Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks (original) (raw)
. Author manuscript; available in PMC: 2012 Jul 14.
SUMMARY
The cellular response to DNA double-strand breaks (DSBs) is mobilized by the protein kinase ATM, which phosphorylates key players in the DNA damage response (DDR) network. A major question is how ATM controls DSB repair. Optimal repair requires chromatin relaxation at damaged sites. Chromatin reorganization is coupled to dynamic alterations in histone posttranslational modifications. Here, we show that in human cells, DSBs induce monoubiquitylation of histone H2B, a modification that is associated in undamaged cells with transcription elongation. We find that this process relies on recruitment to DSB sites and ATM-dependent phosphorylation of the responsible E3 ubiquitin ligase: the RNF20-RNF40 heterodimer. H2B monoubiquitylation is required for timely recruitment of players in the two major DSB repair pathways—nonhomologous end-joining and homologous recombination repair—and optimal repair via both pathways. Our data and previous data suggest a two-stage model for chromatin decondensation that facilitates DSB repair.
INTRODUCTION
Genome stability is essential for prevention of undue cellular death and neoplasia. DNA damage caused by internal or external damaging agents is a major threat to the integrity of the cellular genome. The cellular defense system against this threat is the DNA damage response (DDR)—an elaborate signaling network that activates DNA repair and cell-cycle checkpoints and modulates many physiological pathways (Ciccia and Elledge, 2010). The DDR serves as a barrier against cancer formation by activated oncogenes (Halazonetis et al., 2008). Germline mutations abrogating critical relays in the DDR lead to genomic instability syndromes characterized by sensitivity to genotoxic stresses, tissue degeneration, and excessive cancers.
One of the most powerful activators of the DDR is the DNA double-strand break (DSB). This cytotoxic lesion is induced by ionizing radiation (IR), radiomimetic chemicals, and reactive oxygen species that accompany normal metabolism. DSBs are also formed during meiotic recombination and maturation of the antigen receptor genes (Hiom, 2010). Eukaryotic cells repair DSBs via error-prone nonhomologous end-joining (NHEJ) or high-fidelity homologous recombination repair (HRR), which is preceded by DNA end resection (Holthausen et al., 2010; Lieber, 2010). The vast cellular response to DSBs affects cell-cycle progression, gene expression, protein synthesis, degradation and trafficking, and RNA processing.
The DSB response begins with recruitment of sensor proteins to the damaged sites; these proteins are involved in the initial recognition and processing of the damage and activation of the transducers of the DNA damage alarm (Ciccia and Elledge, 2010). The primary transducer of the DSB response is the nuclear serine-threonine kinase ATM, which phosphorylates a plethora of effectors in various DDR pathways (Derheimer and Kastan, 2010). Loss of ATM causes the genome instability syndrome ataxia-telangiectasia (A-T) (Lavin, 2008).
A major question is how ATM controls a critical branch of the DDR: DSB repair. In laboratory cell lines, timely repair of about 10%–15% of IR-induced DSBs is ATM dependent (Riballo et al., 2004). Like other DNA transactions, DNA repair should be coupled to chromatin reorganization. Chromatin organization is affected by protein-DNA and protein-protein interactions among core histones and nonhistone proteins and posttranslational modifications (PTMs) of these proteins. Dynamic changes in the rich repertoire of histone PTMs indeed accompany chromatin reorganization associated with DNA transactions (Murr, 2010; Zhou et al., 2011).
Here, we show that histone H2B monoubiquitylation, a highly dynamic and mobile histone mark known to be associated with transcription elongation, is induced in human cells after DSB induction in an ATM-dependent manner. The responsible E3 ubiquitin ligase is a heterodimer of the RING-finger proteins RNF20 and RNF40 (Kim et al., 2005; Zhu et al., 2005). Human RNF20 and RNF40 are orthologs of the budding yeast protein Bre1, which together with the ubiquitin-conjugating enzyme Rad6, monoubiquitylates the yeast histone H2B on lysine 123 at sites of transcription elongation (Kao et al., 2004). This histone modification was recently shown to interfere with the compaction of the basic 30 nm chromatin fiber (Fierz et al., 2011). Notably, in yeast and mammalian cells, H2B monoubiquitylation at transcribed chromatin is required for subsequent histone H3 methylations on Lys4 and Lys79 (Dover et al., 2002; Kim et al., 2009; Lee et al., 2007; Schneider et al., 2005).
We find that, similar to transcription-associated H2B monoubiquitylation, the induction of this histone modification after DNA damage is RNF20-RNF40 dependent. Furthermore, upon DNA damage a fraction of RNF20-RNF40 is recruited to DSB sites and undergoes ATM-mediated phosphorylation. ATM’s activity and the presence of its phosphorylation sites on RNF20-RNF40 are required for damage-induced H2B monoubiquitylation. This process is not required for the recruitment of the damage sensors in the very early stage of the DDR, but is essential for timely accumulation of NHEJ and HRR proteins at DSB sites and subsequent optimal repair via both pathways. Collectively, our data and previous data suggest a two-stage process of chromatin relaxation at DSB sites that is essential for timely DSB repair.
RESULTS
A Tight RNF20-RNF40 Heterodimer Is Physically Associated with ATM
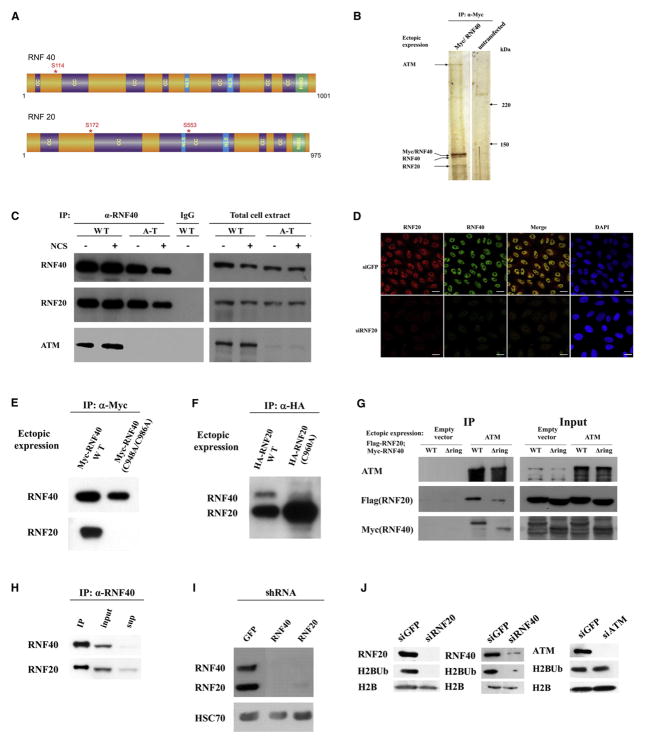
An initial hint that H2B monoubiquitylation might be involved in the DDR came from observation of an ATM-associated protein complex isolated from human cells that contained RNF20 and RNF40 (Figure S1A, available online, and Figure 1A). Coimmunoprecipitation of ATM with ectopic and endogenous RNF40 further demonstrated this interaction (Figures 1B and 1C). Polyclonal and monoclonal antibodies were raised against RNF20 and RNF40 (Figure S1B) and used to show that the two proteins colocalize in the nucleus (Figure 1D) and are bound tightly to each other (Figures 1E–1I). Stability of each protein depended on this mutual interaction, which required the integrity of their RING domains (Figures 1E and 1F). Thus, knockdown of one of them reduced the level of the other (Figures 1D and 1I), as did substitutions of critical residues within their RING domains (Figures 1E and 1F). Deletion of their RING domains reduced but did not abolish completely their individual interaction with ATM (Figure 1G). Importantly, depletion of either RNF20 or RNF40, but not ATM, considerably reduced the level of monoubiquitylated H2B (H2BUb) in unstressed cells (Figure 1J). These results suggest that the functional unit in H2B monoubiquitylation is a tight heterodimer of RNF20 and RNF40.
Figure 1. Characterization of the Human RNF20-RNF40 Heterodimer.
(A) Domain diagrams of RNF20 and RNF40. Locations of serine residues later found to be phosphorylated in an ATM-dependent manner are indicated by asterisks. CC, coiled-coil domain; NLS, nuclear localization signal; RING, RING finger domain.
