the _cis_-regulation underlying nuclear retention of incompletely spliced presynaptic mRNAs.
Our RNAi experiments (Fig. 5; Supplemental Fig. S8) argue that the incompletely spliced Stx1b mRNA retained in the nucleus is cleared by a branch of the nuclear RNA surveillance machinery comprising the exosome complex and Tpr, a mammalian homolog of the yeast nuclear pore basket proteins Mlp1/2 previously implicated in nuclear surveillance of unspliced mRNA in Saccharomyces cerevisiae (Galy et al. 2004; Houseley and Tollervey 2009; Tomecki and Dziembowski 2010; Coyle et al. 2011). The involvement of Tpr in the surveillance of natural intron-retained mRNA species is an important finding, since the role of this protein in the metabolism of incompletely spliced RNAs in the mammalian system has thus far been documented only for recombinant transcripts containing intronic Tap/NXF1-dependent nuclear export signals (Coyle et al. 2011).
Interestingly, of the four exosome subunits analyzed in our study, knockdown of Dis3/Rrp44, Exosc10/Rrp6, and Exosc9/Rrp45 led to comparable up-regulation of the intron-retained transcripts (except for the lack of significant effect of shExosc9 on Vamp2 expression), whereas down-regulation of Exosc1/Csl4 resulted in noticeably less-pronounced effects (Fig. 5; Supplemental Fig. S8). These data suggest that only a subset of exosomal components may function in the nuclear surveillance of incompletely spliced RNAs, consistent with the recently proposed “exozyme” hypothesis (Kiss and Andrulis 2011).
In conclusion, this study suggests that Ptbp1-controlled intron retention synchronizes the expression of critical presynaptic proteins during neuronal differentiation and counters their aberrant expression in nonneuronal cells. We predict that similar mechanisms might contribute to the regulation of other functionally linked gene networks in higher eukaryotes.
Materials and methods
RNAi
The parental CAD cells (Y Li et al. 2007) and their derivatives were transfected with Ptbp1- and/or Ptbp2-specific or control ON-TARGETplus siRNAs (Dharmacon) using Lipofectamine 2000 (Invitrogen) and analyzed 72 h post-transfection. Expression of shRNAs was induced by treating corresponding CAD-A13 cell pools with 2 μg/mL Dox for 72 h as described (Khandelia et al. 2011). To knock down Ptbp1 and Ptbp2 expression in PMEF cells, they were transfected with the corresponding siRNAs and RNAi MAX (Invitrogen) twice over a 36-h interval so that the cells were exposed to siRNA for 72 h in total. To deliver siRNAs into neurosphere cultures, we used the Amaxa mouse NSC nucleofection protocol with minor modifications. Briefly, 4 × 106 cells were nucleofected with 100 pmol of corresponding siRNA and plated onto 6-cm culture dishes coated with polyornithine and fibronectin. The cells were harvested 72 h following the nucleofection. Alternatively, we used two rounds of nucleofection, each with 50 pmol of siRNA. In this case, the cells were maintained as neurospheres for 36 h following the first round and as adherent culture for 36 h following the second round of nucleofection.
RNA-seq
RNA-seq libraries were prepared in principle as described (Mortazavi et al. 2008). Total RNAs were extracted from siRNA-treated CAD cells with Trizol (Invitogen), and the poly(A)+ mRNA fractions were prepared using a Dynabeads mRNA Direct kit (Invitogen). The purified mRNA samples were partially hydrolyzed for 2.5 min at 94°C in the presence of Mg2+, which was followed by the first strand cDNA synthesis with the SuperScript III and random primers (Invitrogen). The second strand cDNA synthesis and adapter ligation steps were carried out as recommended by Illumina. DNA fragments of 350 bp ± 50 bp were then purified with SizeSelect E-gels (Invitrogen) and amplified by 15 cycles of PCR using Phusion DNA polymerase (New England Biolabs) and Illumina primers. Sequencing was carried out using a Genome Analyzer IIx (Illumina). The RNA-seq data were analyzed using the ExpressionPlot pipeline (Friedman and Maniatis 2011). The RNA-seq data have been deposited in NCBI Gene Expression Omnibus (accession no. GSE37933).
RMCE
CAD-A13 cells expressing Stx1b minitransgenes and shRNAs against RNA surveillance components were generated as described (Khandelia et al. 2011). Briefly, 2 × 105 CAD-A13 cells were transfected with a mixture containing 99% of a recombination donor plasmid and 1% of the Cre expression plasmid (pEM784) using Lipofectamine 2000 (Invitrogen). Transgenic cells populations were obtained by treating the transfected cells with 2.5–5 μg/mL puromycin for 7–10 d and pooling the puromycin-resistant colonies. Single-copy integration of BAC-encoded _FRT_-marked Stx1b transgenes was carried out similarly, except five to six individual puromycin-resistant colonies were picked up in this case and propagated independently. Clones with correctly integrated Stx1b transgenes were used in the subsequent experiments. A complete description of the BAC RMCE protocol will be published elsewhere (K Yap, ZQ Lim, and EV Makeyev, in prep.).
