ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize (original) (raw)
Abstract
Teosinte, the progenitor of maize, is restricted to tropical environments in Mexico and Central America. The pre-Columbian spread of maize from its center of origin in tropical Southern Mexico to the higher latitudes of the Americas required postdomestication selection for adaptation to longer day lengths. Flowering time of teosinte and tropical maize is delayed under long day lengths, whereas temperate maize evolved a reduced sensitivity to photoperiod. We measured flowering time of the maize nested association and diverse association mapping panels in the field under both short and long day lengths, and of a maize-teosinte mapping population under long day lengths. Flowering time in maize is a complex trait affected by many genes and the environment. Photoperiod response is one component of flowering time involving a subset of flowering time genes whose effects are strongly influenced by day length. Genome-wide association and targeted high-resolution linkage mapping identified ZmCCT, a homologue of the rice photoperiod response regulator Ghd7, as the most important gene affecting photoperiod response in maize. Under long day lengths ZmCCT alleles from diverse teosintes are consistently expressed at higher levels and confer later flowering than temperate maize alleles. Many maize inbred lines, including some adapted to tropical regions, carry ZmCCT alleles with no sensitivity to day length. Indigenous farmers of the Americas were remarkably successful at selecting on genetic variation at key genes affecting the photoperiod response to create maize varieties adapted to vastly diverse environments despite the hindrance of the geographic axis of the Americas and the complex genetic control of flowering time.
Keywords: genetic diversity, quantitative trait locus
The rapid spread of agriculture from the Fertile Crescent was enabled in part by the East–West axis of Eurasia, permitting crop cultivation to spread across large geographic regions at a common latitude (1). The relatively simple genetic control of flowering time of the key crops domesticated in the Fertile Crescent, wheat and barley (2), coupled with a predominantly self-fertilizing mating system, also facilitated colonization of new environments by rare mutants with large effects on flowering time responses to day length and temperature.
In contrast, the spread of maize from its origin in Southern Mexico 6 to 10,000 y ago was relatively slow (1), hindered by the North–South axis of the Americas. Maize (Zea mays L. subsp. mays) was domesticated from the Mexican native teosinte Zea mays L. subsp. parviglumis (3), a species adapted to day lengths less than 13 h. Under the longer day lengths of higher latitudes, teosinte flowers very late or not at all (4). From its Meso-American origin, maize was spread by early humans to geographically and ecologically diverse environments from Canada to Chile well before the arrival of Columbus to the Americas (5, 6), requiring its adaptation to long day lengths. Thus, although the spread of maize occurred later than that of wheat, it was a remarkable achievement, given its ancestral adaptation to short day lengths, outcrossing mating habit, and complex genetic regulation of flowering time (7).
The complexity of maize flowering time has hindered the identification of genes regulating its initiation of flowering. The few maize flowering time genes identified to date (8–11) appear to control only a small amount of the standing genetic variation for the photoperiod response (12, 13). Here we analyzed the flowering time photoperiod response of the maize nested association mapping (NAM) population, a set of 5,000 mapping lines derived from 25 related crosses that enables powerful, comprehensive linkage analysis of complex traits (7, 14). We used a genome-wide association study (GWAS) based on a dense maize haplotype map (15), taking advantage of the control over population structure and genetic background variation permitted by NAM (16, 17). Targeted high-resolution genetic mapping was used to resolve a key quantitative trait locus (QTL) on chromosome 10 to a single gene, ZmCCT. The expression and phenotypic effect of ZmCCT alleles from teosinte uniformly conferred late flowering under long day lengths. Thus, the spread of maize to higher latitudes appeared to require the selection by prehistoric humans of rare mutations at ZmCCT and a few other critical genes that reduced sensitivity to long day lengths.
Results
Genetic Architecture of Flowering Time Photoperiod Response.
We measured the photoperiod response of the maize NAM recombinant inbred lines (RILs) by evaluating their flowering time in eight long-day length and three short-day length environments. Thermal time [i.e., growing degree days (GDDs)] to flowering was measured within each environment to minimize the effect of temperature differences among environments on the observed flowering times (Dataset S1). Photoperiod response for each RIL was measured as the difference in mean thermal time to flowering between long- and short-day length environments (Dataset S2). Substantial variation for the response to day length was observed among the inbred founders of the NAM population (SI Appendix, Fig. S1). Long day lengths delayed flowering time of inbreds from tropical regions eight times more than inbreds from temperate zones. Photoperiod responses for thermal time to male and female flowering were highly correlated (r = 0.92; P < 0.0001) and had similar QTL positions (Fig. 1 and SI Appendix, Table S1 and Fig. S2); we focus on time to female flowering (i.e., silking) because of its higher heritability (76%). Family main effects and 14 QTLs detected with joint linkage analysis explained 98% of the genetic variation in NAM (Figs. 1 and 2 and SI Appendix, Table S2 and Dataset S3). Prediction of founder photoperiod responses with the additive joint linkage model was as accurate as possible given the heritability of the trait (_r_2 = 75%; SI Appendix, Fig. S3).
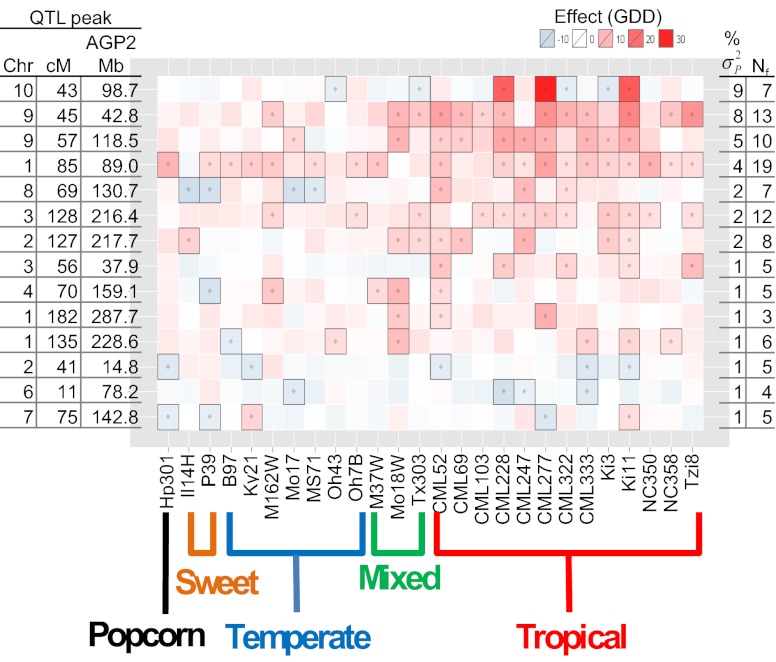
Fig. 1.
Heat map of NAM QTL effects for photoperiod effect on thermal time to silking. Rows of the heat map correspond to the 14 QTLs for silking photoperiod response, ordered by the proportion of variation they explain (in percentage of 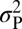 ). Columns correspond to the 25 diverse founders of NAM and the Mo17 founder for IBM population, ordered according to subpopulation assignment. QTL alleles that are significantly (<5% false discovery rate) different from the B73 reference allele are represented with boxed outlines and dots. The number of founders (Nf) segregating for significant allele effects for each QTL are shown on the right.
). Columns correspond to the 25 diverse founders of NAM and the Mo17 founder for IBM population, ordered according to subpopulation assignment. QTL alleles that are significantly (<5% false discovery rate) different from the B73 reference allele are represented with boxed outlines and dots. The number of founders (Nf) segregating for significant allele effects for each QTL are shown on the right.