(B) Validation of the interaction of RNF20 and RNF40 with ATM. Myc-tagged RNF40 was expressed in HEK293 cells and immunoprecipitated with an anti-Myc antibody. The immune complexes underwent gel electrophoresis and protein bands were visualized by silver staining. The band at ~350 kDa was identified as ATM.
(C) Further validation of the interaction between endogenous ATM and RNF20-RNF40. RNF40 was immunoprecipitated from total cell extracts of human lymphoblasts, coimmunoprecipitating RNF20 and ATM. The amount of proteins in the right-hand panel (input) represents 5% of the amount used for immunoprecipitation.
(D) Dependence of RNF40 level on the presence of RNF20 demonstrated in HeLa cells by using immunostaining with the antibodies shown in Figure S1B. Scale bar = 20 μm.
(E) Dependence of RNF20-RNF40 interaction on the integrity of RNF40s RING domain. Myc-tagged RNF40 in wild-type or RING mutant (C984A/C986A) versions was ectopically expressed in HEK293 cells, and an anti-Myc antibody was used to immunoprecipitate the ectopic proteins. Western blotting analysis was employed to demonstrate coimmunoprecipitation of endogenous RNF20 with wild-type, but not the mutant RNF40.
(F) Analogous experiment to that described in (E), demonstrating the reciprocal immunoprecipitation, which now depended on the integrity of the RING domain of RNF20.
(G) Interaction with ATM of RNF40 and RNF20 with RING domain deletion. FLAG-RNF20 and Myc-RNF40 in wild-type and RING deletions spanning aa 922–960 in RNF20 and 948–986 in RNF40 were ectopically expressed in HEK293 cells concomitantly with GFP-tagged ATM. ATM was immunoprecipitated with an anti-GFP antibody. The immune complexes were blotted with the indicated antibodies. The protein amounts in the input gel represent 10% of the amount used for immunoprecipitation.
(H) Interaction between endogenous RNF20 and RNF40 demonstrated by coimmunoprecipitation of RNF20 with RNF40 in HEK293 cell extracts. Note the concomitant depletion of both proteins from the supernatant. The input represents 10% of the protein amount used for immunoprecipitation.
(I) Dependence of the stability of RNF20 and RNF40 on their interaction with each other. Depletion of one of these proteins in HEK293 cells by using stable expression of shRNA leads to the loss of the other.
(J) RNF20-RNF40 dependence and ATM independence of histone H2B monoubiquitylation in unstressed cells. Depletion of endogenous RNF20, RNF40, and ATM was obtained in HeLa cells by using siRNA. H2BUb was detected on western blots of cellular extracts by using a monoclonal antibody against this modification (Minsky et al., 2008).
RNF20-RNF40 and H2B Monoubiquitylation on K120 Are Required for Timely DSB Repair
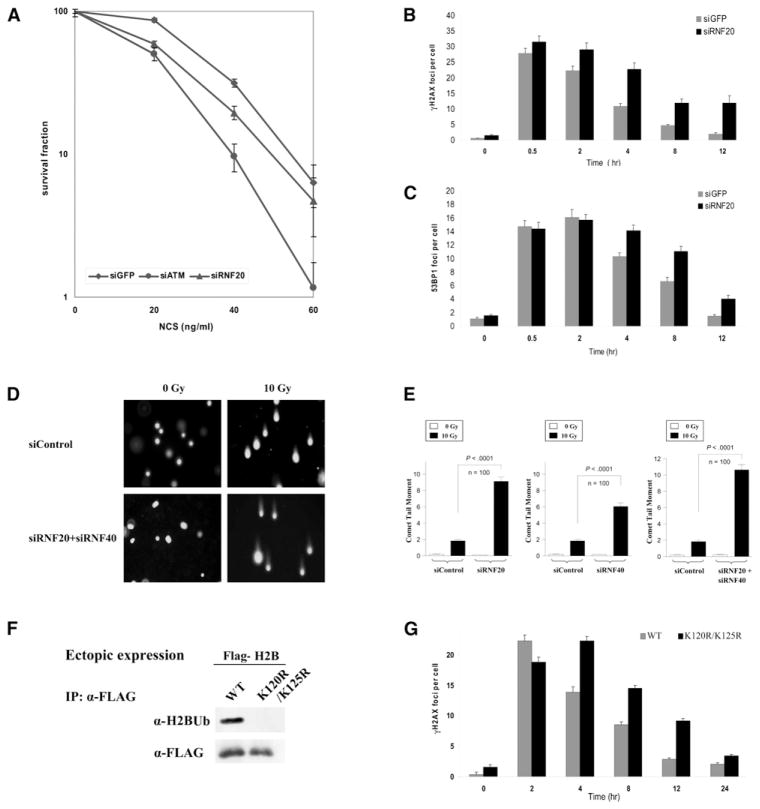
The stable association between ATM and RNF20-RNF40 suggested the involvement of the latter in the DDR. Indeed, reduction in cellular RNF20-RNF40 level obtained by using RNAi increased the sensitivity of human cells to the cytotoxic effect of the radiomimetic drug neocarzinostatin (NCS) (Figure 2A). This phenotype usually points to abrogation of a proximal process in the DNA damage response and is often associated with suboptimal DSB repair. The time course of DSB repair is reflected in the dynamics of DSB hallmarks, such as the local phosphorylation of histone H2AX over large genomic regions flanking DSBs (reviewed by Löbrich et al., 2010) and nuclear foci formed at DSB sites by early damage response proteins such as 53BP1(reviewed by FitzGerald et al., 2009). Indeed, we observed slower disappearance with time of both phosphorylated H2AX (γH2AX) and 53BP1 foci in RNF20-depleted cells (Figures 2B and 2C and Figures S2A and S2B). Direct evidence of an overt repair defect was further obtained by using the neutral comet assay, which monitors the presence of DSBs in single cells by microscopic detection of DNA migration in gel (Dhawan et al., 2009). Importantly, this sensitive assay demonstrated a marked defect in DSB sealing in cells depleted of RNF20-RNF40 (Figures 2D and 2E).
Figure 2. Effect of RNF20 Depletion or Interference with H2B Monoubiquitylation on the DNA Damage Response.
(A) Survival curves based on clonogenic growth of HeLa cells transfected with siRNA against GFP (control), ATM and RNF20 (Figure 1J) and treated with increasing doses of the radiomimetic drug neocarzinostatin (NCS). The experiment was carried out in triplicate. Bars represent standard error of the mean.
(B and C) Counts of γH2AX (B) and 53BP1 foci (C) at various time points after irradiation with 1 Gy of ionizing radiation in U2-OS cells transfected with siRNA against GFP or RNF20 (average of 100 cells). Bars represent standard error of the mean.
(D) Direct observation of DNA damage 4 hr after irradiation with 10 Gy of IR by using the neutral comet assay in U2-OS cells pretreated with various siRNAs. Shown are representative microscopic images of the assay.
(E) Quantitation of the comet data. The length and intensity of DNA tails relative to heads is shown as % of the relative comet tail moment (n = 100). Bars represent standard error of the mean. The statistical differences between siControl and siRNF20-RNF40 are indicated with p values. The IR dose of 10 Gy is equivalent to 200 ng/ml of NCS based on clonogenic survival experiments.
(F) Western blotting analysis showing the monoubiquitylation of ectopic FLAG-tagged wild-type histone H2B expressed in HeLa cells and lack of monoubiquitylation of a mutant (K120R/K125R) version of H2B, under the same conditions. Ectopic H2B was immunoprecipitated with an α-FLAG antibody and the immune complexes were blotted with the indicated antibodies.
(G) Ectopic H2B proteins were expressed in HeLa cells for 48 hr, the cells were irradiated as above, and γH2AX foci were counted specifically in cells expressing ectopic H2B as judged by FLAG staining.
We also examined in RNF20-depleted cells the activation of the cell-cycle checkpoints. Treatment of hTERT-immortalized human fibroblasts with 100 ng/ml of NCS led to a cell-cycle arrest at G1 and G2 and concomitant depletion of the S phase followed by slow recovery, in a roughly similar manner to cells transfected with irrelevant siRNA and cells depleted for RNF20 (not shown). We concluded that RNF20-RNF40 loss might have affected directly DSB repair without having a marked effect on activation of the cell-cycle checkpoints.
