Rethinking dog domestication by integrating genetics, archeology, and biogeography (original) (raw)
Abstract
The dog was the first domesticated animal but it remains uncertain when the domestication process began and whether it occurred just once or multiple times across the Northern Hemisphere. To ascertain the value of modern genetic data to elucidate the origins of dog domestication, we analyzed 49,024 autosomal SNPs in 1,375 dogs (representing 35 breeds) and 19 wolves. After combining our data with previously published data, we contrasted the genetic signatures of 121 breeds with a worldwide archeological assessment of the earliest dog remains. Correlating the earliest archeological dogs with the geographic locations of 14 so-called “ancient” breeds (defined by their genetic differentiation) resulted in a counterintuitive pattern. First, none of the ancient breeds derive from regions where the oldest archeological remains have been found. Second, three of the ancient breeds (Basenjis, Dingoes, and New Guinea Singing Dogs) come from regions outside the natural range of Canis lupus (the dog’s wild ancestor) and where dogs were introduced more than 10,000 y after domestication. These results demonstrate that the unifying characteristic among all genetically distinct so-called ancient breeds is a lack of recent admixture with other breeds likely facilitated by geographic and cultural isolation. Furthermore, these genetically distinct ancient breeds only appear so because of their relative isolation, suggesting that studies of modern breeds have yet to shed light on dog origins. We conclude by assessing the limitations of past studies and how next-generation sequencing of modern and ancient individuals may unravel the history of dog domestication.
Keywords: genomics, phylogeography
Darwin speculated about the origins of several domestic animals and suggested that, given the vast morphological variation across numerous breeds, dogs must have had more than one wild ancestor (1). Recent genetic studies, however, support the notion that dogs are descended exclusively from the gray wolf (Canis lupus) (2).
Beyond questions regarding wild ancestry, geneticists and generations of archeologists have investigated not only how and why dogs were domesticated, but also when, where, and how many times it may have occurred. Unique among all domestic animals, the first unambiguous domestic dogs precede the appearance of settled agriculture in the archeological record by several thousand years. Identifying the earliest dogs is difficult, however, because key morphological characters established by zooarcheologists to differentiate domestic animals from their wild wolf ancestors (e.g., size and position of teeth, dental pathologies, and size and proportion of cranial and postcranial elements) were not yet fixed during the initial phases of the domestication process. Furthermore, the range of natural variation among these characters in ancient wolf populations and the time it took for these traits to appear in dogs are unknown. Free-ranging wolves attracted to the refuse generated by human camps most likely followed a commensal pathway to domestication that was neither deliberate nor directed (3). Because the process was not unidirectional, the telltale traits archeologists use to differentiate wolves and dogs probably took numerous generations to become apparent in the archeological record.
Despite the difficulties associated with the use of archeological evidence to pinpoint the timing of domestication, there is a general consensus that domestic dogs were present in the Levant (including Cyprus), Iraq, Northern China, and the Kamchatka peninsula in Far Eastern Russia by ∼12,000 y ago, and in western Europe a few millennia before that. Recent studies have made claims that domestic (or incipient) dogs were present even earlier during the Late Pleistocene in Belgium (4), the Czech Republic (5), and southwestern Siberia (6). Morphological analyses suggest that although some of the early canid remains possess characteristics broadly similar to those found in modern dogs, it remains possible that the bones represent either wolves going through the initial phases of an incomplete domestication process (6) or a morphologically distinct local, now-extinct population of wolves.
The use of more advanced morphometric analyses is allowing zooarcheologists to have greater confidence in identifying early dogs (7). Given the geographical breadth of these finds, archeologists have (generally) been reluctant to postulate exact locations where dogs may have been domesticated. Instead, they have broadly accepted the plausibility of the existence of numerous, independent centers of dog domestication beginning in the Late Pleistocene (8).
Many genetic studies of modern dogs and wolves have been less circumspect. Armed first with fragments of mitochondrial DNA and molecular clocks, the authors of one study concluded that dogs were domesticated 135,000 y ago (9). A separate study later analyzed a similar mitochondrial fragment sequenced from 654 dogs and, on the basis of regional patterns of modern dog diversity, deduced that dogs were domesticated just once in East Asia (10).
Both of these claims have since been challenged. First, it is highly likely that the use of deep fossil calibrations for molecular clocks has led to a significant overestimation of the timing of dog domestication (11). Second, analyses of African street dogs suggested that a single East Asian origin was too simplistic (12). A study of 48,000 SNPs in 912 dogs and 225 gray wolves concluded that both East Asian and Near Eastern wolf populations contributed DNA to modern dog breeds (13). Other studies that have incorporated nuclear markers also suggest diverse geographic origins of dogs (14), and with the application of a broader, more integrated approach, the genetic and archeological perspectives have become more closely aligned. However, despite the volume of new data, the estimates of when, where, and how many times dogs were domesticated remain disconcertingly imprecise.