Fig. 2.
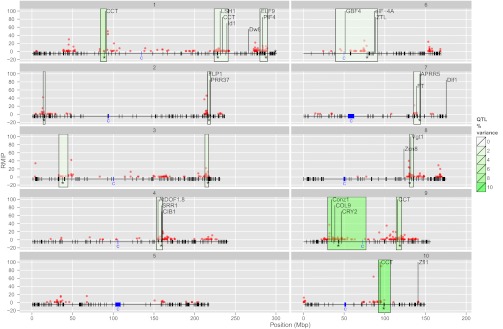
Joint linkage QTL and GWAS of photoperiod sensitivity for thermal time (in GDD) to silking in maize NAM and IBM populations. Each box represents one of the 10 chromosome pairs of maize. Vertical black hashes at bottom of each box represent the 1,106 SNP markers mapped directly on the NAM RILs positioned according to their physical location on the maize AGP v2 reference sequence. Blue boxes and the letter “c” on the physical maps represent centromeres. Green boxes delineate QTL support intervals; the intensity of green color represents the proportion of phenotypic variation associated with each QTL, and asterisks identify the positions of the markers at QTL peaks. Red dots represent HapMap v2 SNPs or CNVs associated with photoperiod response in at least one GWAS subsample analysis. Vertical position of the HapMap variants represents the frequency with which they were detected in GWAS subsample analyses (i.e., RMIP). Selected candidate gene positions are indicated with vertical black lines and gene names.
The 14 photoperiod response QTL largely represent a subset of the 39 flowering time per se QTLs previously identified in NAM under the same long-day length environments (7): 13 of 14 photoperiod response QTL intervals are within 5 cM of a marker at the peak of a long-day length flowering time per se QTL (SI Appendix, Table S1 and Dataset S3). To directly compare the number of QTL for photoperiod response vs. long-day length flowering time without the confounding effects of methodological differences, we reanalyzed the long-day length flowering time data by using the same scale (i.e., GDDs) and statistical threshold, resulting in detection of 29 QTL for flowering time per se, still more than twice the number of photoperiod response QTL (SI Appendix, Table S1). The magnitude of QTL effects for photoperiod response and long-day-length flowering time measured in GDDs was similar (Dataset S4). For example, the strongest allele effect on both traits was conferred by the CML277 allele at chromosome 10 QTL; the effect was similar in magnitude for both traits [27 GDDs or 1.7 d on long-day length flowering time (7) and 31 GDDs on photoperiod response]. The total phenotypic variation was smaller for photoperiod response than for long-day length flowering time; however, therefore, the phenotypic variance associated with individual QTL was greater for photoperiod response (mean 2.7% and maximum 9.0% of variation for photoperiod QTL vs. mean 1.1% and maximum 4.1% for long-day length flowering time QTL; SI Appendix, Table S2).
Whereas flowering time was highly consistent across long-day length environments (7), photoperiod differences substantially increased the ratio of genotype-by-environment interaction to genotypic variation when short day lengths were included in the environment sample (0.18 vs. 0.10; SI Appendix, Table S3). When this ratio was partitioned between families derived from tropical or temperate parents crossed to B73, the results were even more striking (0.35 for tropical families, 0.04 for temperate families; SI Appendix, Table S3). In summary, photoperiod response in maize is under modestly complex genetic control, regulated by at least 14 QTLs, but represents one component of the more complex flowering time per se, with fewer genes controlling the component trait. Dissection of a complex trait into its components aids high-resolution genetic analysis.
Four of five QTL with largest effects (collectively explaining more than 20% of the phenotypic variation) coincided with the most important QTL detected in an independent study (12). The QTL explaining the most variation and expressing the strongest allele effects mapped to chromosome 10 (Figs. 1 and 2); the importance of this region has been observed consistently across maize photoperiod mapping studies (12, 18, 19). Alleles at this locus have highly heterogeneous effects, even within the tropical subgroup of founder lines (Fig. 1 and Dataset S5). Only three tropical lines carry alleles that significantly increase the photoperiod response, and the alleles from tropical founders Ki3 and CML333 reduce the photoperiod effect relative to the temperate reference founder B73 allele (Fig. 1) (20). The allele from temperate founder Ky21 increases photoperiod response for time to anthesis, although not for silking (Fig. 1 and SI Appendix, Fig. S2). Although the allele conferring a strong photoperiod response at this QTL segregates in only three NAM families, its effect in those families is so great that it explains more phenotypic variation than any other QTL (Fig. 1). The distribution of allelic effects in the NAM founders varies widely among the photoperiod QTLs. For example, the rarity of the photoperiod sensitive allele at chromosome 10 contrasts with the more common occurrence of photoperiod-sensitive alleles in tropical lines at key QTLs on chromosomes 1, 8, and 9 (Fig. 1).
The sensitivity of allelic effects at photoperiod-response QTLs can be seen by comparing their effects on flowering time across individual environments (Fig. 3). For example, the tropical founder alleles with strongest photoperiod response at the chromosome 10 QTL had reduced effects under short day lengths and consistently strong positive effects under long day lengths (Fig. 3). Almost all tropical alleles at the chromosome 9 (45 cM) QTL had consistently negative (i.e., shorter flowering time) effects under short day lengths and positive effects under most long day lengths. The two North Carolina environments presented an exception to the pattern, however, with many tropical alleles having negative effects, perhaps because of the slightly shorter day length (14 h) in the early growing season compared with the other long-day length environments (> 15 h day lengths). At the chromosome 8 QTL, tropical alleles had larger positive effects under long than short day lengths, but many temperate alleles had a markedly opposite response, with increasingly negative effects under long day lengths, reducing the photoperiod responses of those lines.
Fig. 3.
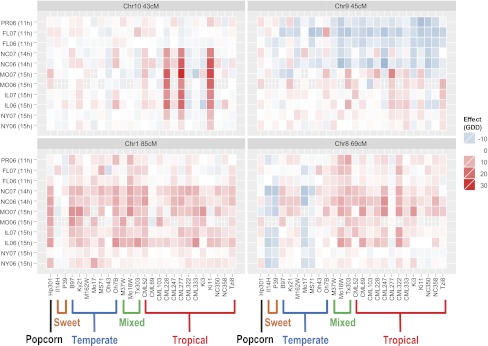
Heat map of allelic effects at four key QTLs for thermal time to silking within each of eight evaluation environments. Within each QTL heat map, rows are environments (ordered from top to bottom as shorter to longer day lengths), and columns are NAM founders.