Notably, loss of RNF20-RNA40 did not affect other early steps in the DDR: initial 53BP1 recruitment (Figure 2C) and the recruitment of two other central players in the early phase of the DDR, MDC1 and RNF8 (reviewed by Ciccia and Elledge, 2010 and Al-Hakim et al., 2010) (Figures S2C and S2D). ATM activation—as evidenced by its autophosphorylation on Ser1981 (Bakkenist and Kastan, 2003) —and the time course and intensity of ATM-mediated phosphorylation of KAP-1, a typical ATM substrate (Ziv et al., 2006), also were not affected by RNF20-RNF40 depletion (not shown). Collectively, our data therefore suggest that RNF20-RNF40 is not required for the early steps of the DDR, but is necessary during the later step of DSB repair.
Assuming that RNF20-RNF40 is involved in the DDR in its capacity as the E3 ligase in histone H2B monoubiquitylation, we examined the role of this process in DSB repair by using ectopic expression of tagged wild-type or mutant (K120R/K125R) H2B. The K120R substitution prevents monoubiquitylation at this position (Figure 2F); the K125R substitution does not allow a secondary monoubiquitylation that may occur in the absence of the primary monoubiquitylation site at K120 (Minsky et al., 2008). Importantly, ectopically expressed histone H2B fused with various tags was heavily incorporated into the chromatin (Figures S2E–S2G). Similarly to RNF20-RNF40 depletion, expression of the mutant H2B slowed the disappearance of the DSB-flagging foci (Figure 2G), indicating that the ectopic mutant H2B exerted a dominant-negative effect on H2B monoubiquitylation. This result suggests that H2B monoubiquitylation is indeed required for timely DSB repair.
DNA Damage Triggers ATM-Dependent H2B Monoubiquitylation
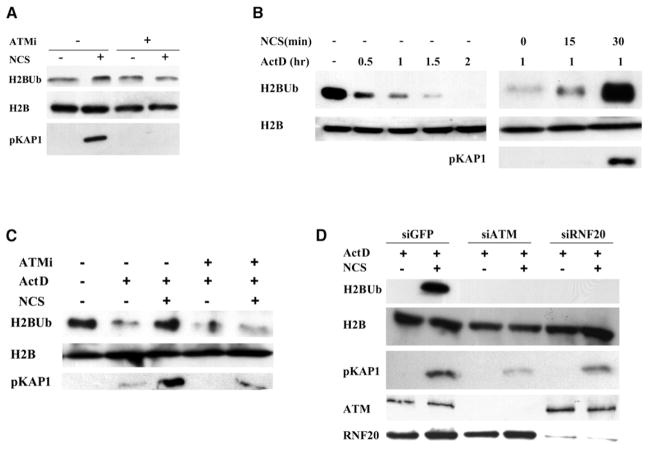
In undamaged cells, RNF20-RNF40 is associated with the ongoing, dynamic H2B monoubiquitylation that accompanies transcription elongation. The above data suggested that, after DSB induction, RNF20-RNF40 might mediate the damage-induced H2B monoubiquitylation that is required for optimal DSB repair. We explored whether DNA damage enhances H2B monoubiquitylation by monitoring the amount of cellular H2BUb with an antibody highly specific for this modification (Minsky et al., 2008). Western blotting analysis of cellular extracts showed transient elevation of total cellular H2BUb after DNA damage, but the background of ongoing, transcription-associated H2BUb made it difficult to discern the incremental damage-induced elevation in this modification (Figure 3A). We therefore attempted to reduce this background by inhibiting transcription. Examination of the effect of several transcription inhibitors on basal H2BUb levels showed that H2BUb was considerably reduced by the transcription inhibitors actinomycin D (Figure 3B), 5,6-dichloro-β-D- ribofuranosylbenzimidazole (DRB), and α-amanitin (Figures S3A and S3B). Damage-induced H2BUb was clearly observed in the presence of each of these inhibitors (Figure 3B and Figures S3A and S3B). Importantly, damage-induced H2BUb exhibited ATM dependence: ATM knockdown or the ATM inhibitor KU-55933 (Hickson et al., 2004) markedly reduced the induction of this modification by DNA damage (Figures 3C and 3D). RNF20-RNF40 depletion had a similar effect (Figure 3D), suggesting that this ubiquitin ligase plays an essential role in damage-induced H2B monoubiquitylation, similar to its role in transcription-associated H2BUb. Notably, this pathway was not noticeably affected by depletion of MDC1 or RNF8 (Figure S3C), suggesting that it was separate from the MDC1-RNF8 axis, which is responsible for extensive protein ubiquitylation at DSB sites, including histone H2A monoubiquitylation (reviewed by Al-Hakim et al., 2010).
Figure 3. ATM- and RNF20-RNF40-Dependent Monoubiquitylation of Histone H2B after DNA Damage.
(A) HeLa cells were treated with 1 μg/ml of NCS for 1 hr, with or without 30 min pretreatment with 20 μM of the ATM inhibitor KU-55933 and cellular extracts were blotted with the indicated antibodies. A slight, ATM-dependent increase in H2BUb is noticed. DNA damage induction was monitored with an antibody against the phosphorylated form of an ATM substrate, KAP-1.
(B) Detection of damage-induced H2BUb in cells treated with actinomycin D. Left panel: HeLa cells were treated with 5 μg/ml of actinomycin D and H2BUb levels were assessed at the indicated time points. Right panel: The cells were incubated with actinomycin D for 1 hr and then with 1 μg/ml of NCS for the indicated time periods.
(C) Effect of the ATM inhibitor KU-55933 on damage-induced H2BUb in transcription-inhibited cells. HeLa cells were pretreated with 20 μM of the inhibitor (ATMi) for 30 min and then with 5 μg/ml of actinomycin D and 1 μg/ml of NCS for 1 hr.
(D) Dependence of damage-induced H2BUb on ATM and RNF20. HeLa cells transfected with siRNA against GFP, ATM, or RNF20 were treated for 1 hr with 5 μg/ml of actinomycin D and 1 μg/ml of NCS.
The RNF20-RNF40 Heterodimer Is an ATM Target
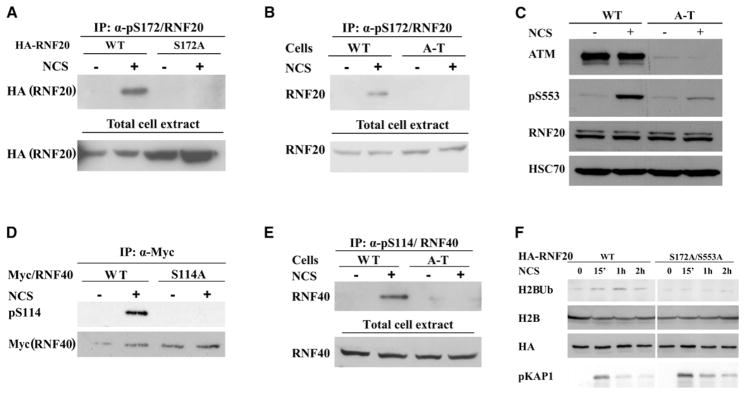
In typical ATM-mediated pathways, pivotal players are usually downstream targets of ATM’s kinase activity, and their phosphorylation modulates the pathway in which they function (Derheimer and Kastan, 2010). ATM phosphorylates its targets preferentially on serine or threonine residues within SQ or TQ motifs. Examination of RNF20 and RNF40 amino acid sequences revealed several potential targets for ATM-mediated phosphorylation; two of them, Ser172 on RNF20 and Ser114 on RNF40 (Figure 1A), were phosphorylated by ATM in vitro (Figures S4A and S4B). A recent phosphoproteomic screen (Matsuoka et al., 2007) identified Ser553 on RNF20 (Figure 1A) as another potential ATM target. Phosphospecific antibodies raised against the three phosphorylation sites confirmed that in response to DSB induction in cells, RNF20 and RNF40 were phosphorylated at these sites in an ATM-dependent manner (Figures 4A–4E). Notably, however, only a fraction of the cellular content of these proteins was phosphorylated in cells (Figure S4C).
Figure 4. ATM-Mediated Phosphorylation of RNF20 and RNF40 in Response to DNA Damage and Its Requirement for Damage-Induced H2B Monoubiquitylation.
(A) Phosphorylation of S172 of RNF20. HA-tagged RNF20 in wild-type and S172A mutant versions was ectopically expressed in HEK293 cells. The cells were treated for 1 hr with 200 ng/ml of NCS. The fraction of the protein that is phosphorylated in these cells was most clearly detected by immunoprecipitation of the phosphorylated protein with the phosphospecific antibody and subsequent western blotting analysis with an anti-HA antibody. The input (total cell extract) represents 10% of the protein amount that was used for immunoprecipitation. Note lack of phosphorylation of the S172A mutant.
