The Salmonella Type III Secretion System Inner Rod Protein PrgJ Is Partially Folded (original) (raw)
Background: The structure of the protein family of the bacterial inner rod proteins is unknown.
Results: CD and NMR were used to determine the structural properties of the Salmonella PrgJ inner rod protein.
Conclusion: PrgJ is a partially folded protein and contains only a short helix.
Significance: Protein flexibility is a hallmark of the inner rod proteins.
Keywords: Bacteria, NMR, Protein Assembly, Protein Secretion, Protein Structure
Abstract
The type III secretion system (T3SS) is essential in the pathogenesis of many bacteria. The inner rod is important in the assembly of the T3SS needle complex. However, the atomic structure of the inner rod protein is currently unknown. Based on computational methods, others have suggested that the Salmonella inner rod protein PrgJ is highly helical, forming a folded 3 helix structure. Here we show by CD and NMR spectroscopy that the monomeric form of PrgJ lacks a tertiary structure, and the only well-structured part of PrgJ is a short α-helix at the C-terminal region from residues 65–82. Disruption of this helix by glycine or proline mutation resulted in defective assembly of the needle complex, rendering bacteria incapable of secreting effector proteins. Likewise, CD and NMR data for the Shigella inner rod protein MxiI indicate this protein lacks a tertiary structure as well. Our results reveal that the monomeric forms of the T3SS inner rod proteins are partially folded.
Introduction
Many Gram-negative bacteria require the type III secretion system (T3SS)4 to infect humans, animals, and plants (reviewed in Ref. 1). The structural component of the T3SS is the needle complex, which is a syringe-like macromolecular assembly that functions to inject effector proteins into eukaryotic cells (reviewed in Ref. 2). Salmonella typhimurium has two T3SSs, the one encoded by the Salmonella pathogenesis island 1 (SPI-1) is assembled from over 14 different proteins and consists of a basal structure that spans the inner and outer bacterial membranes (3–6), the external needle formed by the polymerization of the needle protein PrgI (5, 7, 8), and a tip complex formed by SipD (9) and the translocon proteins SipB and SipC (10). The translocon forms a channel in the host cell membrane to allow the passage of effectors into the host cell to modulate host cell biology for the benefit and survival of bacteria.
Within the T3SS basal structure is the inner rod (3–5), which forms a conduit for protein transport through the periplasm. The inner rod is assembled by the polymerization of PrgJ in S. typhimurium (5, 6), MxiI in Shigella flexneri (11, 12), and YscI in Yersinia pestis (13). Other T3SS inner rod proteins are BsaK (Burkholderia pseudomallei), EprJ and EscI (Escherichia coli), and PscI (Pseudomonas aeruginosa) (Fig. 1). In Salmonella, the assembly of the PrgJ inner rod is important in the assembly of the entire needle complex (4, 5). The inner rod is surrounded by membrane proteins that form ring-like structures within the inner and outer bacterial membranes (reviewed in Ref. 2). PrgJ is critical in virulence as evidenced by the non-invasive phenotype of a PrgJ-null mutant (14). Likewise, MxiI (11, 12) and YscI (13) are essential in the pathogenesis of Shigella and Yersinia, respectively. Additionally, YscI plays a critical role in substrate specificity switching (13), wherein the order of protein export through the needle is regulated (for example, effectors are secreted after the structural proteins).
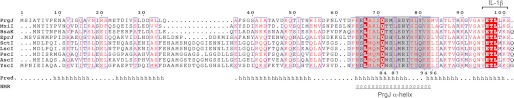
FIGURE 1.
Sequence alignment of the T3SS inner rod proteins. Computer programs (39) predicted (Pred.) that PrgJ is mostly helical (h). NMR, however, reveals that only residues 65–82 form a regular α-helix (shaded gray) in monomeric PrgJ. Miao et al. (15) showed that the seven C-terminal residues of PrgJ are important in activating the inflammatory cytokine interleukin 1β (_IL-1_β). Also denoted are the YscI residues (84, 87, 94–96), that were mutated by Wood et al. (13) and shown to be defective in needle assembly. The proteins used in the alignment are PrgJ S. typhimurium; MxiI, S. flexneri; BsaK, B. pseudomallei; EprJ, E. coli 0157:H7; SctI. Photorhabdus luminescens; LscI. Photorhabdus luminescens; PscI, P. aeruginosa; AscI, Aeromonas salmonicida; and YscI, Y. pestis.
Recently, the inner rod proteins were shown to be potent inducers of the inflammasome (15–19). Miao et al. (15) showed that PrgJ and other inner rod proteins (BsaK, EprJ, EscI, and PscI) interact with the NOD-like receptor NLRC4 to activate the maturation of interleukin-1β (IL-1β), which triggers the host inflammatory response against microbial infection. Specifically, the C-terminal seven residues of PrgJ were identified by deletion mapping to be essential in activating the host immune response (15).
Despite the importance of the inner rod proteins in assembly of the needle complex (3–6), pathogenesis (11–14, 20), and host immune response (15–18), little is known about their biophysical and structural properties. Currently, no CD or NMR data has been reported for any T3SS inner rod proteins. As such, their monomeric structures have been inferred from computer predictions (15). For example, PrgJ is predicted to have a folded structure with three α-helices (15). Here, we show by CD and NMR spectroscopy that monomeric PrgJ is essentially a partially folded protein. Likewise, CD and NMR data for monomeric MxiI suggest this protein lacks a tertiary structure as well.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Full-length and various truncations of PrgJ were subcloned as fusion proteins with a His6-tagged GB1, the B1 immunoglobulin binding domain of Streptococcus protein G, to form the fusion constructs PrgJGB1 (with GB1 attached at the C terminus of PrgJ) and GB1PrgJ (with GB1 attached at the N terminus of PrgJ). The fusion proteins also contained a tobacco etch virus (TEV) protease cleavage site to recover PrgJ (supplemental Fig. S1_A_). Isotopically (15N or 15N, 13C) labeled proteins were overexpressed by freshly transforming E. coli BL21(DE3) with the expression plasmid and growing in 1 L M9 minimal medium. Cells were grown at 37 °C, induced with 1 mm IPTG at _A_600 ∼0.8, and cell growth was continued at 15 °C overnight. Cells were harvested by centrifugation, resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5 mm imidazole, 1 mm phenylmethanesulfonyl fluoride, PMSF), and sonicated. The cell lysate was centrifuged (13,000 × g, 15 min, 4 °C) and for proteins that expressed as inclusion bodies, the cell pellet was resuspended in lysis buffer and centrifuged (13,000 × g, 15 min, 4 °C). The inclusion body pellet was solubilized in 35 ml buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm PMSF, 4 m urea) and loaded on a 5 ml Ni2+-affinity column (Sigma), washed with 35 ml of binding buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 50 mm imidazole, 4 m urea), and eluted with a total of 15 ml elution buffer (500 mm NaCl, 20 mm Tris-HCl, pH 8.0, 250 mm imidazole, 4 m urea). The fusion proteins were dialyzed in 1 liter of buffer (10 mm sodium phosphate, pH 6.9, 10 mm NaCl) with progressively decreasing concentrations of urea (4, 2, 1, 0.5, 0 m urea). Soluble proteins were purified by Ni2+-affinity chromatography as described but using buffers without urea.