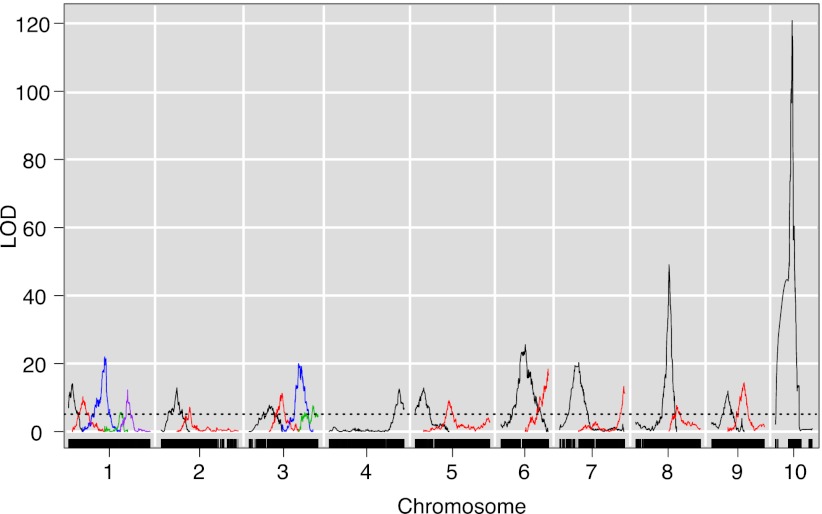
To gain insights into the evolutionary changes that occurred during and following domestication of maize from teosinte, we performed a parallel linkage analysis of flowering time in a population of 866 RILs derived from backcrosses of teosinte to a temperate maize line. We identified 23 QTLs controlling 74% of the phenotypic variation for flowering time under long day lengths (Fig. 4). NAM QTL peaks for days to anthesis under long day lengths were within 5 cM of the support intervals of approximately half (n = 13) of these maize-teosinte QTL (Dataset S6) (7). The most important flowering time QTL in the maize-teosinte population was syntenous to the major chromosome 10 photoperiod QTL identified in NAM (Fig. 4). Briggs et al. (21) demonstrated previously that the teosinte allele at this QTL is highly responsive to photoperiod; its effect is observed only under long day lengths, similar to the CML228/CML277/Ki11 allele at this region in NAM.
Fig. 4.
Whole-genome scan of flowering time under long day lengths in a maize-teosinte mapping population. Black hash marks on bottom axis represent genetic markers, curves represent logarithm of odds (LOD) scores for QTL at each genomic position. LOD curves for distinct QTLs on a single chromosome are plotted with different colors.
Genome-Wide Association Analysis in NAM.
A GWAS of photoperiod response was conducted in the NAM population using 26.5 M SNPs and 1 M copy number variants (CNVs) identified in the maize HapMap version 2 (22). Model resampling techniques were used to identify variants associated with silk photoperiod response and characterize their frequency of association with the phenotype across data samples [i.e., resample model inclusion probability (RMIP)] (23). We identified 118 SNPs and five CNVs associated with silking photoperiod response (at RMIP ≥5%), of which 37% were located within a QTL support interval (Fig. 2 and Dataset S7). This represents an almost fourfold enrichment compared with the 10% of the complete set of 27.5 M HapMap variants within QTL support intervals.
We identified a priori 198 predicted genes in the B73 reference genome with homology to genes shown to be involved in regulation of flowering time in Arabidopsis. We also identified seven loci previously shown to affect flowering time in maize (Dlf1, Dw8, eIF-4A, Id1, Vgt1, Zcn8, and Zfl1) (8, 10, 11, 24–27), as well as 10 homologues of the key rice photoperiod regulatory gene, Ghd7/CCT (28, 29), and three homologues of the most important photoperiod regulatory gene in Sorghum, Ma1/PRR37 (30) (Dataset S8). We tested the hypothesis that candidate genes were related to GWAS associations by comparing the frequency of significant SNP-trait associations within and outside of 10-kb or 100-kb windows around candidate genes. GWAS associations did not occur within 10 kb or 100 kb of candidate gene positions more frequently than expected by chance. Only 25 of the 218 candidate genes mapped within QTL intervals, including maize genes eIF-4A, Id1 and Vgt1, but not Dw8 or Zcn8 (Fig. 2). Although there was no general pattern of agreement between candidate gene positions and GWAS associations, a few interesting candidate gene associations with QTLs were observed. A maize gene with 65% predicted protein homology to the key sorghum photoperiod gene Ma1/PRR37 was located in the photoperiod QTL on the long arm of chromosome 2 (associated with 1% of the variation; Fig. 2). Remarkably, four of the 10 genes with homology to Ghd7/CCT were located inside QTL intervals, including the two genes with highest homology in three of the most important QTLs on chromosomes 1, 9, and 10 (Fig. 2).
Chromosome 10 QTL Resolved to ZmCCT.
Given the importance of the QTL on chromosome 10, we targeted this region for high-resolution linkage mapping in maize and maize-teosinte mapping families. Three maize high-resolution mapping families were created to fine-map this QTL segregating for the reference B73 allele and NAM founder alleles with strongest effects (CML228, CML277, and Ki11; SI Appendix, Fig. S4). Within a mapping family, only the QTL region segregated against a common fixed genetic background (SI Appendix, Fig. S4). Progeny lines homozygous for recombinant chromosome segments within this region were selected using sequence-based markers defining the QTL region, and contrasting pairs of nearly isogenic recombinant and nonrecombinant subfamilies were evaluated for flowering time under long-day length conditions conducive to expression of late flowering by photoperiod-sensitive alleles (SI Appendix, Fig. S5). Substitution mapping based on the breakpoints of the observed recombinant progeny refined the QTL position consistently to a region around 94 Mbp on the chromosome 10 physical map (AGP v2; SI Appendix, Figs. S6–S8). The highest resolution was obtained in the Ki11 population, where the QTL was narrowed to a 443-kbp region containing six predicted genes in the reference sequence, including ZmCCT, a homologue of rice Ghd7 (SI Appendix, Fig. S6) (19).
Fine-mapping of the maize-teosinte chromosome 10 QTL was conducted by using a BC2S3 family segregating for a 51-Mbp region around the QTL. Recombinant chromosomes for the QTL target region were isolated from selfed plants of this family to create a set of homozygous nearly isogenic lines. These lines segregate into discrete early and late classes that flower approximately 9 d apart under long day lengths (Fig. 5). These data allowed us to refine the QTL to a 202-kbp interval. This region includes part of the first exon and first intron of only one gene, ZmCCT, and 200.2 kbp between ZmCCT and the next upstream gene (Fig. 5).
Fig. 5.
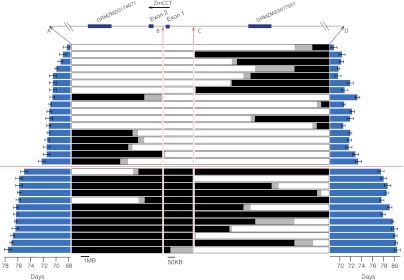
Fine-mapping chromosome 10 QTLs in a maize-teosinte population. Graphical genotypes of homozygous recombinant progeny lines at markers defining intervals within the target QTL region are shown in the center, where white segments indicate markers homozygous for the maize W22 allele, black segments indicate markers homozygous for the teosinte allele, and gray segments indicate unknown genotype. Blue bars represent days to silk (Right) and days to pollen shed (Left) under long day lengths for each recombinant progeny line. Markers “B” and “C” delineate the QTL position and correspond to a reference sequence containing the upstream region and first exon of only one gene, ZmCCT.
Sequence and Association Analysis of ZmCCT in Maize Diversity Panel.