(B) ATM dependence of S172 phosphorylation, demonstrated by similar analysis as in (A), of endogenous RNF20 in wild-type and A-T lymphoblasts. The input (total cell extract) represents 10% of the protein amount that was used for immunoprecipitation.
(C) ATM dependence of phosphorylation of S553 of RNF20. After treatment of human lymphoblasts with 200 ng/ml of NCS for 1 hr, total cellular extracts were blotted with the indicated antibodies.
(D and E) Phosphorylation of S114 of RNF40. The analyses are similar to those in (A) and (B). Ectopic RNF40 is Myc-tagged.
(F) Requirement of ATM phosphorylation sites on RNF20 for damage-induced H2B monoubiquitylation. HA-tagged RNF20 was expressed in wild-type or in S172A/S553A versions by using siRNA-resistant expression constructs in HeLa cells in which endogenous RNF20 had been knocked down by using RNAi. The cells were treated with 1 μg/ml of NCS and H2B monoubiquitylation was monitored at the indicated time points.
Functional Significance of ATM-Mediated Phosphorylation of RNF20-RNF40
A robust method to examine the functional significance of protein phosphorylation entails knocking down the endogenous protein, concomitant expression of ectopic wild-type or non-phosphorylatable versions of the same protein, and measurement of the effect of this manipulation on the cellular phenotype. A technical difficulty of this manipulation of RNF20-RNF40 is the tendency for a large portion of ectopic, overexpressed RNF20-RNF40 to aggregate in inclusion bodies, leaving only a small fraction of the ectopic proteins active. Fine-tuning of the experimental conditions did, however, enable us to detect the activity of free ectopic RNF20-RNF40, which was sufficient for limited but clearly detectable damage-induced H2B monoubiquitylation. Significantly, this activity was not observed with a non-phosphorylatable mutant (S172A/S553A) of RNF20 (Figure 4F). This suggests that the damage-induced ATM-RNF20-RNF40-H2BUb axis is regulated via ATM-mediated phosphorylation of RNF20-RNF40.
RNF20-RNF40 Is Recruited to Damage Sites
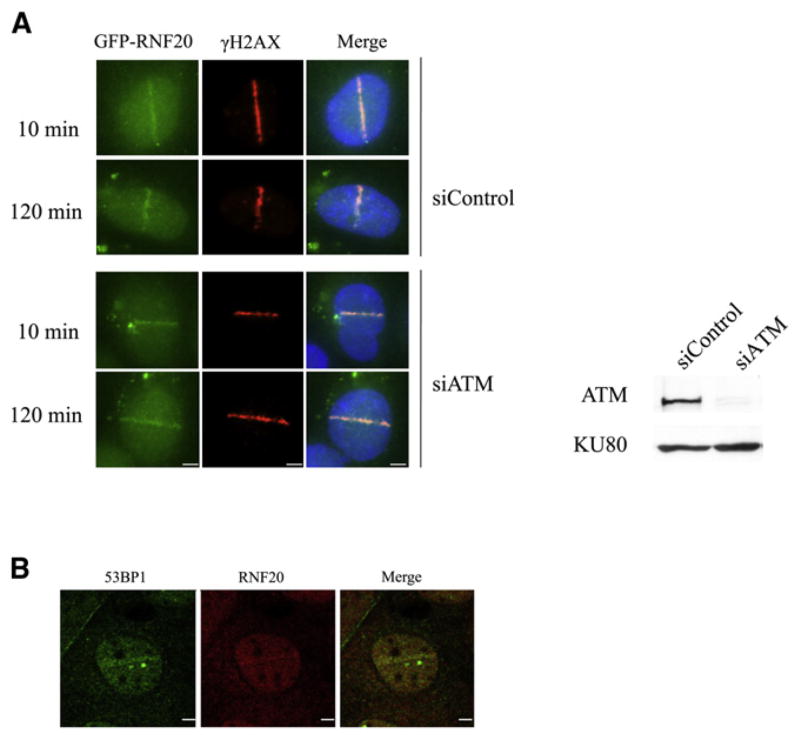
A key question is the location of damage-induced H2BUb. In view of the importance of this modification for DSB repair, it was reasonable to assume that this histone modification occurred at the damaged DNA sites. We asked whether, upon DNA damage, RNF20-RNF40 was recruited to DSB sites. RNF20-RNF40 nuclear foci are not readily observed against the background of total nuclear RNF20-RNF40 after induction of global damage, probably because of the small fraction of cellular RNF20-RNF40 that is involved in damage-induced H2B monoubiquitylation (Figure S4C). We therefore induced spatially localized DSBs in cells by using focused laser microirradiation (Bekker-Jensen et al., 2006). This technique allowed us to detect the recruitment of a small but distinct fraction of ectopic GFP-tagged RNF20 (Figure 5A), as well as endogenous RNF20 (Figure 5B), to the laser-induced DNA damage tracks. Notably, RNF20 recruitment to the damaged sites was ATM independent (Figure 5A).
Figure 5. Recruitment of RNF20 to DSB Sites.
(A) Accumulation of ectopic, GFP-tagged RNF20 at DSBs induced in HeLa cells by focused laser microirradiation. This process is ATM independent. ATM knockdown is shown in the western blot on the right. Scale bar = 5 μm.
(B) Accumulation of a fraction of endogenous RNF20 at laser-induced damage in U2-OS cells 10 min after laser irradiation. The cells were stained with antibodies against RNF20 (red) and the damage sensor 53BP1 (green). Scale bar: 5 μm.
The ATM-RNF20-RNF40-H2BUb Axis Is Required for Recruitment of HRR and NHEJ Proteins
We reasoned that damage-induced H2BUb might be necessary to create the appropriate chromatin microenvironment for DSB repair. We first examined the effect of abrogation of the H2BUb pathway on each of the two major repair mechanisms, NHEJ and HRR. Importantly, we found that the efficiencies of both processes were markedly reduced in RNF20-RNF40-depleted cells (Figures 6A and 6B). This effect could be due to reduced levels of repair proteins which might have resulted from abrogation of transcription because of reduced H2BUb levels. We examined the amount of several DDR and repair proteins in cells depleted for RNF20-RN40, and the results (Figure S5A) did not support this possibility. Importantly, gene expression profiles of RNF20-depleted cells have recently indicated that the expression of genes encoding DSB repair proteins is not affected by RNF20-RNF40 depletion (Shema et al., 2008).
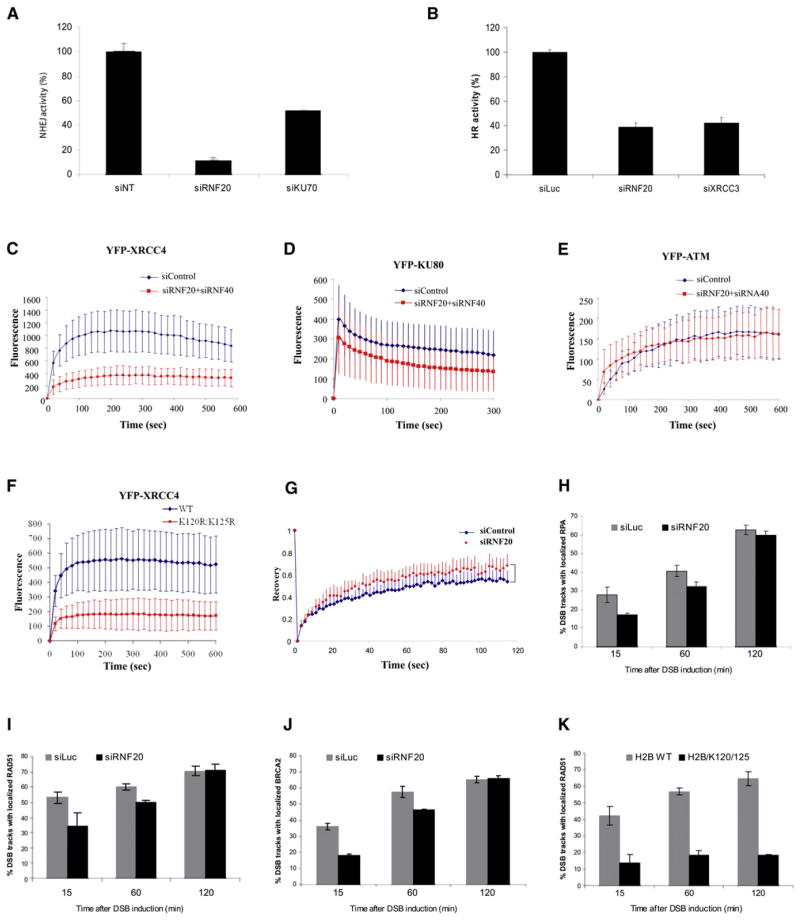
Figure 6. The Role of the RNF20-RNF40-H2BUb Pathway in DSB Repair.