To obtain PrgJ, the GB1PrgJ fusion protein was incubated in 0.02% by volume of 0.1 mm recombinant TEV protease (21) in buffer (50 mm Tris, pH8.0, 20 mm NaCl, 0.5 mm EDTA, 1 mm DTT) at 25 °C overnight, followed by passage through a Ni2+-affinity column and dialysis in low salt buffer (10 mm sodium phosphate, pH 7). For NMR spectroscopy, the fusion proteins (GB1PrgJ and PrgJGB1) were dialyzed up to 10 times in buffer (10 mm sodium phosphate, 10 mm NaCl, pH 6.4).
MxiI was also expressed as a GB1MxiI fusion protein similar to PrgJ as described above. Additionally, a soluble construct lacking the last five residues, MxiICΔ5, was subcloned in pET-21b, which appended the sequence “LEH6” at the C terminus, overexpressed in E. coli and purified by Ni2+-affinity chromatography followed by size exclusion chromatography (GE Healthcare S75 16/60; 150 mm NaCl, 20 mm Tris, pH 7.5).
IL-1β Assay
Mouse bone marrow-derived macrophages (BMDM) were isolated by previously described methods (22). Briefly, bone marrow was flushed from femurs of 8–12 week old C57Bl/6 mice, resuspended in DMEM-10 (Invitrogen) supplemented with 40 ng/ml M-CSF (R&D Systems) and incubated 5 days. Medium was replaced on day 5; and on day 7 the resulting BMDMs were washed with PBS and detached using trypsin-EDTA (Invitrogen). BMDMs were primed using established protocols (23). Briefly, cells were primed using 2 units/ml IFN-γ for 4 h, washed, and triggered using 10 ng/ml LPS for 1 h. After activation, BMDMs were transfected with varying concentrations of each purified protein using Profect P1 transfection reagent (Targeting Systems) per manufacturer's protocol. IL-1β concentration was measured 24 h later by IL-1β-specific ELISA (Invitrogen). The mock transfected control expressed ∼25 pg/ml of IL-1β, and this was considered to represent residual IL-1β that might have been activated by the priming LPS. Thus, IL-1β present in the transfected cultures would represent protein induced by the added bacterial proteins. In each case, bacterial proteins were transfected at 8, 16, or 31 ng/ml of culture. The transfected proteins obtained after the Ni2+-column purification harbored between 25 and 125 EU/ml (∼2.5 - 12.5 ng/ml) LPS as measured using serial dilutions and the ToxinSensor Gel Clot Endotoxin Assay Kit (GenScript, Piscataway N.J.). Each stock protein (1 ml) was diluted to deliver 8, 16, or 31 ng/ml to be transfected. Total (minimal) dilution factor was 1:250,000 for PrgJ, delivering a maximal concentration of LPS to the BMDM of 63 fg/ml in culture. As one example of potency, a study several years ago (24) determined that for an average preparation of LPS from E. coli, 100 ng was stimulatory for macrophages whereas 10 ng was not.
Functional Assays
A S. typhimurium prgIJ deletion strain served as the background for all cultures. For wild type and site-specific substitutions, a plasmid carrying PrgI and PrgJ with the desired mutation was provided in trans. Cultures were grown in 0.3 m LB to late logarithmic phase and culture supernatant and pellet fractions were collected and screened for the presence of T3SS-specific substrates using antibodies specific for SipB, SptP, InvJ, and PrgJ.
Needle Complex Assembly
Needle complexes were purified as described elsewhere (5, 25) and probed with antibodies specific to PrgH, PrgK, InvG, and PrgJ. Purified needle complexes were applied to glow discharged carbon coated Cu-grids and stained with 2% phosphotungstic acid, pH 7.0 and imaged by electron micrscopy. Images were acquired using a Tecnai Biotwin TEM (FEI Company, Hillsboro) at 80 kV using Morada Soft Imaging system and 6 m pixel CCD camera (Olympus, Munster, Germany) (25).
CD Spectroscopy
Samples for circular dichroism (CD) spectroscopy contained 5–10 μm protein in buffer (10 mm NaPO4, pH 7.0). CD spectra were acquired in triplicate using a JASCO J-815 Spectropolarimeter. Wavelength scans were collected at 25 °C at a scan rate of 50 nm/min and thermal denaturation curves were monitored at 222 nm at a temperature ramp rate of 15 °C/h. For MxiICΔ5, CD was performed using a sample at 9 μm protein in buffer (10 mm NaPO4, pH 7.5, 150 mm NaF). Spectra were acquired four times using a JASCO J-715 Spectropolarimeter. Wavelength scans were collected at 25 °C over the range 195–260 nm.
NMR Spectroscopy
NMR data for PrgJ and MxiI were acquired at 25 °C using a Bruker Avance 800 MHz spectrometer equipped with a cryoprobe, processed with NMRPipe (26), and analyzed with NMRView (27). Typical samples contained 0.1 mm PrgJ or 0.3 mm PrgJGB1 or GB1PrgJ in buffer (10 mm NaPO4, pH 7.0, 10 mm NaCl 10% D2O). For MxiICΔ5, 2D 1H-15N HSQC were collected from a 96 μm sample (150 mm NaCl, 20 mm Tris pH 7.5, 5% D2O) at 25 °C using a Bruker 500 MHz spectrometer equipped with a cryoprobe, processed with TopSpin (Bruker) and analyzed with Sparky (28).
