A MatP–divisome interaction coordinates chromosome segregation with cell division in E. coli (original) (raw)
Abstract
Initiation of chromosome segregation in bacteria is achieved by proteins acting near the origin of replication. Here, we report that the precise choreography of the terminus region of the Escherichia coli chromosome is also tightly controlled. The segregation of the terminus (Ter) macrodomain (MD) involves the structuring factor MatP. We characterized that migration of the Ter MD from the new pole to mid-cell and its subsequent persistent localization at mid-cell relies on several processes. First, the replication of the Ter DNA is concomitant with its recruitment from the new pole to mid-cell in a sequential order correlated with the position on the genetic map. Second, using a strain carrying a linear chromosome with the Ter MD split in two parts, we show that replisomes are repositioned at mid-cell when replication of the Ter occurs. Third, we demonstrate that anchoring the Ter MD at mid-cell depends on the specific interaction of MatP with the division apparatus-associated protein ZapB. Our results reveal how segregation of the Ter MD is integrated in the cell-cycle control.
Keywords: divisome, macrodomain, MatP, replication factory, ZapB
Introduction
The development of fluorescence microscopy together with the advances of genetical and biochemical investigations has considerably improved our understanding of cellular organization. Studies performed in different bacterial models have revealed how cell morphology is controlled by cytoskeletal structures and how cell division is spatially orchestrated and coordinated within the cell cycle (for review see Shapiro et al, 2009). In Escherichia coli, cell division involves septation at mid-cell. The divisome consists of a complex machinery (>10 proteins) assembled around the FtsZ ring that is used as a scaffold (Adams and Errington, 2009). The assembly of the FtsZ ring is a key step of the cell cycle targeted by several systems that monitor the integrity and segregation of the chromosome. One of these processes, called nucleoid occlusion (Woldringh et al, 1991; Wu and Errington, 2004; Bernhardt and de Boer, 2005), inhibits FtsZ ring assembly if a certain level of chromosome segregation is not achieved (Cho et al, 2011; Tonthat et al, 2011). The accurate distribution of the sister chromatids into each daughter cell imposes a necessary synchronization of chromosome segregation and division processes. Because of the control of cell division and chromosome segregation, optimal nucleoid folding is essential to avoid interference with division ring assembly (Sawitzke and Austin, 2000).
Nucleoid folding and compaction relies on several processes: unrestrained DNA supercoiling, formation of a chromatin-like structure through the interaction of nucleoid-associated proteins with DNA, DNA condensation by structural maintenance of chromosome (SMC)-like proteins, and macromolecular crowding (Thanbichler et al, 2005). Because replichores tend to be localized into distinct compartments of cell, it is proposed that E. coli chromosome presents a transversal subcellular organization (Nielsen et al, 2006; Wang et al, 2006). This replichore localization bias is generated by the non-random segregation of the leading and lagging strands, respectively, towards the old and the new pole of the cell (White et al, 2008). The chromosome is organized into large regions called macrodomains (MDs) and non-structured (NS) regions (Niki et al, 2000; Valens et al, 2004). MDs have been defined as large regions in which DNA interactions occurred preferentially and DNA interactions between the different MDs are highly restricted; in NS regions, DNA sites can interact with both flanking MDs (Valens et al, 2004). The E. coli chromosome contains four MDs: the Ori MD contains oriC while the opposite Ter MD contains the replication terminus and the chromosome dimer resolution dif site. The Ter MD is flanked by the left and right MDs, whereas the Ori MD is flanked by the two NS (NSRight and NSLeft) regions. Each MD occupies a specific territory inside the nucleoid and is segregated with specific properties (Espeli et al, 2008). The binding of the MatP protein to 23 matS sites distributed in the 800-kb Ter region promotes the structuring of the Ter MD; this interaction provokes the condensation of the Ter region in a small territory. The MatP focus forms the centre of the Ter MD territory. The binding of MatP to matS sites constrains DNA mobility of markers in the Ter region. Inactivation of MatP results in severe segregation defects at fast growth rates; anucleated and filamentous cells with unsegregated nucleoids are observed. As a consequence of the chromosome segregation defects noticed in the matP mutants, alterations of the division process are observed (Mercier et al, 2008).
The understanding of bacterial chromosome segregation has improved considerably in recent years. In several species, parABS partition systems are involved in driving the segregation process (reviewed by Possoz et al, 2012). In E. coli, no such mitotic apparatus has been identified and several proposals have been put forth, among them extrusion of the DNA by the replication machinery, _migS_-driven segregation or entropic exclusion of sister chromosomes (reviewed by Toro and Shapiro, 2010). In Bacillus subtilis, the replication itself has been proposed to contribute to the segregation process. In the capture extrusion model, a static replication machinery (called the DNA factory) is involved in the capture of chromosomes at mid-cell for replication; thereafter, newly replicated sister chromatids would be segregated into the future daughter cells (Lemon and Grossman, 2001). The recent observations in E. coli that replisomes are actually mobile in the cell suggested that the replication machinery followed the chromosome rather than constraining its localization (Bates and Kleckner, 2005; Reyes-Lamothe et al, 2008a). This observation made the capture extrusion model less likely in E. coli.
The segregation of the Ter region of the E. coli chromosome has been described in several studies. The frequent localization of any FROS tagging the Ter MD or of the MatP focus at mid-cell has been repeatedly observed (Li et al, 2002; Bates and Kleckner, 2005; Adachi et al, 2008; Espeli et al, 2008; Mercier et al, 2008). The localization pattern of the Ter MD is lost in the absence of MatP (Mercier et al, 2008). The choreography of the Ter MD in the E. coli wt strain is tightly controlled and includes three steps: (i) migration from the pole to the mid-cell in the new-born cells; (ii) a prolonged localization at mid-cell and finally (iii) duplication of the sister Ter MD foci around mid-cell a short time before septation.
In this study, we analysed the role played by the division apparatus and the replication machinery in the segregation of the Ter region. Simultaneous observation of the assembly of the septation ring and migration of MatP, allowed us to analyse how MatP interacts with cellular components at mid-cell. A new assay indicated the extended localization of MatP at mid-cell, and tests of the implication of divisome proteins in this localization revealed that MatP interacts directly with the early septal protein ZapB. The anchoring of MatP to ZapB contributes to the maintenance of the Ter MD in a condensed territory. By using a strain carrying a linearized chromosome with the Ter MD split in two parts (Cui et al, 2007), we have shown that MatP associated to ZapB can hold together the two halves of the Ter MD even if they are physically separated. Finally, we have shown that MatP is not responsible for the migration of the Ter MD from the pole to the mid-cell. Instead, we present strong evidence that Ter DNA is recruited to mid-cell during the process of replication with replisomes localized at mid-cell.
Results
Localization pattern of MatP
We previously reported that, during a persistent step of the cell cycle, the Ter MD is observed at mid-cell (Espeli et al, 2008). Time-lapse experiments revealed that during a cell cycle with overlapping replication and division, new-born cells presented one MatP focus localized near the new pole and, as cells progressed in the cell cycle, the MatP focus was found to mid-cell where it remained for a long period. A few minutes before division, the MatP focus duplicates into two and the two sister foci remain juxtaposed for a few minutes before moving away from mid-cell (Mercier et al, 2008). We have constructed a MatP–mCherry in order to refine this pattern of localization (Figure 1A–C). As observed previously for MatP–GFP, every cell presented at least one MatP focus; new-born cells displayed one MatP–mCherry focus near the pole of the cell, larger cells presented one MatP–mCherry focus localized at mid-cell, and two separated foci were observed in the largest cells before division (Figure 1A–C). About 80% of the cells presented one MatP focus at mid-cell (Figure 1C), confirming that the Ter MD remains localized at mid-cell for a large part of the cell cycle. Because of a greater signal-to-noise ratio for the MatP–mCherry fusion compared with the MatP–GFP, we were able to observe a significant number (≈5–8% of the population) of small cells with two MatP–mCherry foci. This corresponds to cells presenting one focus near the pole and one focus near mid-cell. Usually the two foci were not of equal intensity (Figure 1A, arrow; Supplementary Figure S1). We measured the fluorescence intensity in each focus in 26 cells and observed an inverse relationship between the amount of fluorescence in the polar foci and the central foci (Supplementary Figure S1).
Figure 1.
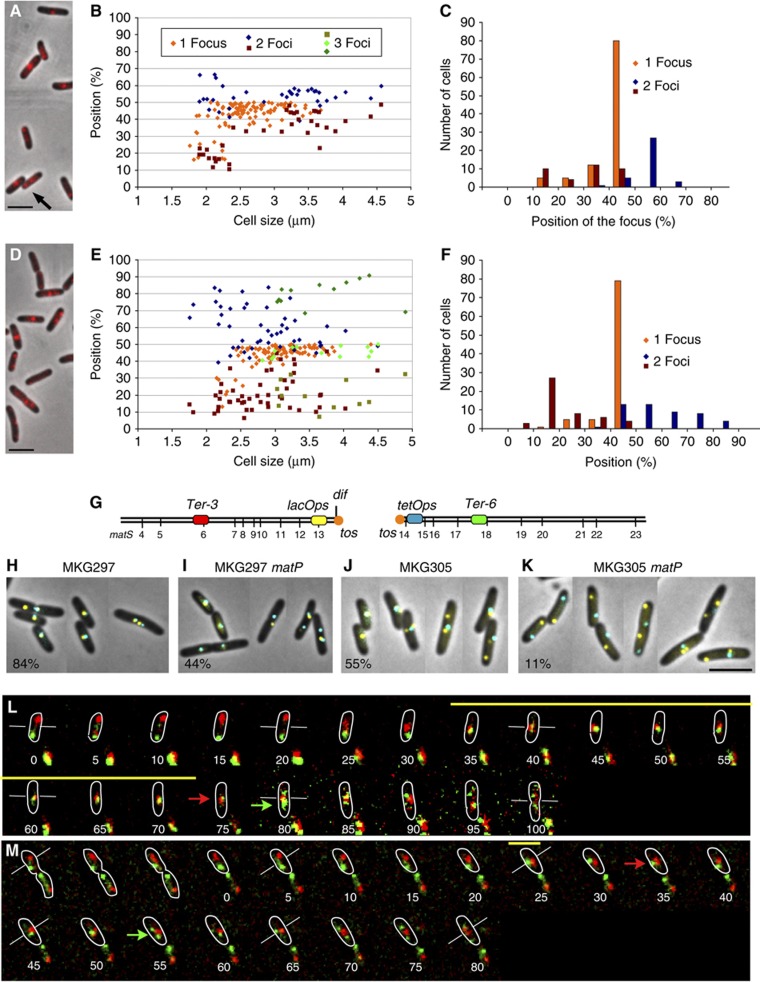
MatP holds the Ter MD at mid-cell. (A) Merged picture of MatP–mCherry (red) and phase-contrast micrographs (grey) of MG1655 cells with a circular chromosome. (B) Histogram of the relative positions from the pole of the cell of MatP–mCherry foci observed in (A) according to cell size. Cells with one focus or two foci are indicated. (C) Histogram representing the number of cells with 1, 2 or 3 MatP–mCherry foci observed in (A) according to the position of the focus. (D) Merge of MatP–mCherry (red) and phase-contrast micrographs (grey) in MG59 cells with a linearized chromosome. Cells with one focus or two foci are indicated. (E) Histogram of the relative positions from the pole of the cell of the MatP–mCherry foci according to cell size. (F) Histogram representing the number of cells with 1, 2 or 3 MatP–mCherry foci according to the position of the focus. (G) Map of the Ter MD with the position of the matS sites and the FROS tags used in the following panels. (H–K) localization of the lacOps and tetOps FROS tags in strains with circular or linear chromosome. (H) wt MKG297 strain. (I) MKG297_matP_. (J) MKG305 linear chromosome. (K) MKG305_matP._ The percentage of cells presenting at least one lacOps and one tetOps colocalized at mid-cell is indicated. (L) Merged pictures of a time-lapse experiment with 5 min intervals for the segregation of the Ter-3 (red) and Ter-6 (green) loci in strain MG59 (linear chromosome). Because photobleaching grey levels in the montage were not homogeneously set, starting at the time point 75 min a narrow grey scale was used for both fluorescence channels. The yellow bar represents the extent of colocalization of the red and green foci, the white bars tag the mid-cell for specific stages, the red and green arrows represent, respectively, the duplication of the Ter-3 and Ter-6 focus. Outlines of the cells were drawn according to phase-contrast images. The red and green foci at the bottom of each image correspond to foci from adjacent cells. (M) Merged pictures of a time-lapse experiment with 5 min intervals for the segregation of the Ter-3 (red) and Ter-6 (green) loci in strain MG59_matP_ (linear chromosome). Same representation as in (L). Scale bars represent 3 μm.
MatP is required to maintain the Ter MD at mid-cell
To evaluate the potential of MatP to organize and target the Ter MD at mid-cell, we analysed its ability to gather the two separated halves of the Ter MD split apart. To do this, we used a strain where the Ter region was split into two parts by linearization in the middle of the chromosome near the dimer resolution dif site. This strain was constructed using the linearization module of the N15 prophage (Cui et al, 2007). MatP–mCherry formed foci in the cells with a linear chromosome (Figure 1D–F). The new-born cells frequently displayed two foci close to each pole and larger cells presented one focus at mid-cell. More than 60% of the cells displayed a single MatP–mCherry focus at mid-cell. The central MatP–mCherry was sometimes flanked by one or two polar foci. Altogether, these results suggested that the two halves of the Ter MDs merged at mid-cell at a defined step of the cell cycle, mostly in cells of median size (Figure 1C). To rule out the possibility that MatP–mCherry would only tag one of the chromosome ends, we used FROS tags (lacO and tetO) (Lau et al, 2003) to simultaneously observe both ends of the linear chromosome (Figure 1G–K; Supplementary Figure S2). We observed a frequent colocalization of lacOps and tetOps at mid-cell in wt cells with a circular chromosome (84%), in matP cells with a circular chromosome (44%) and in wt cells with a linear chromosome (55%). However, in the absence of MatP only a small percentage of cells with a linear chromosome (11%) presented a mid-cell colocalization of both operators. These results revealed that MatP has the ability to maintain together the two separated halves of the Ter MD.
To follow the dynamics of chromosomal tags inserted in the Ter region of a linear chromosome in the presence and absence of MatP, we performed time-lapse experiment using parS/ParB-P1 (_parS_P1) and parS/ParB-pMT1 (parS pMT1) FROS (Nielsen et al, 2006). E. coli grown in minimal medium supplemented with casamino acids and glucose is characterized by overlapping replication and division cycles (Espeli et al, 2008). Time-lapse experiments were performed over a complete cell cycle (120 min) as described before (Espeli et al, 2008; Mercier et al, 2008). The Ter-3 (parS P1) and Ter-6 (parS pMT1) tags were inserted 200 kb from the tos site on each chromosome arm of a linear chromosome (Figure 1G). The montage presented in Figure 1L and M and Supplementary Figure S3 are representative of the population observed. In the new-born matP+ cells (Figure 1L), one red (Ter-3) and one green (Ter-6) focus were distant and localized close to each pole. About 30 min after birth, the foci moved to mid-cell, where they remained colocalized for 40–50 min. After this colocalization period, each focus duplicated, and the colocalization of the chromosome ends did not persist. In the absence of MatP (Figure 1M), the first part of the cell cycle was similar to that observed in MatP+ cells. The tagged chromosome arms were localized to distinct cell halves in the new-born cells. As in the wt cells, ∼30 min after birth, both tags moved to mid-cell. However, the two foci rapidly (<10 min of colocalization) separated at mid-cell and were promptly separated from each other (Figure 1M; Supplementary Figure S3). Our results indicated that Ter MD migration to mid-cell is independent of MatP, whereas MatP is required for an extended colocalization of chromosome ends at mid-cell.
The termini regions of linear chromosomes are replicated at mid-cell
The migration of the two ends of the linear chromosome to mid-cell occurs 30–40 min after birth (Figure 1L and M), a period that corresponds to the estimated timing of replication of the loci under consideration (Espeli et al, 2008). To analyse more precisely the timing of the Ter MD migration to mid-cell during the cell cycle, we followed the dynamics of the replisomes together with that of chromosomal markers. First, using a ssb–ypet fusion, we analysed the dynamics of the replisomes in strains with a circular or a linear chromosome (Figure 2A). In strain with circular chromosome, new-born cells were finishing the replication rounds initiated in the mother cell; they presented two SSB–YPet foci corresponding to the forks that replicated the right and left replichores. Later, the two foci merged at mid-cell. We assumed that merging corresponded to the replication of the terminus region. After a brief period without replication (cells lacking SSB–YPet focus), the replication was initiated simultaneously on two chromosomes (large cells with two SSB–YPet foci). A few minutes before division, the replication forks separated, giving rise to cells with 4 SSB–Ypet foci (Figure 2A; Supplementary Figure S4A). Remarkably, this pattern is maintained with little modification in the cells with a linear chromosome (Figure 2A; Supplementary Figure S4A). Moreover, MatP plays no role in the replisome choreography since similar values were obtained during replication of either circular or a linear chromosome, in both wt and matP backgrounds. The most remarkable result was the observation, with a linear or a circular chromosome, of a single SSB–YPet focus near mid-cell at the predicted moment of replication of the two termini (Figure 2A).
Figure 2.
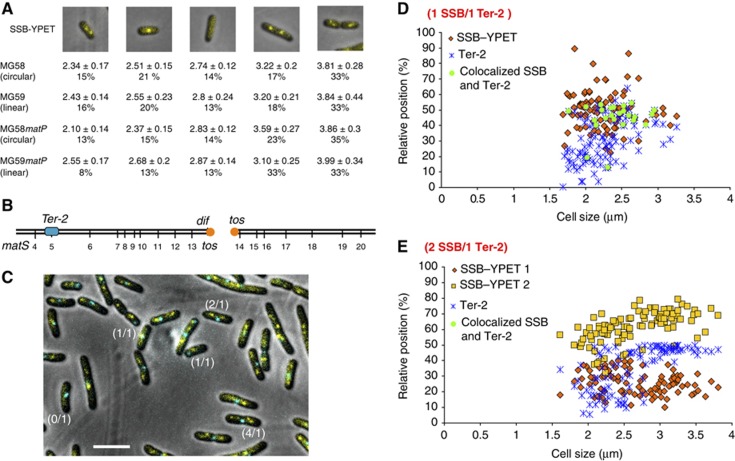
The Ter MD is replicated at mid-cell in strains with circular or linear chromosome. (A) Localization of the SSB–YPet fusion in MG58, MG58_matP_, MG59 and MG59_matP_ strains. Average size (mm) and the percentage of cells in each category are indicated. (B) Position of the Ter-2 (parS P1) tag inserted in the chromosome of the MG59 strain (MG1655 with a linear chromosome). (C) Localization of SSB–Ypet and the of Ter-2 parS P1 tag in the MG59 cells. Examples of cells with 0 SSB focus and 1 Ter-2 focus (0/1), 1 SSB and 1 Ter-2 (1/1), 2 SSB and 1 Ter-2 (2/1) and 4 SSB and 1 Ter-2 (4/1) are highlighted. The cells were grown to O.D.=0.2 in liquid medium, spread on agar pads and observed immediately. Scale bar represents 3 μm. (D) Histogram of the localization of SSB–YPet and the Ter-2 tag in MG59 cells containing one focus for each. (E) Histogram of the localization of SSB–YPet and the Ter-2 tag in MG59 cells containing one focus of Ter-2 and two SSB–Ypet foci. Scale bars represent 3 μm.
Second, we simultaneously followed the dynamics of the replisomes with that of chromosomal Ter markers. We tagged a locus in the terminus region of a linear chromosome (Ter-2 parS P1; Figure 2B) in a strain expressing a ssb–ypet fusion (Figure 2B–E; Supplementary Figure S4B and C). On a linear chromosome, replication is forced to finish at the tos site, ensuring that replication of the Ter-2 site is achieved by the rightward replication fork 250 kb before it ends. As expected, we observed cells with 0, 1, 2 and 4 SSB–Ypet foci and cells with 1 or 2 Ter-2 foci (Figure 2C). At the moment of its replication, the Ter-2 locus must be colocalized with an SSB focus (interfocal distance<100 nm). These events were rare (≈3% of the population) and solely observed in the context of cells presenting 1 Ter focus and 1 SSB–YPet focus (Figure 2C and D). Strikingly, none of the cells with two SSB–YPet foci displayed a colocalized Ter-2 focus (Figure 2C and E; Supplementary Figure S4). Colocalized single SSB and Ter-2 foci were found near mid-cell in most cells (around 80%). These observations demonstrated that the replication of the Ter-2 locus and likely that of a large terminus region (Ter-2 is localized 250 kb before dif) is achieved at mid-cell for most of the cells with a linear chromosome.
The migration of the Ter MD to mid-cell is coordinated with its replication
The ends of each chromosome’s arm of a linear chromosome were brought to mid-cell at the time of their replication. This observation suggests that in cells with a circular chromosome, the merging of the replisome foci in one focus may not solely be the consequence of two forks running in the opposite orientation on a circular track and meeting at the terminus already localized at mid-cell. An alternative hypothesis stipulates that the two replisomes could meet at mid-cell before replicating the Ter MD and therefore could provoke Ter MD migration towards mid-cell for its replication. A consequence of this hypothesis would be that the loci from the terminus region would migrate to mid-cell in sequential order according to their genetic distance from oriC. To test this hypothesis, we analysed the dynamic behaviour of different FROS tags inserted in the Ter region. Using a triple-labelled strain with MatP–mCherry, Ter-7 parS pMT1 (2000, kb from oriC) and Ter-6 parS P1 (2200, kb from oriC) tags (Figure 3A), we observed that the Ter-7 tag was localized at mid-cell in the smallest cells, while the Ter-6 and the majority of the MatP–mCherry signals were still near the pole of the cell (Figure 3B, picture 1). In larger cells (picture 2), most of the MatP–mCherry signal observed at mid-cell colocalized with Ter-7, while the Ter-6 tag remained at the pole and colocalized with a faint second MatP focus. These two categories of cells (pictures 1 and 2) only represented 4% of the population, suggesting that splitting of the ter MD loci is a transient event during the cell cycle. Later (picture 3) the Ter-6 tag migrated to mid-cell and merged with the Ter-7 tag and MatP–mcherry (60% of the population). In every further step of the cell cycle, at least one Ter-6, one Ter-7 and one MatP focus were colocalized near mid-cell (pictures 4–7). This experiment showed that the Ter MD migrates progressively to mid-cell starting from its oriC proximal part. Together with the results showing an inverse relationship between the amount of fluorescence in the polar and central MatP foci (Figure 1; Supplementary Figure S1), these results indicated that the MatP focus does not migrate as a whole from the pole to mid-cell but rather that MatP molecules dissociated progressively from the Ter region at the pole and reassembled at mid-cell after migration of the Ter DNA.
Figure 3.
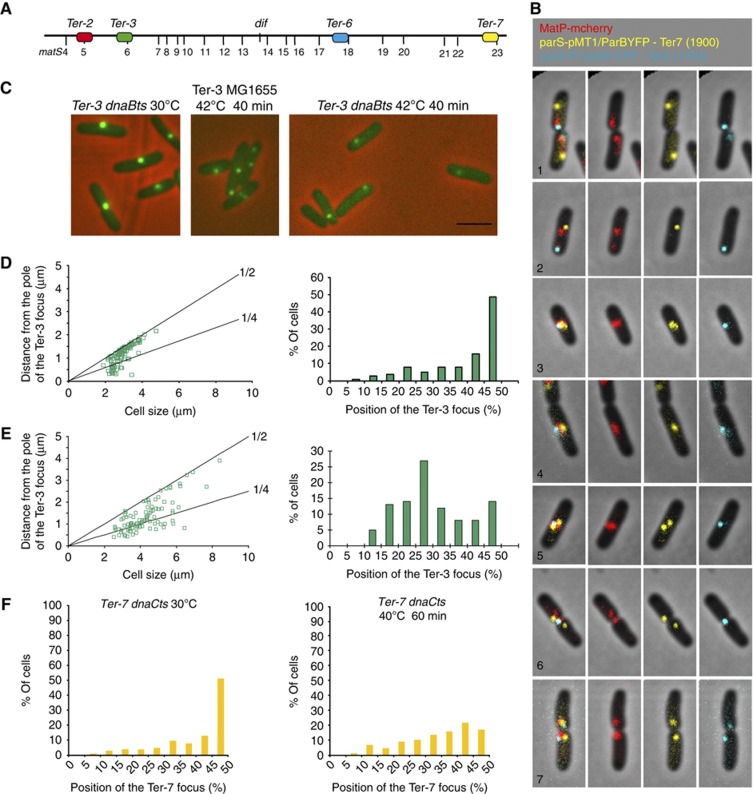
The replication machinery is involved in recruiting the Ter DNA to mid-cell. (A) Map of the Ter MD indicating the position of the matS sites and the parS tags used in the following panels. (B) Examples of representative cells tagged with parS pMT1/ParBYFP at Ter-7 (yellow), parS P1/ParBCFP at Ter-6 (blue) and MatP–mCherry (red). The pictures are arranged from new-born cells (1) to predivisional cells (7) according to cell size. Together, groups 1 and 2 represent 4% of the population, groups 3–5 represent 60% of the population, group 6 represents 20% and group 7 represents 11% of the population. The left panel is a montage of the merge pictures presented in the right panels. (C) Localization of the Ter-3 parS P1 tag in MG1655 and MG1655 dnaBts cells during a 30–42°C shift. Scale bar represents 3 μm. (D) Histogram of the position of the Ter-3 focus in MG1655 dnaBts cells according to cell size at 30°C (Left). Only cells with one focus were included. Histogram of the percentage of MG1655 dnaBts cells present in each category defined by the Ter-3 focus position at 30°C (Right). (E) Histogram of the position of the Ter-3 focus in MG1655 dnaBts cells according to cell size at 42°C (Left). Only cells with one focus were included. Histogram of the percentage of MG1655 dnaBts cells present in each category defined by the Ter-3 focus position at 42°C (Right). (F) Histograms of the percentage of MG1655 dnaCts cells present in each category defined by the Ter-3 focus position at 30°C or after 60 min at 40°C.
To test directly the influence of replication on the migration of the Ter MD to mid-cell, we analysed a locus from the terminus region (Ter-3 parS P1) following replication arrest mediated by the inactivation of the DnaB helicase (Figure 3C). At 42°C, a dnaB ts mutation inhibits the progression of DNA replication forks. At 30°C, most MG1655_dnaBts_ cells presented one central Ter-3 focus (Figure 3C and D). After 40 min at 42°C, wt cells presented 1 central focus or 2 foci localized at the mid-cell and quarter positions (Figure 3C). Cells from the dnaBts allele, presented mostly one focus near the quarter position (Figure 3C and E) suggesting that in the absence of replication, the Ter-3 migration from the new pole to mid-cell was blocked. To confirm this observation, we chose to block replication at the initiation step with the dnaCts allele (Figure 3F). As expected, we observed a strong decrease of the number of cells with a mid-cell localized Ter-7 parS pMT1 tag following a 1-h initiation block at 40°C (Figure 3F). In contrast from the dnaBts block, the dnaCts allele did not provoke the accumulation of cells with a focus near the quarter position; this could reflect a different organization of chromosomes frozen during replication compared with fully replicated chromosomes.
We used chromosome rearrangements to test directly the influence of the position on the genetic map on the migration of Ter loci. In the FBGT2 strain where the Ter MD is interrupted by the right MD, that is, a large Ter MD region is present at an ectopic position replicated 600 kb earlier (strain FBGT2 in Supplementary Figure S5A–C). Experiments showed that arrival at mid-cell for a locus in the terminus region (Ter-3 tag) can be observed earlier when it is replicated earlier. Using another strain carrying unbalanced replication arms (Esnault et al, 2007), we observed that a delay in the replication of a locus from the terminus region (Ter-3 tag) also delays its timing of migration toward mid-cell (Supplementary Figure S5D and E). Migration to mid-cell of a locus in the terminus region is therefore coordinated with its replication. Altogether, our results (Figures 1, 2, 3; Supplementary Figures S3–S5) showed that replication has direct consequences on the subcellular positioning of the terminus region during the cell cycle.
Anchoring of the Ter MD to the septal ring
We observed that the Ter MD is maintained at mid-cell for a long time (Figures 1A, L and 3B) following its replication (Figure 2E) in a MatP-dependent manner (Figure 1L and M). To determine whether the division apparatus that assembles at mid-cell is involved in the precise localization of MatP and of the Ter region, we analysed more precisely the assembly of the septal ring and the migration of MatP to mid-cell. The septal ring is formed of several proteins that assemble progressively at mid-cell around the tubulin-like polymer of FtsZ (Adams and Errington, 2009). MatP–mCherry and FtsZ–CFP fusions were constructed (Figure 4A). A snapshot analysis of MatP–FtsZ colocalization was performed with a strain expressing simultaneously MatP–mCherry and FtsZ–CFP fusions. As expected, FtsZ–CFP formed a mid-cell ring in the majority of cells. Only the smallest cells failed to present a distinct Z ring (Figure 4A and B). MatP–mCherry foci were present in every cell. They were frequently found under the FtsZ ring. Interestingly, MatP followed FtsZ dynamics, that is, small cells without an FtsZ ring presented a MatP focus near the pole, cells with a mid-cell MatP focus always presented a colocalized FtsZ ring and the largest cells presented two MatP foci flanking the FtsZ ring (Figure 4B). We noticed that the duplication of the MatP focus into two adjacent foci is concomitant with septum constriction; that is, 96% of cells with a distinct septum displayed two MatP–mCherry foci. MatP also frequently colocalized with the FtsZ ring in a strain with a linear chromosome (Supplementary Figure S6). Because simultaneous localization of MatP–mCherry and FtsZ–CFP could rely on independent but synchronous processes, we altered the amount of FtsZ polymers in the cell by over expressing FtsZ–CFP (Figure 4C). The over expression provoked the accumulation of heterogeneous and mislocalized FtsZ–CFP signals. We distinguished eight categories of cells that testify to the profound alteration of cell physiology observed in this context. Only a small percentage of the cells have a wt pattern (category 1, 18% of the population). MatP foci were frequently observed to be colocalized with aberrant FtsZ rings (categories 3, 4, 5 and 7). Aberrant MatP localization can also be observed without FtsZ colocalization (category 6 and category 8 (filaments)). Overall, these experiments indicate that the division machinery can be targeted by MatP and that FtsZ is not the only determinant controlling this process.
Figure 4.
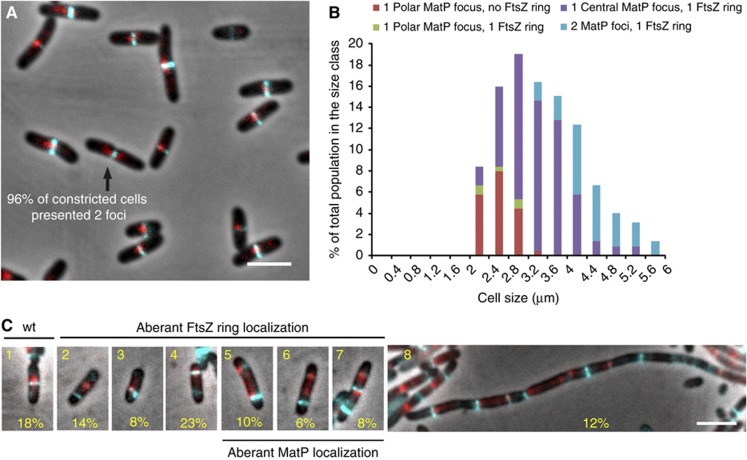
MatP is colocalized with the septal ring. (A) Merged pictures of MatP–mCherry (red), FtsZ–CFP (cyan) and phase-contrast micrographs (grey) of the MG1655 cells grown in minimal medium supplemented with casamino acids and glucose. (B) Histograms representing the appearance of the FtsZ ring and the positioning of MatP–mCherry near the pole (distance from the pole<30% of the cell size) or at mid-cell (position of the focus comprised between 45 and 50% of the cell size) according to the cell size. (C) Merged picture of cells expressing MatP–mCherry (red) and FtsZ–CFP (cyan) with phase-contrast microscopy (grey), grown under FtsZ–CFP overexpression conditions. Eight categories of cells were defined; the percentage of cells in each category is indicated. Scale bars represent 3 μm.
ZapB is the target of MatP at mid-cell
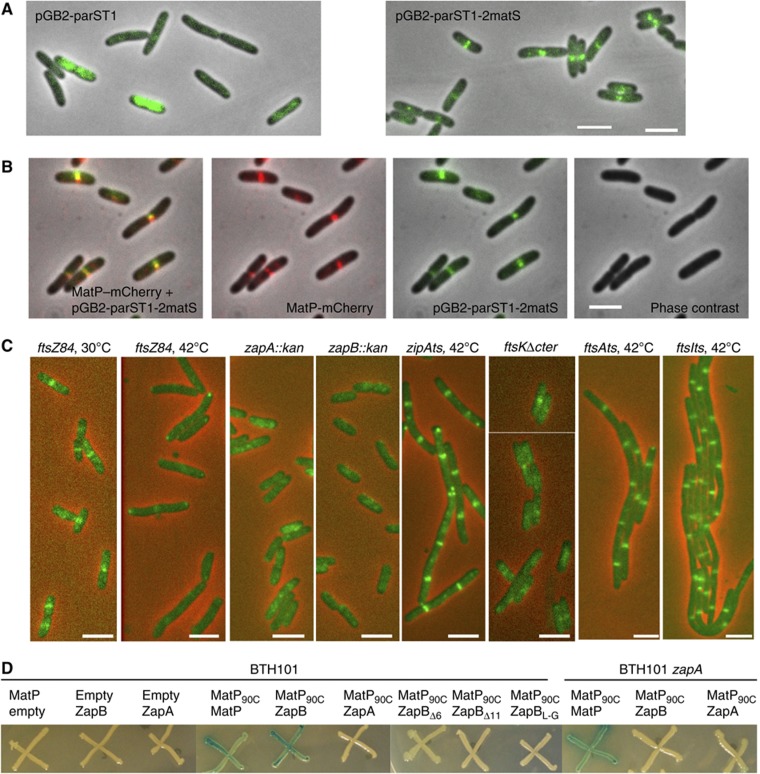
To identify the factors involved in anchoring of MatP to mid-cell, it was necessary to develop an assay capable of directly revealing this association. We anticipated that MatP could anchor _matS_-containing DNA to mid-cell. We therefore used a reporter plasmid containing a parS _pM_T1 and 2 matS sites. A dynamic diffused signal was detected using the control plasmid devoid of a matS site, indicating random distribution of the plasmids moving rapidly in the cellular space (Figure 5A; Supplementary Figure S7A). In contrast, when matS sites were present on the plasmid, a very distinct pattern of localization was observed. Plasmids with matS sites accumulated at mid-cell in a very bright and stable ring-like structure (Figure 5A; Supplementary Figure S7B). The ring-like structure is also observed in strains with different genetic backgrounds than MG1655 such as AB1157 (Supplementary Figure S7C). The matS plasmid localization to mid-cell is dependent on MatP since no accumulation of fluorescence was detected at mid-cell in matP cells (Supplementary Figure S7D). Using a strain containing the matS plasmid and expressing MatP–mCherry, MatP was simultaneously found associated to these ring-like structures (Figure 5B). Remarkably, in this context, most MatP–mCherry formed ring-like structures rather than foci (compare Figures 1A, 3B and 4A with 5B).
Figure 5.
MatP interacts with ZapB. (A) Merged pictures of the pGB_parS_ T1 or the pGB_parS_ T1_-2matS plasmid (green) and phase-contrast micrographs (grey) in MG1655 cells. (B) Montage of merged pictures of the MG1655_matP–mCherry strain containing the pGB_parS_ T1_-2matS plasmid. pGB_parS T1_-2matS (green), MatP–mCherry (red) and phase-contrast microscopy (grey) are shown. (C) Localization of the pGB_parS T1_-2matS plasmid (green) in MG1655_ftsZ84, zapA, zapB, ftsk_Δ_C strains grown at 30°C and the MG1655_ftsZ84, zipAts_, ftsAts and ftsIts strains grown at 42°C for 1 h. Scale bars represent 3 μm. (D) Bacterial two-hybrid assay; the BTH101 and BTH101_zapA_ strains containing the pKT25, pKT25MatP, pKT25ZapA, pKT25ZapB, pKT25ZapBΔ6, pKT25ZapBΔ11 or pKT25ZapBL-G and pUT18c or pUT18CMatP90C plasmids were grown in LB supplemented with kanamycine, ampicilin, IPTG and Xgal at 30°C; pictures were taken after 2 days of incubation at 30°C.
We took advantage of the robust signal of the system described above to survey the involvement of various septal ring proteins in MatP-dependent anchoring to mid-cell of plasmids carrying matS sites. The septal ring is composed of multiple proteins, both essential and accessory, which localize to the septum. To probe for the involvement of these different proteins, we tested the accumulation of the reporter matS plasmid in deletion (zapA, zapB, ftsK_Δ_C) or thermo sensitive (ftsZ, zipA, ftsA, ftsI) mutants. At non-permissive temperatures, regularly spaced bands of the matS plasmids were observed in the filaments after inhibition of ftsI, zipA, ftsA, or deletion of 3′ end of ftsK. In contrast, inhibition of ftsZ and the deletion of zapA and zapB genes profoundly altered the band pattern (Figure 5C; Supplementary Figure S7E). Because ZapA and ZapB septal ring associations are dependent on FtsZ, and nearly normal FtsZ rings are produced in zapA or zapB mutants, it is likely that FtsZ is not the direct target of MatP. The ZapA and ZapB proteins have been shown to stabilize FtsZ rings (Gueiros-Filho and Losick, 2002; Ebersbach et al, 2008). It was demonstrated that ZapA interacts directly with FtsZ, while ZapB requires ZapA to associate with the septum (Galli and Gerdes, 2010). Direct evidence for a ZapB–MatP interaction was obtained by a bacterial two-hybrid assay (Figure 5D). ZapB interacts strongly with the C-terminus half of MatP (Figure 5D). No interaction between ZapA and MatP was detected by the two-hybrid assay, indicating that ZapB is the main target of MatP among the candidates tested above. Interestingly, the interaction between MatP and ZapB, revealed by the two-hybrid assay, is abolished in the absence of ZapA (Figure 5D), suggesting that MatP and ZapB only interact when ZapB is attached to the septal ring. The interaction of ZapB with ZapA requires the N-terminal end of ZapB but not its C-terminal end (Galli and Gerdes, 2012). Remarkably, ZapB did not interact with MatP after deletion or alteration of its C-terminal domain (Figure 5D) confirming the specificity of the ZapB–MatP interaction.
ZapA and ZapB are required for the mid-cell anchoring of MatP
As expected, MatP–mCherry was perfectly colocalized with ZapB–GFP (Figure 6A) or ZapA–GFP (Figure 6B). As reported previously (Galli and Gerdes, 2010), the absence of ZapB did not alter the localization of ZapA–GFP, confirming that ZapA did not require ZapB for proper anchoring to the septal ring (Figure 6C). In a zapB mutant strain, about half of the cells displayed two MatP–mCherry foci that were distant from the ZapA–GFP ring (Figure 6C–E; Supplementary Figure S8), confirming that a ZapA was not the target of MatP and was not sufficient to promote accumulation of MatP at mid-cell. The absence of ZapA profoundly altered the localization of ZapB–GFP and MatP–mCherry (Figure 6F). ZapB–GFP formed bright polar foci that could reflect an aggregation of the protein (Galli and Gerdes, 2010). MatP foci were not colocalized with the ZapB–GFP foci. Moreover, in this context, MatP foci presented a localization pattern similar to that observed in the zapB mutant (Figure 6D and E; Supplementary Figure S8). This is in agreement with the two-hybrid experiments, indicating that ZapA is required for the MatP–ZapB interaction. Altogether, these results indicated that MatP interacts with ZapB when associated in a larger complex comprising at least ZapB, ZapA and FtsZ.
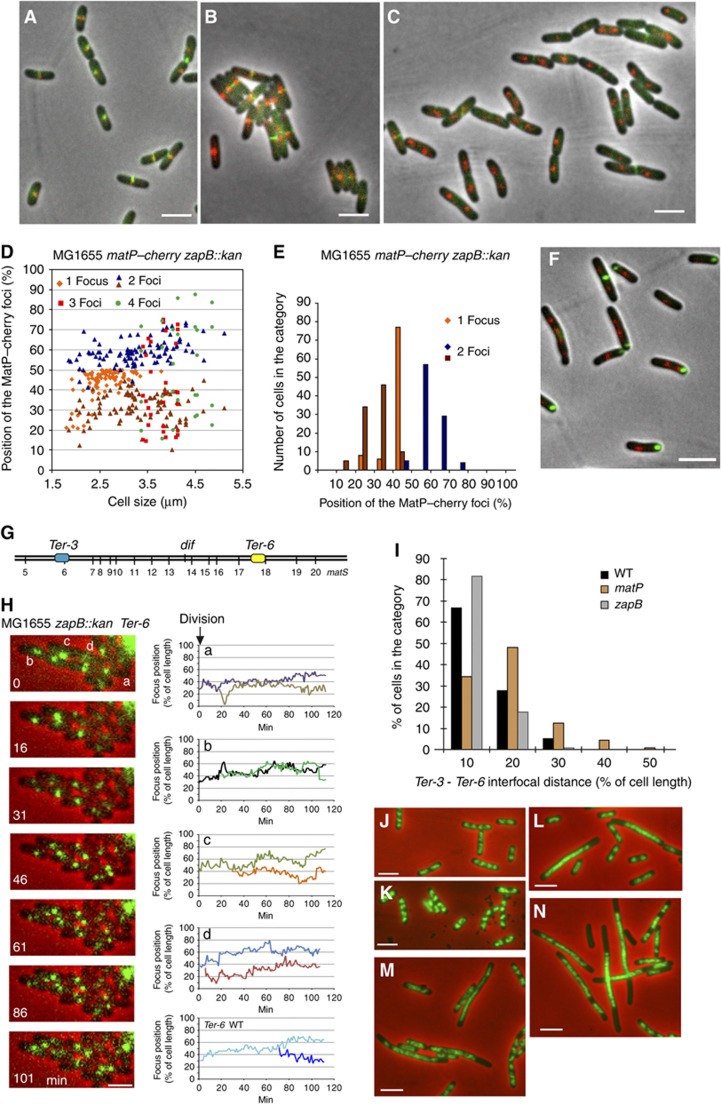
Figure 6.
Influence of ZapB-mediated anchoring on the Ter MD and chromosome dynamics. (A) MatP–mCherry (red) and ZapB–GFP (green) are colocalized in wild-type cells. (B) MatP–mCherry (red) and ZapA–GFP (green) are colocalized in wild-type cells. (C) MatP–mCherry (red) and ZapA–GFP (green) are not colocalized in the MG1655_zapB_ mutants. (D) Histogram of the relative position of MatP–mCherry foci according to MG1655_zapB_ cell size. Cells with 1, 2, 3 or 4 foci are indicated. (E) Histogram of the number of cells in each category of MatP–mCherry position (180 cells were counted). (F) MatP–mCherry (red) and ZapB–GFP (green) are not colocalized in the MG1655_zapA_ strain. (G) Map of the tags inserted in the chromosome of the MG1655 zapB strain that were used in (H–J), parS P1 (Ter-3) and parS pMT1 (Ter-6) positions are represented. (H) Segregation of Ter-6 (parS pMT1) tag. MG1655_zapB_ cells were followed during a 120-min time lapse at 1-min intervals. One picture every 16 min was represented on the montage. Traces representing the segregation of the Ter-6 tag in the MG1655_zapB_ cells annotated in the montage (a–d) and in wt MG1655 cells. (I) Measure of the interfocal distance between Ter-3 (parS P1) and Ter-6 (parS pMT1) loci in the MG1655, MG1655_matP_ and MG1655_zapB_ strains. (J) DAPI staining (green) of the nucleoid of the MG1655_zapA_ strain grown in rich medium (LB). (K) DAPI staining of the nucleoid of the MG1655_zapB_ strain. (L) DAPI staining of the nucleoid of the MG1655_matP_ strain. (M) DAPI staining (green) of the nucleoid of the MG1655_zapAmatP_ strain. (N) DAPI staining of the nucleoid of the MG1655_zapBmatP_ strain. Cells presented on the panels (J–N) were grown in rich LB medium to O.D.=0.3. Scale bars represent 3 μm.
Deletion of zapB alters the segregation of the Ter MD
To analyse the effect of the ZapB inactivation on the segregation of the Ter MD, we monitored the localization of either MatP–mCherry foci in zapB and zapA mutants. The localization pattern of MatP–mCherry was altered in the absence of ZapB (Figure 6D and E) compared with the wild-type strain (Figure 1B and C). A larger number of cells with two MatP–mCherry foci were observed whatever the cell size, and a broader distribution of the MatP foci was observed. Also, a significant number of cells displayed three or four MatP–mCherry foci. Similar alterations of the MatP pattern were observed in the zapA mutant. The pattern of segregation of the Ter-6 parS pMT1 tag (Figure 6G and H) was also altered in the absence of ZapB. The Ter-6 focus duplicated earlier and segregated in a less precise manner than observed in the wt cells (Espeli et al, 2008). The global pattern of localization of the Ter-6 tag was also altered in the absence of ZapB (Supplementary Figure S9) compared with wt; more cells with two foci were detected (11 and 35% in the wt and the zapB mutant, respectively). Further, the youngest cells with two foci were smaller than in wt (2.2 μm in the zapB strain compared with 3.5 μm in the wt), and the localization appeared to be less precise. Altogether, these observations suggest that MatP and ZapB are required for the prolonged localization of the Ter MD at mid-cell. Since the chromosome replication programme was not significantly altered in the absence of ZapB (data not shown), this indicates that sister Ter MD colocalization at mid-cell is reduced in the absence of ZapB.
Deletion of zapB does not influence the condensation of the Ter MD
The absence of ZapB did not alter the colocalization of MatP with any parS tag tested in the terminus region (Supplementary Figure S9C), showing that MatP remained associated with the matS sites in the absence of ZapB. MatP inactivation affected the condensation of the Ter region (interfocal distance increased) (Mercier et al, 2008). To determine whether the association of the Ter region with the septal ring was responsible for the condensation of the region, we measured interfocal distances between two tags of the Ter MD separated by 255 kb in zapB mutant cells. As previously observed (Mercier et al, 2008), the interfocal distance increased in the absence of matP. In contrast, the absence of ZapB did not provoke an increase in the interfocal distance (Figure 6I). This observation suggested that the ZapB-mediated anchoring of the newly replicated Ter MDs to the septal ring has no measurable effect on the condensation per se of the Ter MD.
Nucleoid segregation is dramatically altered in the matP zapA or matP zapB strains
We constructed double-deletion mutants for matP and zapA or zapB. On LB agar plates, the colonies were small and heterogeneous in size. Nevertheless, growth in rich medium was possible, allowing microscopic observation of the nucleoid following DAPI staining (Figure 6J–N). Both matP–zapA and matP–zapB double mutants displayed many filamentous cells with unsegregated nucleoids and anucleated cells. Respectively, 48 and 54% of the cells were abnormal in the matP–zapA and the matP–zapB strains. The combination of deletions had synergistic effects because in the matP deletion strain, no more than 20% of the cells were abnormal, and in the zapA or zapB deletion strains, <4% of the cells were filamentous, and none displayed major nucleoid segregation defects. The synergistic effects observed with the absence of ZapA or ZapB and MatP suggest that each protein may have a yet uncharacterized function that is only revealed in the absence of its partner (see Discussion).
Discussion
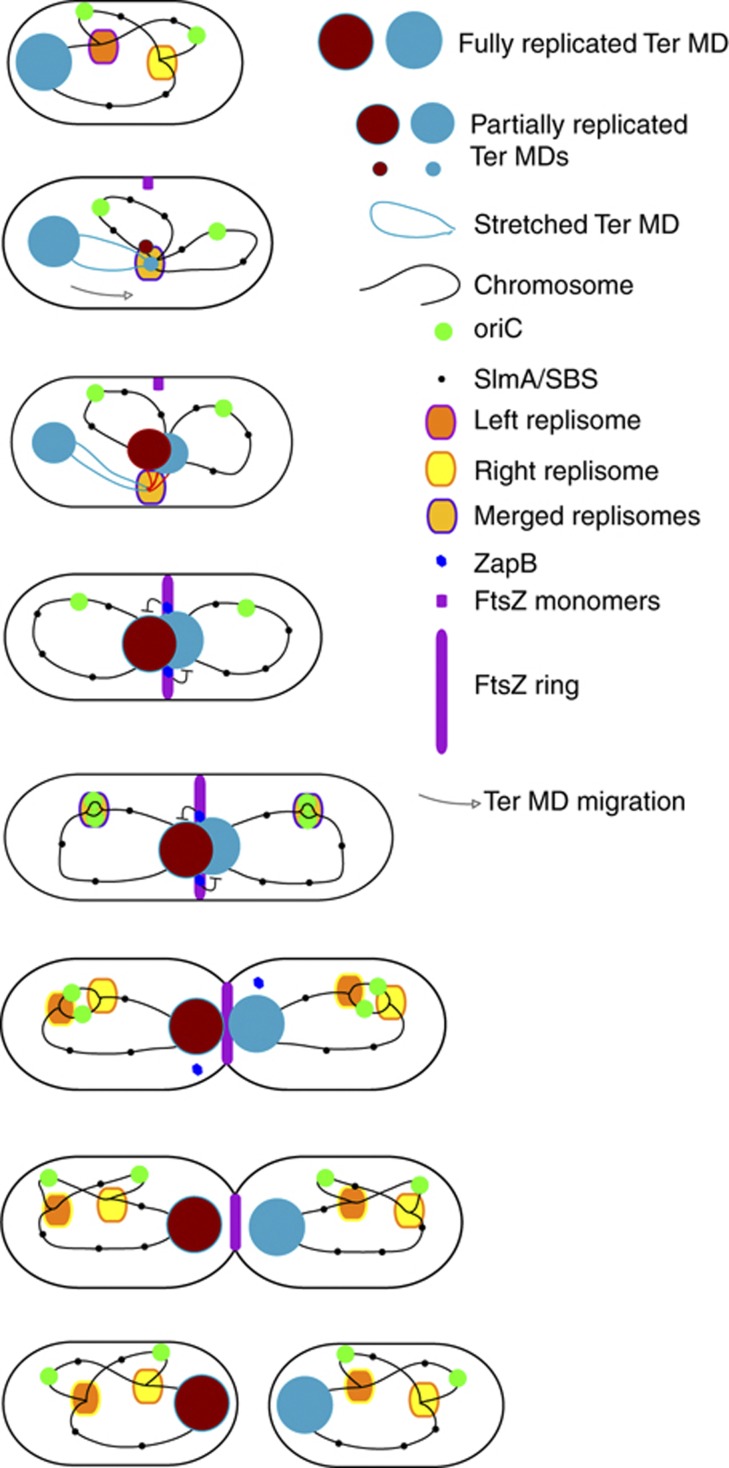
The organization of the Ter MD relies on the site-specific binding protein MatP. Interaction of MatP with matS sites affects several observable parameters, that is, mobility constraint of DNA markers, colocalization of sister markers, DNA condensation inside the Ter MD, anchoring of the Ter MD to mid-cell (Mercier et al, 2008). Here, we analysed the role of MatP in two different steps of the Ter MD choreography: (i) the migration of the terminus region from the pole to mid-cell and (ii) the prolonged step of mid-cell localization of the Ter MD. We demonstrated that replication of the Ter MD by a centrally localized replisome is coordinated with its migration to mid-cell where the MatP–ZapB interaction stabilized the association of the Ter MD sister chromatids with the septal ring (Figure 7). MatP and ZapB are thus essential for the Ter MD choreography during the cell cycle.
Figure 7.
Model for the Ter MD dynamics during the cell cycle. Schematic diagram of a complete cell cycle, from birth to the next division. Steps of ∼10 min are described. The dynamics of the Ter MD (red–brown ball) is proposed according to the observations described above. The migration of the Ter DNA is illustrated with an arrow. ZapB-mediated anchoring of the sister Ter MDs to the septal ring is illustrated.
The migration of the Ter MD to mid-cell is coordinated with its replication
Migration towards mid-cell of the loci of the Ter MD in a circular or a linear chromosome was independent of MatP. Nonetheless, it was temporally controlled. We demonstrated that migration to mid-cell was initiated by the oriC proximal part of the Ter MD and progressed gradually until the migration of the distal loci occurred. This correlated with a progressive disappearance of the polar MatP focus and the reciprocal appearance of a mid-cell focus after migration of the Ter DNA. Ter DNA migration preceded its colocalization with the SSB–YPet focus and likely its replication. As observed for the circular chromosome, a centrally localized replication factory performed the replication of the Ter region in the context of the linear chromosome. This observation does not support a model proposing that replisomes progress only on the chromosome as trains on a track. Together with previous studies reporting the choreography of replisomes in E. coli cells (Reyes-Lamothe et al, 2008b), our results are consistent with a model in which the replisomes assemble at oriC, then separate to replicate left and right portions of the chromosome and are finally brought together, independently of the circular or linear chromosome configuration, for the completion of the replication process of the Ter region. This model requires that replisome choreography is directed by a different superstructure than the chromosome itself; this superstructure remains to be characterized. A consequence of this replisome choreography is a revival, in E. coli, of the capture part of the ‘capture and extrusion model’ (Lemon and Grossman, 2001). We attempted to directly test this capture step of the Ter MD mid-cell by the replication factory in three ways. We have provided evidence suggesting that delay of replication from the Ter MD induces a delay in its migration towards mid-cell, and reciprocally, that an earlier replication induces earlier migration. Finally, we have shown that following a replication blockage, the position of the Ter MD to mid-cell was lost. These elements indicate that sequential migration of the loci from the Ter MD to mid-cell is coordinated with their replication and likely involves their recruitment by the replication machinery. Alternatively, it was proposed that newly replicated sister regions might push one another apart, thus bringing unreplicated regions to mid-cell (Bates and Kleckner, 2005); such process could participate in the migration of the Ter MD to the vicinity of a mid-cell localized replication factory.
An interaction between MatP and the septal ring mediates the anchoring of the sister chromatids at mid-cell by their Ter MD
Sister chromatid colocalization for extensive periods of time has been observed in E. coli in several studies (Bates and Kleckner, 2005; Adachi et al, 2008; Espeli et al, 2008). The colocalization period appears to be dependent on the locus tested and follows the general rule that colocalization is long in the MDs and short in the NS regions (Espeli et al, 2008). Recently, an analysis at high density of the Ori MD revealed that some regions, called SNAPs, displayed longer cohesion than others (Joshi et al, 2011). In the terminus region, the colocalization of sister chromatids could be particularly long and is dependent on the MatP/matS interaction (Mercier et al, 2008). Because Ter MD migrated towards mid-cell to be replicated early in the cell cycle (10–20 min after division at 30°C in a 70-min cell cycle, 30–40 min after division at 25°C in a 120-min cell cycle) and remained localized at mid-cell until septum constriction (20–30 min before division), we deduce that sister Ter MDs colocalize at mid-cell for 30–50 min. This prolonged step is strongly dependent on MatP (Mercier et al, 2008) but almost unaffected by the linearization of the chromosome (Figure 1). We observed that the MatP–mCherry focus that clustered the two sister Ter MDs associated with the division machinery at mid-cell. This colocalization is regulated because constriction or alteration of the FtsZ regulation allowed separation of FtsZ and MatP at mid-cell. ZapA and ZapB were required for an extended localization of the Ter MD at mid-cell and the bacterial two-hybrid assay revealed a ZapB–MatP interaction. ZapA and FtsZ are required to observe the interaction, indicating that MatP interacts with ZapB when it is associated with the divisome. Because the two-hybrid assay revealing the MatP–ZapB interaction was performed in E. coli, we cannot exclude that they do not interact directly but are brought together by a third, yet uncharacterized partner. ZapB dissociation from the septal ring (Galli and Gerdes, 2010) could be the trigger for segregation of the Ter MDs outside of the division plane as soon as constriction is initiated. However, plasmids with matS sites behaved differently; they remained associated to the septal ring until scission. This suggests that ZapB in the presence of matS plasmid remained associated with the constricting septal ring. An interesting possibility would be that other DNA metabolism process(es), for example, segregation in the oriC proximal region or nucleoid splitting (Bates and Kleckner, 2005), would trigger the loss of the Ter MD–septal ring association.
Alteration of MatP/septal ring anchoring perturbs nucleoid segregation
We analysed the behaviour of MatP–mCherry and parS tags under conditions in which the association with the septal ring was altered. zapA and zapB mutants altered the segregation pattern of the Ter MD during growth at a slow growth rate. As published previously (Ebersbach et al, 2008), these mutations had no major consequences on nucleoid segregation, even at a fast growth rate. In contrast to what was observed in the matP mutant, we did not observe a significant increase in the interfocal distance between the Ter loci in the zapB mutant, which indicated that internal organization of the Ter MD was not altered. Therefore, MatP interaction with the septal ring is not critical for its role as a MD organizer. In both MC1000 (Ebersbach et al, 2008) and MG1655 (this work) strains, inactivation of zapB resulted in a mild phenotype of elongated cells with no effect on mass doubling time. The most striking defects consisted in a small proportion of filamenting cells and the morphology of the FtsZ ring that formed arc-like and helix-like structures in the zapB strain. The interaction of MatP with ZapB is not essential to achieve chromosome segregation. In the absence of ZapB, many cells still presented a MatP focus at mid-cell. This may reflect other possibilities for MatP to persist at mid-cell and influence chromosome segregation, either by itself after binding Ter DNA or by interacting with another partner. As observed with other chromosome organization/segregation systems (i.e., ParABS systems), the anchoring of MatP to ZapB might appear to be essential when combined with other mutation(s) or specific growth conditions.
Remarkably, combining matP with zapB or zapA deletion has consequences on cell physiology and nucleoid segregation stronger than expected if the role of ZapB was simply to anchor the Ter MD at the septum. These results revealed a synergistic effect of the double mutation, indicating a putative role of ZapB in chromosome segregation that is important in the absence of MatP or a role of MatP in cytokinesis that is important in the absence of ZapB. We have not yet identified any direct involvement of MatP in cytokinesis. A challenge now is to identify whether either ZapB alone or the properly configured Z ring play a direct role in chromosome segregation.