(A) Effect of RNF20-RNF40 depletion on DSB repair via NHEJ. The experimental system is based on cells in which reporter plasmids containing recognition sites of the rare cutter restriction endonuclease I-SceI were incorporated into the cellular genome. The repair of I-SceI-induced DSBs via NHEJ activates expression of GFP which is monitored by using a FACS-based fluorescence readout (Mansour et al., 2008; Schulte-Uentrop et al., 2008). The system was originally developed in Chinese hamster cells and its adaptation to human HeLa cells was used here. Cells depleted of the KU70 protein, a major NHEJ player, served as positive control. Error bars represent the standard error of the mean. The figure represents three independent experiments.
(B) Effect of RNF20-RNF40 depletion in U2-OS cells on DSB repair via HRR. This extensively documented system (Pierce et al., 1999) too is based on following the repair of I-SceI-induced breaks, but is engineered to specifically monitor the HRR pathway. A recent modification of this system (Puget et al., 2005) was used. Knockdown of XRCC3, a central HRR player, served as positive control. Error bars represent the standard error of the mean. The figure represents three independent experiments.
(C) Recruitment of YFP-XRCC4 to DSBs was monitored by using live cell imaging in HeLa cells transfected with control siRNA or with a combination of siRNAs against RNF20 and RNF40. YFP-XRCC4 was transiently expressed in the cells 24 hr after siRNA treatment, and 72 hr later they were microirradiated. Images were obtained at 10 s intervals at the indicated times. To compensate for nonspecific photobleaching, we subtracted the background fluorescence from the accumulation spot. Each data point is the average of ten independent measurements and the experiment was repeated three times. Error bars represent standard deviation. The same statistical analysis was applied to the data in (D)–(F).
(D) Accumulation kinetics of YFP-KU80 at DSB sites.
(E) Recruitment of YFP-ATM to DSBs was examined in an A-T cell line (AT5BIVA) devoid of endogenous ATM, in which YFP-ATM was ectopically expressed. RNF20-RNF40 was depleted in these cells and they were treated and analyzed as described above.
(F) Recruitment of XRCC4 to laser-induced damage in cells expressing ectopic, mCherry-tagged histone H2B in wild-type and mutant (K120R/K125R) versions. Cells positive for RFP fluorescence (i.e., expressing the H2B proteins) were analyzed.
(G) Dynamics of XRCC4 at DSB sites studied by using fluorescence recovery after photobleaching assay (FRAP). HeLa cells were microirradiated 96 hr after transfection with siRNAs and 10 min later, when XRCC4 accumulation at DSB sites reached its maximum, the DSB regions were photobleached. Fifty images were then acquired in 1.5 s intervals. Shown are FRAP curves. Each data point is the average of 18 independent normalized measurements. Error bars represent standard deviation. Prebleach intensity levels were normalized to 1, and postbleach intensity levels were normalized to 0. The data indicate that the immobile XRCC4 fraction at DSBs is reduced in RNF20-RNF40-depleted cells compared to RNF20-RNF40-proficient cells.
(H) Quantification of the accumulation of RPA at DSB sites in the presence or absence of RNF20. HeLa cells were transfected with siRNAs against luciferase (siLuc, control) or RNF20, cultured for 48 hr, and microirradiated with α particles (Stap et al., 2008). At the indicated time points the cells were stained for DNA (DAPI), γH2AX, and RPA34. The columns represent average percentage of cells in which RPA and γH2AX foci were colocalized along DSB tracks. One hundred cells containing DSB tracks were scored per experiment. Error bars represent the range of percentages obtained from three independent experiments.
(I) Similar analysis of RAD51 accumulation at DSB tracks. Statistical analysis is as in (H).
(J) Similar analysis of BRCA2 accumulation at DSB tracks. Statistical analysis is as in (H).
(K) Similar analysis as in (H), carried out in cells expressing wild-type of mutant RFP-tagged histone H2B. Cells showing RFP fluorescence were scored. Fifty to one hundred cells were scored per experiment. Error bars represent the range of percentages obtained from three independent experiments. Note the profound, sustained effect on RAD51 accumulation at damage sites of the mutant H2B.
This general effect on both DSB repair pathways with no effect on repair protein levels suggested that abrogation of damage-induced H2BUb might diminish the access of repair proteins to the damage sites, which would be reflected in their reduced accumulation at these sites. By using live cell imaging of fluorophore-tagged proteins and immunostaining of endogenous proteins, we followed the accumulation of major players in the NHEJ and HRR pathways at localized DNA damage induced by microirradiation. Depletion of RNF20-RNF40 led to reduced accumulation of YFP-tagged XRCC4 and KU80, major components of the NHEJ pathway, at laser-induced damage (Figures 6C and 6D). YFP-ATM, on the other hand, showed normal accumulation at DSB sites in RNF20-RNF40-deficient cells (Figure 6E) in agreement with our previous findings that lack of RNF20-RNF40 affected neither the recruitment of the early damage response proteins nor ATM activation. Significantly, XRCC4 accumulation was markedly affected by the expression of the mutant H2B (Figure 6F), indicating again the dominant-negative effect of the mutated protein on H2B monoubiquitylation. We further studied the dynamics of XRCC4 at DSB sites by using fluorescence recovery after photobleaching (FRAP). When XRCC4 accumulation at DSBs reached its maximum, the DSB regions were photobleached and images were acquired at timed intervals. Both control and RNF20-RNF40-depleted cells showed rapid recovery of fluorescence, demonstrating a dynamic exchange between bound and free XRCC4 at DSB sites (Figure 6G). Even though both cell lines exhibited similar recovery rates (p = 0.79891), there was a significant difference in maximum value of recovery: at 115 s it reached 53.9% of the prebleached intensity in control cells and 68.2% in RNF20-RNF40-depleted cells (p = 0.0018469). That the XRCC4 fraction recruited to DSB sites is more readily exchangeable in RNF20-RNF40-depleted cells indicates RNF20-RNF40 is required for retention of XRCC4 at these sites.
Unlike the NHEJ proteins, recruitment of HRR players to DSBs occurs mainly at the late S and G2 phases of the cell cycle. The HRR mechanism is functional when sister DNA molecules are available for recombination, and the levels of these proteins are low at G1 (Holthausen et al., 2010). This repair pathway is preceded by deep 5′ to 3′ end resection, which creates the long 3′ single-strand overhangs necessary for HRR (Huertas, 2010). We studied the recruitment to damage sites of HRR proteins in HeLa cells, in which normal cell-cycle distribution is maintained after depletion of RNF20-RNF40 (Figure S5B) (Shema et al., 2008). DSB tracks were induced by α-particle irradiation (Stap et al., 2008) or laser microirradiation (Bekker-Jensen et al., 2006). The recruitment to the damage sites of the replication protein A (RPA) complex (Oakley and Patrick, 2010), which indicates that end resection has taken place, and two key HRR proteins—RAD51 and BRCA2—was monitored by following their colocalization with the 53BP1 protein or γH2AX foci along the damage tracks by using immunostaining. In RNF20-RNF40-depleted cells a transient but considerable delay was observed in the recruitment to DSBs of RPA (Figure 6H), BRCA2 (Figure 6J), and RAD51 (Figure 6I and Figures S5C and S5D). Contrary to the transient effect of RNF20-RNF40 depletion on the recruitment of these proteins, expression of the K120R/K125R histone H2B led to a profound, sustained effect on RAD51 recruitment (Figure 6K), indicating again that optimal kinetics of this process indeed require H2B monoubiquitylation. Notably, throughout this study, the dominant-negative effect of the ectopic mutated H2B on the monoubiquitylation of endogenous H2B was stronger than RNF20-RNF40 knockdown, whose efficiency probably varies, while the incorporation into the chromatin of mutant H2B should strictly block H2B monoubiquitylation and consequently, H2BUb-driven processes. It cannot be ruled out, however, that the mutant H2B protein exerts its effect also by inactivating the catalytic activity of RNF20-RNF40.