One significant insight that genetic studies have yielded, using both microsatellites (15) and SNPs (13), is the identification of several genetically divergent modern dog breeds in well-supported basal positions on phylogenetic trees. This early-branching pattern has been used to designate these breeds as “ancient” (13). To avoid conflating genetic differentiation with presumed ancient heritage (16), we will instead refer to these lineages as “basal.”
The term “breed” is also problematic. The focus on general classes of dogs (e.g., sight hounds, scent hunters, shepherd dogs, and giant dogs) likely has prehistoric roots and led to the development of broadly distinct forms of dogs. For example, three differently sized dog types have been recorded at the 8,000-y-old Svaerdborg site in Denmark (17). Modern breeding practices, focused on distinct breeds with strict aesthetic requirements and closed bloodlines, only emerged in the 19th century, and claims for the antiquity (and long-term continuity) of modern breeds are based upon little or no historical or empirical evidence. In fact, recent historical records clearly demonstrate that most modern breeds experienced significant population fluctuations within the past 100 y (Table S1). Here, we only use the term “breed” when referring to modern dog breeds recognized by kennel clubs.
To test the branching pattern of the previously identified basal breeds and to assess the status of unstudied breeds (Table 1 and Table S1), we used 49,024 SNPs typed in 19 wolves and 1,375 dogs from 35 breeds. In addition, we compiled a broad temporal and geographic survey of dog domestication by undertaking a global examination of the archeological record (Tables S2 and S3). By comparing the zooarcheological evidence with the geographical origins of the total set of modern breeds, we established a framework for understanding why some breeds have retained basal signatures and why most have not.
Table 1.
A list of 16 breeds that were either labeled “ancient” in previous publications or were identified as basal in this study
| Breed | Parker et al. (15) | Vonholdt et al. (13) | Present study |
|---|---|---|---|
| Afghan Hound1 | y | y | |
| Akita2 | y | y | y |
| Alaskan Malamute3 | y | y | |
| American Eskimo (recent) | y* | ||
| Basenji4 | y | y | y |
| Canaan5 | y | ||
| Chow Chow6 | y | y | |
| Dingo7 | y | ||
| Eurasier (recent) | y | ||
| Finnish Spitz8 | y | ||
| New Guinea singing dog9 | y | ||
| Saluki10 | y | y | y |
| Samoyed11 | n | y | |
| Shar-Pei12 | y | y | y |
| Shiba Inu13 | y | ||
| Siberian Husky14 | y | y |
Results and Discussion
Genetic History of Modern Breeds.
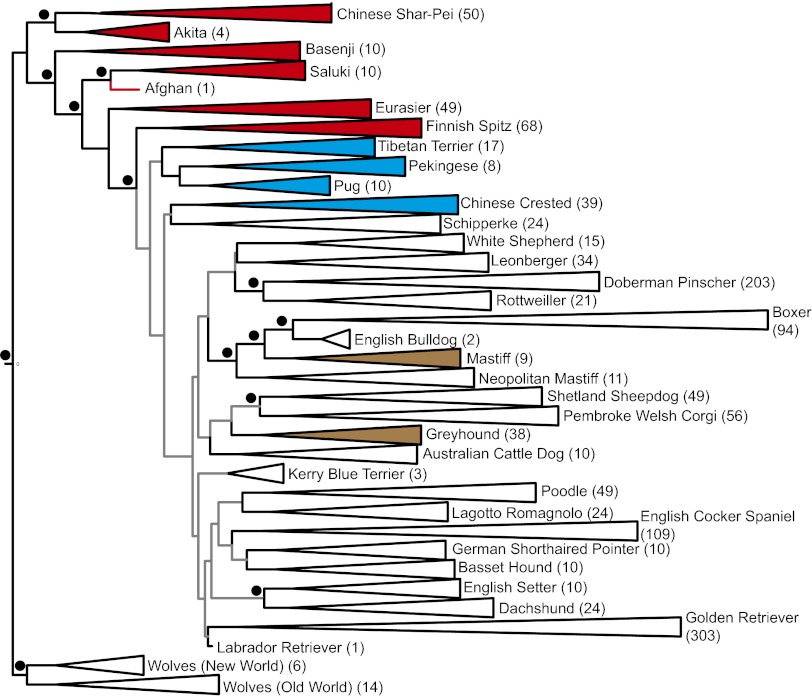
A neighbor-joining phylogenetic tree inferred using our data (Fig. 1) was broadly similar to those described previously (13, 15). A deep genetic split is evident between Old World and New World wolves (Table S4) at the base of the tree. From there, high bootstrap values (>95%) support the basal position and genetic distinctiveness of the so-called ancient (basal) breeds: the Akita, Basenji, Eurasier, Finnish Spitz, Saluki, and Shar-Pei (Fig. 1 and Table 1). Although the relationships between numerous breeds that have been crossed recently (e.g., Dachshunds) are well supported, and although each of nonbasal breeds is strongly monophyletic, the relationships between them are poorly resolved (Fig. 1).