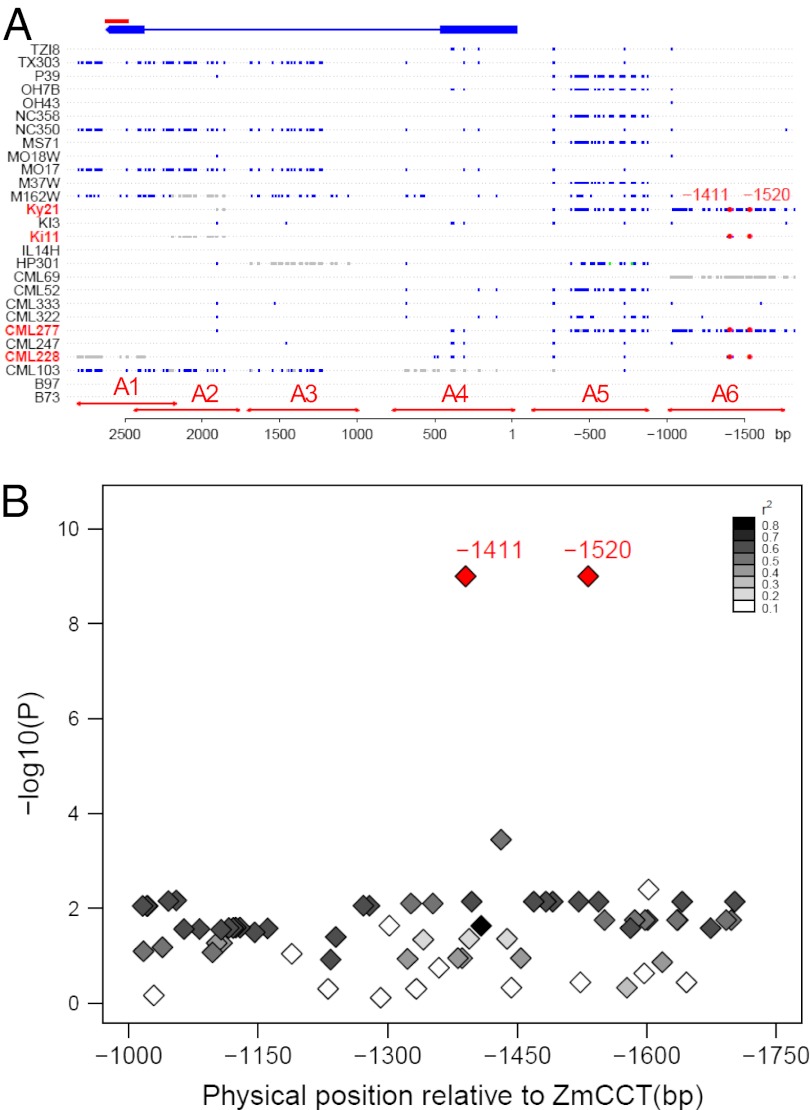
To discover the potential functional variants at ZmCCT region, we sequenced ZmCCT and its upstream region in the 27 NAM founders and 16 teosinte inbreds (SI Appendix, Table S4). A total of 227 variants (SNPs and indels) were discovered across a 4.5-kb region (Fig. 6). Only 11 sequence variants are located in the coding region, none in the conserved CCT domain (Fig. 6 and SI Appendix, Fig. S9). Overall, we did not find evidence for a selective sweep at this gene, as the ratio of diversity in maize to teosinte ranged from 41% to 67% for the six sequenced regions (SI Appendix, Table S5), in line with the genome average of 57% (31). Two SNPs, located at 1,411 bp (SNP1411) and 1,520 bp (SNP1520) upstream of ZmCCT, are consistent with the pattern of QTL effects in this genomic region (with positive effects resulting from CML277, CML228, Ki1, and Ky21 alleles; Fig. 1 and SI Appendix, Fig. S2). All seven teosinte inbreds successfully sequenced in this region also have the same alleles as those four maize lines at SNP1411 and SNP1520.
Fig. 6.
(A) Haplotype around ZmCCT across 27 NAM founders. ZmCCT is 2,809 bp long and consists of two exons, indicated in blue boxes at the top. The CCT domain is located at exon 2 and indicated with a red bar. Six primers (A1–A6 from left to right), shown in red arrows in the bottom, were used to sequence a 4.5-kb region around ZmCCT including 1.8 kb of its upstream region. All polymorphic variants including SNPs and indels relative to B73 allele are shown in blue boxes. The missing genotypes and the third alleles are indicated in gray and green boxes, respectively. All positions are shown relative to the start codon of ZmCCT. Two SNPs, located at 1411 and 1520 bp upstream of ZmCCT respectively, are consistent with NAM QTL mapping results for this region at which four lines—CML277, CML228, Ki11, and Ky21, indicated in red in the graph—had significant or nearly significant photoperiodic response allele effects relative to B73 allele in NAM joint linkage mapping. (B) Association analysis of SNPs in upstream region (A6) of ZmCCT for photoperiod response for GDDs to anthesis date in a panel of 282 diverse maize lines. SNP1411 and SNP1520 showed the most significant (P = 10−9) associations with photoperiod response. SNP1411 and SNP1520 are in complete LD in the population. Intensity of gray shading indicates the level of LD (_r_2) between each SNP and SNP1411 or SNP1520.
The exclusive sharing of alleles at SNP1411 and SNP1520 upstream of ZmCCT by teosintes and those NAM founders with chromosome 10 QTL alleles conferring increased photoperiod sensitivity suggests that they are in strong linkage disequilibrium (LD) with the causal sequence variants. Therefore, we sequenced a ∼750-bp region containing these SNPs (region A6 in Fig. 6) in a larger panel of 282 diverse maize lines (32), and tested the discovered SNPs for association with photoperiod response in this population. By using a mixed model that corrects for large-scale population structure and pairwise relatedness among individuals (33), the two SNPs associated with the pattern of QTL effects in the founders, SNP1411 and SNP1520, also showed the most significant (P = 10−9) associations with photoperiodic response in diverse lines (Fig. 6). Five obvious haplotypes are observed at this region in the association panel (Table 1). Lines carrying photoperiod response alleles of SNP1411 and SNP1520 split into four haplotypes, which maintained a typical level of sequence diversity compared with teosinte (SI Appendix, Table S6). In contrast, the sixth haplotype group, which contained B73 and most temperate lines, had a 10-fold reduction in diversity in this region, suggesting selection acting on this locus within certain maize lineages (SI Appendix, Table S6). Of the four NAM founders showing positive photoperiod responses, CML277 and Ky21 have haplotype I and CML228 and Ki11 have haplotype IV. Haplotype groups II and IV are predominantly composed of tropical lines and had significantly (P < 0.01) greater mean photoperiod responses effects than the B73 allele group haplotype V (Table 1). The strong association between SNP1411/1520 and photoperiod response in the diversity panel does not imply that they are the causal variants, but does indicate that they are at least in strong LD with the causal variants of increased photoperiod response, which may exist even further upstream of ZmCCT than the regions sequenced. Furthermore, NAM founders whose chromosome 10 QTL alleles significantly reduced photoperiod response (CML322, Ki3, Oh43, and Tx303; Fig. 1) were not distinguishable from B73 based on haplotypes in this region (Fig. 6), suggesting that additional sequence variation that affects photoperiod response is harbored within more complex haplotype structure in this region, or that there is a closely linked gene that segregates for the earlier flowering effect.
Table 1.
Haplotypes at A6 region of ZmCCT in maize association panel
| GDD to anthesis photoperiod response† | |||||||
|---|---|---|---|---|---|---|---|
| Haplotype | No. of lines | NAM founders included | SNP1411 and SNP1520* | Lines carrying MITE at vgt1 vgt1) | Mean | SD | Tropical origin, % |
| I | 16 | CML277, Ky21 | 1 | 5 | 49.0 | 14.0 | 61.4 |
| II | 2 | 1 | 0 | 126.1‡ | 36.0 | 97.9 | |
| III | 6 | 1 | 5 | 13.7 | 20.7 | 69.0 | |
| IV | 9 | CML228, Ki11 | 1 | 0 | 129.3‡ | 18.1 | 95.1 |
| V | 222 | B73, other NAM founders | 0 | 56 | 24.3 | 6.2 | 23.2 |
We further discovered a significant interaction in the maize diversity panel between SNP1411 or SNP1520 at ZmCCT and the flowering time gene Vgt1 (11) on chromosome 8 (SI Appendix, Fig. S10). SNP1411 and SNP1520 at ZmCCT had significant effects for photoperiod response only in lines not containing the MITE insertion at Vgt1.