These results suggest that the H2BUb-driven pathway functions upstream of both major DSB repair pathways. H2B monoubiquitylation is required also for the DNA end resection that precedes the HRR pathway, as suggested by its requirement for RPA recruitment. Collectively, the data place the ATM-RNF20-RNF40-H2BUb axis at a late stage of the early phase of the DSB response after and independent of the recruitment of the early damage sensors and before the entry of the repair machineries into the DDR cascade.
Histone H3 Methylations at Damage Sites
H2B monoubiquitylation at transcribed chromatin is required for di- and trimethylation of Lys4 and Lys79 of histone H3 (reviewed by Smith and Shilatifard, 2010) and H2BUb was shown to stimulate DOT1L, the Lys79 methylase (Chatterjee et al., 2010). We examined the possibility that H2BUb primes H3 methylations on Lys4 and Lys79 at damage sites. Immunoblotting analysis with antibodies against H3K79Me2, H3K79Me3, H3K4Me2, and H3K4Me3 disclosed no damage-induced increase in the amounts of these histone marks (Figure S6A). However, because this analysis might be insensitive to small differences, we used the antibodies against H3K79Me3 and H3K4Me2 in an attempt to observe possible changes in the density of these histone marks along damage tracks induced by microirradiation with a focused laser beam. No such change was observed for H3K4Me2 (Figure S6C), but there was increased staining along the damage tracks corresponding to H3K79Me3. Importantly, however, this increase was not affected by RNF20-RNF40 depletion or overexpression of the dominant-negative mutant H2B (Figures S6B and S6C), suggesting that it was independent of the H2BUb pathway.
DISCUSSION
An ATM-Dependent Pathway Linking DSB Repair and Chromatin Organization
Information about the interfaces between the DDR and chromatin organization on one hand and the DDR and ubiquitin-driven mechanisms on the other hand is rapidly accumulating (Al-Hakim et al., 2010; van Attikum and Gasser, 2009). We describe here an arm of the ATM-mediated DDR that represents a meeting point of the three arenas: it leads to monoubiquitylation of histone H2B by a fraction of RNF20-RNF40 that is recruited to DSB sites and phosphorylated by ATM. This pathway, presumably mediating chromatin decondensation, is required for timely recruitment of NHEJ and HRR proteins and subsequent end resection and optimal DSB repair via both pathways. Abrogation of this pathway leaves a fraction of unrepaired DSBs for extended periods of time. The critical nature of DSBs makes this delay in repair sufficient to cause discernible cellular sensitivity to DSB-inducing agents—a severe phenotype with potential dire consequences at the organismal level. This phenotype flags the importance of this pathway in the vast ATM-mediated network.
The ATM-dependent, H2BUb-driven pathway is added to the previously documented ATM-dependent pathway that facilitates DSB repair via chromatin reorganization, mediated by the ATM substrate KAP-1 (Goodarzi et al., 2008; Ziv et al., 2006). Recent data from our lab indicate that ATM facilitates DSB repair also by directly targeting repair enzymes (H. Segal-Raz and Y.S., unpublished data). Parallel modulation of several pathways that converge at the same biological endpoint is typical of manyATM-mediated processes, presumably allowing fine-tuning of these processes (Shiloh, 2006).
H2B monoubiquitylation was previously implicated in the DDR in the budding yeast (Game and Chernikova, 2009; Game et al., 2006; Giannattasio et al., 2005) and recently depletion of RNF20 or RNF40 (also called Bre1a and Bre1b) was shown to interfere with HRR in human cells (Chernikova et al., 2010). However, the HRR-mediated pathway is responsible for the repair of only a fraction of DSBs in mammalian cells, while the majority of breaks are repaired by NHEJ (Holthausen et al., 2010; Lieber, 2010). Our data show that the H2BUb pathway is important for timely action of wholesale DSB repair and is embedded in the ATM-mediated signaling web.
The DDR is a complex and highly structured process. The precise and strict temporal order of the recruitment of the DDR players to damage sites probably reflects a functional order and hierarchy (Bekker-Jensen and Mailand, 2010; Huen and Chen, 2010). Our data suggest that the H2BUb-driven pathway might be independent of the early damage sensors, but ATM dependent, and activated prior to the entry of the repair machineries into the DDR cascade.
In the transcription context, H2B monoubiquitylation in yeast and mammals is dependent on the early steps in transcription initiation and elongation and the corresponding E3 ligase closely interacts with many proteins that take part in this process, some of which associate directly with RNA Pol II. This ongoing spatio-temporal dynamic reflects the mobility of the corresponding ubiquitylating system on the one hand and a delicate balance between ubiquitylation and deubiquitylation on the other (reviewed by Weake and Workman, 2008). Our data suggest that in the DDR context, H2B monoubiquitylation is confined to the damage sites and is retained at those sites for extended periods of time. Thus, upon DNA damage the H2B ubiquitylating module is probably taken out of its regular context to carry out the same reaction in conjunction with different proteins, most notably ATM. This might explain why we did not observe H2BUb-dependent methylations of Lys4 and Lys79 on histone H3, which are dependent on this modification in the transcription context.
The Mechanism of H2BUb Effect on Chromatin Organization
A major question is how H2BUb facilitates transcription elongation and recruitment of repair proteins to DSB sites. A plausible mechanism would be based on the binding of proteins that contain a ubiquitin-binding domain and assist in these processes. However, because H2BUb is required for two very different DNA transactions—transcription elongation and DSB repair—a unifying mechanism based on local chromatin relaxation as a result of the mere histone ubiquitylation might be attractive. Such a mechanism was recently proposed by Fierz et al., 2011, who showed that just the addition of a ubiquitin moiety to histone H2B is sufficient to interfere with compaction of the 30 nm chromatin fiber and lead to an open, biochemically accessible fiber conformation. This mechanism might explain how H2BUb locally enhances the accessibility of DNA embedded in chromatin to the corresponding enzymes in various DNA transactions including DNA repair.
Differences between the Roles of H2AUb and H2BUb in Chromatin Reorganization
An important example of the involvement in the DDR of another histone mark that is normally associated with gene regulation is histone H2A monoubiquitylation, which was linked to transcription silencing (reviewed by Higashi et al., 2010 and Weake and Workman, 2008). This process was recently implicated in facilitating DSB repair and was associated with three DDR players, the RING finger proteins RNF8 and RNF168 and the HERC2 protein (reviewed by Al-Hakim et al., 2010). It was suggested that in mouse embryo fibroblasts RNF8 might be involved in histone H2B monoubiquitylation as well (Wu et al., 2009); our data indicate, however, that in human cells RNF20-RNF40 is the major E3 ligase in DSB-induced H2BUb, as in transcription-associated H2BUb. Interestingly, unlike with H2BUb, the H2AUb ubiquitin ligases that function in the gene regulation context (Vidal, 2009) were not implicated in DNA damage-induced H2A monoubiquitylation. There are additional differences in the appearance of the two apparently analogous histone marks in DNA damage sites, such as the dependence of the RNF8-mediated pathway on MDC1, and its requirement for 53BP1 recruitment, contrary to what we see in damage-induced H2BUb. Importantly, H2AUb was recently shown to be involved in ATM-mediated transcription silencing and prevention of RNA Pol II elongation-dependent chromatin decondensation at regions distal to DSBs (Shanbhag et al., 2010). Thus, the two histone monoubiquitylations play opposing roles in the dynamics of chromatin organization, H2BUb in the immediate vicinity of DSBs and H2AUb further away from these lesions. Indeed, despite the apparent analogy between the two modifications, H2AUb, at the opposing side of the nucleosomal surface, does not lead to chromatin fiber decompaction (Fierz et al., 2011).