Fig. 1.
A neighbor-joining tree depicting the relationships between 35 breeds (with sample sizes) and rooted with New and Old World Wolves. All clades have been collapsed. Gray branches are poorly supported, whereas black branches and black circles indicate bootstrap values >95%. Clade colors depict breeds that retain a basal signature (red), non-European breeds that are not basal (blue), and European breeds that are rumored to have deep histories but are not basal (brown). The well-supported relationships between Rottweilers and Doberman Pinschers, Neapolitan Mastiffs, Mastiffs, English Bulldogs, Boxers, Shetland Sheepdogs, and Pembroke Welsh Corgis are the result of known or suspected recent admixture between these breeds. The well-support relationship between Dachshunds and English Setters reflects a recent interbreeding between the Dachshund individuals used in this study with English Setters.
When our results are combined with those from the two previous studies (13, 15), the total number of basal breeds increases to 16. Two of these basal breeds have shallow histories. The American Eskimo breed was deliberately created by crossing Keeshonds, Volpinos, and Pomeranians, and after World War II, Japanese Spitzes may also have been incorporated (18). The name “American Eskimo” was derived from the kennel that originally began breeding them, despite the fact that the breed never had an association with Inuits. The highly mixed heritage of the breed is evident from its position on the phylogenetic tree, which is dependent on the choice of analytical technique. The American Eskimo appears alongside the basal Samoyed in trees estimated using 10-SNP windows; however, it is positioned next to Pomeranians on a tree inferred using individual SNPs (13).
The Eurasier is also a recently created breed, developed deliberately and fixed in the 1960s by mixing Chow Chows with Keeshonds and a single Samoyed (18). Because the majority of the breeds used to create Eurasiers possess basal signatures (13, 15), Eurasiers also appear basal, although they are the only breed whose monophyly is weakly supported (33% bootstrap value).
The remaining 14 basal breeds [including Samoyeds, which do not appear basal on the phylogenetic tree inferred from microsatellite data (15), but are basal when using SNPs (13)] have generally avoided admixture with other breeds (Table S1). This avoidance is probably the only reason why they retain a genetic legacy that extends beyond the age of modern breeding and the establishment of kennel clubs during the second half of the 19th century (19).
Despite the long history of human selection for specific dog forms, there is a major disconnect between truly ancient dogs and modern breeds. For example, unsubstantiated claims have been made for the antiquity of the modern Irish Wolfhound. Wolfhound-type dogs were used to hunt wolves across Europe. In Ireland, wolves were exterminated by 1786 (20), after which the demand for Wolfhounds plummeted, and by 1840 the type was either extinct or all but extinct. George Augustus Graham revitalized (or recreated) the form by breeding one possible wolfhound to Scottish Deerhounds, and then incorporated Borzois and Great Danes to create the modern breed that retained the aesthetic of the original form, but not the genetic ancestry (18).
The story of the Irish Wolfhound is not unusual. Although the origin myths of the Cardigan and Pembroke Welsh Corgis state that their respective introductions to England differed by 2,000 y (21), both types were allowed to interbreed for centuries before being split into two modern breeds in the 1920s (18). Whatever their deeper history, these breeds form strongly supported sister clades on phylogenetic trees (13), meaning that their preadmixture heritage is invisible even with the resolving power of tens of thousands of SNPs.
Both World Wars had a major impact on the genetic diversity of the domestic dog. In the United Kingdom, English Mastiffs were reduced to 14 individuals (18), Sussex Spaniels to 10 (22), and Manchester Terriers to 11 (18). Bernese Mountain Dogs (18) and Italian Greyhounds (22) vanished completely and many other breeds suffered significant bottlenecks (Table S1). Bolstering or recreating these breeds was accomplished by crossing numerous other breeds, a practice that obscured whatever genetic signatures of their early heritage that existed before the World Wars, and ultimately led to highly inbred modern populations (23). Interestingly, the recent genetic homogenization has occurred despite the increase in phenotypic disparity as breeders have simultaneously closed breeding lines and selected for extreme morphological traits (24).