Phenotypic Effects and Gene Expression of ZmCCT Alleles from Diverse Teosintes.
Maize NAM mapping results indicated that ZmCCT alleles have phenotypic effects functionally similar to the ancestral teosinte allele in only a subset of tropical maize lines (Fig. 1). To determine if photoperiod-sensitive alleles at ZmCCT are frequent in teosinte, we measured the effects of ZmCCT QTL alleles from eight teosinte donors representing the genetic and geographic variability within extant populations of the progenitor species in crosses with the temperate inbred W22. Segregating families from each cross between W22 and a teosinte donor were evaluated in the field under long day lengths and genotyped with flanking markers to identify which lines carried the teosinte alleles in the genome region encompassing ZmCCT and to estimate the effects of those alleles on flowering time. Across all donors, the additive effect (half the difference between homozygous classes) of a teosinte allele ranged from +3.3 to +5.6 d, suggesting that all teosintes carry a late flowering allele (Fig. 7).
Fig. 7.
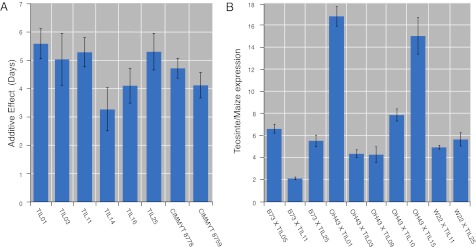
(A) Effect of eight distinct teosinte alleles in ZmCCT region on flowering time under long day lengths measured in backcrosses to maize. The mean difference in number of days to flowering for progenies homozygous for the teosinte allele vs. those homozygous for the maize allele are shown for each family. (B) Allele specific expression of ZmCCT in 10 F1 crosses between photoperiod-insensitive maize inbreds and eight distinct teosintes. The relative expression of teosinte vs. maize alleles is plotted for each F1.
To relate the phenotypic effects of teosinte QTL alleles in the ZmCCT region to the expression of the gene itself, we measured allele-specific ZmCCT transcript abundance in F1 crosses between eight different teosintes and three temperate maize lines grown under long day lengths. We hypothesized that teosinte ZmCCT alleles would have higher expression levels than maize alleles if ZmCCT acts to repress flowering time in maize as Ghd7 does in rice. Our allele-specific expression analysis revealed that the teosinte allele was expressed at higher levels than the maize allele in all cases (range, 2.9–12.6 times greater; Fig. 7). Thus, all teosintes tested carried _cis_-acting regulatory elements that increased the expression of ZmCCT relative to the maize allele, consistent with the fine-mapping and association experiments that identified the upstream region of ZmCCT within the causal QTL interval (Figs. 5 and 6 and SI Appendix, Figs. S6–S8). If ZmCCT functions in a manner homologous to its rice orthologue, higher expression of the teosinte allele under long day lengths is expected to repress expression of the maize florigen required to initiate flowering.
Discussion
The genetic architecture of maize photoperiod response demonstrates the general trend of more polygenic control for flowering time in maize relative to self-fertilizing species. For example, in the self-fertilizing tropical cereal crop sorghum, a handful of genes control most of the variation for photoperiod response in cultivated varieties, and allelic variation at the Ma1 locus alone can change flowering time by 60 d under long day lengths (30). In contrast, we discovered 14 maize QTLs for photoperiod response at which the strongest allele changes flowering time by only approximately 3 d when homozygous under long day lengths (7). Nevertheless, among quantitative traits of maize, a spectrum of complexity for genetic architecture exists (7, 16, 17, 34, 35) on which photoperiod response exhibits simpler genetic control than flowering time per se and other quantitative traits previously analyzed, as evidenced by the smaller number of photoperiod response QTLs, each contributing to a larger proportion of genetic variation. It appears that photoperiod response genes in maize represent a subset of genes affecting flowering time under long day lengths. Furthermore, we found that QTL alleles from teosinte and inbred line CML254 had stronger effects than those of any NAM parent (Figs. 5 and 7) (20) (perhaps because of selection of founder lines for NAM capable of reproducing under long day lengths), which could further simplify the inheritance of photoperiod response in specific segregating populations.
The maize NAM and maize-teosinte mapping results are congruent regarding the importance of a photoperiod response QTL on chromosome 10, and further, to the localization of its causal sequence variant to near ZmCCT. The difference between effects of the photoperiod-sensitive teosinte allele and the maize photoperiod-insensitive allele appears greater than the difference between sensitive and insensitive alleles within NAM (Figs. 2, 5, and 7). Nevertheless, photoperiod-sensitive alleles at ZmCCT appear to play a critical role in conferring day length adaptation in teosinte and some tropical maize varieties.
The identification of ZmCCT as the most important photoperiod response gene in maize is consistent with previous maize fine-mapping studies that suggested this gene as the probable causal gene of the photoperiod response QTL on chromosome 10 (19, 20). ZmCCT is homologous to Ghd7, a key regulator of photoperiod response in rice (28) that encodes a CO, CO-LIKE, and TIMING OF CAB1 (CCT) domain protein and is maximally expressed only in certain photoperiod-sensitive lines grown under long day lengths (29). The expression of Ghd7 represses the transcription of the B-type regulator Ehd1, which up-regulates the rice florigen gene Hd3a (29). Thus, Ghd7 is a major contributor to the photoperiod sensitivity of rice, and variation at this locus is associated with latitude of origin of rice cultivars (28, 29).
Fine-mapping and gene expression results demonstrate that sequence variation within or very near ZmCCT causes phenotypic variation for photoperiod response in maize-teosinte populations. Fine-mapping results in maize were consistent with this result, but not definitive, as five other predicted genes exist within the narrowest interval defined for the chromosome 10 photoperiod QTL in maize (SI Appendix, Figs. S6–S8). The larger allele effect observed between maize and teosinte at this QTL facilitated mendelization of the gene and consequently better mapping resolution (Fig. 5). The strongest association with photoperiod response in the NAM GWAS analysis was a SNP located at 95.7 Mbp on the chromosome 10 reference sequence, approximately 1.5 Mbp from ZmCCT itself, located at 94.2 Mbp (Fig. 2). Surprisingly, another SNP with polymorphisms in the same founders but located less than 4 kb upstream of ZmCCT was selected as an associated SNP in only 4% of resampling analyses (Dataset S7). These two SNPs are within the same 1-cM genetic interval on the NAM map, and, therefore, their imputation on the NAM RILs depends on the same flanking marker information, but differs in the weighting given to the two flanking markers in RILs that have a recombination in this interval. In the absence of information on how recombination rate varies within the interval, we used physical distance between HapMap SNPs and flanking markers to weight their influence on the imputation. To investigate the possibility that inaccuracies in linkage-based imputations caused the GWAS association signal to move away from the location of the causal variants, we used the recently released genotyping-by-sequencing (GBS) markers (36) assayed directly on the NAM RILs to identify haplotypes centered around these two markers that tracked their inheritance in the NAM lines. By reimputing genotypes at each of these two SNPs based on the GBS markers, we found that the SNP marker closest to ZmCCT had a slightly stronger association with photoperiod response than the more distant SNP marker (_R_2 value greater by 0.1%; Dataset S9), but their effects were not easily separated. Thus, our results demonstrate that GWAS in NAM may have limited resolution in cases in which recombination rates vary within a genetic interval and among families, and when the number of families segregating for a causal variant is low, limiting the number of recombination events that are needed to separate the effects of closely linked SNPs. In such cases, complementary analysis of the diverse maize association panel, characterized by low LD, can provide higher resolution, as observed in this case.