Integration of chromosome management in the control of the cell cycle
Eukaryotic cells utilize a number of quality control systems, also called checkpoints, that block the cell cycle when one of the steps is not completed properly (Elledge, 1996; Tyson and Novak, 2008). Equivalent systems are not well characterized in bacteria. The SulA checkpoint is among the best unravelled. Following DNA damage and as a part of the SOS response, the SulA protein will favour the depolymerization of FtsZ and therefore block cell division (Huisman et al, 1984). The nucleoid occlusion system (Woldringh et al, 1991) has been molecularly characterized in B. subtilis and E. coli (Wu and Errington, 2004; Bernhardt and de Boer, 2005); in E. coli, it involves the SlmA protein, which binds to 24 SBS sites along the chromosome and blocks FtsZ polymerization when SBS sites remain in the division plane. Remarkably, SBS sites are excluded from the Ter MD (Cho et al, 2011; Tonthat et al, 2011). Presumably, the selection of matS sites in the Ter region and SBS sites outside the Ter region ensure coordination of chromosome segregation with cell division: SlmA act as a negative regulator of cell division if the non-Ter region is found at mid-cell whereas matS sites promote the anchoring of the Ter MD near the division machinery. While SlmA acts in a checkpoint controlling chromosome segregation and if necessary blocks cytokinesis, MatP acts as an organizer that anchors the Ter MD to mid-cell to get the chromosome ready for segregation. An association between the septal ring and the terminus region of the chromosome has already been described for decatenation of entangled chromosomes (Hojgaard et al, 1999; Espeli et al, 2003) and chromosome dimer resolution (Blakely et al, 1991; Steiner et al, 1999). A challenge now is to determine whether the MatP–ZapB interaction influences these transactions or another as yet uncharacterized process.
Materials and methods
Strains and growth conditions
Strains and plasmids are described in Supplementary Table S1. MG1655, MG58, MG59 and their derivatives were grown in minimal medium A supplemented with casamino acids (0.2%) and glucose (0.5%) unless specified. Strains MKG297 and MKG305 were grown in minimal medium A supplemented with casamino acids (0.2%) and succinate (0.5%), and 30 min before observation, arabinose (0.1%), IPTG (5 μM) and anhydro-tetracyclin (40 ng/ml) were added (Lau et al, 2003). The ZapA–GFP and ZapB–GFP fusion proteins were observed following a brief induction of the PBAD promoter during 10 min in the presence of arabinose (0.2%) (Ebersbach et al, 2008). The FBGT2 strain has been obtained by a transposition event of a segment of the Right MD in the Ter MD (Thiel et al, 2012).
Microscopy
Bacteria were grown at 30°C (unless specified) in the indicated medium until O.D.=0.2, washed in fresh medium and concentrated 10 times. Then, 2 μl were spread on agarose and immediately observed using a Leica DM6000 microscope, a coolsnap HQ CCD camera (Roper) and Metamorph software or a Zeiss axio observer Z1 microscope, an Evolve EM-CCD camera (Roper) and Axiovision software. The analysis was performed with ImageJ software. The Stackreg (Thevenaz et al, 1998) and the MtrackJ (Meijering et al, 2012) plugins were used for the analysis of time-lapse images; the Pointpicker plugin was used for snapshot analyses. Custom-made Excel sheets were used to monitor the cell size, focus positions, interfocal distance, septum constriction, fluorescence intensity and DNA mobility. Septum constriction was monitored, thanks to a line scan analysis of the phase-contrast images. We empirically determined that a 20% reduction of the phase signal, measured at mid-cell, was the threshold to consider invagination. For each snapshot experiment, at least 180 cells were counted.
Two-hybrid assay
BTH101 or BTH101 ΔzapA cells were cotransformed by the pUT18c or the pUT18cmatPΔN30 and the pKT25, pKT25ΔN30matP, pKT25zapA, pKT25zapB, pKT25zapBΔ6, pKT25zapBΔ11, pKT25zapBL-G plasmids. Transformed cells were streaked on LB plates supplemented with Xgal (20 μg/ml) and IPTG (500 μM) and incubated for 2 days at 30°C.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank K Gerdes, T Horiuchi, D Ladent, B Michel and D Sherratt for the kind gift of strains and plasmids; S Duigou, B Michel, L Sperling, M Valens and I Vallet-Gely for critically reading the manuscript and thoughtful discussions. This work was supported by CNRS and ANR (Grant ANR 08-Blan-0119 2009-2012 to FB and ANR JCJC 2010-2014 SISTERS to OE).
Author contributions: OE, RB, PD, AT and EG performed the experiments; OE, RB, PD, RM and FB designed the experiments; FB and OE wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi S, Fukushima T, Hiraga S (2008) Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells 13: 181–197 [DOI] [PubMed] [Google Scholar]
- Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 [DOI] [PubMed] [Google Scholar]
- Bates D, Kleckner N (2005) Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121: 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PA (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over Chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakely G, Colloms S, May G, Burke M, Sherratt D (1991) Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol 3: 789–798 [PubMed] [Google Scholar]
- Cho H, McManus HR, Dove SL, Bernhardt TG (2011) Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA 108: 3773–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui T, Moro-oka N, Ohsumi K, Kodama K, Ohshima T, Ogasawara N, Mori H, Wanner B, Niki H, Horiuchi T (2007) Escherichia coli with a linear genome. EMBO Rep 8: 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebersbach G, Galli E, Moller-Jensen J, Lowe J, Gerdes K (2008) Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol 68: 720–735 [DOI] [PubMed] [Google Scholar]
- Elledge SJ (1996) Cell cycle checkpoints: preventing an identity crisis. Science 274: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Esnault E, Valens M, Espeli O, Boccard F (2007) Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Lee C, Marians KJ (2003) A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J Biol Chem 278: 44639–44644 [DOI] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Galli E, Gerdes K (2010) Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol 76: 1514–1526 [DOI] [PubMed] [Google Scholar]
- Galli E, Gerdes K (2012) FtsZ-ZapA-ZapB interactome of Escherichia coli. J Bacteriol 194: 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueiros-Filho FJ, Losick R (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16: 2544–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojgaard A, Szerlong H, Tabor C, Kuempel P (1999) Norfloxacin-induced DNA cleavage occurs at the dif resolvase locus in Escherichia coli and is the result of interaction with topoisomerase IV. Mol Microbiol 33: 1027–1036 [DOI] [PubMed] [Google Scholar]
- Huisman O, D'Ari R, Gottesman S (1984) Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci USA 81: 4490–4494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi MC, Bourniquel A, Fisher J, Ho BT, Magnan D, Kleckner N, Bates D (2011) Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci USA 108: 2765–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ (2003) Spatial and temporal organization of replicating Escherichia coli chromosomes. InMol Microbiol 49: pp731–743England [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD (2001) The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev 15: 2031–2041 [DOI] [PubMed] [Google Scholar]
- Li Y, Sergueev K, Austin S (2002) The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol 46: 985–996 [DOI] [PubMed] [Google Scholar]
- Meijering E, Dzyubachyk O, Smal I (2012) Methods for cell and particle tracking. InMethods Enzymol 504: pp183–200United States [DOI] [PubMed] [Google Scholar]
- Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135: 475–485 [DOI] [PubMed] [Google Scholar]
- Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S (2006) Progressive segregation of the Escherichia coli chromosome. Mol Microbiol 61: 383–393 [DOI] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223 [PMC free article] [PubMed] [Google Scholar]
- Possoz C, Junier I, Espeli O (2012) Bacterial chromosome segregation. InFront Biosci 17: pp1020–1034United States [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ (2008a) Independent positioning and action of Escherichia coli replisomes in live cells. Cell 133: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Wang X, Sherratt D (2008b) Escherichia coli and its chromosome. Trends Microbiol 16: 238–245 [DOI] [PubMed] [Google Scholar]
- Sawitzke JA, Austin S (2000) Suppression of chromosome segregation defects of Escherichia coli muk mutants by mutations in topoisomerase I. Proc Natl Acad Sci USA 97: 1671–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R (2009) Why and how bacteria localize proteins. Science 326: 1225–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner W, Liu G, Donachie WD, Kuempel P (1999) The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol 31: 579–583 [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Wang SC, Shapiro L (2005) The bacterial nucleoid: a highly organized and dynamic structure. J Cell Biochem 96: 506–521 [DOI] [PubMed] [Google Scholar]
- Thiel A, Valens M, Vallet-Gely I, Espéli O, Boccard F (2012) Long-range chromosome organization in E. coli: a site-specific system isolates the Ter macrodomain. PLoS Genet 8: e1002672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7: 27–41 [DOI] [PubMed] [Google Scholar]
- Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA (2011) Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30: 154–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro E, Shapiro L (2010) Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol 2: a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Novak B (2008) Temporal organization of the cell cycle. Curr Biol 18: R759–R768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F (2004) Macrodomain organization of the Escherichia coli chromosome. EMBO J 23: 4330–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu X, Possoz C, Sherratt DJ (2006) The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev 20: 1727–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR (2008) Non-random segregation of sister chromosomes in Escherichia coli. Nature 455: 1248–1250 [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142: 309–320 [DOI] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File