Even the basal breeds identified in this and other studies experienced recent and significant demographic change. The Shiba Inu faced extinction in World War II and the modern breed is an amalgamation of three isolated and distinct Japanese lineages (18). The Finnish Spitz, supposedly used for millennia by Finno-Ugric people, was nearly extinct by 1880. A single breeder, Hugo Roos, set out to rescue the type by traveling to remote villages and collecting the few remaining individuals least likely to have been crossed (accidentally or purposely) with other breeds (18). The fact that Finnish Spitzes retain a basal genetic signature is testament to the success of Roos’s efforts to obtain uncrossed individuals.
With the exception of the Alaskan Malamute, all 14 basal breeds have geographic origins in the Old World (Table S1); this is despite the fact that dogs were an integral part of the human occupation of the New World and that several modern breeds, including the Chihuahua, are thought to have been at least partly derived from domestic dogs native to the New World. The general lack of basal lineages in the Americas is likely because of the fact that European breeds, initially introduced only 500 y ago, have overwhelmed the native lineages. This finding was demonstrated by a recent study of mitochondrial variation among street dogs in South America, which concluded native maternal lineages were almost entirely absent in New World dogs (25).
Finally, numerous widely geographically distributed dog populations share identical mutations responsible for specific phenotypes. Chinese and Mexican breeds both possess the same hairless gene (26), sub-Saharan African and Thai breeds possess a ridged line of hair on their backs caused by the same genetic mutation (27), and at least 19 different breeds possess the identical mutation for foreshortened limbs (28). These mutations are unlikely to have arisen multiple times independently, implying a significant degree of gene flow between breeds. This evidence, combined with known demographic fluctuations in numerous breeds, suggests that throughout history global dog populations experienced numerous episodes of diversification and homogenization. Each successive round further reduced the power of genetic data derived from modern breeds to infer the early history of dog domestication.
Dogs in the Archeological Record.
Identifying dog remains in the archeological record is not always straightforward. First, it can be difficult to discriminate between dogs and wolves, because dogs were still morphologically wolf-like at the earliest stages of domestication. In addition, and in contrast to their modern patchy distribution, wolves were once dispersed across the Northern Hemisphere (29) (Fig. 2). As a result, zooarcheologists cannot establish the wild or domestic status of dog remains based solely on geographic location as they can for sheep and goats, the native wild ranges of which were much more restricted.
Fig. 2.
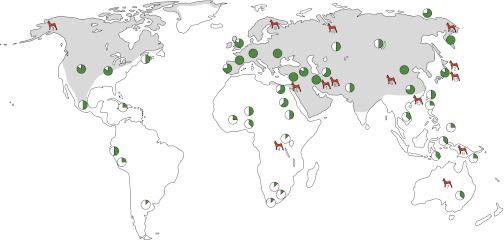
A world map in which the approximate maximal range of gray wolves (Canis lupus) is shaded in gray (based on ref. 29). Green circles represent regions where confidently dated remains of domestic dogs have been described in at least one archeological site (Table S3). Circles are divided into eight segments, each of which represents 1,500 y, visually depicting the age of the oldest remains at sites in the region over which the circle sits. Filled circles represent remains older than 10,500 y. Each red dog represents a basal breed. The number under each dog refers to the breeds in Table 1; their locations are based upon their suspected geographic origins, described in Table S1.
Second, identifying dogs can be confounded by the presence of several other extant and extinct species of similar-sized canids, including foxes (Vulpes spp.) and maned wolves (Chrysocyon brachyurus) in South America, dholes (Cuon spp.) in Europe and Asia, jackals in Africa and Asia (Canis aureus, Canis adustus, and Canis mesomelas), and African wild dogs (Lycaon pictus) (30). Recent efforts have been made to differentiate dogs from these canid species using shape analyses (7), and numerous early claims for domestic dogs have since been rejected because reanalyses have revealed contradictory designations (Table S2). This is often the case when preserved specimens are relatively scarce or fragmented, reducing the presence of specific distinguishing features necessary to discriminate between closely related forms.
Third, a variety of factors can introduce biases against the preservation of certain vertebrate taxa in the archeological record. These include taphonomic processes [particularly in humid tropical settings (31)], and the general paucity of canid remains relative to other prey and domestic animals in the fossil record. In addition, the absence of archeological excavations in many parts of the world biases our interpretation of domestication history. The universal human propensity to bury dogs either on their own or within human burials (32), however, has significantly enhanced the archeological visibility of dogs.
Finally, even when zooarcheologists can confidently attribute remains to Canis familiaris, dating can prove problematic. The earliest dogs in North America were originally reported from the Jaguar Cave site in Idaho with an associated date of 10,400 y cal B.P. (33). Subsequent direct dating of the bones revealed that two Jaguar Cave dogs are ∼3,500 and ∼1,000 y old (34).
An interesting pattern emerges when directly dated and confidently identified dog specimens (Table S3) are mapped onto the historic distribution of wolves across the Old and New Worlds (Fig. 2). First, remains ∼12,000 y or older are present in numerous sites in Europe, the Levant, Iraq, Northern China, and the Kamchatka peninsula in the Russian Far East. Dogs appear in contexts older than 8,000 y everywhere else within the maximal distribution of wolves, suggesting independent domestications of local populations of wolves, migration of humans possessing dogs, or the secondary acquisition of dogs by groups that were not involved in the domestication process.
Dogs appear south of the original wolf distribution in the Old and New Worlds almost always with the arrival of agriculture. For example, despite the fact that human remains are present in much older contexts at Coxcatlan Cave in Mexico, dogs first appear only ∼5,200 B.P. alongside the appearance of agricultural communities (35). The same is true in sub-Saharan Africa, where dogs appear after the advent of the Sudanese Neolithic ∼5600 B.P. (36), in Peninsular Southeast Asia ∼4,200 B.P. (37), and in Island Southeast Asia ∼3,500 B.P. (38). Dogs only arrived in South Africa ∼1,400 y ago following the arrival of cows, sheep, and goats a few hundred years before (39), and in southern South America ∼1,000 y ago with the arrival of sedentary societies (40).
Biogeographical Perspective.
Mapping the geographic location of the 14 basal dog lineages onto the maximal wolf distribution and the archeological data reveals several counterintuitive patterns. First, although domestic dogs were present in numerous European archeological sites ∼15,000 y ago, and despite the fact that textual references or depictions superficially suggest temporally deep origins for 13 European breeds including the Pharaoh and Ibizan Hounds (Table S1), only the Finnish Spitz retains a basal signature. Second, although dogs reached Island Southeast Asia ∼3,500 y ago and southern Africa ∼1,400 y ago, the branches leading to three breeds from these regions (Basenjis, New Guinea Singing Dogs, and Dingoes) are located in basal positions on the tree (Fig. 1). This pattern confounds the expectation that basal breeds should originate from the regions that possess the oldest archeological dog remains, or at least the regions that possess the deepest historical records of types recognizable in modern breeds.
The two breeds closest to central Europe that retain basal signatures (the Finnish Spitz and the Israeli Canaan Dog), are both known to have been isolated from their European counterparts. Efforts to create modern breed standards included a policy of avoiding those individuals that had been bred with foreign, recently introduced breeds (18). Most basal breeds have hybridized with other lineages. If those breeds have either been crossed with other basal breeds (e.g., the Shiba Inu) or if a few of the least introgressed individuals are retained and bred [e.g., the Finnish Spitz or the Dingo; though at least 80% of wild dingoes have interbred with European breeds (41)], then a basal genetic signal is retained.
As discussed above, many basal breeds have also experienced severe bottlenecks that have exaggerated their unique genetic signatures. The extant captive population of the New Guinea Singing Dog is descended from only eight individuals (42), European Afghans went extinct during the World Wars and were re-established using just three imported dogs, and the modern European Basenji stock was initiated with just a handful of individuals collected in 1936 and supplemented with dogs acquired from central Africa in 1988 (21). The combination of introgression and bottlenecks suggests that basal breeds have little or no genetic connections to their ancestral populations, and that genetic distinctiveness alone cannot be used as a proxy to signify an ancient heritage.
The most predictive factor in determining whether a breed retains a basal signature is a lack of gene flow, or at least a lack of introgression with breeds that do not possess basal signatures. Thus, the unifying characteristic among the 14 basal dog lineages (Table 1) is geographic or cultural isolation from the primary center of dog breeding in Europe that began in the 19th century. If geography alone determined basal status, however, then the Africanis, Chihuahua, Chinese Crested, Lhasa Apso, Pekingese, Pug, Rhodesian Ridgeback, Shih Tzu, and Tibetan Terrier should also be basal. In these cases, however, a significant degree of introgression with European breeds is recorded or strongly suspected (Table S1). Although there is pictorial, written (43), and zooarcheological (44) evidence for toy dogs spanning at least the last 2,000 y, no toy breeds possess a basal signature, probably a result of the ease with which they can be transported and interbred with local dogs.
Populations of numerous taxa that live at isolated peripheries, including the Falkland Islands Wolf (45), Homo floresiensis (46), and woolly mammoths (47), often either outlived or appear different from their continental relatives. Island populations of dogs (both real and metaphorical) are more likely to retain their genetic integrity not because related populations on the mainland have gone extinct, however, but because peripheral populations have avoided amalgamation into a larger group that, as a consequence, has lost its genetic distinctiveness.
