Plexins Are GTPase Activating Proteins for Rap and Are Activated by Induced Dimerization (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 7.
Published in final edited form as: Sci Signal. 2012 Jan 17;5(207):ra6. doi: 10.1126/scisignal.2002636
Abstract
Plexins are cell surface receptors that bind to semaphorins and transduce signals that regulate neuronal development, immune responses, and other processes. Signaling through plexins has been proposed to rely on specific GTPase activating protein (GAP) activity for R-Ras and M-Ras. Activation of this GAP activity appears to require simultaneous binding of semaphorin to the plexin extracellular domain and of a RhoGTPase to the cytoplasmic region. However, GAP activity of plexins has eluded detection in several recent studies. We show that the purified cytoplasmic region of plexin uses a non-canonical catalytic mechanism to act as a GAP for Rap, but not for R-Ras or M-Ras. The RapGAP activity of plexins was normally autoinhibited and could be activated by induced dimerization. Our biochemical and crystallographic analyses demonstrate that binding of the RhoGTPases did not directly contribute to activation of plexin RapGAP activity. Semaphorin stimulated the RapGAP activity of full-length plexin in cells, which was required for plexin-mediated neuronal growth cone collapse. These findings together define a pathway for plexin signaling and provide new insights into the mechanism for semaphorin-induced activation of plexin.
Introduction
Plexins are a large family of cell surface receptors for the axon guidance molecules semaphorins (1, 2). Semaphorin-plexin signaling is essential for the regulation of neuronal development and other processes. Dysfunction in plexin pathways has been implicated in various diseases including neurological disorders and cancer (3, 4).
Semaphorins are secreted or membrane bound multi-domain proteins, containing an N-terminal Sema domain that is responsible for both dimerization and binding to plexin (5, 6). Dimerization of some semaphorins is further stabilized by an inter-chain disulfide bond (7, 8). Plexins are transmembrane proteins with a multi-domain extracellular region that also contains an N-terminal Sema domain. Binding of semaphorin to the Sema domain of plexin leads to activation of its cytoplasmic region, which relays the signal to downstream pathways. Several lines of evidence suggest that plexin is activated by semaphorin-induced dimerization or oligomerization (9, 10). Three structural studies have revealed how one semaphorin dimer binds two plexin Sema domains simultaneously (11–13). However, the mechanism by which this interaction leads to activation of the plexin cytoplasmic region remains unclear, partly due to the absence of the multiple membrane proximal domains of plexin in these structures. The distance between the C-termini of the two plexin extracellular segments in the structures could be bridged by the missing membrane proximal domains, allowing dimerization of the transmembrane and cytoplasmic regions for triggering activation (12, 13). On the other hand, these structures do not exclude the possibility that in the semaphorin-plexin complex the cytoplasmic regions from the two monomers remain separated. This would be consistent with a mechanism in which the cytoplasmic region of plexin forms a pre-formed autoinhibitory dimer, and is activated by semaphorin-induced separation of this dimer (5, 14).
The cytoplasmic region of plexin contains two segments that show sequence similarity to Ras guanosine triphosphatase (GTPase) activating proteins (RasGAPs) (15, 16), which fold together to form an intact GAP domain with an overall fold virtually identical to canonical RasGAPs such as p120GAP (17, 18). The segment between the two GAP homology regions forms a RhoGTPase-binding domain (RBD) that resides at one side of the GAP domain. A juxtamembrane segment that connects the transmembrane region to the GAP domain adopts a helix-loop-helix-loop conformation and wraps around the GAP domain. The GAP domain is essential for plexin signaling because mutating the conserved arginine residue corresponding to the catalytic “arginine finger” in p120GAP abolishes plexin-mediated changes in cell morphology (15, 19–21). The RBD binds to a group of Rho family GTPases, such as Rac1 and Rnd1, in their GTP-bound active state (10, 15, 22, 23). Plexin activation appears to require both binding of a RhoGTPase to the RBD and semaphorin to the extracellular region (19, 24, 25). There is indirect evidence indicating that the RhoGTPases contribute to activation by inducing a conformational change in the GAP domain (24).
Plexins has been reported to act as GAPs for the Ras homologues R-Ras and M-Ras and inactivate them by catalyzing hydrolysis of the bound GTP to GDP (19, 20, 24, 25). Inactivation of R-Ras by plexin GAP domains may contribute to neuronal axon growth cone collapse by decreasing integrin-mediated adhesion (26). Intriguingly, although binding between plexins and R-Ras has been observed (18, 21), detection of the R-Ras GAP activity of plexins has proven to be challenging (17, 18, 21). Therefore plexins have been suggested to inhibit R-Ras by sequestering it from downstream effectors, rather than converting it to the GDP-bound inactive state (21).
In search of the enzymatic substrates of the plexin GAP domain, we noticed that SynGAP and the GAP1 family proteins, whose GAP domains are structurally similar to those in plexins, display GAP activity towards both Ras and its homologue Rap (27, 28). As exemplified by p120GAP, most GAPs for Ras, R-Ras and M-Ras catalyze GTP hydrolysis for these Ras homologues by using the “arginine finger” residue, which stabilizes the leaving γ-phosphate group in GTP. (29). Canonical GAPs for Rap, however, are structurally distinct and use a fundamentally different catalytic mechanism that requires an “asparagine thumb” residue for orienting a catalytic water molecule. (30). Although the dual specific GAPs apparently use the same mechanism as p120GAP for Ras GTP hydrolysis, they can also act on Rap through a non-canonical mechanism that is yet to be structurally characterized (31).
These analyses prompted us to test whether plexins possess RapGAP activity. Our results demonstrate that purified plexin cytoplasmic regions exhibit GAP activity for Rap, but not for R-Ras or M-Ras. RapGAP activity could be stimulated by induced dimerization of plexin, suggesting an allosteric activation mechanism exerted by inter-molecular interactions. We also showed that RBD-binding RhoGTPases did not affect the plexin RapGAP activity in solution. Our crystal structure of the PlexinA1-Rac1 complex demonstrated that Rac1 does not induce any obvious conformational change in plexin, nor is it likely to facilitate binding of Rap to plexin by interacting with Rap directly. We further showed that semaphorin stimulated plexin RapGAP activity in cells, which was required for plexin-mediated neuronal growth cone collapse, supporting the notion that Rap is a critical downstream transducer in plexin signaling pathways.
Results
Purified plexin cytoplasmic regions exhibit GAP activity for Rap but not for R-Ras or M-Ras
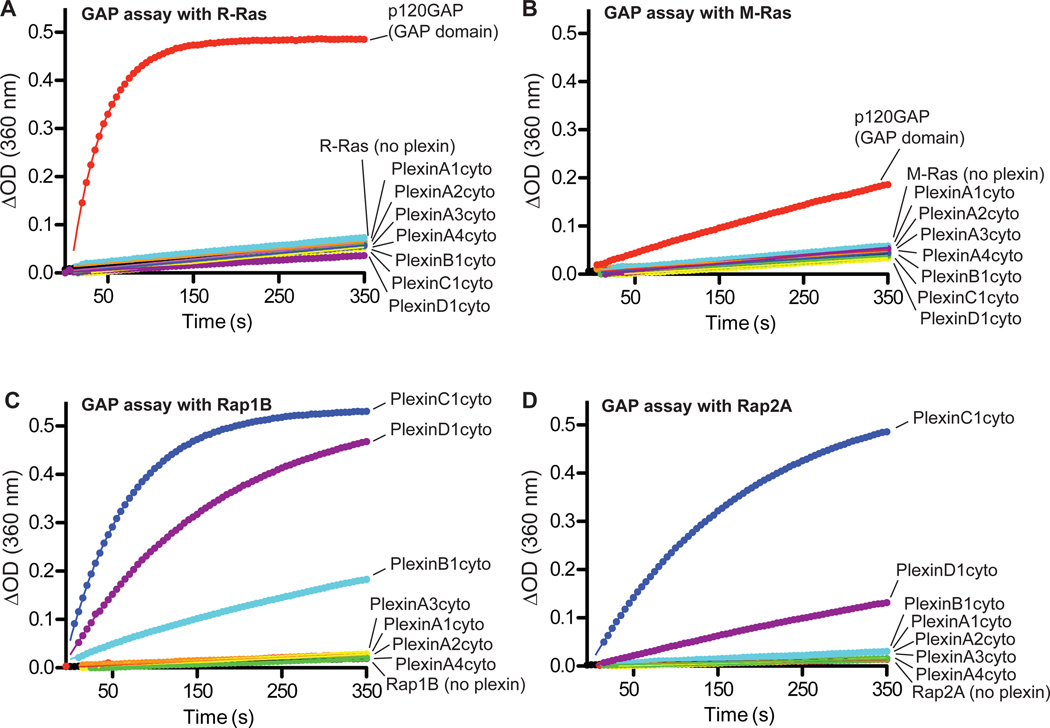
To investigate the GAP activity of plexin family members, we purified the cytoplasmic regions (referred to as plexinscyto) of Plexin A1 to A4, B1, C1 and D1. The GAP activity of these proteins towards purified, GTP-loaded R-Ras and M-Ras was measured with a continuous photometric assay (32). GTP hydrolysis of either R-Ras or M-Ras was accelerated by the GAP domain of p120GAP (0.05 µM) but not by the plexincyto proteins (10 µM) (Fig. 1, A and B).
Fig. 1. Purified plexinscyto display GAP activity for Rap but not for R-Ras or M-Ras.
(A to D) None of the tested plexinscyto accelerate GTP hydrolysis for R-Ras (A) or M-Ras (B).
Plexinscyto show GAP activity towards both Rap1B (C) and Rap2A (D).
Data shown are representative of at least 3 independent experiments.