Across the whole genome, we have evidence that a number of candidate photoperiod response genes are associated with QTL (Fig. 2), but, in general, SNPs associated with photoperiod response do not represent variants at candidate flowering or photoperiod genes. Perhaps homologues of important photoperiod pathway genes from Arabidopsis do not tend to share similar functions in maize, or perhaps a high proportion of the causal genetic variation occurs in noncoding regions, and so they are not detected as an enrichment of GWAS associations within candidate genes. This result also may reflect an inherent limitation of GWAS based on single SNPs to resolve allelic series as a result of more complex haplotypic variation.
The consistently high levels of expression and phenotypic effects on photoperiod response of teosinte alleles at ZmCCT suggests that the mutation(s) that reduced the effect of ZmCCT on photoperiod response were rare or absent in teosinte, but increased to high frequencies following the initial domestication of maize. The effect of reduced ZmCCT expression on fitness in maize is highly dependent on environments, so it is possible that these alleles were not selected for until humans attempted to move maize culture to higher latitudes, which succeeded only in the most recent 2,000 y (5, 37). The relatively common occurrence of ZmCCT alleles with limited or no effect on photoperiod response among tropical inbred lines could be a result of much earlier selection sweep related to adaptation of maize to rainfall patterns and other climate differences between the domestication zone and other tropical environments (e.g., highland areas) where the effect of ZmCCT might have hindered flowering under day lengths greater than 12 h but less than 13 h. Alternatively, Ghd7 in rice is known to affect inflorescence architecture, so ZmCCT may have been selected first in tropical maize for its effects on ear morphology, helping to move alleles that were preadapted to longer day lengths to a high frequency in tropical maize. More recent exchange of maize germplasm among geographic regions may also be involved, but, in any case, ZmCCT alleles with reduced effects on photoperiod response appear to be selectively neutral in many tropical environments. Standing variation at photoperiod response loci provided early American peoples the raw material with which to select maize with adaptation to increasingly higher latitudes. Although the North–South spread of maize from its center of origin to cover vast distances in latitudes and ecologies was slower than the East–West spread of temperate cereals in Eurasia, it was accomplished before the arrival of Columbus to the Americas and before the discovery of the scientific principles of plant breeding, highlighting the skill of early agriculturalists and the power of artificial selection to shape unique genomic variation rapidly on an evolutionary time scale.
Materials and Methods
Maize Population Development and Genotyping.
The maize NAM population was created by crossing 25 diverse inbred lines to the common reference parent B73 and deriving 200 RILs from each cross to form 5,000 mapping lines (14). The NAM population was evaluated in 11 environments, including eight environments with mean day lengths greater than 13 h during the growing season (long-day length environments) and three environments with mean day lengths less than 13 h (short-day length environments) (38). A sample of 200 intermated B73 × Mo17 (IBM) RILs (39) and a panel of 281 lines encompassing much of the global diversity of public maize inbreds (“maize association panel”) (32) were also included alongside the NAM lines in field evaluations. The total number of inbred lines evaluated in field experiments was 5,481. Details of experimental design and phenotype data analysis are provided in the work of Hung et al. (38) and in SI Appendix, SI Materials and Methods. Each NAM and IBM line was genotyped with a common set of 1,106 SNP markers chosen to have high informativeness in these crosses (14). Linkage map and genotype scores are available at www.panzea.org (40). Lines identified with greater than 8% heterozygosity or putative contamination based on SNP analysis were excluded from QTL and GWAS analyses, resulting in a sample size of 4,699 NAM and 162 IBM (total of 4,861) RILs used for genetic analysis.
Maize Fine-Mapping Chromosome 10 QTLs.
Three maize fine-mapping populations were created to resolve the position of the chromosome 10 QTLs by selecting lines carrying recombinations in the QTL region between chromosomes from B73 and CML277, CML228, or Ki11 in homogenized genetic backgrounds (SI Appendix, Figs. S4–S8). The substitution mapping procedure widely used in fine mapping (41) was used to delimit the causal QTL region as detailed in SI Appendix, SI Materials and Methods.
Sequencing ZmCCT Alleles and Association Analysis in a Maize Diverse Panel.
Six primers were used to sequence a 4.5-kb region around ZmCCT including 1.8 kb of its upstream region in the 27 NAM and IBM founders and in 16 teosinte (Z. mays subsp. parviglumis) inbred lines (Fig. 6, Dataset S10, and SI Appendix, Table S4). Sequencing reactions were performed on PCR products in both directions with BigDye v3.1 on an Applied Biosystems 3700 automated sequencer. PHRED and PHRAP (42) were used for base calling, quality checks, and sequence assembly. Biolign (http://en.bio-soft.net/dna/BioLign.html) was used for multiple sequence alignments. Region A6, which includes SNP1411 and SNP1520 (Fig. 6), was further sequenced in a panel of 282 diverse maize lines (32). A mixed linear model that corrects for broad-scale population structure and pairwise relationships among lines (33) was used to conduct association testing for each variant identified at region A6. The population structure (i.e., Q) and relative kinship (i.e., K) matrices were calculated based on 89 microsatellites and 553 random SNPs, respectively, as described previously (33).
Teosinte Population Development and Genotyping.
To compare the QTL controlling differences in anthesis date between maize and teosinte to NAM QTLs, we mapped flowering time QTLs in 866 maize-teosinte BC2S3 lines genotyped at 19,838 GBS markers. The large effect QTL for days to anthesis on chromosome 10 was fine-mapped by using a single maize-teosinte BC2S3 family that segregated for a 50.77-Mb chromosomal segment including the QTL region introgressed from teosinte into the W22 genetic background. By using two marker loci that flanked the QTL, we isolated 74 lines homozygous for chromosomal recombinations around the QTL. These recombinant chromosome nearly isogenic lines were genotyped at 31 SNP and indel markers across the region (SI Appendix, Table S7) and phenotyped for flowering time under long day lengths. To assess whether late flowering alleles are prevalent in teosinte, we evaluated long-day length flowering time of BC1S1 families derived from crosses between temperate maize line W22 and six teosinte inbred lines. For each of the six families, 190 plants were genotyped at the ZmCCT locus and phenotyped for days to anthesis (SI Appendix, Table S7) to estimate the effect of substituting the maize allele with a teosinte allele around this gene. A more detailed description of maize-teosinte mapping experiments is provided in SI Appendix, SI Materials and Methods).
Allele-Specific Expression Assay.
We used an allele-specific assay of ZmCCT transcript accumulation in the F1 hybrids of inbred maize and teosinte parents. The hybrids were created by crossing eight teosinte inbred lines to three temperate maize inbred lines creating a total of 10 F1s (SI Appendix, Table S9). The plants were grown in long-day conditions in the field (summer 2011, West Madison Agricultural Research Station, Madison, WI) or in growth chambers (set to a 17:7-h light:dark daily cycle). Total cellular RNA was isolated from fully expanded leaf tissue from the third or fourth leaf of each plant. We assayed three to five biological replicates per cross and two to four technical replicates per biological replicate. Leaf tissue for RNA extraction was collected in the morning after the plants had experienced approximately 3 to 4 h of light. Aliquots of each RNA sample were DNase treated and reverse-transcribed using a polyT primer and SuperScript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. We used actin primers (wh182, 5′-CCAAGGCCAACAGAGAGAAA-3′; WH183, 5′-CCAAACGGAGAATAGCATGAG-3′) to check the integrity of the cDNA and genomic DNA contamination. We found no evidence of genomic DNA contamination.