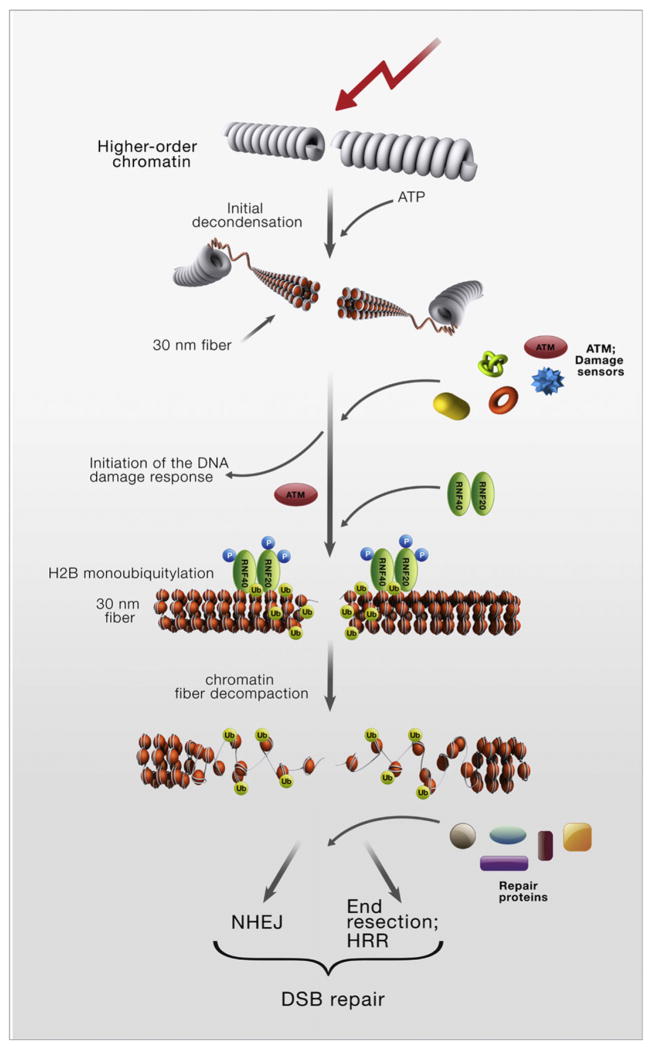
A Two-Stage Model for Chromatin Relaxation at DSB Sites
Nussenzweig and colleagues previously showed an initial, very rapid chromatin relaxation at DSB sites that was ATP dependent and ATM independent and was required for the recruitment of the damage sensors (Kruhlak et al., 2006). Our data suggest that the ATM-RNF20-RNF40-H2BUb pathway functions independently and later and is required for the actual DSB repair process. The role of H2B monoubiquitylation in the decompaction of the basic chromatin fiber (Fierz et al., 2011) fits the requirement of this modification for the repair process, which takes place on exposed DNA. Collectively, the data suggest a model of chromatin relaxation at DSB sites as a two-stage process consisting of a rapid, ATM-independent chromatin de-condensation that facilitates the recruitment of sensors and an ATM-dependent stage at the level of the 30 nm chromatin fiber, which is mediated by ATM’s substrate, RNF20-RNF40 (Figure 7). This ubiquitin ligase is temporarily called into action for the DDR, outside its normal working environment: transcribed chromatin. Further investigation of the role of histone PTMs in shaping chromatin organization in response to DNA damage will reveal the fine structural aspects of the DNA damage response.
Figure 7.
A Scheme Depicting a Two-Stage Model of DNA Damage-Induced Chromatin Relaxation at DSB Sites
EXPERIMENTAL PROCEDURES
Expression Constructs
Full-length cDNA clones of RNF20, RNF40, and histone H2B were obtained from the I.M.A.G.E. Consortium and subcloned into pcDNA3.0 (RNF20) and pcDNA4.0 (RNF40) vectors. HA-tag was added at the amino terminus of RNF20 by using PCR and appropriate adaptor primers. Site-directed mutagenesis was carried out by using the QuikChange Site-Directed Mutagenesis System (Stratagene, La Jolla, CA). Wild-type and mutant (K120R/K125R) H2B open reading frames previously cloned in pcDNA3.1 (Minsky et al., 2008) were excised from the vector by using BamH1 and Xba1 and subcloned in a pMCherry-C1 vector by using recognition sites of the same restriction enzymes. The expressed protein was fused to red fluorescent protein (RFP) at its amino terminus.
RNA Interference
siRNA oligonucleotides (siGenome OnTarget Plus pools) were obtained from Thermo Scientific (Dharmacon RNAi Technologies, Lafayette, CO). The following siRNA sequence served for construction of shRNA against RNF20: CCAAUGAAAUCAAGUCUAAUU.
DNA Damage Responses
Clonogenic survival of cell lines, immunoblotting, immunostaining, and the ATM in vitro kinase assay were carried out as previously described (Ziv et al., 2006). Microscopic photography equipment and data acquisition were described in Ziv et al., 2006.
Comet Assay
MCF7 cells were transfected with siRNAs against RNF20 or RNF40, or both, and irradiated 48 hr later with 10 Gy of IR. After 4 hr, a neutral comet assay was performed as described before (Tsai et al., 2008). Briefly, cells were harvested and mixed with low-melting agarose. After lysis, electrophoresis was performed at 1 V/cm and 15 mA for 40 min. After staining the slides with SYBG Green dye for 10 min, images of 100 randomly selected cells per sample were captured by using a Zeiss fluorescent microscope and digital fluorescent images were obtained by using the AxioVision software. In this assay, the relative length and intensity of DNA tails relative to heads is proportional to the amount of DNA damage in individual nuclei. These parameters were measured by Olive tail moment with TriTek Comet Score software (TriTek Corp., Sumerduck, VA).
NHEJ Assay
HeLa cells with the plasmid pEJSSA (Mansour et al., 2008) stably integrated into their genome were transfected with siRNAs targeting RNF20 or KU70 and/or with nontargeting siRNA, and 36 hr later were transfected with an I-SceI-expressing vector or an empty vector. Parallel samples were transfected with a GFP-expressing plasmid to control for transfection efficiencies. Thirty-six hours after the second transfection, cells were harvested and suspended in phosphate-buffered saline supplemented with 10% fetal bovine serum, and the fraction of GFP-positive cells was determined by cell sorting with FACSort flow cytometer (Becton Dickinson, Franklin Lakes, NJ) at 50,000 events/sample. The results were processed with the Cyflogic data analysis software (http://www.cyflogic.com/).
HRR Assay
Site-specific recombination after DSB induction by I-SceI was measured as described (Puget et al., 2005). An siRNA against the HRR player XRCC3 was used as a control.
Recruitment and Accumulation of DNA Damage Response Proteins at DSBs
Live cell imaging combined with laser microirradiation was described previously (Uematsu et al., 2007; Yano et al., 2008). Briefly, fluorescence data were acquired with an Axiovert 200 M microscope (Carl Zeiss MicroImaging, Thornwood, NY). A 365 nm pulsed nitrogen laser (Spectra-Physics, Mountain View, CA) was directly coupled to the epifluorescence path of the microscope. DSBs were introduced in the nucleus by microirradiation with 365 nm laser. Time-lapse images were taken with an AxioCam HRm camera and fluorescence intensities of microirradiated areas were determined by using Axiovision Software, version 4.5 (Carl Zeiss). During microirradiation and time-lapse imaging, cells were maintained in CO2-free medium (Invitrogen) at 37°C. To compensate for nonspecific photobleaching, we subtracted the background fluorescence from the values recorded at the accumulation spots. When DSBs were induced by using α particles, irradiation was carried out as previously described (Stap et al., 2008). Briefly, HeLa cells were transfected with siRNA targeting luciferase mRNA (CGUACGCGGAAUACUUCGAdTdT) or with RNF20 and 24 hr later cells were plated on a 1.8-μm-thick polyester membrane and transfection was repeated. After irradiation with α particles, cells were fixed and stained for immunofluorescence as previously described (Aten et al., 2004).
FRAP Analysis
The analysis was carried out by using an LSM 510 Meta confocal microscope as described before (Uematsu et al., 2007). Briefly, cells expressing YFP- XRCC4 were maintained in CO2-free medium (Invitrogen) at 37°C and micro-irradiated with the 365 nm laser as described above. Ten minutes later, when YFP-XRCC4 accumulation reached its maximum, the accumulated spots were photobleached with a 514 nm argon laser. Subsequently, 20 images were acquired at 1.5 s intervals. To compensate for nonspecific photo-bleaching, we subtracted the background fluorescence from the accumulation spot. Normalized intensity (NI) was calculated according to the formula: NI (t) = It/Ipre, where Ipre represents the prebleach value of the intensity light. The normalized curves were plotted by using SigmaPlot and recovery curves were fitted to a single-order exponential equation, Y = Ymax(1−exp(−KX), in order to obtain the maximum recovery (Ymax) and half-time (t1/2) values (Schmiedeberg et al., 2004).