See the Supplemental Material for additional details.
Acknowledgments
We thank Neal Copeland, Nancy Jenkins, and Robin Reed for reagents; Rick Myers's laboratory for the help with RNA-seq; and Tom Maniatis and Snezhka Oliferenko for valuable discussions. We also thank the NTU Protein Production Platform for the help with protein purification. This work was supported by the National Research Foundation grant NRF-RF2008-06 (to E.V.M.).
Footnotes
Supplemental material is available for this article.
References
- Ameur A, Zaghlool A, Halvardson J, Wetterbom A, Gyllensten U, Cavelier L, Feuk L 2011. Total RNA sequencing reveals nascent transcription and widespread co-transcriptional splicing in the human brain. Nat Struct Mol Biol 18: 1435–1440 [DOI] [PubMed] [Google Scholar]
- Averbeck N, Sunder S, Sample N, Wise JA, Leatherwood J 2005. Negative control contributes to an extensive program of meiotic splicing in fission yeast. Mol Cell 18: 491–498 [DOI] [PubMed] [Google Scholar]
- Ballas N, Mandel G 2005. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol 15: 500–506 [DOI] [PubMed] [Google Scholar]
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ 2010. Deciphering the splicing code. Nature 465: 53–59 [DOI] [PubMed] [Google Scholar]
- Bell TJ, Miyashiro KY, Sul JY, Buckley PT, Lee MT, McCullough R, Jochems J, Kim J, Cantor CR, Parsons TD, et al. 2010. Intron retention facilitates splice variant diversity in calcium-activated big potassium channel populations. Proc Natl Acad Sci 107: 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M Jr, Black DL 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev 21: 1636–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley PT, Lee MT, Sul JY, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J 2011. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron 69: 877–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Sharp PA 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795 [DOI] [PubMed] [Google Scholar]
- Coyle JH, Bor YC, Rekosh D, Hammarskjold ML 2011. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. RNA 17: 1344–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona N, Potter K, Wise JA 2010. A meiotic gene regulatory cascade driven by alternative fates for newly synthesized transcripts. Mol Biol Cell 22: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR 2003. Nuclear mRNA export: Insights from virology. Trends Biochem Sci 28: 419–424 [DOI] [PubMed] [Google Scholar]
- Davidson EH. 2006. The regulatory genome.Gene regulatory networks in development and evolution. Academic Press, San Diego. [Google Scholar]
- DeBoer SR, You Y, Szodorai A, Kaminska A, Pigino G, Nwabuisi E, Wang B, Estrada-Hernandez T, Kins S, Brady ST, et al. 2008. Conventional kinesin holoenzymes are composed of heavy and light chain homodimers. Biochemistry 47: 4535–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. 2005. Escaping the nuclear confines: Signal-dependent pre-mRNA splicing in anucleate platelets. Cell 122: 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu DE, Chanfreau G 2011. Proofreading and spellchecking: A two-tier strategy for pre-mRNA splicing quality control. RNA 17: 383–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman BA, Maniatis T 2011. ExpressionPlot: A Web-based framework for analysis of RNA-seq and microarray gene expression data. Genome Biol 12: R69 doi: 10.1186/gb-2011-12-7-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galante PA, Sakabe NJ, Kirschbaum-Slager N, de Souza SJ 2004. Detection and evaluation of intron retention events in the human transcriptome. RNA 10: 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2009. The many pathways of RNA degradation. Cell 136: 763–776 [DOI] [PubMed] [Google Scholar]
- Isken O, Maquat LE 2008. The multiple lives of NMD factors: Balancing roles in gene and genome regulation. Nat Rev Genet 9: 699–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Srivastava D 2010. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell 7: 36–41 [DOI] [PubMed] [Google Scholar]
- Keene JD 2007. RNA regulons: Coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Khandelia P, Yap K, Makeyev EV 2011. Streamlined platform for short hairpin RNA interference and transgenesis in cultured mammalian cells. Proc Natl Acad Sci 108: 12799–12804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss DL, Andrulis ED 2011. The exozyme model: A continuum of functionally distinct complexes. RNA 17: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE 2007. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 623: 190–211 [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M 1989. Some _cis_- and _trans_-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57: 573–583 [DOI] [PubMed] [Google Scholar]
- Lemieux C, Marguerat S, Lafontaine J, Barbezier N, Bahler J, Bachand F 2011. A pre-mRNA degradation pathway that selectively targets intron-containing genes requires the nuclear poly(A)-binding protein. Mol Cell 44: 108–119 [DOI] [PubMed] [Google Scholar]
- Li Y, Bor YC, Misawa Y, Xue Y, Rekosh D, Hammarskjold ML 2006. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 443: 234–237 [DOI] [PubMed] [Google Scholar]
- Li Q, Lee JA, Black DL 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 8: 819–831 [DOI] [PubMed] [Google Scholar]
- Li Y, Hou LX, Aktiv A, Dahlstrom A 2007. Studies of the central nervous system-derived CAD cell line, a suitable model for intraneuronal transport studies? J Neurosci Res 85: 2601–2609 [DOI] [PubMed] [Google Scholar]
- Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, et al. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat Struct Mol Biol 17: 1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T 2007. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 27: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla A, Lopez-Sanchez C, de la Rosa EJ, Garcia-Martinez V, Martinez-Salas E, de Pablo F, Hernandez-Sanchez C 2005. Developmental regulation of a proinsulin messenger RNA generated by intron retention. EMBO Rep 6: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinescu V, Loomis PA, Ehmann S, Beales M, Potashkin JA 2007. Regulation of retention of FosB intron 4 by PTB. PLoS ONE 2: e828 doi: 10.1371/journal.pone.0000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldon A, Malapeira J, Gabrielli N, Gogol M, Gomez-Escoda B, Ivanova T, Seidel C, Ayte J 2008. Promoter-driven splicing regulation in fission yeast. Nature 455: 997–1000 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ 2009. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Munding EM, Igel AH, Shiue L, Dorighi KM, Trevino LR, Ares M Jr 2010. Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev 24: 2693–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson P, Muhlemann O 2010. Cutting the nonsense: The degradation of PTC-containing mRNAs. Biochem Soc Trans 38: 1615–1620 [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR 2010. Expansion of the eukaryotic proteome by alternative splicing. Nature 463: 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ 2008. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40: 1413–1415 [DOI] [PubMed] [Google Scholar]
- Parenteau J, Durand M, Morin G, Gagnon J, Lucier JF, Wellinger RJ, Chabot B, Abou Elela S 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331 [DOI] [PubMed] [Google Scholar]
- Pereira S, Massacrier A, Roll P, Verine A, Etienne-Grimaldi MC, Poitelon Y, Robaglia-Schlupp A, Jamali S, Roeckel-Trevisiol N, Royer B, et al. 2008. Nuclear localization of a novel human syntaxin 1B isoform. Gene 423: 160–171 [DOI] [PubMed] [Google Scholar]
- Rain JC, Legrain P 1997. In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J 16: 1759–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer P, Nehrbass U 2005. Quality control of messenger ribonucleoprotein particles in the nucleus and at the pore. Curr Opin Cell Biol 17: 294–301 [DOI] [PubMed] [Google Scholar]
- Spellman R, Llorian M, Smith CW 2007. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol Cell 27: 420–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC 2004. The synaptic vesicle cycle. Annu Rev Neurosci 27: 509–547 [DOI] [PubMed] [Google Scholar]
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR 2010. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 17: 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M 2011. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells 16: 1035–1049 [DOI] [PubMed] [Google Scholar]
- Taniguchi I, Masuyama K, Ohno M 2007. Role of purine-rich exonic splicing enhancers in nuclear retention of pre-mRNAs. Proc Natl Acad Sci 104: 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki R, Dziembowski A 2010. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA 16: 1692–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM 2009. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet 10: 252–263 [DOI] [PubMed] [Google Scholar]
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S 2011. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 147: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites CL, Craig AM, Garner CC 2005. Mechanisms of vertebrate synaptogenesis. Annu Rev Neurosci 28: 251–274 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik SM, Brose N 2007. Regulation of membrane fusion in synaptic excitation-secretion coupling: Speed and accuracy matter. Neuron 55: 11–24 [DOI] [PubMed] [Google Scholar]
- Wollerton MC, Gooding C, Wagner EJ, Garcia-Blanco MA, Smith CW 2004. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol Cell 13: 91–100 [DOI] [PubMed] [Google Scholar]
- Xu Q, Walker D, Bernardo A, Brodbeck J, Balestra ME, Huang Y 2008. Intron-3 retention/splicing controls neuronal expression of apolipoprotein E in the CNS. J Neurosci 28: 1452–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. 2009. Genome-wide analysis of PTB–RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 36: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL 2012. PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2. Nat Neurosci 15: 381–388, S381 [DOI] [PMC free article] [PubMed] [Google Scholar]