To distinguish maize vs. teosinte transcripts in an allele-specific expression assay, we used an indel (PZD00131.indel) that is polymorphic between maize and teosinte in ZmCCT (SI Appendix, Table S10). We performed PCR on the cDNAs from each cross by using fluorescently labeled primers (GHD7EP1 forward, FAM-5′-TCATCACCGTCGTCATGAGT-3′; GHD7DEP1 reverse, 5′-CGCTTGCTTCTGCTGTCTC-3′; TAQ Core Kit; Qiagen). PCR products were assayed on an ABI 3700 fragment analyzer (Applied Biosystems), and areas under the peaks for the maize and teosinte transcripts were determined by using Gene Marker version 1.70. Relative expression was calculated as the ratio of the area under the peak for teosinte allele to the area under the peak for the maize allele. The calculated relative expression ratios were corrected for biased PCR amplification of the maize vs. teosinte allele by using the observed ratio of teosinte to maize PCR product for genomic DNA from each cross. Mean relative expression across biological replicates and SEs are reported (Fig. 7).
Supplementary Material
Supporting Information
Acknowledgments
This work was supported by National Science Foundation (NSF) Division of Biological Infrastructure Grant 0321467 and NSF Integrative Organismal Systems Grant 0820619, and by the US Department of Agriculture-Agricultural Research Service.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10 reported in this paper are deposited on the Panzea Web site, http://www.panzea.org/lit/publication.html#2012.
See Author Summary on page 11068 (volume 109, number 28).
References
- 1.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 2.Cockram J, et al. Control of flowering time in temperate cereals: genes, domestication, and sustainable productivity. J Exp Bot. 2007;58:1231–1244. doi: 10.1093/jxb/erm042. [DOI] [PubMed] [Google Scholar]
- 3.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf PC. Corn: Its Origin, Evolution, and Improvement. Cambridge, MA: Harvard Univ Press; 1974. [Google Scholar]
- 5.Crawford GW, Saunders D, Smith DG. Pre-contact maize from Ontario, Canada: Context, chronology, variation, and plant association. In: Staller J, Tykot R, Benz B, editors. Histories of Maize: Multidisciplinary Approaches to the Prehistory, Linguistics, Biogeography, Domestication, and Evolution of Maize. San Diego: Academic; 2006. pp. 549–559. [Google Scholar]
- 6.Weatherwax P. Indian Corn in Old America. New York: McMillan; 1954. [Google Scholar]
- 7.Buckler ES, et al. The genetic architecture of maize flowering time. Science. 2009;325:714–718. doi: 10.1126/science.1174276. [DOI] [PubMed] [Google Scholar]
- 8.Thornsberry JM, et al. Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet. 2001;28:286–289. doi: 10.1038/90135. [DOI] [PubMed] [Google Scholar]
- 9.Colasanti J, et al. The maize INDETERMINATE1 flowering time regulator defines a highly conserved zinc finger protein family in higher plants. BMC Genomics. 2006;7:158–175. doi: 10.1186/1471-2164-7-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muszynski MG, et al. delayed flowering1 Encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006;142:1523–1536. doi: 10.1104/pp.106.088815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salvi S, et al. Conserved noncoding genomic sequences associated with a flowering-time quantitative trait locus in maize. Proc Natl Acad USA. 2007;104:11376–11381. doi: 10.1073/pnas.0704145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics. 2010;184:799–812. doi: 10.1534/genetics.109.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camus-Kulandaivelu L, et al. Maize adaptation to temperate climate: Relationship between population structure and polymorphism in the Dwarf8 gene. Genetics. 2006;172:2449–2463. doi: 10.1534/genetics.105.048603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McMullen MD, et al. Genetic properties of the maize nested association mapping population. Science. 2009;325:737–740. doi: 10.1126/science.1174320. [DOI] [PubMed] [Google Scholar]
- 15.Gore MA, et al. A first-generation haplotype map of maize. Science. 2009;326:1115–1117. doi: 10.1126/science.1177837. [DOI] [PubMed] [Google Scholar]
- 16.Tian F, et al. Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet. 2011;43:159–162. doi: 10.1038/ng.746. [DOI] [PubMed] [Google Scholar]
- 17.Kump KL, et al. Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population. Nat Genet. 2011;43:163–168. doi: 10.1038/ng.747. [DOI] [PubMed] [Google Scholar]
- 18.Wang CL, et al. Genetic analysis of photoperiod sensitivity in a tropical by temperate maize recombinant inbred population using molecular markers. Theor Appl Genet. 2008;117:1129–1139. doi: 10.1007/s00122-008-0851-y. [DOI] [PubMed] [Google Scholar]
- 19.Ducrocq S, et al. Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics. 2009;183:1555–1563. doi: 10.1534/genetics.109.106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coles ND, Zila CT, Holland JB. Allelic effect variation at key photoperiod response QTL in maize. Crop Sci. 2011;51:1036–1049. [Google Scholar]
- 21.Briggs WH, McMullen MD, Gaut BS, Doebley J. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics. 2007;177:1915–1928. doi: 10.1534/genetics.107.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chia J-M, et al. Maize HapMap2 identifies extant variation from a genome in flux. Nat Genet. 2012 doi: 10.1038/ng.2313. 10.1038/ng.2313. [DOI] [PubMed] [Google Scholar]
- 23.Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics. 2009;182:1263–1277. doi: 10.1534/genetics.109.100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coneva V, Zhu T, Colasanti J. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J Exp Bot. 2007;58:3679–3693. doi: 10.1093/jxb/erm217. [DOI] [PubMed] [Google Scholar]
- 25.Meng X, Muszynski MG, Danilevskaya ON. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell. 2011;23:942–960. doi: 10.1105/tpc.110.081406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazakis CM, Coneva V, Colasanti J. ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J Exp Bot. 2011;62:4833–4842. doi: 10.1093/jxb/err129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand E, et al. Flowering time in maize: Linkage and epistasis at a major effect locus. Genetics. 2012;190:1547–1562. doi: 10.1534/genetics.111.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 29.Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- 30.Murphy RL, et al. Coincident light and clock regulation of pseudoresponse regulator protein 37 (PRR37) controls photoperiodic flowering in sorghum. Proc Natl Acad USA. 2011;108:16469–16474. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright SI, et al. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- 32.Flint-Garcia SA, et al. Maize association population: A high-resolution platform for quantitative trait locus dissection. Plant J. 2005;44:1054–1064. doi: 10.1111/j.1365-313X.2005.02591.x. [DOI] [PubMed] [Google Scholar]
- 33.Yu JM, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- 34.Brown PJ, et al. Distinct genetic architectures for male and female inflorescence traits of maize. PLoS Genet. 2011;7:e1002383. doi: 10.1371/journal.pgen.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cook JP, et al. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol. 2012;158:824–834. doi: 10.1104/pp.111.185033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elshire RJ, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. 2011;6:e19379. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith BD. Origins of agriculture in eastern North America. Science. 1989;246:1566–1571. doi: 10.1126/science.246.4937.1566. [DOI] [PubMed] [Google Scholar]
- 38.Hung HY, et al. The relationship between parental genetic or phenotypic divergence and progeny variation in the maize nested association mapping population. Heredity (Edinb) 2012;108:490–499. doi: 10.1038/hdy.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharopova N, et al. Development and mapping of SSR markers for maize. Plant Mol Biol. 2002;48:463–481. doi: 10.1023/a:1014868625533. [DOI] [PubMed] [Google Scholar]
- 40.Canaran P, et al. Panzea: An update on new content and features. Nucleic Acids Res. 2008;36(database issue):D1041–D1043. doi: 10.1093/nar/gkm1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paterson AH, DeVerna JW, Lanini B, Tanksley SD. Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics. 1990;124:735–742. doi: 10.1093/genetics/124.3.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
Proc Natl Acad Sci U S A. 2012 Jul 10;109(28):11068–11069.