In the RasGAP family, SynGAP and three proteins of the GAP1 group exhibit dual specificity towards both Ras and Rap (27, 28). The high degree of structural similarity of the plexin GAP domain to that of SynGAP (17) led us to speculate whether plexins also possess GAP activity towards Rap. Rap1B and Rap2A, two members of the Rap subfamily, were used as substrates to measure the GAP activity of plexinscyto. The photometric assays showed that the plexincyto proteins accelerated GTP hydrolysis of both Rap1B and Rap2A (Fig. 1, C and D). These results were confirmed by an HPLC-based assay that assesses the ratio of GTP and GDP bound to Rap after incubation with plexins (fig. S1). Among the plexins we tested, PlexinC1cyto displayed the highest activity (first order rate constant k = 0.013 s−1 for Rap1B). Class A plexins were less active (k =1.4~1.6×10−4 s−1 for Rap1B), but showed higher activity than the intrinsic activity of Rap (Rap1B: k = 9.1×10−5 s−1).
Dimerization stimulates the RapGAP activity of plexinscyto
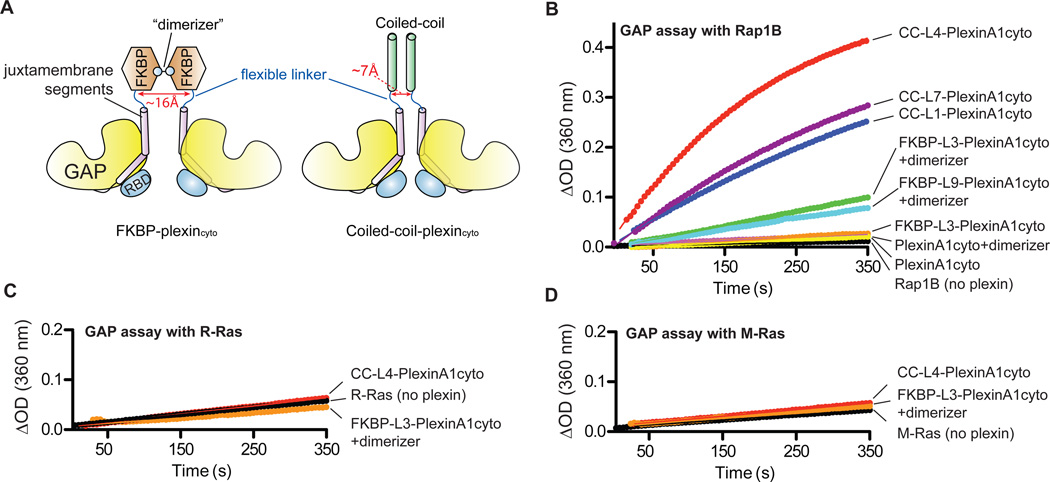
Plexinscyto are predominantly monomeric in solution (fig. S2B) (17). To test whether their RapGAP activity is regulated by dimerization, we fused PlexinA1cyto to the C-terminus of a modified FKBP (FK506-binding protein), which can be induced to homodimerize upon binding to the bivalent compound AP20187 (the dimerizer) (33). We placed FKBP at the N-terminus rather than the C-terminus of PlexinA1cyto, because this topology is likely to better mimic semaphorin-induced dimerization of full-length plexins (13). A flexible linker (3 or 9 residues) was included between FKBP and PlexinA1cyto to avoid imposing constraints on the distance and orientation between the two plexincyto molecules in the induced dimer (Fig. 2A). These proteins are referred to as FKBP-L3-PlexinA1cyto and FKBP-L9-PlexinA1cyto, respectively.
Fig. 2. Effect of dimerization on the GAP activity of plexinscyto.
(A) Schematic diagrams of the FKBP or coiled-coil plexincyto fusions to enable induced or constitutive dimerization.
(B) The RapGAP activity of PlexinA1cyto is stimulated by dimerization. “+dimerizer” indicates pre-incubation of the FKBP fusions with the dimerizer.
(C to D) CC-L4-PlexinA1cyto and dimerized FKBP-PlexinA1cyto do not show GAP activity for R-Ras (C) or M-Ras (D).
Data shown are representative of at least 3 independent experiments.
Similar to native PlexinA1cyto, the FKBP-PlexinA1cyto fusion proteins were predominantly monomeric in solution (fig. S2A). Their RapGAP activities were also similar to that of the native protein (Fig. 2B). As expected, adding the dimerizer induced robust dimerization of the FKBP-PlexinA1cyto fusions (fig. S2A). Dimerized FKBP-PlexinA1cyto proteins showed higher RapGAP activity (Fig. 2B). The dimerizer did not alter the activity of native PlexinA1cyto (Fig. 2B), supporting the notion that the stimulated activities of the FKBP-PlexinA1cyto proteins can be attributed to formation of an activating dimer. To test the generality of this dimerization-driven activation mechanism, we constructed a FKBP fusion of PlexinC1cyto with the 3-residue linker, which showed three-fold higher activity when dimerized (fig. S2C) compared to monomeric PlexinC1cyto (Fig. 1C). We also tested the dimerization-induced activation of FKBP-L3-PlexinC1cyto in cells by using a GST-RalGDS pull-down assay (34). Although degradation of the protein after treatment with the dimerizer obscured the expected increase in RapGAP activity, the basal activity of PlexinC1 was readily detected by this cell-based assay (fig. S3).
FKBP-L3-PlexinA1cyto was activated by dimerization to a greater extent (4-fold) than was FKBP-L9-PlexinA1cyto (3-fold) (Fig. 2B), suggesting that closer proximity of the two plexincyto monomers favors formation of the activating dimer. To further explore the effect of the inter-monomer distance on plexin activation, we fused the PlexinA1cyto to the C-terminus of the coiled-coil dimerization motif of the transcription factor GCN4 (general control non-repressed 4) to enforce constitutive dimerization on plexins (35). The distance between the two C-termini in the GCN4 coiled-coil helix dimer was ~7 Å (PDB ID: 2ZTA), smaller than that in the FKBP dimer (~16Å) (PDB ID: 1BL4). We also included a flexible linker of various lengths (1, 4 and 7 residues) between the coiled-coil and PlexinA1cyto to enable sampling of a wider range of inter-monomer distances. The 1-residue linker essentially restrains the two N-termini in plexinscyto within the range of direct contacts, whereas the 7-residue linker allows more than 40 Å distance between the two N-termini (assuming ~3 Å span per linker residue, 7 × 3 × 2 = 42 Å).
Insertion of the coiled-coil motif induced robust dimerization of PlexinA1cyto (fig. S2). The coiled-coil PlexinA1cyto dimer was more active than the dimerized FKBP-PlexinA1cyto fusion (Fig. 2B). The construct with the 4-residue linker (CC-L4-PlexinA1cyto) displayed the highest RapGAP activity (k = 3.9 × 10−3 s−1 for Rap1B), which was 28-fold and 7-fold higher than the native protein and FKBP-L3-PlexinA1cyto, respectively. In contrast, none of the dimerized FKBP- and coiled-coil-PlexinA1cyto stimulated GTP hydrolysis for either R-Ras or M-Ras under similar conditions (Fig. 2, C and D).
We also generated and purified coiled-coil fusions of PlexinA2 and A4. They both formed constitutive dimers and displayed higher RapGAP activity than their respective native proteins (fig. S2D). The degree of activation varied among different plexin family members, which may be due to differences in their intrinsic catalytic capacity or the degree of activating dimer formation induced by the coiled-coil motif.
These results together suggest that the RapGAP activity of monomeric plexins is autoinhibited, and it can be activated by formation of the activating dimer. Previous studies have suggested that SynGAP and other dual specificity GAPs interact with Rap in a binding mode similar to that of the p120GAP-Ras complex (27, 31). Assuming that plexins also bind Rap in the same way, the Rap binding sites in monomeric PlexinA3 and B1 structures are clearly narrower than that in SynGAP (17, 18). This might prevent Rap binding and serve as the autoinhibition mechanism. Higher activity of the coiled-coilplexincyto dimers compared to the FKBP-plexincyto dimers suggests that close proximity of the two juxtamembrane segments in plexin monomers promotes formation of the activating dimer. These findings support the idea that semaphorin activates plexin by inducing dimerization of the plexin cytoplasmic region, as opposed to separation of a preformed inhibitory dimer.
Plexins use a non-canonical mechanism for catalyzing GTP hydrolysis for Rap
Although Rap and Ras are close homologues, their respective canonical GAPs are structurally unrelated and use different catalytic mechanisms. RasGAPs such as p120GAP catalyze GTP hydrolysis for Ras, R-Ras, or M-Ras by using an arginine finger residue (29), which neutralizes the developing negative charge of the leaving g-phosphate group in GTP. The arginine residue also stabilizes Gln61 in Ras, which orients a nucleophilic water molecule for catalysis. Rap lacks the ability to position the nucleophilic water because it contains a threonine residue at the equivalent position (Thr61). Canonical RapGAPs facilitate GTP hydrolysis for Rap with an asparagine residue (“Asn thumb”), which plays a role equivalent to that of Gln61 in Ras (30).
The dual specificity GAPs, such as SynGAP, can act as RapGAPs without an “Asn thumb” (31, 36). They are structurally related to RasGAPs and also possess the arginine finger. This arginine residue is required for GTP hydrolysis for Rap, although its precise role is unknown (31). Mutation of Gln63 in Rap to either alanine (Gln63→Ala; Q63A) or glutamate (Gln63→Glu; Q63E) renders Rap insensitive to the dual specificity GAPs, but only slightly affects GTP hydrolysis catalyzed by canonical RapGAPs (31). These observations have led to the model that the dual specificity GAPs induce a conformational change in Rap, which pulls Gln63 into the active site where it functions in catalysis in a manner similar to that of Gln61 in Ras (31).
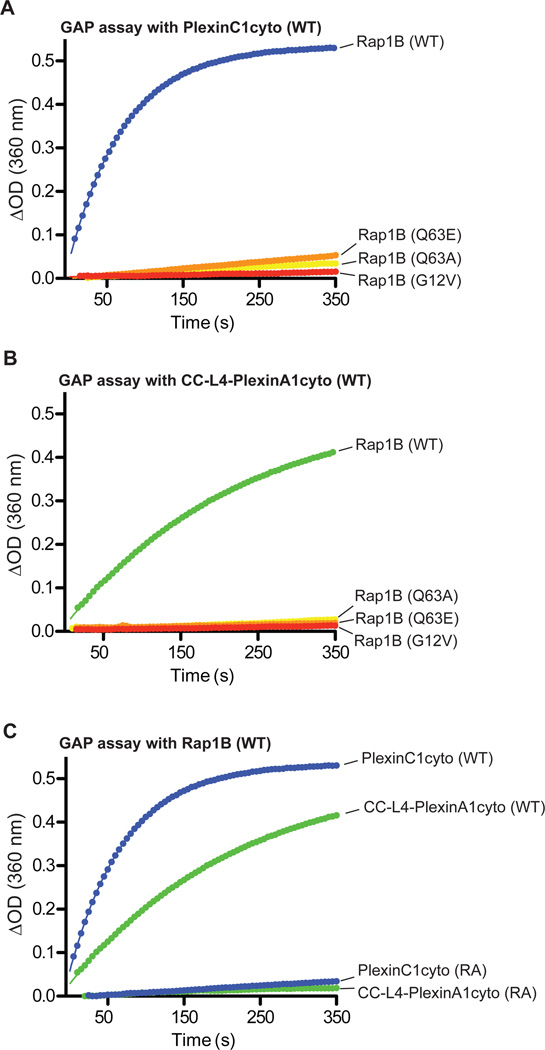
The structural similarity of plexins to SynGAP suggests that they use the same alternative mechanism for catalyzing GTP hydrolysis for Rap. To test this idea, we measured the activity of plexins towards the Rap1B Q63E and Q63A mutants. Both of these two mutations inhibited GTP hydrolysis catalyzed by either monomeric or dimeric plexins (Fig. 3A and B). In addition, we constructed the Rap G12V mutant, which is also sensitive to the canonical RapGAPs but not to SynGAP (27). This mutation impaired plexin-catalyzed GTP hydrolysis. Similar to other dual specificity GAPs, mutating the arginine finger residue (Arg1429) in PlexinA1 to alanine rendered both monomeric and dimeric plexinscyto catalytically inactive (Fig. 3C).
Fig. 3. Mutational analysis of the RapGAP activity of plexinscyto.
(A to B) The G12V, Q63A or Q63E mutation in Rap1B inhibits GTP hydrolysis catalyzed by PlexinC1cyto (A) or CC-L4-PlexinA1cyto (B).
(C) Mutation of the arginine finger (RA) abolishes the RapGAP activity of both PlexinC1cyto and CC-L4-PlexinA1cyto.
Data shown are representative of at least 3 independent experiments.
Rac1 does not induce notable conformational changes in plexincyto
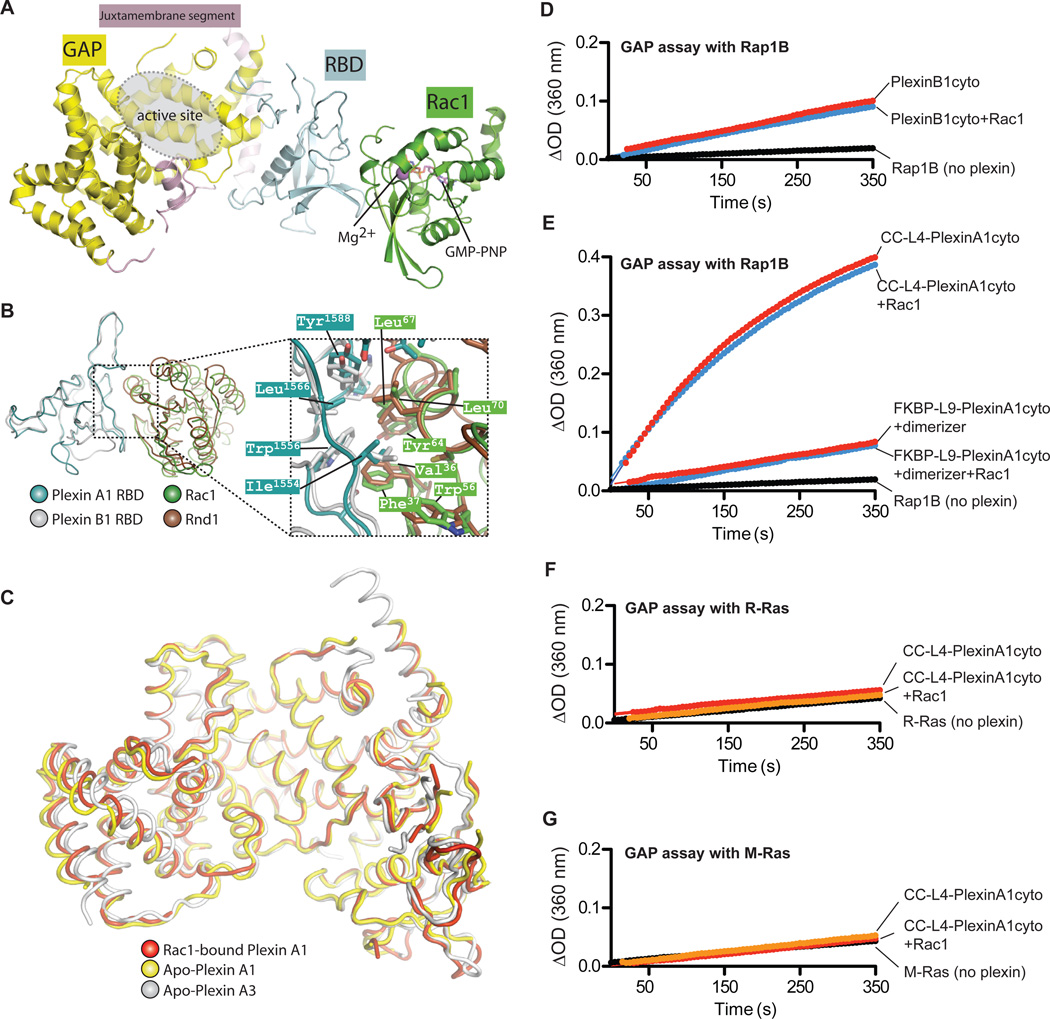
The mechanism by which the RhoGTPase-RBD interaction contributes to plexin activation remains largely unclear. Our previous structural comparison of PlexinA3cyto and the Rnd1-bound RBD of PlexinB1 indicated that RhoGTPases do not induce a substantial conformational change in plexin (17). To validate this notion, we determined the crystal structure of full-length PlexinA1cyto bound to the RhoGTPase Rac1 loaded with the nonhydrolyzable GTP analogue GMP-PNP. The Rac1 protein contained the Gln61→Leu (Q61L) mutation, which renders Rac1 constitutively active but does not affect plexin binding (14). The asymmetric unit of the crystal contained one Rac1(GMP-PNP) and two PlexinA1 molecules. One of the PlexinA1 molecules was bound to Rac1 in a mode similar to that of the PlexinB1 RBD-Rnd1 complex. The RhoGTPase-binding site on the other plexin monomer was empty (fig. S4B). The presence of both apo- and Rac1-bound plexins in the same crystal provides an opportunity to directly compare these two states of plexin.
The structure shows that the switch I and II regions of Rac1, which are the structural elements for binding both guanine nucleotides and effector proteins, interact with the hydrophobic patch on the plexin RBD that is distal to the GAP domain (Fig. 4, A and B). A superimposition of this structure with the PlexinB1 RBD-Rnd1 complex based on the RBDs leads to good alignment between Rac1 and Rnd1, especially in regions that interact with the RBDs (Fig. 4B) (14). Regions distal to the interface deviate to larger extents due to both the structural difference between the two proteins and a minor misalignment, which can be described as a small rotation of Rac1 relative to Rnd1 along the axis of the switch II helix. Many of the residues in the PlexinA1 RBD-Rac1 interface overlap with their counterparts in the PlexinB1 RBD-Rnd1 structure, underscoring the similarity between the two binding interfaces (Fig. 4B).
Fig. 4. Binding of Rac1 does not affect the structure or the GAP activity of plexinscyto.
(A) Overall structure of the PlexinA1cyto-Rac1(Q61L) complex. The second plexin molecule in the asymmetric unit, which is not bound to Rac1, is not shown.
(B) Comparison of the PlexinA1cyto-Rac1 complex and the PlexinB1 RBD-Rnd1 complex (PDB ID: 2REX). The superimposition is based on the RBDs in the two structures. The juxtamembrane segment and the GAP domain of PlexinA1 are omitted for clarity.
(C) Superimposition of Apo-PlexinA3 (PDB ID: 3IG3), Apo-PlexinA1 and Rac1-bound PlexinA1.
(D) Active Rac1 does not affect the RapGAP activity of PlexinB1cyto. For GAP assays with Rac1, plexin proteins were incubated with GTP-loaded Rac1(Q61L) before the assays.
(E) Active Rac1 does not affect the RapGAP activity of dimeric PlexinA1cyto.
(F to G) CC-L4-PlexinA1cyto in the presence of Rac1 remains inactive towards R-Ras (F) and M-Ras (G).
Data shown are representative of at least 3 independent experiments.
A superimposition of the Rac1-bound PlexinA1 molecule with the apo-PlexinA1 in the crystal shows that they adopt the same conformation, with a root mean square deviation (RMSD) of 0.89 Å for the Ca atoms of the 519 residues that are ordered in both molecules (Fig. 4C). They are also similar to the previously determined apo PlexinA3 structure (17). The RMSD between Rac1-bound PlexinA1 and apo PlexinA3 is 1.36 Å for 529 aligned Ca atoms, and that between apo-PlexinA1 and A3 is 1.82 Å for 531 aligned Ca atoms. The only obvious difference among the three structures resides in the last two helices in the proteins, which are located on the side of the GAP domain that is distal to both the GAP active site and the RBD. They are largely solvent exposed and likely flexible as indicated by their high B-factors.
A structural study of the PlexinB1-Rac1 complex shows that in addition to the binding site shown here, Rac1 simultaneously engages a different site on a second plexin molecule with much lower affinity (37). These bivalent interactions lead to a trimeric arrangement of the plexin-Rac1 complex in the crystal, which has been proposed to contribute to plexin activation by facilitating its higher order oligomerization. Nevertheless, the plexin molecules in this structure adopt the same inactive conformation as in our structure (37). The high degree of structural similarity between apo- and Rac1-bound plexins suggest that Rac1, and likely other RhoGTPases, do not induce either substantial local structural changes in the RBD or large-scale reorientation between different domains in plexin. The large distance between the bound Rac1 and the GAP active site suggests that Rac1 is unlikely able to facilitate catalysis by binding to Rap directly (Fig. 4A).
Rac1 does not stimulate the RapGAP activity of plexinscyto in solution
To validate the structural observations, we tested whether Rac1 modulates the RapGAP activity of plexincyto. The experiments were designed to measure the effect of the plexin-Rac interaction as shown in the structure in the present study and previous structures (18), but not the new potential secondary interaction in the PlexinB1-Rac1 structure, which is weak and does not form stably in solution (37). We used PlexinB1cyto to test the effect of Rac1 on plexin monomers because it has relatively high basal activity and its binding to Rac1 has been well characterized (14, 18, 37). To ensure sufficient binding between Rac1 and plexin in the assay, the concentration of GTP-loaded Rac1 Q61L was 10-fold higher than that of PlexinB1cyto and ~2-5-fold higher than the dissociation constant (Kd) value (~20–43 µM) (18, 37). Rac1 did not alter the RapGAP activity of PlexinB1cyto (Fig. 4D) or the activities of the FKBP or coiled-coil dimer of PlexinA1cyto (Fig. 4E). These results together with the crystal structure demonstrate that Rac1 does not directly contribute to activation of the RapGAP activity of plexin in solution. This likely holds true for the other plexin-interacting RhoGTPases, which bind to plexin in the same mode with similar affinities (14, 18).
Biochemical assays have suggested that induced clustering of RhoGTPase-bound plexinscyto elicits their GAP activity to R-Ras (24, 25). We used R-Ras or M-Ras as the substrate in GAP assays and found that dimerized plexinscyto remained inactive to both RRas and M-Ras in the presence of Rac1(GTP) (Fig. 4, F and G).
RapGAP activity of full-length plexins in cells is stimulated by semaphorin
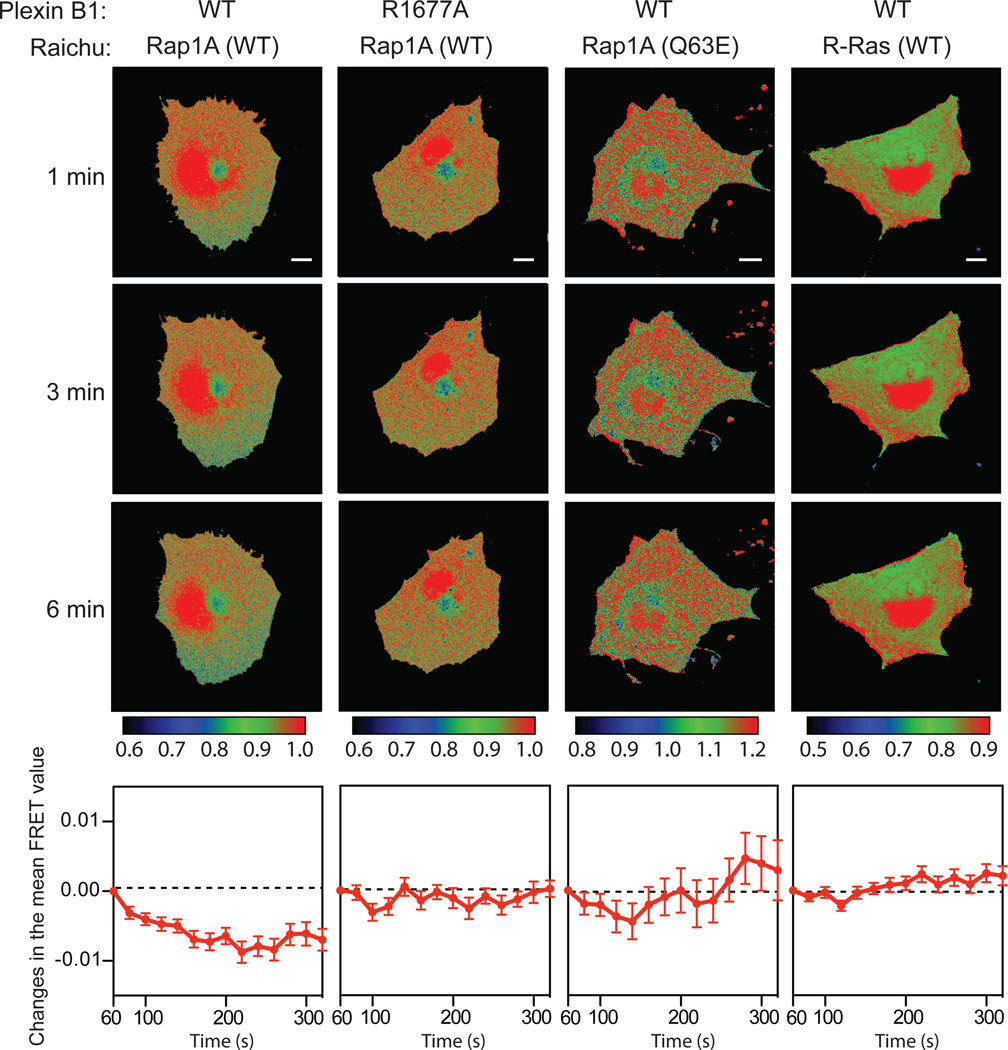
To examine whether full-length plexins function as RapGAPs when activated by their semaphorin ligands, we employed the Raichu-Rap reporter system to monitor the activity of Rap in living cells (38). In this system, an increase in the amount of GTP-bound Raichu-Rap leads to stronger fluorescence resonance energy transfer (FRET) between the reporter fluorescent proteins CFP and YFP, and vice versa. We used PlexinB1 for these assays because its activation by the Sema4D ligand has been well characterized, and unlike the interaction between some class A plexins and class 3 semaphorins its interaction with Sema4D does not require the presence of neuropilin as the co-receptor (1, 2).
COS7 cells cotransfected with full-length PlexinB1 and the Raichu-Rap reporter were stimulated with Sema4D and subjected to time-lapse FRET imaging. The Raichu-Rap protein is not localized exclusively at the cell membrane where plexin is activated by semaphorin (38, 39), consequently plexin could only act on a fraction of the Raichu-Rap reporter in the cell. In particular, we observed strong fluorescence signals in the perinuclear region of the cell, consistent with previous reports that a large amount of Rap resides on the Golgi apparatus and late endosomes (40–42). This pool of Rap is not accessible to plexin and we therefore excluded it from FRET analyses. The mean FRET value of the peripheral regions of cells transfected with wild-type PlexinB1 and the Raichu-Rap reporter decreased within 5 minutes after Sema4D treatment (Fig. 5). PlexinB1 bearing the arginine finger mutation R1677A, which inactivates RapGAP activity, did not decrease FRET of the Raichu-Rap reporter (Fig. 5). In addition, cells transfected with the Raichu-Rap reporter bearing the plexin-resistant Q63E mutations exhibited no FRET decrease after Sema4D treatment (Fig. 5). We also performed the same FRET assays using the Raichu reporter for R-Ras. FRET in cells expressing the Raichu-R-Ras reporter and PlexinB1 did not decrease after Sema4D treatment (Fig. 5).
Fig. 5. Semaphorin activates the RapGAP activity of full-length PlexinB1 in cells.
FRET images were recorded after semaphorin treatment of the cells transfected with the combinations of plasmids as indicated at the top panel. Each column contains three timelapse images of one representative cell for each transfection combination. Scale bars: 10 µm. The images are pseudo-colored based on the FRET value of each pixel. The calibration bars for the coloring schemes are shown below each column. In the bottom panels, the changes in the mean FRET value of all the imaged cells in each group are plotted as a function of time. The plots are combined data from at least 3 independent experiments (n = 8~12 cells). Error bars represent standard error of the mean.
These results suggest that plexins can not only act as Rap-specific GAPs in solution, but also in cells. We propose that semaphorins stimulates this activity, likely by inducing dimerization of the plexin cytoplasmic regions in a mode similar to that in the FKBP and coiled-coil dimerization systems, in which the juxtamembrane helices in the two plexin monomers are brought close to each other.
Rap inactivation is essential in plexin-mediated neuronal growth cone collapse
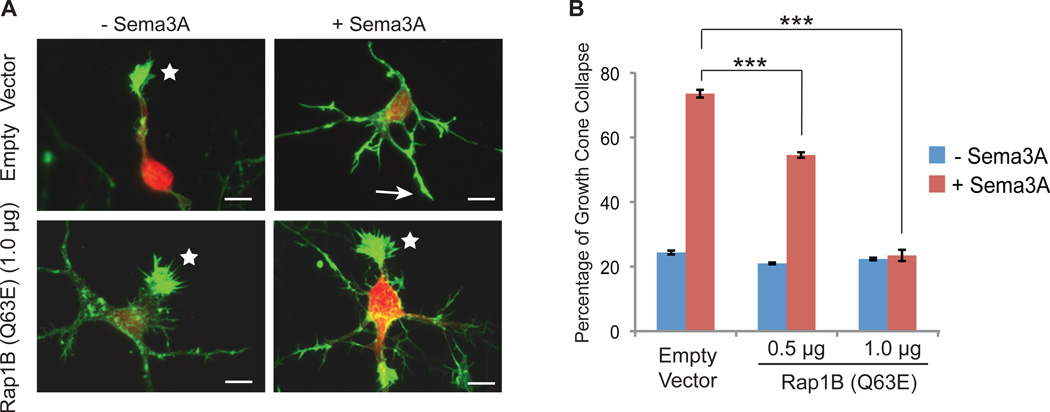
To determine whether the plexin RapGAP activity is required for semaphorininduced repulsive axon guidance, we cultured primary rat cortical neurons, which have endogenous PlexinA1 and its co-receptor Neuropilin1 (43, 44), and analyzed Sema3A-induced growth cone collapse. Sema3A promotes growth cone repulsion through the PlexinA1-Neuropilin1 holo-receptor complex (1). Under basal conditions, addition of Sema3A to the culture medium induced a robust collapse (~70%) of the growth cones by 1 hour (Fig. 6, A and B). If the plexin RapGAP activity is important for semaphorin-induced growth cone repulsion, expression of Rap1B (Q63E), which is resistant to plexins but remains sensitive to canonical RapGAPs (31), would be expected to reduce or block Sema3A-induced growth cone collapse. Ectopically expressed Rap1B (Q63E) inhibited Sema3A-induced growth cone collapse in a DNA concentration-dependent manner compared to vector only-transfected neurons (Fig. 6, A and B). These findings indicate that inactivation of Rap by plexin plays an essential role in semaphorin-induced growth cone collapse. Consistently, mutating the arginine finger in plexin, which abrogates its RapGAP activity, abolishes plexin-induced cell morphological changes (15, 19–21).
Fig. 6. The RapGAP activity of plexin is required for semaphorin induced neuronal growth cone collapse.
(A) Representative images of semaphorin-induced growth cone collapse of rat cortical neurons. Growth cone morphology was visualized by staining of F-actin with Alexa-554 conjugated phalloidin (green). Transfected neurons were identified based on the mCherry fluorescence signal (red). Uncollapsed and collapsed growth cones are indicated by stars and arrows, respectively. Scale bars: 10 µm.
(B) Quantification of growth cone collapse of the neurons that are transfected with empty vector or various amounts of Rap1B Q63E mutant. Error bars represent standard error of the mean. Asterisks (***) indicate significant differences (p<0.001) determined by analysis of variance (ANOVA) followed by Bonferroni post-tests.
Discussion
Identification of the RasGAP domain in plexins over a decade ago raised the critical question which Ras family member they act on for signal transduction (15, 16). We show here that plexins, although structurally homologous to the RasGAPs, possess GAP activity specific for Rap. It appears that the RasGAP fold has evolved a spectrum of specificity for the Ras family members. Although p120GAP and other proteins with the RasGAP fold are only active to the Ras, M-Ras, and R-Ras group, SynGAP and some GAP1 proteins are dual specificity GAPs that can also act on Rap. Plexins likely reside at the other end of the spectrum, because they are active towards Rap but not Ras, R-Ras, or M-Ras under the conditions examined here. Our mutational analysis suggests that plexin catalyzes GTP hydrolysis for Rap by using the alternative mechanism proposed recently for the dual specificity GAPs (31). Although plexins may also display GAP activity towards R-Ras and M-Ras as reported earlier under certain conditions, it is possible that they inactivate R-Ras and M-Ras through an indirect mechanism or by sequestering them from their downstream effectors (21).
Together with previous analyses of the arginine finger mutants of plexins, our growth cone collapse assays with the Rap Q63E mutant suggest that inactivation of Rap by plexin is essential for plexin-mediated neuronal growth cone collapse and other changes in cell morphology (15, 19–21). This inactivation of Rap is likely limited to the plasma membrane regions where plexin is activated by semaphorin. The RapGAP activity of plexins as shown by our kinetic assays is low when compared to other RapGAPs (45). This localized, low RapGAP activity may allow plexin to trigger localized cell morphological changes without perturbing other functions of Rap inside the cells such as vesicular transport (46). The EphA4 receptor induces growth cone collapse by inactivating Rap, although EphA4 itself does not possess GAP activity and does so by recruiting a canonical RapGAP called spine-associated RapGAP (47). Conversely, activation of Rap by the Rap-specific guanine nucleotide exchange factor Epac leads to axon growth (48, 49). It appears that multiple signaling pathways converge on Rap, and its activity plays a key role in determining whether the axon undergoes collapse or outgrowth. This is consistent with several studies showing that active Rap promotes neurite outgrowth by regulating cell adhesion and cytoskeleton dynamics through integrin and RhoA, respectively (47, 50, 51).
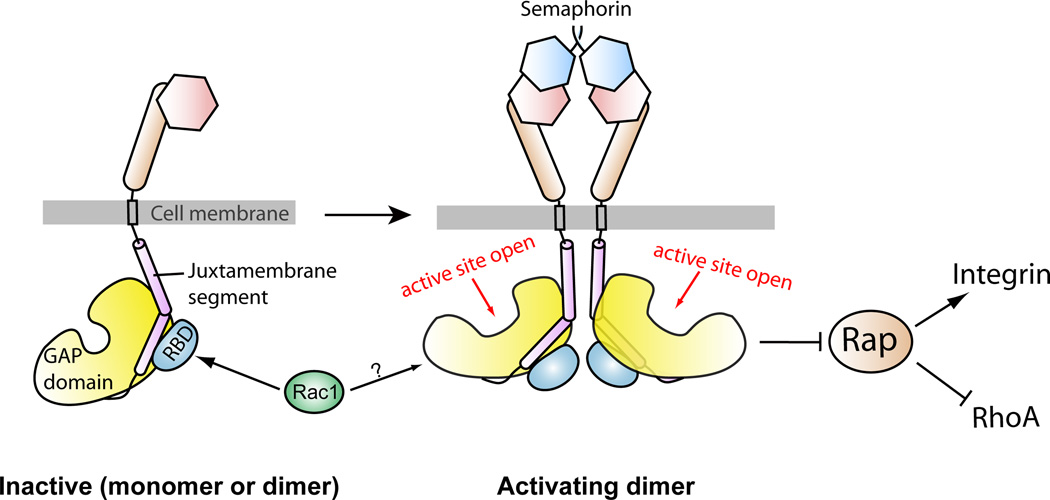
Structural studies of the plexin ectodomains have elucidated the binding mode between plexin and semaphorin, but have not addressed how this binding triggers activation of the plexin cytoplasmic region (11–13). We show that the RapGAP activity of purified plexin cytoplasmic region can be activated by dimerization and the extent of activation increases as the distance between the two monomers decreases. These results suggest an activation mechanism that involves formation of an activating dimer (Fig. 7). Given that the GAP activity does not modify plexins themselves, plexins cannot employ an activation mechanism similar to that used by most receptor tyrosine kinases in which two kinases activate each other by cross phosphorylation (52). Instead, activation of the plexin GAP domain could be achieved by a specific inter-monomer interaction that triggers an activating conformational change in the GAP domain (Fig. 7). The dualspecificity GAP CAPRI has been suggested to be regulated by dimerization (53). The dimerization mode and the conformational change required for activation await structural analyses of dimeric plexins and related GAPs.
Fig. 7. A schematic model for the regulation of the RapGAP activity of plexin.
Prior to semaphorin binding, plexin is an inactive monomer or dimer, in which the RapGAP activity is autoinhibited. Semaphorin-induced dimerization of the plexin extracellular region promotes formation of the activating dimer of the cytoplasmic region, which converts the GAP domain to the active state through an allosteric mechanism. Rac1 or other activator RhoGTPases are not involved in this allosteric activation process, and their roles in plexin activation are not clear. Plexin inactivates Rap to promote neuronal growth cone collapse and other changes in cell morphology, which may rely on the ability of Rap to regulate cell adhesion and cytoskeleton dynamics through integrin and RhoA, respectively.
This activation mechanism involving the activating dimer does not rule out the possibility of a preformed, inactive dimer of plexin on the cell surface (5, 14). In this case, semaphorin binding would trigger the switch from the inactive to the activating dimer. The structure of the “head-on” dimer of PlexinA2 extracellular region is of particular interest in this regard (13). The two C-termini in this dimer point to opposite directions, which may impose spatial and orientational restraints that prevent formation of the activating dimer of the cytoplasmic region.
The role of the RBD-binding RhoGTPases is another intriguing issue in plexin signaling. Our biochemical and structural analyses suggest that the RhoGTPases are not involved directly in the dimerization-driven activation mechanism of plexin. Given that RhoGTPases, plexins and Rap are all associated with the plasma membrane, RhoGTPases may contribute to plexin activation in the context of the membrane surface. Binding of RhoGTPases may help restrain the plexin cytoplasmic region in the orientation that favors formation of the activating dimer or increase the accessibility of the GAP active site to Rap. Structural analysis of the PlexinB1-Rac1 complex suggests a mechanism in which Rac1 facilitates higher order oligomerization of plexins on the cell surface by binding to two plexin molecules through two distinct binding sites (37). Alternatively, RhoGTPases may destabilize the inactive dimer of plexin formed on the cell surface as discussed above and shift the equilibrium towards the active state. Moreover, the RBD-RhoGTPase interaction has been suggested to sequester RhoGTPases from their downstream effectors, thereby contributing to signaling without altering the activation state of plexins (54).
Materials and Methods
Protein expression and purification
The coding sequence for the cytoplasmic domain of mouse PlexinA1 (residues 1269–1894), A2 (1264–1894), A3 (residues 1247–1872), A4 (1263–1893), C1 (residues 975–1571), D1 (residues: 1297–1916) (provided by Dr. Masahiko Taniguchi) and human PlexinB1 (1515–2315) were amplified by PCR. These cDNAs were sub-cloned into either a modified pET28 vector (Novagen) that contains a recognition site for the human rhinovirus C3 protease following the His6-tag (His-tagged), or another modified pET28 vector that expresses the target protein with an N-terminal His6-Sumo tandem tag (His-sumo-tagged). The constructs of FKBP-plexinscyto with either one of the two linkers (3-residue linker: GSG; 9-residue linker: GSSGSGSSG) were generated by PCR by using the coding sequence for FKBP in the pC4-Fv1E plasmid (ARIAD) and those of plexins. The GCN4 coiled-coil sequence (VKQLEDKVEELLSKNAHLENEVARLKKLV) with three different linkers (1-residue linker: G; 4 residue linker: GSSG; 7-residue linker: GSSGSSG) were fused with plexinscyto by PCR and subcloned into the same expression vectors. Mutants of plexins were generated by Quickchange (Stratagene). Expression and purification of these proteins were conducted as described previously (17). The oligomerization states of the proteins were assessed by gel filtration chromatography using Superdex 200 10/300 GL columns (GE healthcare).
The GAP domain of p120GAP (residues 714–1047) was expressed by using the His-tagged vector in the bacteria strain BL21(DE3) and purified with a 1 ml Histrap column (GE Healthcare) followed by ion exchange chromatography (1 ml resource Q, GE Healthcare). The small GTPases including Rac1 (residues 2–177), Rap1B (residues 2–167), Rap2A (residues 2–167), R-Ras (residues 22–201) and M-Ras (residues 10–178) were all expressed and purified through similar procedures. Mutations were generated by Quickchange. Guanine nucleotide exchange was performed by incubating the purified small GTPases with GTP or the nonhydrolyzable analogue guanylyl imidodiphosphate (GMP-PNP) at 40-fold excess in the presence of 4 mM EDTA on ice for 2 hours. The reaction was terminated by 10 mM MgCl2, and excess nucleotide was removed by gel filtration using a Superdex 75 10/300 GL column (GE Healthcare).
Crystallization and structure determination
PlexinA1 and GMP-PNP loaded Rac1 Q61L were mixed at final concentrations of 3 mg/ml and 1.3 mg/ml respectively (~1:1.5 molar ratio) and subjected to crystallization trials in 96-well sitting drop plates. Crystals grew initially in 0.1 M SPG buffer, pH 7.0 and 25% PEG1500 at 20°C. Larger crystals were grown by hanging drop vapor diffusion in the same condition. Crystals were further improved by streak seeding in similar conditions with lower concentrations of PEG1500 (12–20%). Crystals were flash frozen in liquid nitrogen in the crystallization condition supplemented with 20% glycerol. Diffraction data were collected at Beamline 19ID at the Advanced Photon Source (Argonne National Laboratory) at 100 K. Data were indexed and scaled with the HKL2000 package (55). The data are consistent with the space group P43212 and extend to 3.6 Å resolution. The detailed statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics.
| Data collection | |
|---|---|
| Space group | P43212 |
| Unit cell (Å) | a=b=110.78, c=265.63 |
| Resolution (Å) | 50-3.60 (3.73-3.60)a |
| Number of reflections | 103,082 |
| Number of unique reflections | 20,090 |
| Completeness (%) | 99.7 (98.8) |
| I/σ | 17.6 (2.4) |
| Rsym (%)b | 10.3 (64.7) |
| Refinement | |
| Rwork/Rfree (%) | 26.7/33.7 |
| Protein molecules/asymmetric unit | Plexin: 2; Rac1: 1 |
| Number of protein atoms | 9,083 |
| Non-protein atoms | Mg2+ and GMP-PNP |
| RMSD bond length (Å) | 0.003 |
| RMSD bond angle (°) | 0.490 |
| Ramachandran plot (favored, allowed, disallowed) (%) | 82.9, 17.1, 0 |
The structure of PlexinA3 (PBD ID: 3IG3) was used as the search model for molecular replacement in the Phenix package (56), which located two plexin molecules in the asymmetric unit. An electron density map calculated using phases from these two plexin molecules showed clear density for Rac1 bound to one of the plexin molecules, allowing Rac1 to be placed manually with the program Coot (57). The other plexin monomer was not bound to Rac1, and its RhoGTPase-binding site was partially blocked by two symmetry-related plexin molecules (fig. S4B). Iterative model building and refinement were performed by using Coot and Phenix, respectively. Due to the low resolution of the data, tight restraints on the geometry and B-factors were applied in the refinement steps to prevent over-fitting. Noncrystallographic symmetry restrains were not applied to avoid artificial suppression of the differences between the two PlexinA1 molecules in the asymmetric unit. Kicked maps generated by the Phenix refinement program were used in model building to detect and correct for model bias. The GMPPNP molecule was not included in the model until Rfree decreased below 37%. Maps calculated prior to including GMP-PNP in the model showed strong difference density at the nucleotide binding site in Rac1 (fig. S4A), confirming the validity of the structure. The detailed statistics of refinement is included in Table 1.
In solution GAP activity assays
The photometric GAP assays were performed as described with minor modifications (32). Briefly, GTP-loaded small GTPases were added to the reaction solution containing 50 mM Tris-HCl pH 7.6, 20 mM NaCl, 2 mM MgCl2, 10% glycerol, 4 unit/ml purine nucleoside phosphorylase (Sigma) and 200 µM 7-methyl-6-thioguanosine (Berry & Associates). The final concentration of all the small GTPases in the reactions was 50 µM. The absorbance at 360 nm was monitored continuously in a 1 cm path-length cuvette. In these conditions, an increase of 1 O.D. unit was calibrated to be equivalent to release of 95 µM inorganic phosphate from the GTP hydrolysis reaction
After 20–30 s of baseline reading with small GTPases in the reaction solution, plexins (10 µM) or p120GAP (0.05 µM) was added and mixed, and GAP-catalyzed reactions were monitored for 400 s in total. For assays involving dimerized FKBP-plexinscyto, 10 µM FKBP-plexinscyto were first incubated with 5 µM AP20187 at room temperature for 5 min. Then 50 µM small GTPase was added and mixed to start the assay. To test the effect of Rac1 on the GAP activity of plexinscyto, 100 µM GTP-loaded Rac1 (Q61L) was incubated with 10 µM plexinscyto at room temperature for 5 min before the assay. The data were fit to the single exponential decay model to obtain the rate constants k using the program Prism5.
The HPLC-based GAP assay was performed similar to described in (58). Briefly, aliquots of Rap were heat denatured to release the bound guanine nucleotides. Denatured proteins were removed by centrifugation and following ultrafiltration using a 10 kD cut-off concentrator (Amicon). The nucleotide containing flow-through was loaded onto an anion exchange column (AX300,100 × 4.6mm, SynChropak), and eluted with a buffer gradient of KH2PO4 (pH 7.0) from 0 mM to 700 mM. Elution of the nucleotides was monitored by the absorbance at 254 nm. Standard elution profiles of GTP and GDP were obtained by running analytical grade GTP and GDP from Sigma on the same column.
RalGDS pull-down assay for the RapGAP activity of FKBP-PlexinC1cyto in cells
The FKBP-L3-PlexinC1cyto in the pIRES-EGFP2 vector (Clontech) expressed in cells failed to reduce the abundance of GTP-bound Rap by this assay, likely due to cytosolic localization of the protein which cannot act efficiently on the Rap substrate localized on the membrane surface (59). FKBP-PlexinC1cyto was therefore targeted to the membrane by including the myristoylation signal sequence from the kinase Yes (residue 1–11: MGCIKSKENKS) at the N-terminus (fig. S3A). A C-terminal Myc-tag was also included for detection by Western blot. The assay was performed as described previously (34). Briefly, HEK293T cells in 6-well plates were transfected with Myr-FKBPPlexinC1cyto alone or together with Myc-tagged Rap1B in pcDNA3.1 (Invitrogen) using Fugene HD (Promega). Twenty-four hours after transfection, cells were washed once in PBS and treated with 100 nM AP20187. Cells were washed with cold PBS and then lysed in the lysis buffer (50 mM HEPES, pH 7.3, 200 mM NaCl, 2 mM MgCl2, 1% Triton X100, 5% glycerol, 2 mM DTT with a protease inhibitor cocktail (Roche)). The lysates were cleared by centrifugation, and the supernatant was incubated with 5 µl pre-equilibrated GST beads bound to 5 µg purified GST-RalGDS at 4°C for 45 min. GST-beads were washed 4 times and the bound GTP-loaded Rap were eluted with the SDS-PAGE sample buffer (50 mM Tris-HCl, pH 6.8, 1% SDS, 10% glycerol, 5% β mercaptoethanol, 0.02% bromophenol blue) and boiled for 1 min before SDS-PAGE and Western blot analyses with an anti-Myc antibody (Cell Signaling) for transfected Myc-Rap1B or an anti-Rap1 antibody (BD Transduction) for endogenous Rap1. Loading amounts were normalized by the total protein concentrations as determined with the Bio-Rad protein assay. A fraction of the lysates was analyzed by Western blot directly to probe the total amounts of Rap and Myr-FKBP-PlexinC1cyto.
FRET-based RapGAP activity assays in cells
Full-length VSV-tagged PlexinB1 in pcDNA3 (Invitrogen) was obtained from Dr. Kun-liang Guan at UCSD. The Raichu-Rap and Raich-R-Ras constructs were provided by Dr. Michiyuki Matsuda at Kyoto University. Various mutations of these constructs were generated by Quickchange.
COS7 cells (~1.0 × 105) in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal bovine serum (FBS) were plated onto 35mm glass bottom dishes (MatTek). Cells were transfected with different combinations of the plexin constructs and Raichu reporters on the next day with Fugene6 (Roche) following the manufacturer’s instruction with a Fugene: plexin: Raichu ratio of 15:4:1. Two days after transfection, cells were washed with 1 ml phosphate buffered saline (PBS) and then supplied with 600 µl DMEM/F-12(1:1) medium (GIBCO) containing 25 mM HEPES but without serum or phenol red. Live imaging was performed on a DeltaVisionRT microscope equipped with a 37°C chamber and a CoolSNAP HQ CCD camera. Images were acquired using a 436/10 excitation filter together with 465/30 or 535/30 emission filters for the CFP or YFP channel, respectively. Cells with net fluorescent signal intensities over one hundred for both channels were chosen for FRET measurements. For each dish, 4–6 cells were imaged using a 40×/1.35 oil lens and 10% neutral density filter. Images were first acquired for 2–3 min to ensure stability of the focus and fluorescence signals. Cells were then treated with Sema4D at the final concentration of ~5 nM, followed by refocusing and subsequent image acquisition. All the acquisitions were performed at 20-s interval for 6 min, with the exposure time set to 400 ms. Images were processed with the program ImageJ (60).
After background subtraction, an intensity threshold was chosen to define the boundary of the cell. Pixels outside of this region were set to zero intensity. For each pixel the intensity ratio between the YFP (FRET acceptor) and CFP (FRET donor) channels was calculated and taken as the FRET value (61). FRET images were pseudo-colored according to the FRET values. For quantification of the FRET data for each transfection combination, all the cells in each group (8–12 cells) that remained in focus during the imaging process were included. For each cell, we randomly selected 2–4 regions in the peripheral areas of the cell for quantification. Strong fluorescence signals were observed in the perinuclear regions, which likely represent Raichu-Rap localized on perinuclear membrane compartments (39). This pool of Raichu-Rap is not accessible to plexin and was therefore excluded from the FRET analyses. The mean FRET values of the regions of interest were calculated using ImageJ. The changes in the mean FRET values with respect to that of the first time point were plotted as a function of time.
Neuron growth cone collapse assay
Dissociated rat cortical neurons were generated from E18 embryos using standard methods (62). Dissociated neurons were cultured on poly-D-lysine- and laminin-coated glass coverslips in Neurobasal medium (Invitrogen) plus B27 supplement (Invitrogen), penicillin/streptomycin and glutamine. After dissociation in papain, and before plating, the neurons were cotransfected with pcDNA3.1-Rap1B (Q63E) or vector control together with a plasmid expressing mCherry using Lipofectamine 2000. The total amount of DNA transfected was held at 2 mg, and the titrated amount of Rap1B plasmid was balanced with empty vector (pcDNA3.1). Two days after plating (100,000 neurons/well in a 24- well plate), neurons were stimulated with Sema3A at 7.5 nM or conditioned media for 1 hour at 37°C and 5% CO2. The neurons were then fixed in PBS supplemented with 4% paraformaldehyde and 2% sucrose for 8 min, F-actin was visualized using partial permeabilization and phalloidin-Alexa-554 (Invitrogen) as described (62). Transfected neurons were identified by the mCherry fluorescent signal. The relative percentage of collapsed growth cones was determined under experimenter-blinded conditions using standard criteria as described (62). Over 450 growth cones were counted for each experimental condition. Two-way ANOVA showed a significant interaction between Rap1B (Q63E) transfection and semaphorin 3A stimulation (F2,12=287.2, p<0.0001). Simple main effects were determined by multiple one-way ANOVA and independent samples t-tests. One-way ANOVA of the stimulated group alone showed a significant effect of Rap1B (Q63E) transfection (F2,6=337.2, p<0.0001). Bonferroni post hoc analyses showed significant differences between vector-only and 0.5 µg Rap1B (Q63E) transfection (p<0.001), as well as between vector-only and 1.0 µg Rap1B (Q63E) transfection (p<0.001). One-way ANOVA of the unstimulated group alone also showed that Rap1B (Q63E) transfection affected growth cone collapse (F2,6=10.17, p<0.05); Bonferroni analysis indicated this effect was significant between vector and 0.5 µg Rap1B (Q63E) (p<0.05). Individual t-tests performed at each level of Rap1B (Q63E) transfection showed that semaphorin stimulation significantly induced growth cone collapse when cells were transfected either with vector (t4=33.09, p<0.0001) or 0.5 µg Rap1B (Q63E) (t4=35.36, p<0.0001).
Supplementary Material
Supplemental Data
Acknowledgements
We thank Hongtao Yu, Yuh Min Chook, Laura Smith, Jonathan Terman and members in the Zhang laboratory for discussions and technical assistance. We also thank the Live Cell Imaging Core at UTSW for assistance with FRET imaging, and the staff at beamline 19ID at the Advanced Photon Source for assistance with X-ray data collection. X. Z. is a Virginia Murchison Linthicum Scholar in Medical Research at UTSW.
Funding: The work is supported in part by grants to X.Z. from NIGMS (5R01GM088197), the Welch foundation (I-1702) and the American Heart Association (10GRNT3430013). C.W.C is supported by grants from NEI (EY018207 and EY020799) and the Whitehall Foundation. J.J. is supported by grants from NIH (GM61269) and the Welch foundation (I-1603). Use of the beamline at the Advanced Photon Source was supported by the United States DOE under contract DE-AC02-06CH11357.
Footnotes
**
This manuscript has been accepted for publication in Science Signaling. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencesignaling.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supplementary Materials
Fig. S1. HPLC-based assay for the plexin RapGAP activity.
Fig. S2. The RapGAP activity of plexinscyto is stimulated by induced dimerization.
Fig. S3. GST-RalGDS pull-down assays for the RapGAP activity of plexin in cells.
Fig. S4. Crystal structure of the PlexinA1cyto-Rac1 complex.
Author contributions X.Z, Y.W. and H.H. conceived the project. X.Z. and S.V. performed cloning. Y.W. and H.H. conducted protein purification and in vitro GAP assays. X.Z. and H.H. determined the crystal structure. Y.W. performed the FRET experiments with assistance from Y.C. and J.J. N.S. and C.W performed the neuronal growth cone collapse assays.
Competing interests: The authors declare that they have no competing interests.
Accession code The structure of the PlexinA1-Rac1 complex has been deposited to the Protein Data Bank with accession Code 3RYT.
References
- 1.Tran TS, Kolodkin AL, Bharadwaj R. Semaphorin regulation of cellular morphology. Annual review of cell and developmental biology. 2007;23:263–292. doi: 10.1146/annurev.cellbio.22.010605.093554. [DOI] [PubMed] [Google Scholar]
- 2.Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nature reviews. 2005;6:789–800. doi: 10.1038/nrm1740. [DOI] [PubMed] [Google Scholar]
- 3.Capparuccia L, Tamagnone L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment--two sides of a coin. Journal of cell science. 2009;122:1723–1736. doi: 10.1242/jcs.030197. [DOI] [PubMed] [Google Scholar]
- 4.Rujescu D, Meisenzahl EM, Krejcova S, Giegling I, Zetzsche T, Reiser M, Born CM, Moller HJ, Veske A, Gal A, Finckh U. Plexin B3 is genetically associated with verbal performance and white matter volume in human brain. Mol Psychiatry. 2007;12:190–194. 115. doi: 10.1038/sj.mp.4001903. [DOI] [PubMed] [Google Scholar]
- 5.Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, Rajashankar KR, Lu M, Hoemme C, Puschel AW, Nikolov DB. Structure of the semaphoring-3A receptor binding module. Neuron. 2003;39:589–598. doi: 10.1016/s0896-6273(03)00502-6. [DOI] [PubMed] [Google Scholar]
- 6.Love CA, Harlos K, Mavaddat N, Davis SJ, Stuart DI, Jones EY, Esnouf RM. The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nature structural biology. 2003;10:843–848. doi: 10.1038/nsb977. [DOI] [PubMed] [Google Scholar]
- 7.Koppel AM, Raper JA. Collapsin-1 covalently dimerizes, and dimerization is necessary for collapsing activity. The Journal of biological chemistry. 1998;273:15708–15713. doi: 10.1074/jbc.273.25.15708. [DOI] [PubMed] [Google Scholar]
- 8.Klostermann A, Lohrum M, Adams RH, Puschel AW. The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. The Journal of biological chemistry. 1998;273:7326–7331. doi: 10.1074/jbc.273.13.7326. [DOI] [PubMed] [Google Scholar]
- 9.Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. The Journal of biological chemistry. 2002;277:43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- 10.Driessens MH, Hu H, Nobes CD, Self A, Jordens I, Goodman CS, Hall A. Plexin-B semaphorin receptors interact directly with active Rac and regulate the actin cytoskeleton by activating Rho. Curr Biol. 2001;11:339–344. doi: 10.1016/s0960-9822(01)00092-6. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Juo ZS, Shim AH, Focia PJ, Chen X, Garcia KC, He X. Structural basis of semaphorin-plexin recognition and viral mimicry from Sema7A and A39R complexes with PlexinC1. Cell. 2010;142:749–761. doi: 10.1016/j.cell.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen BJC, Robinson RA, Pérez-Brangulí F, Bell CH, Mitchell KJ, Siebold C, Jones EY. Structural basis of semaphorin–plexin signalling. Nature. 2010;467:1118–1122. doi: 10.1038/nature09468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nogi T, Yasui N, Mihara E, Matsunaga Y, Noda M, Yamashita N, Toyofuku T, Uchiyama S, Goshima Y, Kumanogoh A, Takagi J. Structural basis for semaphorin signalling through the plexin receptor. Nature. 2010;467:1123–1127. doi: 10.1038/nature09473. [DOI] [PubMed] [Google Scholar]
- 14.Tong Y, Chugha P, Hota PK, Alviani RS, Li M, Tempel W, Shen L, Park HW, Buck M. Binding of Rac1, Rnd1, and RhoD to a Novel Rho GTPase Interaction Motif Destabilizes Dimerization of the Plexin-B1 Effector Domain. The Journal of biological chemistry. 2007;282:37215–37224. doi: 10.1074/jbc.M703800200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohm B, Rahim B, Kleiber B, Hovatta I, Puschel AW. The semaphorin 3A receptor may directly regulate the activity of small GTPases. FEBS letters. 2000;486:68–72. doi: 10.1016/s0014-5793(00)02240-7. [DOI] [PubMed] [Google Scholar]
- 16.Hu H, Marton TF, Goodman CS. Plexin B mediates axon guidance in Drosophila by simultaneously inhibiting active Rac and enhancing RhoA signaling. Neuron. 2001;32:39–51. doi: 10.1016/s0896-6273(01)00453-6. [DOI] [PubMed] [Google Scholar]
- 17.He H, Yang T, Terman JR, Zhang X. Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration. Proc Natl Acad Sci U S A. 2009;106:15610–15615. doi: 10.1073/pnas.0906923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tong Y, Hota PK, Penachioni JY, Hamaneh MB, Kim S, Alviani RS, Shen L, He H, Tempel W, Tamagnone L, Park HW, Buck M. Structure and function of the intracellular region of the plexin-b1 transmembrane receptor. The Journal of biological chemistry. 2009;284:35962–35972. doi: 10.1074/jbc.M109.056275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oinuma I, Ishikawa Y, Katoh H, Negishi M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- 20.Saito Y, Oinuma I, Fujimoto S, Negishi M. Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO reports. 2009;10:614–621. doi: 10.1038/embor.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakurai A, Gavard J, Annas-Linhares Y, Basile JR, Amornphimoltham P, Palmby TR, Yagi H, Zhang F, Randazzo PA, Li X, Weigert R, Gutkind JS. Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Molecular and cellular biology. 2010;30:3086–3098. doi: 10.1128/MCB.01652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanata SM, Hovatta I, Rohm B, Puschel AW. Antagonistic effects of Rnd1 and RhoD GTPases regulate receptor activity in Semaphorin 3A-induced cytoskeletal collapse. J Neurosci. 2002;22:471–477. doi: 10.1523/JNEUROSCI.22-02-00471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vikis HG, Li W, He Z, Guan KL. The semaphorin receptor plexin-B1 specifically interacts with active Rac in a ligand-dependent manner. Proc Natl Acad Sci U S A. 2000;97:12457–12462. doi: 10.1073/pnas.220421797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oinuma I, Katoh H, Negishi M. Molecular dissection of the semaphorin 4D receptor plexin-B1-stimulated R-Ras GTPase-activating protein activity and neurite remodeling in hippocampal neurons. J Neurosci. 2004;24:11473–11480. doi: 10.1523/JNEUROSCI.3257-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toyofuku T, Yoshida J, Sugimoto T, Zhang H, Kumanogoh A, Hori M, Kikutani H. FARP2 triggers signals for Sema3A-mediated axonal repulsion. Nature neuroscience. 2005;8:1712–1719. doi: 10.1038/nn1596. [DOI] [PubMed] [Google Scholar]
- 26.Oinuma I, Katoh H, Negishi M. Semaphorin 4D/Plexin-B1-mediated R-Ras GAP activity inhibits cell migration by regulating beta(1) integrin activity. J Cell Biol. 2006;173:601–613. doi: 10.1083/jcb.200508204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pena V, Hothorn M, Eberth A, Kaschau N, Parret A, Gremer L, Bonneau F, Ahmadian MR, Scheffzek K. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO reports. 2008;9:350–355. doi: 10.1038/embor.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kupzig S, Deaconescu D, Bouyoucef D, Walker SA, Liu Q, Polte CL, Daumke O, Ishizaki T, Lockyer PJ, Wittinghofer A, Cullen PJ. GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. The Journal of biological chemistry. 2006;281:9891–9900. doi: 10.1074/jbc.M512802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 30.Scrima A, Thomas C, Deaconescu D, Wittinghofer A. The Rap-RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. Embo J. 2008;27:1145–1153. doi: 10.1038/emboj.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sot B, Kotting C, Deaconescu D, Suveyzdis Y, Gerwert K, Wittinghofer A. Unravelling the mechanism of dual-specificity GAPs. EMBO J. 2010;29:1205–1214. doi: 10.1038/emboj.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb MR, Hunter JL. Interaction of GTPase-activating protein with p21ras, measured using a continuous assay for inorganic phosphate release. The Biochemical journal. 1992;287(Pt 2):555–559. doi: 10.1042/bj2870555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clackson T, Yang W, Rozamus LW, Hatada M, Amara JF, Rollins CT, Stevenson LF, Magari SR, Wood SA, Courage NL, Lu X, Cerasoli F, Jr, Gilman M, Holt DA. Redesigning an FKBP-ligand interface to generate chemical dimerizers with novel specificity. Proc Natl Acad Sci U S A. 1998;95:10437–10442. doi: 10.1073/pnas.95.18.10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwartkruis FJ, Wolthuis RM, Nabben NM, Franke B, Bos JL. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. EMBO J. 1998;17:5905–5912. doi: 10.1093/emboj/17.20.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 36.Kupzig S, Bouyoucef-Cherchalli D, Yarwood S, Sessions R, Cullen PJ. The ability of GAP1IP4BP to function as a Rap1 GTPase-activating protein (GAP) requires its Ras GAP-related domain and an arginine finger rather than an asparagine thumb. Molecular and cellular biology. 2009;29:3929–3940. doi: 10.1128/MCB.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bell CH, Aricescu AR, Jones EY, Siebold C. A dual binding mode for RhoGTPases in plexin signalling. PLoS Biol. 2011;9:e1001134. doi: 10.1371/journal.pbio.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mochizuki N, Yamashita S, Kurokawa K, Ohba Y, Nagai T, Miyawaki A, Matsuda M. Spatio-temporal images of growth-factor-induced activation of Ras and Rap1. Nature. 2001;411:1065–1068. doi: 10.1038/35082594. [DOI] [PubMed] [Google Scholar]
- 39.Ohba Y, Kurokawa K, Matsuda M. Mechanism of the spatio-temporal regulation of Ras and Rap1. Embo J. 2003;22:859–869. doi: 10.1093/emboj/cdg087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beranger F, Goud B, Tavitian A, de Gunzburg J. Association of the Ras-antagonistic Rap1/Krev-1 proteins with the Golgi complex. Proc Natl Acad Sci U S A. 1991;88:1606–1610. doi: 10.1073/pnas.88.5.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pizon V, Desjardins M, Bucci C, Parton RG, Zerial M. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. Journal of cell science. 1994;107(Pt 6):1661–1670. doi: 10.1242/jcs.107.6.1661. [DOI] [PubMed] [Google Scholar]
- 42.Nomura K, Kanemura H, Satoh T, Kataoka T. Identification of a novel domain of Ras and Rap1 that directs their differential subcellular localizations. The Journal of biological chemistry. 2004;279:22664–22673. doi: 10.1074/jbc.M314169200. [DOI] [PubMed] [Google Scholar]
- 43.Murakami Y, Suto F, Shimizu M, Shinoda T, Kameyama T, Fujisawa H. Differential expression of plexin-A subfamily members in the mouse nervous system. Dev Dyn. 2001;220:246–258. doi: 10.1002/1097-0177(20010301)220:3<246::AID-DVDY1112>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Chen G, Sima J, Jin M, Wang KY, Xue XJ, Zheng W, Ding YQ, Yuan XB. Semaphorin-3A guides radial migration of cortical neurons during development. Nature neuroscience. 2008;11:36–44. doi: 10.1038/nn2018. [DOI] [PubMed] [Google Scholar]
- 45.Brinkmann T, Daumke O, Herbrand U, Kuhlmann D, Stege P, Ahmadian MR, Wittinghofer A. Rap-specific GTPase activating protein follows an alternative mechanism. The Journal of biological chemistry. 2002;277:12525–12531. doi: 10.1074/jbc.M109176200. [DOI] [PubMed] [Google Scholar]
- 46.Gloerich M, Bos JL. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011;21:615–623. doi: 10.1016/j.tcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 47.Richter M, Murai KK, Bourgin C, Pak DT, Pasquale EB. The EphA4 receptor regulates neuronal morphology through SPAR-mediated inactivation of Rap GTPases. J Neurosci. 2007;27:14205–14215. doi: 10.1523/JNEUROSCI.2746-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray AJ, Shewan DA. Epac mediates cyclic AMP-dependent axon growth, guidance and regeneration. Mol Cell Neurosci. 2008;38:578–588. doi: 10.1016/j.mcn.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Murray AJ, Tucker SJ, Shewan DA. cAMP-dependent axon guidance is distinctly regulated by Epac and protein kinase A. J Neurosci. 2009;29:15434–15444. doi: 10.1523/JNEUROSCI.3071-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jeon CY, Kim HJ, Lee JY, Kim JB, Kim SC, Park JB. p190RhoGAP and Rap-dependent RhoGAP (ARAP3) inactivate RhoA in response to nerve growth factor leading to neurite outgrowth from PC12 cells. Exp Mol Med. 2010;42:335–344. doi: 10.3858/emm.2010.42.5.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamada T, Sakisaka T, Hisata S, Baba T, Takai Y. RA-RhoGAP, Rap-activated Rho GTPase-activating protein implicated in neurite outgrowth through Rho. The Journal of biological chemistry. 2005;280:33026–33034. doi: 10.1074/jbc.M504587200. [DOI] [PubMed] [Google Scholar]
- 52.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 53.Dai Y, Walker SA, De Vet E, Cook S, Welch HC, Lockyer PJ. Ca2+-dependent monomer and dimer formation switches capri between RasGAP and RapGAP activities. The Journal of biological chemistry. 2011 doi: 10.1074/jbc.M110.201301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vikis HG, Li W, Guan KL. The plexin-B1/Rac interaction inhibits PAK activation and enhances Sema4D ligand binding. Genes & development. 2002;16:836–845. doi: 10.1101/gad.966402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta crystallographica. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Biddlecome GH, Berstein G, Ross EM. Regulation of phospholipase C-beta1 by Gq and m1 muscarinic cholinergic receptor. Steady-state balance of receptor-mediated activation and GTPase-activating protein-promoted deactivation. The Journal of biological chemistry. 1996;271:7999–8007. doi: 10.1074/jbc.271.14.7999. [DOI] [PubMed] [Google Scholar]
- 59.Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 60.Abramoff MD, Magelhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 61.Nakamura T, Matsuda M. In vivo imaging of signal transduction cascades with probes based on Forster Resonance Energy Transfer (FRET) Curr Protoc Cell Biol. 2009:10. doi: 10.1002/0471143030.cb1410s45. Chapter 14, Unit 14. [DOI] [PubMed] [Google Scholar]
- 62.Cowan CW, Shao YR, Sahin M, Shamah SM, Lin MZ, Greer PL, Gao S, Griffith EC, Brugge JS, Greenberg ME. Vav family GEFs link activated Ephs to endocytosis and axon guidance. Neuron. 2005;46:205–217. doi: 10.1016/j.neuron.2005.03.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data