Backbone assignments were obtained from 2D 1H-15N HSQC (29) and 3D HNCA (30), CBCA(CO)NH (30), HNCACB (31), and HNCO (32). Secondary structures were identified from the Cα, Cβ, C′, and Hα chemical shifts (33). Side chain assignments were obtained from 2D 1H-13C HMQC (34), 3D HBHA(CO)NH (35), and 3D 13C-edited HMQC-NOESY (36) (_t_mix = 120 ms). NOE (nuclear Overhauser effect) crosspeaks were identified from 3D 15N-edited NOESY-HSQC (37) (_t_mix = 120 ms), and 3D 13C-edited HMQC-NOESY (36) (_t_mix = 120 ms). NOE distance restraints were classified into upper bounds of 2.7, 3.5, 4.5, and 5.5 Å and lower bound of 1.8 Å based on peak volumes. Backbone dihedral angles in the α-helical region (Lys-65 to Ser-82) were constrained to ϕ (−60 ± 20°) and ψ (−40 ± 40°). Structure calculations were done using CYANA (38). The heteronuclear {1H}-15N NOE was acquired with 2048 (1H) × 128 (15N) complex points, 32 scans per point, and a 5 s recycle delay. Triplicate data were acquired to estimate the error bars, and overlapped peaks were excluded in determining the {1H}-15N NOE.
RESULTS
Protein Expression
The inner rod proteins show sequence conservation at their C termini (Fig. 1), and computer programs (39) predicted that PrgJ (15) and other inner rod proteins are likely to be highly α-helical in secondary structure (Fig. 1). Additionally, computer programs that predict disordered regions in proteins such as DISOPRED2 (40) and PONDR (41), predicted that PrgJ is a folded protein. Nevertheless, full-length PrgJ or MxiI when expressed without any tags in E. coli formed inclusion bodies that required solubilization in 8 m urea. Thus, PrgJ was expressed in E. coli as an N- or C-terminal fusion protein with GB1, a 56-residue domain serving as a solubility enhancer tag for NMR characterization (originally used as such in Ref. 42 and recently reviewed in Ref. 43). The GB1PrgJ, PrgJGB1, and GB1MxiI fusion proteins expressed as inclusion bodies and purification required solubilization in 4 m urea followed by exhaustive dialysis to remove the urea and refold the fusion proteins. The refolding of the fusion protein was monitored by CD spectroscopy and two-dimensional NMR spectroscopy. TEV protease digestion of GB1PrgJ and GB1MxiI followed by dialysis in low salt buffer (10 mm sodium phosphate pH 7) yielded monomeric PrgJ and MxiI at concentrations below 0.1 mm (supplemental Fig. S1_B_), which was sufficient for CD and 2D NMR spectroscopy. There was a difference in the solubility of PrgJGB1 and GB1PrgJ. PrgJGB1 (with GB1 attached at the C terminus of PrgJ) was more soluble and can be concentrated to 1 mm whereas GB1PrgJ (with GB1 attached to the N terminus of PrgJ) can only be concentrated to 0.3 mm. Thus, less sensitive NMR experiments such as 3D NMR and heteronuclear {1H}-15N NOE that require higher protein concentrations were acquired using PrgJGB1.
Truncations, which have been successfully used in solubilizing the T3SS needle proteins (7, 44) were also tried to obtain soluble inner rod proteins. Deletion of 20 C-terminal PrgJ residues in GB1PrgJ yielded a soluble protein, PrgJCΔ20, however, other truncations (NΔ22, NΔ38, CΔ5, and CΔ15) yielded inclusion bodies. On the other hand, truncation of the last five residues of MxiI yielded a protein (MxiICΔ5) that was soluble and was purified under non-denaturing condition.
Because full-length PrgJ and MxiI were refolded from inclusion bodies, their bioactivity was assayed by their ability to activate the host inflammatory response. Our purified PrgJ and MxiI triggered an increased level of IL-1β when transfected into mouse bone marrow-derived macrophages (Fig. 2), similar to the results shown by others that the inner rod proteins were able to increase the amount of IL-1β (15–17).
FIGURE 2.
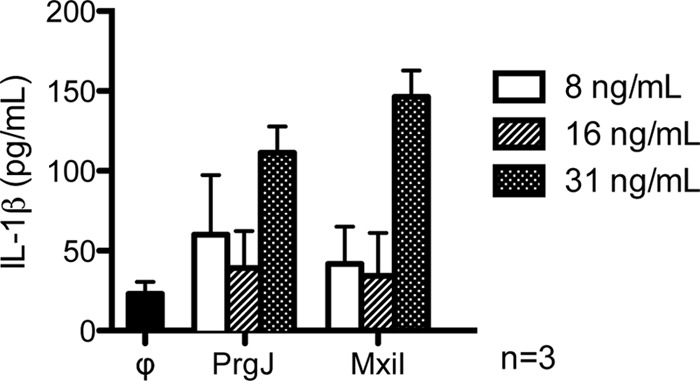
Purified recombinant PrgJ and MxiI were transfected into mouse bone marrow-derived macrophages and probed for IL-1β by ELISA 24 h post-transfection. Control (ϕ) was transfection buffer.
CD Spectroscopy
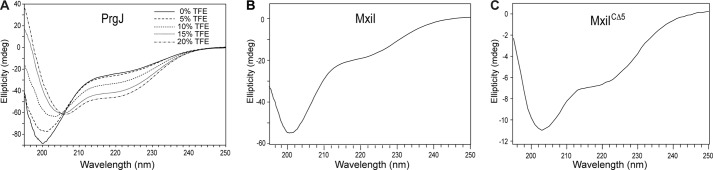
The CD spectrum of PrgJ showed a prominent peak at 200 nm (Fig. 3A) and a slight dip at 220 nm, which indicated that recombinant PrgJ contained some helical content but was largely unfolded. Increasing amounts of trifluoroethanol (TFE), which induces α-helical formation in proteins, increased the helical content of PrgJ (Fig. 3A) as shown by the decreased ellipticity at 220 nm. Likewise, the CD thermal denaturation plot of PrgJ monitored at 222 nm (data not shown) showed no transition or melting temperature, indicating that PrgJ lacked a tertiary structure. The CD spectra of GB1PrgJ and PrgJGB1 indicated that the fusion proteins were folded with peak minima at 200 and 220 nm (supplemental Fig. S1_C_). However, the CD melting curves for GB1PrgJ and PrgJGB1 showed only one transition temperature at 73 °C, which can be superimposed with the melting curve for GB1 (supplemental Fig. S1_D_). The results of CD thermal denaturation showed that PrgJ did not contribute in the thermal stability of the fusion proteins and confirmed that PrgJ lacked a tertiary structure. The CD spectra of the two protein constructs tested for the Shigella inner rod protein, full-length MxiI (Fig. 3B) and the C-terminal truncation MxiICΔ5 (Fig. 3C) suggested that MxiI was largely unfolded as well as indicated by the dominant peak near 200 nm but contained a small amount of helicity as indicated by the slight dip at 222 nm.
FIGURE 3.
CD spectra of recombinant PrgJ with increasing amount of trifluoroethanol (TFE). A, CD spectra of full-length MxiI (B) and MxiICΔ5 a construct where the C-terminal five residues were truncated (C).
NMR Spectroscopy
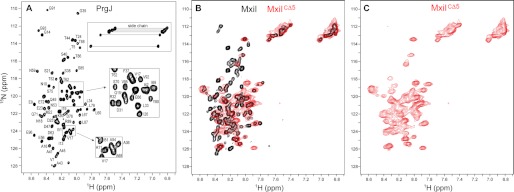
Full-length PrgJ in low salt buffer yielded a 1H-15N HSQC spectrum (Fig. 4A) that was indicative of a monomeric protein based on the sharp HSQC peaks. The narrow backbone proton chemical shift range of about 1 ppm centered around 8.2 ppm suggested that monomeric PrgJ was unfolded or highly α-helical in secondary structure. Likewise, MxiI was soluble at sub-0.1 mm concentration and yielded a 2D 1H-15N HSQC spectrum with a narrow proton chemical shift range suggesting MxiI behaved similarly to PrgJ (Fig. 4B). MxiICΔ5 yielded a 2D 1H-15N HSQC with a narrow proton chemical shift range (Fig. 4C). However, of the 96 peaks expected for MxiICΔ5, only 50 peaks can be seen in Fig. 4C indicating that about half of the MxiICΔ5 peaks were exchanged broadened. However, majority of the backbone peaks and all of the side chain peaks of MxiICΔ5 (Fig. 4C) were in identical position with that of the refolded full-length MxiI (Fig. 4B), suggesting that the refolding protocol yielded the native conformation of MxiI. Based on the results of 2D NMR, PrgJ was deemed better suited for further 3D NMR characterization because the PrgJ peaks were sharper and more uniform in intensities compared with the broader MxiI peaks (Fig. 4). However, at concentrations higher than 0.1 mm, PrgJ precipitated in solution, which prevented further 3D NMR characterization. Thus, 3D NMR was carried out using the more soluble PrgJGB1 fusion proteins.
FIGURE 4.
Two-dimensional 1H-15N HSQC spectra of PrgJ (A), shown with backbone assignments; overlay of the 1H-15N HSQC spectra of full-length MxiI (black peaks) and the construct lacking the C-terminal five residues, MxiICΔ5 (red peaks) (B); and the 1H-15N HSQC spectra of MxiICΔ5 (C).
We first established that tethering PrgJ to GB1 did not alter the native conformation of PrgJ and that NMR characterization of PrgJ using the fusion proteins PrgJGB1 and GB1PrgJ was valid. Both GB1PrgJ and PrgJGB1 also yielded sharp and distinct 1H-15N HSQC peaks (supplemental Fig. S2). The HSQC spectrum of free GB1 can be superimposed with the HSQC spectra of GB1PrgJ and PrgJGB1 (supplemental Fig. S2), thus identifying the PrgJ HSQC peaks. Further, the PrgJ peaks in both GB1PrgJ and PrgJGB1 HSQCs can be superimposed with each other and the vast majority of PrgJ peaks were nearly in identical positions (supplemental Fig. S3_A_), suggesting that the structure and local chemical environment of PrgJ in both fusion proteins were nearly identical. The differences in the peaks between GB1PrgJ and PrgJGB1 HSQCs were due to the different linkers when GB1 was tethered to the N- or C terminus of PrgJ. Likewise, majority of the peaks in the HSQC of PrgJ without the GB1 tag (Fig. 4) were in identical positions with the PrgJGB1 HSQC peaks (supplemental Fig. S2), indicating that the structure of free PrgJ was similar to that of PrgJ in the fusion proteins.
The GB1PrgJ CΔ20 sample was prepared under non-denaturing condition, and the HSQC of GB1PrgJ CΔ20 confirmed that the resolubilization of PrgJGB1 and GB1PrgJ did not alter the overall structural property of the PrgJ (supplemental Fig. S3_B_). Many of the PrgJ CΔ20 peaks were in near identical positions with the PrgJ peaks in GB1PrgJ (supplemental Fig. S3_B_).
PrgJ Backbone Assignments
The millimolar solubility of PrgJGB1 allowed the acquisition of 3D NMR datasets that led to the completion of the backbone resonance assignments of PrgJ. Of the 97 backbone amides of PrgJ, 90 were assigned. The backbone amides of Ser-2, Ile-3, Asp-30, Asn-59, Val-84, Leu-99, and Arg-100 were in rapid exchange with the solvent and could not be assigned. The secondary Cα, Cβ, and Hα chemical shifts suggested the presence of a short helix from residues 30–35 and another helix from residues 65–82 (Fig. 5A). 15N-edited NOESY indicated that majority of PrgJ residues showed mostly intraresidue NOEs (Fig. 5B). There were sequential NOEs at the N terminus and short-range interresidue helical NOEs in residues 65–82 but not in residues 30–35 (Fig. 5B). Further, there were no long range NOEs in PrgJ. The secondary chemical shifts (Fig. 5A) in combination with the helical NOEs (Fig. 5B) confirmed that PrgJ was largely an unfolded protein with a short α-helix at the C-terminal region spanning residues 65–82. Structure calculations using CYANA with 176 interproton NOE distance restraints and 36 dihedral angle restraints confirmed there was only one structured region in PrgJ, from residues Lys-65 to Ser-82, which formed a short α-helix.
FIGURE 5.
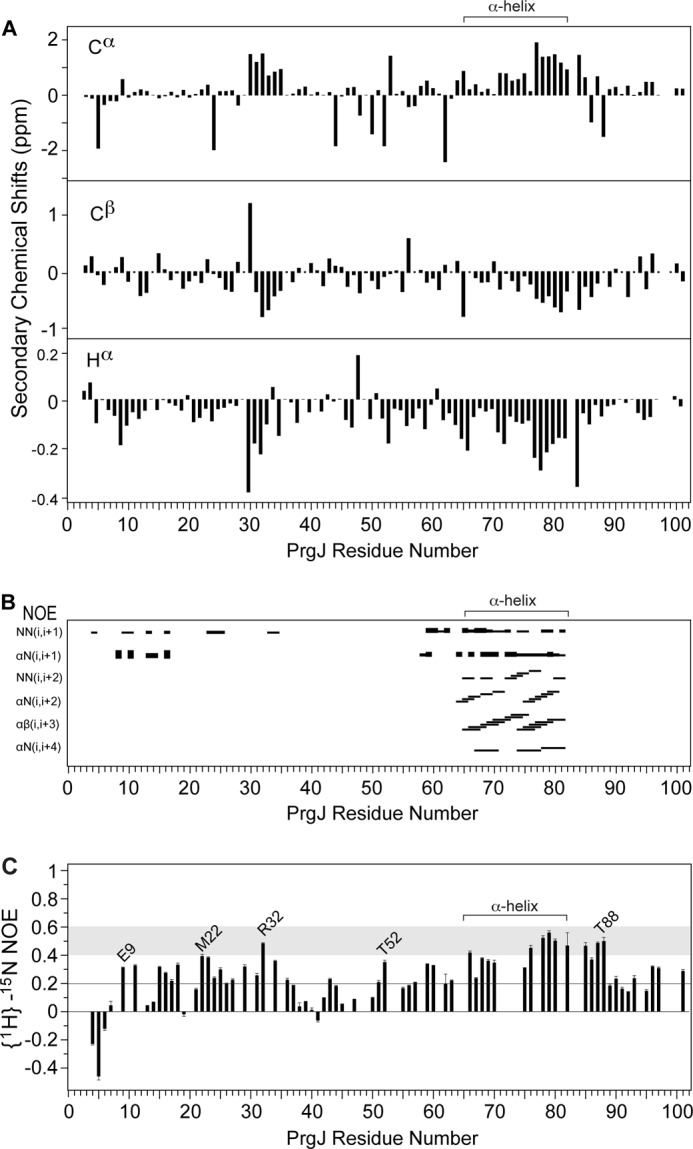
Secondary chemical shifts (Cα, Cβ, and Hα) suggest two potential α-helical regions in PrgJ spanning residues 30–35 and 65–82 (A). PrgJ contains mostly sequential interproton NOEs at the N-terminal region, however, residues 65–82 show α-helical NOEs (B). Heteronuclear {1H}-15N NOE values between 0.4–0.6 (shaded) indicate PrgJ residues have backbone flexibility between that of random coil and well-defined secondary structures (C).
Heteronuclear {1H}-15N NOE
The hetero-nuclear {1H}-15N NOEs provide insight into protein backbone dynamics at the nanosecond to picosecond timescales. Residues with {1H}-15N NOEs values above 0.6 are usually found in well-structured regions of proteins and can be described qualitatively as having rigid flexibility, whereas {1H}-15N NOEs below 0.2 indicate random coil flexibility. The first seven residues of PrgJ were in random coil flexibility as shown by the negative {1H}-15N NOEs for residues 4–6 and the 0.5 {1H}-15N NOE for residue 7 (Fig. 5C). Many residues in the α-helix region (Lys-65 to Ser-82) showed {1H}-15N NOEs between 0.4–0.6. Likewise, several residues outside the α-helix (i.e. Arg-32, Ser-85, Leu-87, and Thr-88) also showed {1H}-15N NOEs between 0.4–0.6, hence these regions were not completely random coil in flexibility but had flexibility approaching that of well-structured secondary structural elements in proteins. Finally, toward the C terminus, for Glu-96, Thr-97, and Ser-101, the region found to be important in activating the host immune response (15), the {1H}-15N NOEs were between 0.2 to 0.4 signifying the C terminus was not a random coil but had flexibility in-between that of random coil and well-structured secondary structures. Additionally, many residues in other regions (i.e. Glu-9, Met-22, Thr-52 in Fig. 5C) outside the α-helix showed {1H}-15N NOEs between 0.2 and 0.4. Thus, although PrgJ lacked extensive secondary structures, many regions were not completely random coil either. Monomeric PrgJ is therefore best described as a partially folded protein.
Mutagenesis and Functional Assays
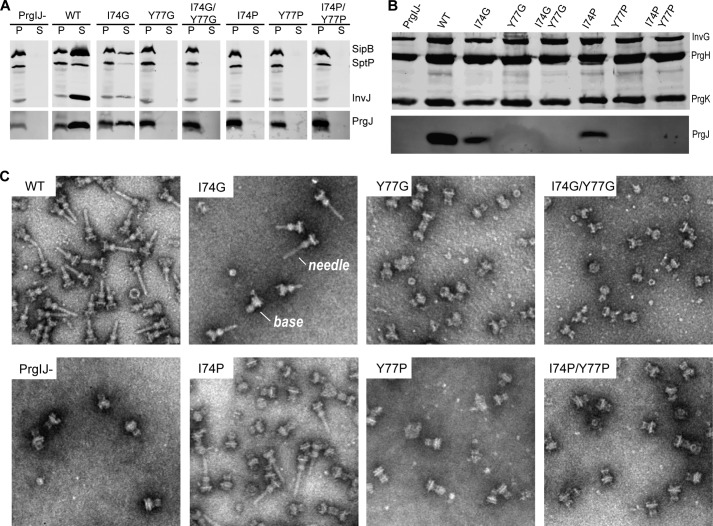
The biological implications of the NMR results showing that the only well-structured part of monomeric PrgJ is an α-helix from residues 65–82 was tested by mutagenesis of residues in the middle of the α-helix. Point mutations that introduced helix destabilizing residues (glycine and proline) were generated at Ile-74 and Tyr-77 of PrgJ. The point mutants (I74G, Y77G, I74P, and Y77P) and the double mutants (I74G/Y77G and I74P/Y77P) were expressed in trans in a Salmonella prgJ deletion strain. The mutations did not significantly alter the level of PrgJ expression (Fig. 6A). The T3SS secretes substrates in a pre-defined order. Therefore we tested the effect of introducing PrgJ helix-destabilizing mutations in the secretion of early (InvJ), middle (SipB), and late (SptP) T3SS substrates. We found that all of the mutants (Y77G, I74P, Y77P, Y77G) but one (Y74G) completely abolished secretion of all the tested T3SS substrates (Fig. 6A). The Y74G mutant exhibit significantly reduced but measurable secretion (Fig. 6A). Combination of different mutations did not significantly alter the observed secretion phenotypes other than the Y74G mutant, that when combined with other mutations, exhibit a loss of function phenotype.
FIGURE 6.
Functional analysis of PrgJ I74 and Y77 glycine and proline mutations. A S. typhimurium PrgIJ deletion strain was transformed with a PrgJ plasmid and grown to late logarithmic phase. Culture pellet (P) and supernatant (S) fractions were probed with antibodies for T3SS proteins SipB, SptP, InvJ and PrgJ (A). Purified needle complexes were probed with antibodies for proteins that form the T3SS basal structure, InvG, PrgH, PrgK, and PrgJ (B). Electron micrographs of purified needle complexes (C).
We also examined isolated needle complexes obtained from strains carrying the different prgJ mutations for the presence of PrgJ in the needle complex structures (Fig. 6B). We found that complexes isolated from the Y77G and Y77P (alone or in combination with others) lacked PrgJ. These results indicate that these mutations abolished the ability of PrgJ to be incorporated into the needle complex base substructure. In contrast, complexes isolated from the I74G and I74P mutants showed reduced but significant levels of PrgJ incorporated into the needle complex base substructure. The I74G mutant was competent for secretion so its incorporation into the base substructure is consistent with this phenotype. The I74P mutant, on the other hand, was secretion incompetent suggesting that the mutation may affect other aspects of PrgJ function.
We examined isolated needle complexes obtained from the different prgJ mutant strains and examined them under the electron microscope (Fig. 6C). As expected, needle complexes isolated from strains expressing PrgJ mutants that were not incorporated into the needle complex base (i.e. Y77G and Y77P) lack the needle substructure. In contrast, needle complexes isolated from strains expressing PrgJ mutants that were incorporated into the needle complex base (i.e. I74G and I74P) exhibited reduced but detectable levels of the needle substructure. Taken together these results demonstrate that helix-destabilizing mutations in Y77 are defective in secretion and failed to assemble the inner rod within the needle complex base (Fig. 6B).
DISCUSSION
The inner rod proteins are important in pathogenesis (11–14, 20) and host-pathogen interaction (15–19), however, little is known about their biophysical properties. The recombinant full-length PrgJ and MxiI used in this study were bioactive and activated the host inflammatory response when transfected in mouse bone-marrow derived macrophages (Fig. 2). The CD (Fig. 3) and NMR results (Figs. 4 and 5) presented here are currently the only available CD and NMR data for any member of this protein family and show unexpectedly that monomeric PrgJ is essentially a partially folded protein lacking a tertiary structure. The only structured portion of PrgJ is an 18-residue α-helix at the C-terminal region spanning residues Lys-65 to Ser-82 and the rest of PrgJ lacks a well-defined secondary structure. The sequence conservation (Fig. 1), CD (Fig. 3) and 2D NMR data for MxiI (Fig. 4) suggest that monomeric MxiI is partially folded as well.
Our observation that purified PrgJ lacks tertiary structure raises issues about the biological relevance of experiments in which either the purified protein or the protein expressed by itself in mammalian cells lead to inflammasome activation and binding to inflammasome components (15–19). It is unclear whether the conformation of PrgJ after its assembly into inner rods (presumably the form that would be eventually presented to the innate immune system after infection) would be capable of activating the same responses as those activated by the purified protein. More experiments will be necessary to ascertain the biological significance of the effects of purified PrgJ on the innate immune system.
Despite being highly polar (PrgJ has theoretical pI of 4.5), recombinant PrgJ and MxiI tend to aggregate in solution at concentrations needed for 3D NMR studies, making solution-based biophysical characterization challenging. This poor solution behavior of the inner rod proteins is likely due to their self-polymerizing nature during the assembly of the inner rod. Nevertheless, monomeric PrgJ can be solubilized in low salt buffer at concentrations below 0.1 mm, which allowed the acquisition of CD (Fig. 3) and 2D NMR spectra (Fig. 4). To acquire low-sensitivity NMR data, higher solubility of PrgJ was afforded by fusion with the GB1 domain, a small highly soluble protein that has been successfully used to solubilize many difficult proteins for NMR studies (42, 43).
Mutagenesis of residues in the PrgJ α-helix followed by functional assays show a critical role for Tyr-77 in the assembly of the Salmonella inner rod (Fig. 6). The phenotype associated with the Tyr-77 mutants can be explained as resulting mainly from the failure of Salmonella to secrete PrgJ and assemble the inner rod (Fig. 3B), which then affected the assembly of the external needle (Fig. 6C) and subsequent substrate switching. Without substrate switching, effectors or other structural proteins that must pass through the needle complex are thereby not secreted. These results suggest that Tyr-77 is important in the PrgJ protein-protein interactions required for inner rod assembly. Ile-74 mutants on the other hand can be found in purified needle complexes albeit at significantly reduced level (Fig. 6B), suggesting that Ile-74 mutations affected PrgJ function to a lesser degree.
Wood et al. (13) performed an extensive mutagenesis analysis of YscI, the inner rod protein of Y. pestis, and showed that YscI plays a critical role in substrate specificity switching, confirming previous observations in Salmonella implicating PrgJ in substrate switching (4). Wood et al. showed that the YscI mutants Q84A, L87A, E94K, E95K, and L96A are defective in needle assembly (13). These residues are located at the C-terminal region, which shows the highest degree of sequence conservation among the inner rod proteins (Fig. 1). The overall sequence similarity between YscI and PrgJ is 25%, however, for C-terminal 40 residues, the sequence similarity is 40%. Thus, we expect that the YscI residues (Q84A, L87A, E94K, E95K, and L96A) to be in an α-helix as well. We suggest the YscI mutants introduced by Wood et al. (13) altered the surface property of the YscI C-terminal helix, which then affected the protein-protein interactions of YscI that manifested in the phenotypes observed by Wood et al. (13). In Salmonella, Marlovits et al. (5) showed by cryoEM that the positioning of the inner rod within the basal structure suggests that PrgJ may interact with other components of the T3SS basal structure (5). The exact nature of the pairwise protein-protein interactions of PrgJ has not been worked out in detail and will be complicated because some of the expected binding partners are membrane proteins.
The atomic structure of the assembled PrgJ inner rod is currently unknown. We hypothesize that the predicted propensity of PrgJ to be highly α-helical reflects a conformational state that is different from what we have seen in solution. Perhaps, upon assembly of the inner rod, regions of PrgJ outside of the C-terminal α-helix (Lys-65 to Ser-82) will also form regular α-helices that pack together to assemble the inner rod - similar to the mechanism of assembly of the external needle itself (8, 45, 46). As monomers, about half the length of the needle proteins are flexible and do not form well-defined secondary structures (7, 44). Upon needle assembly, the flexible regions of the needle monomers become ordered into α-helices that pack together (46). Others have also shown that needle assembly involves folding transitions of needle monomers into short β-strands (8, 47). Protein folding transitions in T3SS (8, 46, 47) are not only confined to the structural proteins as others have reported that chaperone-effector interaction also involves folding transitions (48). Our results show that protein flexibility is an important facet of the inner rod proteins.
Supplementary Material
Supplemental Data
Supplemental Data
Acknowledgments
We thank Thenmalarchelvi Rathinavelan (IIT-Hyderabad) for helpful discussion, Janet Deane (Oxford University) for deriving the MxiICΔ5 construct, and Thomas Yankee (Kansas University Medical Center) for mouse bone marrow.
*
This work was supported, in whole or in part, by National Institutes of Health Grant AI074856 (to R. N. D.), National Institutes of Health Grant AI030492 (to J. G.), KU Diabetes Institute (to S. B.), and Wellcome Trust Studentship (to M. A. M).
4
The abbreviations used are:
T3SS
type III secretion system
NMR
nuclear magnetic resonance
NOE
nuclear Overhauser effect
NOESY
NOE spectroscopy
HSQC
heteronuclear single quantum coherence
PRE
paramagnetic relaxation enhancement
BMDM
bone marrow-derived macrophage.
REFERENCES
- 1.Galán J. E., Wolf-Watz H. (2006) Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573 [DOI] [PubMed] [Google Scholar]
- 2.Moraes T. F., Spreter T., Strynadka N. C. (2008) Piecing together the type III injectisome of bacterial pathogens. Curr. Opin. Struct. Biol. 18, 258–266 [DOI] [PubMed] [Google Scholar]
- 3.Schraidt O., Marlovits T. C. (2011) Three-dimensional model of Salmonella's needle complex at subnanometer resolution. Science 331, 1192–1195 [DOI] [PubMed] [Google Scholar]
- 4.Marlovits T. C., Kubori T., Lara-Tejero M., Thomas D., Unger V. M., Galán J. E. (2006) Assembly of the inner rod determines needle length in the type III secretion injectisome. Nature 441, 637–640 [DOI] [PubMed] [Google Scholar]
- 5.Marlovits T. C., Kubori T., Sukhan A., Thomas D. R., Galán J. E., Unger V. M. (2004) Structural insights into the assembly of the type III secretion needle complex. Science 306, 1040–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimbrough T. G., Miller S. I. (2000) Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. U.S.A. 97, 11008–11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Ouellette A. N., Egan C. W., Rathinavelan T., Im W., De Guzman R. N. (2007) Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J. Mol. Biol. 371, 1304–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poyraz O., Schmidt H., Seidel K., Delissen F., Ader C., Tenenboim H., Goosmann C., Laube B., Thünemann A. F., Zychlinsky A., Baldus M., Lange A., Griesinger C., Kolbe M. (2010) Protein refolding is required for assembly of the type three secretion needle. Nat. Struct. Mol. Biol. 17, 788–792 [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee S., Zhong D., Nordhues B. A., Battaile K. P., Lovell S., De Guzman R. N. (2011) The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 20, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lara-Tejero M., Galán J. E. (2009) Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 77, 2635–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blocker A., Jouihri N., Larquet E., Gounon P., Ebel F., Parsot C., Sansonetti P., Allaoui A. (2001) Structure and composition of the Shigella flexneri “needle complex”, a part of its type III secreton. Mol. Microbiol. 39, 652–663 [DOI] [PubMed] [Google Scholar]
- 12.Magdalena J., Hachani A., Chamekh M., Jouihri N., Gounon P., Blocker A., Allaoui A. (2002) Spa32 regulates a switch in substrate specificity of the type III secreton of Shigella flexneri from needle components to Ipa proteins. J. Bacteriol. 184, 3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood S. E., Jin J., Lloyd S. A. (2008) YscP and YscU switch the substrate specificity of the Yersinia type III secretion system by regulating export of the inner rod protein YscI. J. Bacteriol. 190, 4252–4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein J. R., Fahlen T. F., Jones B. D. (2000) Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68, 3368–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao E. A., Mao D. P., Yudkovsky N., Bonneau R., Lorang C. G., Warren S. E., Leaf I. A., Aderem A. (2010) Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. U.S.A. 107, 3076–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warren S. E., Duong H., Mao D. P., Armstrong A., Rajan J., Miao E. A., Aderem A. (2011) Generation of a Listeria vaccine strain by enhanced caspase-1 activation. Eur. J. Immunol. 41, 1934–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lightfield K. L., Persson J., Trinidad N. J., Brubaker S. W., Kofoed E. M., Sauer J. D., Dunipace E. A., Warren S. E., Miao E. A., Vance R. E. (2011) Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 79, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y., Yang J., Shi J., Gong Y. N., Lu Q., Xu H., Liu L., Shao F. (2011) The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 [DOI] [PubMed] [Google Scholar]
- 19.Kofoed E. M., Vance R. E. (2011) Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gong H., Vu G. P., Bai Y., Yang E., Liu F., Lu S. (2010) Differential expression of Salmonella type III secretion system factors InvJ, PrgJ, SipC, SipD, SopA, and SopB in cultures and in mice. Microbiology 156, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geisbrecht B. V., Bouyain S., Pop M. (2006) An optimized system for expression and purification of secreted bacterial proteins. Protein Expr. Purif. 46, 23–32 [DOI] [PubMed] [Google Scholar]
- 22.Davies J. Q., Gordon S. (2005) Isolation and culture of murine macrophages. Methods Mol. Biol. 290, 91–103 [DOI] [PubMed] [Google Scholar]
- 23.Meltzer M. S. (1981) Macrophage activation for tumor cytotoxicity: characterization of priming and trigger signals during lymphokine activation. J. Immunol. 127, 179–183 [PubMed] [Google Scholar]
- 24.Zughaier S. M., Ryley H. C., Jackson S. K. (1999) Lipopolysaccharide (LPS) from Burkholderia cepacia is more active than LPS from Pseudomonas aeruginosa and Stenotrophomonas maltophilia in stimulating tumor necrosis factor α from human monocytes. Infect Immun 67, 1505–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kubori T., Matsushima Y., Nakamura D., Uralil J., Lara-Tejero M., Sukhan A., Galán J. E., Aizawa S. I. (1998) Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science 280, 602–605 [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 27.Johnson B. A. (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 28.Goddard T., Kneller T. (2000) SPARKY NMR Analysis Software, University of California, San Francisco, CA [Google Scholar]
- 29.Grzesiek S., Bax A. (1993) The importance of not saturating H2O in protein NMR. Application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 30.Grzesiek S., Döbeli H., Gentz R., Garotta G., Labhardt A. M., Bax A. (1992) 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma. Biochemistry 31, 8180–8190 [DOI] [PubMed] [Google Scholar]
- 31.Wittekind M., Mueller L. (1993) HNCACB, a high sensitivity 3D NMR experiment to correlate amide proton and nitrogen resonances with the alpha-carbon and beta-carbon resonances in proteins. J. Magn. Reson. 101B, 201–205 [Google Scholar]
- 32.Muhandiram D. R., Kay L. E. (1994) Gradient-enhanced triple-resonance three-dimensional NMR experiments with improved sensitivity. J. Magn. Reson. 103, 203–216 [Google Scholar]
- 33.Wishart D. S., Nip A. M. (1998) Protein chemical shift analysis: a practical guide. Biochem. Cell Biol. 76, 153–163 [DOI] [PubMed] [Google Scholar]
- 34.Tolman J. R., Chung J., Prestegard J. H. (1992) Pure-phase heteronuclear multiple-quantum spectroscopy using field gradient selection. J. Magn. Reson. 98, 462–467 [Google Scholar]
- 35.Grzesiek S., Bax A. (1993) Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J. Biomol. NMR 3, 185–204 [DOI] [PubMed] [Google Scholar]
- 36.Fesik S. W., Zuiderweg E. R. P. (1998) Heteronuclear three-dimensional NMR spectroscopy. A strategy for the simplification of homonuclear two-dimensional NMR spectra. J. Magn. Reson. 78, 588–593 [Google Scholar]
- 37.Marion D., Driscoll P. C., Kay L. E., Wingfield P. T., Bax A., Gronenborn A. M., Clore G. M. (1989) Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1β. Biochemistry 28, 6150–6156 [DOI] [PubMed] [Google Scholar]
- 38.Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 39.Combet C., Blanchet C., Geourjon C., Deléage G. (2000) NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 [DOI] [PubMed] [Google Scholar]
- 40.Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 41.Li X., Romero P., Rani M., Dunker A. K., Obradovic Z. (1999) Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Informatics Workshop on Genome Informatics 10, 30–40 [PubMed] [Google Scholar]
- 42.Huth J. R., Bewley C. A., Jackson B. M., Hinnebusch A. G., Clore G. M., Gronenborn A. M. (1997) Design of an expression system for detecting folded protein domains and mapping macromolecular interactions by NMR. Protein Sci. 6, 2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou P., Wagner G. (2010) Overcoming the solubility limit with solubility-enhancement tags: successful applications in biomolecular NMR studies. J. Biomol. NMR. 46, 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang L., Wang Y., Picking W. L., Picking W. D., De Guzman R. N. (2006) Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei. J. Mol. Biol. 359, 322–330 [DOI] [PubMed] [Google Scholar]
- 45.Galkin V. E., Schmied W. H., Schraidt O., Marlovits T. C., Egelman E. H. (2010) The structure of the Salmonella typhimurium type III secretion system needle shows divergence from the flagellar system. J. Mol. Biol. 396, 1392–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deane J. E., Roversi P., Cordes F. S., Johnson S., Kenjale R., Daniell S., Booy F., Picking W. D., Picking W. L., Blocker A. J., Lea S. M. (2006) Molecular model of a type III secretion system needle: Implications for host-cell sensing. Proc. Natl. Acad. Sci. U.S.A. 103, 12529–12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fujii T., Cheung M., Blanco A., Kato T., Blocker A. J., Namba K. (2012) Structure of a type III secretion needle at 7-Å resolution provides insights into its assembly and signaling mechanisms. Proc. Natl. Acad. Sci. U.S.A. 109, 4461–4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodgers L., Gamez A., Riek R., Ghosh P. (2008) The type III secretion chaperone SycE promotes a localized disorder-to-order transition in the natively unfolded effector YopE. J. Biol. Chem. 283, 20857–20863 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data
Supplemental Data