Conclusion
Though clear signs of the dog domestication process are visible 15,000 y ago, dogs were not present across every habitable continent until they reached South Africa and southern South America <1,400 y ago. The number of differentiated, isolated dog populations has since been reduced through human movement and trade that subsequently led to increased gene flow and population homogenization, and through warfare, which often resulted in extreme demographic fluctuations (including extinction). Each time a lineage that had been evolving in isolation came into contact with introduced dogs, the resulting descendant admixture blurred the genetic signature, making it more difficult to deduce their origins before the assimilation.
This pattern is not unique to dogs. When human populations transported domesticates into new regions, the most common result has been an admixed population of introduced and local varieties, many of which arrived during previous expansion episodes. Examples of this phenomenon include European domestic grapes (48), Central American maize (49), and Western Eurasian sheep (50).
Basal dog lineages fall outside the large, poorly supported clade that includes most modern dog breeds (Fig. 1). This result is not because they more closely approximate the earliest domestic dogs, but because they have mostly avoided recent admixture with other breeds that themselves possess a merged genetic heritage from dogs that evolved in a wide variety of geographic regions. It is far easier to avoid introgression by existing at the periphery, beyond landscape and cultural barriers. This theory explains why numerous basal lineages are from those regions where dogs only recently arrived, outside the natural range of wolves, and why no central European breeds retain an ancient signature despite the ∼15,000-y history of domestic dogs. The vast majority of modern breeds were only created in the past 150 y, emerging from what was a relatively homogeneous gene pool formed as a result of millennia of human migration and the subsequent merging of multiple, previously independently evolving dog lineages. This history, along with the closed gene pools and small effective population sizes associated with recent breed formation, also explains the strongly supported genetic monophyly of individual breeds and the lack of resolved relationships between them.
The shallow history of breed formation has eased the process of correlating known breed-specific phenotypes with, in some cases, their causal mutations (51). Unfortunately, our understanding of dog origins has been hampered by our reliance on limited marker sets that type a small portion of the 2.4 billion DNA bases that make up the dog genome (2). Even in datasets that type numerous individuals, methods that use mitochondrial sequences or even tens of thousands of SNPs are only capable of recovering signatures that have resulted from the effects of bottlenecks and reticulate evolution that took place during 19th and 20th century breed formation. As a result, our ability to investigate the deeper history of dog domestication has been severely hampered.
The advent of rapid and inexpensive DNA sequencing technology has made it possible to significantly increase the volume and commensurate resolving power of genetic data, thus allowing a greater time depth to be accessed. In humans, dense genotyping (millions of SNPs) and complete genomes of both ancient and modern individuals have revealed a far more complex history (including inter- and intraspecies admixture) than was previously available using sparser datasets (52). Comparable genetic analyses of modern and ancient domestic dog genomes and the resolving power they possess will soon yield equally complex insights into their domestication and subsequent evolution, thus revealing our deep, shared history with dogs.
Materials and Methods
Genetics.
DNA was isolated from 1,375 domestic dogs (Table S1) and 19 wolves (Table S4) and genotyped for 49,664 SNPs on the Affymetrix canine v2 arrays using the snp5-geno-qc software package, with subsequent QC done using PLINK (53). SNPs on chromosome X and SNPs with genotyping rates <95%, were removed, yielding a dataset of 49,024 SNPs. Duplicate samples were identified and merged based on genome-wide average identity-by-state pairwise identity higher than 98%. Breed assignment was confirmed using principal component analysis with smartpca (part of the EIGENSOFT software package) (54). All dogs included in the analysis had genotyping rates > 75% (median of 98% in dogs and 96% in wolves).
To construct phylogenetic trees, pairwise identity-by-state genetic distances between samples were first estimated across all SNPs that passed quality filters using PLINK (53). The distances were then used to construct a neighbor-joining tree using Phylip (55). The dataset was bootstrapped 1,000 times to obtain support values for each node.
Archeology.
The survey of the archeological literature revealed numerous reports of remains, the details of which (species designation, status determination, and dating) the authors were confident. Many other claims were contentious. We created two tables. The first (Table S2) lists reports of domestic dogs and the rationales for not including them in Table S3, which lists all of the locations, sites, and elements used in Fig. 2. We applied a conservative approach when deciding whether or not to accept individual claims for remains that were identified as domestic dogs. The specific criteria and rationales are discussed in the SI Results and Discussion.
Supplementary Material
Supporting Information
Acknowledgments
We thank April McMahon, Alan de Quieroz, Matthew Breen, Gary Johnson, and Hannes Lohi for comments on the manuscript. G.L. is currently a Research Councils United Kingdom Academic Fellow and was supported by a European Molecular Biology Organization postdoctoral fellowship; K.L.-T. is a European Young Investigator award recipient funded by the European Science Foundation, and was supported by grants from the Swedish Research Council; and A.P. was supported by the British Association for Japanese Studies.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.B. is a guest editor invited by the Editorial Board.
References
- 1.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 2.Lindblad-Toh K, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 3.Zeder MA. Pathways to animal domestication. In: Gepts P, et al., editors. Biodiversity in Agriculture: Domestication, Evolution and Sustainability. Cambridge: Cambridge Univ Press; 2012. [Google Scholar]
- 4.Germonpré M, et al. Fossil dogs and wolves from Palaeolithic sites in Belgium, the Ukraine and Russia: Osteometry, ancient DNA and stable isotopes. J Archaeol Sci. 2009;36:473–490. [Google Scholar]
- 5.Germonpré M, Lázničková-Galetová M, Sablin MV. Palaeolithic dog skulls at the Gravettian Předmostí site, the Czech Republic. J Archaeol Sci. 2011;39:184–202. [Google Scholar]
- 6.Ovodov ND, et al. A 33,000-year-old incipient dog from the Altai Mountains of Siberia: Evidence of the earliest domestication disrupted by the Last Glacial Maximum. PLoS ONE. 2011;6:e22821. doi: 10.1371/journal.pone.0022821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pionnier-Capitan M, et al. New evidence for Upper Palaeolithic small domestic dogs in South-Western Europe. J Archaeol Sci. 2011;38:2123–2140. [Google Scholar]
- 8.Crockford SJ. A commentary on dog evolution: Regional variation, breed development and hybridisation with wolves. In: Crockford SJ, editor. Dogs Through Time: An Archaeological Perspective. Oxford, UK: Archaeopress; 2000. pp. 295–312. [Google Scholar]
- 9.Vilà C, et al. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- 10.Savolainen P, Zhang YP, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- 11.Ho SYW, Larson G. Molecular clocks: When times are a-changin’. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Boyko AR, et al. Complex population structure in African village dogs and its implications for inferring dog domestication history. Proc Natl Acad Sci USA. 2009;106:13903–13908. doi: 10.1073/pnas.0902129106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vonholdt BM, et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray MM, Sutter NB, Ostrander EA, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker HG, et al. Genetic structure of the purebred domestic dog. Science. 2004;304:1160–1164. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 16.Miklósi Á. Dog Behaviour, Evolution, and Cognition. New York: Oxford Univ Press; 2007. [Google Scholar]
- 17.Henriksen BB, Sørensen KA, Sørensen I. Sværdborg I: Excavations 1943–44: A Settlement of the Maglemose Culture. Köbenhavn: Akademisk; 1976. [Google Scholar]
- 18.Morris D. Dogs: The Ultimate Dictionary of Over 1,000 Dog Breeds. North Pomfret, VT: Trafalgar Square; 2002. [Google Scholar]
- 19.Arman K. A new direction for kennel club regulations and breed standards. Can Vet J. 2007;48:953–965. [PMC free article] [PubMed] [Google Scholar]
- 20.Hickey KR. A geographical perspective on the decline and extermination of the Irish wolf Canis lupus: An initial assessment. Ir Geogr. 2000;33:185–198. [Google Scholar]
- 21.American Kennel Club. The Complete Dog Book. 20th Ed. New York: Ballantine; 2006. [Google Scholar]
- 22.Wilcox B, Walkowicz C. Atlas of Dog Breeds of the World. 4th Ed. Neptune, NJ: TFH Publications; 1993. [Google Scholar]
- 23.Calboli FCF, Sampson J, Fretwell N, Balding DJ. Population structure and inbreeding from pedigree analysis of purebred dogs. Genetics. 2008;179:593–601. doi: 10.1534/genetics.107.084954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drake AG, Klingenberg CP. Large-scale diversification of skull shape in domestic dogs: Disparity and modularity. Am Nat. 2010;175:289–301. doi: 10.1086/650372. [DOI] [PubMed] [Google Scholar]
- 25.Castroviejo-Fisher S, Skoglund P, Valadez R, Vilà C, Leonard JA. Vanishing native American dog lineages. BMC Evol Biol. 2011;11:73. doi: 10.1186/1471-2148-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drögemüller C, et al. A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science. 2008;321:1462. doi: 10.1126/science.1162525. [DOI] [PubMed] [Google Scholar]
- 27.Salmon Hillbertz NH, et al. Duplication of FGF3, FGF4, FGF19 and ORAOV1 causes hair ridge and predisposition to dermoid sinus in Ridgeback dogs. Nat Genet. 2007;39:1318–1320. doi: 10.1038/ng.2007.4. [DOI] [PubMed] [Google Scholar]
- 28.Parker HG, et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nowak RM. Wolf evolution and taxonomy. In: Mech L, Boitani L, editors. Wolves: Behavior, Ecology, and Conservation. Chicago: Univ of Chicago Press; 2003. pp. 239–258. [Google Scholar]
- 30.Macdonald DW, Sillero-Zubiri C, editors. The Biology and Conservation of Wild Canids. Oxford: Oxford Univ Press; 2004. [Google Scholar]
- 31.Van Neer W. Domestic animals from archaeological sites in Central and West-Central Africa. In: Blench R, MacDonald K, editors. The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics and Ethnography. New York: Routledge; 2000. pp. 163–190. [Google Scholar]
- 32.Morey DF. Burying key evidence: The social bond between dogs and people. J Archaeol Sci. 2006;33(2):158–175. [Google Scholar]
- 33.Lawrence B. Antiquity of large dogs in North America. Tebiwa. The Journal the Idaho State University Museum. 1968;11(2):43–48. [Google Scholar]
- 34.Gowlett JAJ, Hedges R, Law I, Perry C. Radiocarbon dates from the Oxford AMS system: Archaeometry datelist 5. Archaeometry. 1987;29(1):125–155. [Google Scholar]
- 35.Flannery KV. Vertebrate fauna and hunting patterns. In: Byers DS, editor. The Prehistory of the Tehuacan Valley. Austin: Univ of Texas Press; 1967. pp. 132–177. [Google Scholar]
- 36.Gautier A. The evidence for the earliest livestock in North Africa: Or adventures with large bovids, ovicaptrids, dogs and pigs. In: Hassan FA, editor. Droughts, Food and Culture: Ecological Change and Food Security in Africa’s Later Prehistory. New York: Kluwer Academic/Plenum; 2002. pp. 195–207. [Google Scholar]
- 37.Higham CFW. Archaeology, linguistics and the expansion of the East and Southeast Asian Neolithic. In: Blench R, Spriggs M, editors. Expansion of East and Southeast Asian Neolithic in Archaeology and Language II: Archaeological Data and Linguistic Hypotheses. New York: Routledge; 1998. pp. 103–114. [Google Scholar]
- 38.Bellwood P. The archaeology of Papuan and Austronesian prehistory in the Northern Moluccas, Eastern Indonesia. In: Blench R, Spriggs M, editors. Expansion of East and Southeast Asian Neolithic in Archaeology and Language II: Archaeological Data and Linguistic Hypotheses. New York: Routledge; 1998. pp. 128–140. [Google Scholar]
- 39.Plug I, Voigt EA. Archaeozoological studies of Iron Age communities in southern Africa. Advances in World Archaeology. 1985;4:189–238. [Google Scholar]
- 40.Prates L, Prevosti FJ, Berón M. First records of prehispanic dogs in southern South America (Pampa-Patagonia, Argentina) Curr Anthropol. 2010;51:273–280. [Google Scholar]
- 41.Corbett LK. The Dingo in Australia and Asia. Ithaca, NY: Comstock/Cornell; 1995. [Google Scholar]
- 42.Koler-Matznick J, Yates BC, Bulmer S, Brisbin IL. The New Guinea singing dog: Its status and scientific importance. Aust Mammal. 2007;29:47. [Google Scholar]
- 43.Toynbee JMC, Oost SI. Animals in Roman life and art. History: Reviews of New Books. 1973;2(2):34–34. [Google Scholar]
- 44.Peters J. Roman Animal Keeping and Breeding: A Synthesis Based on Zooarchaeological Analysis and the Written and Pictorial Record (in German) Rahden, Germany: Leidorf; 1998. [Google Scholar]
- 45.Slater GJ, et al. Evolutionary history of the Falklands wolf. Curr Biol. 2009;19:R937–R938. doi: 10.1016/j.cub.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 46.Brown P, et al. A new small-bodied hominin from the Late Pleistocene of Flores, Indonesia. Nature. 2004;431:1055–1061. doi: 10.1038/nature02999. [DOI] [PubMed] [Google Scholar]
- 47.Vartanyan SL, Arslanov KA, Karhu JA, Possnert G, Sulerzhitsky LD. Collection of radiocarbon dates on the mammoths (Mammuthus primigenius) and other genera of Wrangel Island, northeast Siberia, Russia. Quat Res. 2008;70(1):51–59. [Google Scholar]
- 48.Myles S, et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA. 2011;108:3530–3535. doi: 10.1073/pnas.1009363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108:1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chessa B, et al. Revealing the history of sheep domestication using retrovirus integrations. Science. 2009;324:532–536. doi: 10.1126/science.1170587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlsson EK, et al. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 52.Stoneking M, Krause J. Learning about human population history from ancient and modern genomes. Nat Rev Genet. 2011;12:603–614. doi: 10.1038/nrg3029. [DOI] [PubMed] [Google Scholar]
- 53.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5 c. University of Washington, Seattle: Joseph Felsenstein; 1993. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information