Author Summary
Maize was domesticated from a wild grass, teosinte, that grows only in tropical environments in Mexico (1). Maize agriculture spread from its Meso-American center of origin to Canada and Chile before the arrival of Columbus. The transition to flowering in teosinte is highly sensitive to small changes in day lengths that occur in its native range, perhaps as a way to coordinate flowering with rainfall patterns. In contrast, reproductive fitness for maize of the higher latitudes depends on its capacity to flower under much longer summer day lengths (Fig. P1). Thus, the spread of maize from its tropical center of origin to the higher latitudes of the Americas required selection for adaptation to longer day lengths. Genetic analysis of the differences in photoperiod response among teosinte, tropical maize, and temperate maize revealed moderately complex inheritance involving more than 10 genes, one of which, ZmCCT, is similar to an important regulator of day-length response in rice.
Fig. P1.
Reaction of temperate maize and teosinte to long day lengths. Under short day lengths, this teosinte population flowers earlier than temperate maize line B73, but under long day lengths shown here, it flowers much later or never. The figure shows temperate inbred B73, teosinte plants, and their F1 hybrid growing in the field in Clayton, NC, under long day lengths. The temperate inbred line flowered several weeks before this photo was taken. The teosinte plant never flowered before the field was abandoned in September. ZmCCT and several other genes contribute to this differential response to photoperiod. (Photo taken July 14, 2008; courtesy of J.B. Holland.)
To understand the genetic basis of adaptations to different day lengths, we evaluated nearly 5,000 mapping lines derived from crosses of 27 diverse maize inbred lines that compose the maize nested association mapping (NAM) population. The parents of NAM are a geographically and genetically diverse sample of maize inbreds selected by plant breeders for adaptation to a range of latitudes. The photoperiod responses of the NAM lines and a set of 281 diverse inbreds (representing even more of the global diversity of maize) were measured by comparing their time to flowering in eight long-day length (>13 h) environments to three short (< 12 h)-day length environments after adjusting for temperature differences among environments.
Genetic linkage analysis of the NAM families revealed 14 quantitative trait loci (QTLs) associated with most of the observed variation for flowering time photoperiod response. These photoperiod response QTLs were mostly a subset of the more than twice as many QTLs detected for flowering time under long day lengths. The QTL with strongest effects mapped to a region on chromosome 10 previously identified as carrying a strong photoperiod response QTL in maize and teosinte mapping studies (2–4). Surprisingly, only three tropical NAM founders carry variants (alleles) of this QTL that significantly increase the photoperiod response, and two tropical parent lines have QTL alleles with even less sensitivity to photoperiod than the standard temperate allele. In contrast, most tropical founders carry photoperiod-sensitive alleles at other important QTLs. Patterns of environment-dependent effects on flowering time varied substantially among the four key QTLs, demonstrating that each gene has a unique response to photoperiod.
We conducted a genome-wide association study of photoperiod response in the NAM population by using 26.5 M SNPs and 1 M copy-number variants identified in the maize HapMap version 2. We identified 118 SNPs and five CNVs associated with photoperiod response, which mapped within QTL support intervals much more frequently than expected by chance. Genome-wide association study associations were not enriched near 218 candidate genes identified a priori based on their effects on flowering time in maize or homology to flowering-time genes of Arabidopsis, sorghum, or rice, however. These results suggest that casting a wide net for candidate genes to test for associations with maize photoperiod response was largely unsuccessful.
We also performed a parallel whole-genome linkage mapping scan of flowering time under long days in a population of 866 lines derived from backcrosses of teosinte to a temperate maize line. We identified 23 QTLs controlling variation for flowering time under long day lengths, approximately half of which coincided with NAM flowering-time QTLs. The flowering-time QTL with the largest effect in the maize/teosinte population was syntenic to the major chromosome 10 photoperiod QTL identified in NAM. Briggs et al. (2) demonstrated that the teosinte allele at this QTL is highly responsive to photoperiod and is detected only under long day lengths.
Given the strong allelic effects observed at the QTL on chromosome 10, we targeted this region for high-resolution linkage mapping in maize and maize/teosinte mapping families. Results from all four fine-mapping populations were congruent in identifying a region on chromosome 10 that includes ZmCCT, a homologue of rice Ghd7. Highest resolution was obtained in the maize/teosinte mapping population, which identified ZmCCT as the only predicted gene within the QTL interval. Analysis of the sequences of ZmCCT and its upstream region in the maize 281 line global diversity panel revealed two SNPs in the promoter of this gene that were highly associated with photoperiod response in that panel after correction for population structure.
To determine if photoperiod-sensitive alleles at ZmCCT are frequent in teosinte, we measured the effects of ZmCCT QTL alleles from eight teosinte lines representing the genetic and geographic variability of the progenitor species in crosses with temperate maize. All teosinte alleles in this region have a strong effect on delaying flowering time. We also measured allele-specific ZmCCT transcript abundance in F1 crosses between eight different teosintes and three temperate maize lines grown under long day lengths. The teosinte allele was expressed at higher levels than the maize allele in all cases. Thus, all teosintes tested carried _cis_-acting regulatory elements that increased the expression of ZmCCT relative to the maize allele. If ZmCCT functions in a manner homologous to its rice orthologue (5), higher expression of the teosinte allele under long day lengths is expected to repress expression of the maize florigen required to initiate flowering.
These results suggest that the mutation(s) that reduce the effect of ZmCCT on photoperiod response were rare or nonexistent in teosinte, but increased to high frequencies following the initial domestication of maize. The relatively common occurrence of photoperiod-insensitive ZmCCT alleles in tropical inbred lines could be a result of a selection sweep related to adaptation of maize to tropical environments outside of the range of teosinte. Alternatively, Ghd7 in rice is known to affect inflorescence architecture so ZmCCT may have first been selected in tropical maize for its effects on ear morphology, helping to move alleles that were preadapted to longer day lengths to higher frequency in tropical maize. Standing variation at photoperiod response loci provided early Americans the raw material with which they could select maize with adaptation to increasingly higher latitudes.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets S1, S2, S3, S4, S5, S6, S7, S8, S9, and S10 reported in this paper are deposited on the Panzea Web site, http://www.panzea.org/lit/publication.html#2012.
See full research article on page E1913 of www.pnas.org.
References
- 1.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad USA. 2002;99:6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs WH, McMullen MD, Gaut BS, Doebley J. Linkage mapping of domestication loci in a large maize teosinte backcross resource. Genetics. 2007;177:1915–1928. doi: 10.1534/genetics.107.076497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coles ND, McMullen MD, Balint-Kurti PJ, Pratt RC, Holland JB. Genetic control of photoperiod sensitivity in maize revealed by joint multiple population analysis. Genetics. 2010;184:799–812. doi: 10.1534/genetics.109.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducrocq S, et al. Fine mapping and haplotype structure analysis of a major flowering time quantitative trait locus on maize chromosome 10. Genetics. 2009;183:1555–1563. doi: 10.1534/genetics.109.106922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
Supplementary Materials
Supporting Information