Western Blotting and Immunofluorescence Analysis
These procedures were carried out as previously described (Ziv et al., 2006) by using the antibodies listed in the Supplemental Experimental Procedures.
Coimmunoprecipitation
Coimmunoprecipitation of ectopic or endogenous RNF20-RNF40 with ATM was carried out by using HEK293 cells or lymphoblasts, respectively. Cell pellets were suspended in 120 mM NaCl, 50 mM TRIS (pH 7.5), 2 mM EDTA, 10% glycerol, and 1% NP40 and left on ice for 30 min. After centrifugation the supernatant was mixed with GFP-TRAP conjugated beads (ChromoTek, Martinsried, Germany) to precipitate YFP-ATM or anti-RNF40 polyclonal antibody to precipitate endogenous RNF20-RNF40 and the mixture was left at slow shaking for 16 hr at 4°C. RNF20-RNF40 immune complexes were then mixed with protein A-preblocked sepharose beads for 1 hr at 4°C. The beads were washed five times and resuspended in loading buffer, the suspension was boiled for 10 min and centrifuged, and the supernatant underwent SDS-PAGE.
Supplementary Material
supplementary data
Acknowledgments
We thank Ruth Shiloh for critical advice, Yaron Pereg and Rami Khosravi for experimental contributions and advice, Yael Ziv for useful discussions and comments on the manuscript, J. Lukas for cell lines stably expressing GFP-tagged RNF8 and MDC1, M. Lavin for the C3ABR cell line, M. Huen for anti-RNF8 antibody, N. Mailand for the anti-RNF8 antibody and siRNA, M. Goldberg for anti-MDC1 antibody and siRNA, and T. Halazonetis for the α-53BP1 antibody. This work was supported by research grants from The Israel Science Foundation, the A-T Medical Research Foundation, The A-T Children’s Project and the Israel Cancer Research Fund (to Y.S.), a core grant from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to Y.S., A.C., and M.O.), grants from The National Institutes of Health (CA50519 and PO1-CA92584, to D.J.C), the National Cancer Institute (R01 CA113859 to M.C.-T.H. and R37 CA40099 to M.O.), the Netherlands Genomic Initiative/Netherlands Organization for Scientific Research (NWO), Chemical Sciences TOP, and the Dutch Cancer Society (KWF) (to R.K.), and a grant from the German Federal Ministry of Education and Research (BMBF 02S8427) to J.D.-D. Y.S. is a Research Professor of the Israel Cancer Research Fund.
Footnotes
Supplemental Information includes six figures and Supplemental Experimental Procedures and can be found with this article online at doi:10.1016/j.molcel. 2011.02.015.
References
- Al-Hakim A, Escribano-Diaz C, Landry MC, O’Donnell L, Panier S, Szilard RK, Durocher D. The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 2010;9:1229–1240. doi: 10.1016/j.dnarep.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Mailand N. Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228. doi: 10.1016/j.dnarep.2010.09.010. [DOI] [PubMed] [Google Scholar]
- Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J Cell Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat Chem Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- Chernikova SB, Dorth JA, Razorenova OV, Game JC, Brown JM. Deficiency in Bre1 impairs homologous recombination repair and cell cycle checkpoint response to radiation damage in mammalian cells. Radiat Res. 2010;174:558–565. doi: 10.1667/RR2184.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derheimer FA, Kastan MB. Multiple roles of ATM in monitoring and maintaining DNA integrity. FEBS Lett. 2010;584:3675–3681. doi: 10.1016/j.febslet.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan A, Bajpayee M, Parmar D. Comet assay: a reliable tool for the assessment of DNA damage in different models. Cell Biol Toxicol. 2009;25:5–32. doi: 10.1007/s10565-008-9072-z. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald JE, Grenon M, Lowndes NF. 53BP1: function and mechanisms of focal recruitment. Biochem Soc Trans. 2009;37:897–904. doi: 10.1042/BST0370897. [DOI] [PubMed] [Google Scholar]
- Game JC, Chernikova SB. The role of RAD6 in recombinational repair, checkpoints and meiosis via histone modification. DNA Repair (Amst) 2009;8:470–482. doi: 10.1016/j.dnarep.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Game JC, Williamson MS, Spicakova T, Brown JM. The RAD6/BRE1 histone modification pathway in Saccharomyces confers radiation resistance through a RAD51-dependent process that is independent of RAD18. Genetics. 2006;173:1951–1968. doi: 10.1534/genetics.106.057794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. The DNA damage checkpoint response requires histone H2B ubiquitination by Rad6-Bre1 and H3 methylation by Dot1. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Löbrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Higashi M, Inoue S, Ito T. Core histone H2A ubiquitylation and transcriptional regulation. Exp Cell Res. 2010;316:2707–2712. doi: 10.1016/j.yexcr.2010.05.028. [DOI] [PubMed] [Google Scholar]
- Hiom K. Coping with DNA double strand breaks. DNA Repair (Amst) 2010;9:1256–1263. doi: 10.1016/j.dnarep.2010.09.018. [DOI] [PubMed] [Google Scholar]
- Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst) 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Huen MS, Chen J. Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci. 2010;35:101–108. doi: 10.1016/j.tibs.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Müller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löbrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- Mansour WY, Schumacher S, Rosskopf R, Rhein T, Schmidt-Petersen F, Gatzemeier F, Haag F, Borgmann K, Willers H, Dahm-Daphi J. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 2008;36:4088–4098. doi: 10.1093/nar/gkn347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- Oakley GG, Patrick SM. Replication protein A: directing traffic at the intersection of replication and repair. Front Biosci. 2010;15:883–900. doi: 10.2741/3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puget N, Knowlton M, Scully R. Molecular analysis of sister chromatid recombination in mammalian cells. DNA Repair (Amst) 2005;4:149–161. doi: 10.1016/j.dnarep.2004.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riballo E, Kühne M, Rief N, Doherty A, Smith GC, Recio MJ, Reis C, Dahm K, Fricke A, Krempler A, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–724. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Schmiedeberg L, Weisshart K, Diekmann S, Meyer Zu Hoerste G, Hemmerich P. High- and low-mobility populations of HP1 in heterochromatin of mammalian cells. Mol Biol Cell. 2004;15:2819–2833. doi: 10.1091/mbc.E03-11-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell. 2005;19:849–856. doi: 10.1016/j.molcel.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Schulte-Uentrop L, El-Awady RA, Schliecker L, Willers H, Dahm-Daphi J. Distinct roles of XRCC4 and Ku80 in non-homologous end-joining of endonuclease- and ionizing radiation-induced DNA double-strand breaks. Nucleic Acids Res. 2008;36:2561–2569. doi: 10.1093/nar/gkn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, Raver-Shapira N, Minsky N, Pirngruber J, Tarcic G, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes Dev. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402–410. doi: 10.1016/j.tibs.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. The chromatin signaling pathway: diverse mechanisms of recruitment of histone-modifying enzymes and varied biological outcomes. Mol Cell. 2010;40:689–701. doi: 10.1016/j.molcel.2010.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stap J, Krawczyk PM, Van Oven CH, Barendsen GW, Essers J, Kanaar R, Aten JA. Induction of linear tracks of DNA double-strand breaks by alpha-particle irradiation of cells. Nat Methods. 2008;5:261–266. doi: 10.1038/nmeth.f.206. [DOI] [PubMed] [Google Scholar]
- Tsai WB, Chung YM, Takahashi Y, Xu Z, Hu MC. Functional interaction between FOXO3a and ATM regulates DNA damage response. Nat Cell Biol. 2008;10:460–467. doi: 10.1038/ncb1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu N, Weterings E, Yano K, Morotomi-Yano K, Jakob B, Taucher-Scholz G, Mari PO, van Gent DC, Chen BP, Chen DJ. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J Cell Biol. 2007;177:219–229. doi: 10.1083/jcb.200608077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–217. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Vidal M. Role of polycomb proteins Ring1A and Ring1B in the epigenetic regulation of gene expression. Int J Dev Biol. 2009;53:355–370. doi: 10.1387/ijdb.082690mv. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol Cell Biol. 2009;29:849–860. doi: 10.1128/MCB.01302-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano K, Morotomi-Yano K, Wang SY, Uematsu N, Lee KJ, Asaithamby A, Weterings E, Chen DJ. Ku recruits XLF to DNA double-strand breaks. EMBO Rep. 2008;9:91–96. doi: 10.1038/sj.embor.7401137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary data