Identification of novel NRF2-regulated genes by ChIP-Seq: influence on retinoid X receptor alpha (original) (raw)
Abstract
Cellular oxidative and electrophilic stress triggers a protective response in mammals regulated by NRF2 (nuclear factor (erythroid-derived) 2-like; NFE2L2) binding to deoxyribonucleic acid-regulatory sequences near stress-responsive genes. Studies using Nrf2-deficient mice suggest that hundreds of genes may be regulated by NRF2. To identify human NRF2-regulated genes, we conducted chromatin immunoprecipitation (ChIP)-sequencing experiments in lymphoid cells treated with the dietary isothiocyanate, sulforaphane (SFN) and carried out follow-up biological experiments on candidates. We found 242 high confidence, NRF2-bound genomic regions and 96% of these regions contained NRF2-regulatory sequence motifs. The majority of binding sites were near potential novel members of the NRF2 pathway. Validation of selected candidate genes using parallel ChIP techniques and in NRF2-silenced cell lines indicated that the expression of about two-thirds of the candidates are likely to be directly NRF2-dependent including retinoid X receptor alpha (RXRA). NRF2 regulation of RXRA has implications for response to retinoid treatments and adipogenesis. In mouse, 3T3-L1 cells’ SFN treatment affected Rxra expression early in adipogenesis, and knockdown of Nrf2-delayed Rxra expression, both leading to impaired adipogenesis.
INTRODUCTION
NRF2 [nuclear factor (erythroid-derived 2)-like 2; _NFE2L2_] is a key transcriptional mediator of antioxidant and Phase II detoxification genes, which collectively protect the cellular milieu from oxidative and electrophilic stress (1,2). In response to stimuli, NRF2 dissociates from KEAP1, translocates to the nucleus and dimerizes with other bZIP proteins such as small Maf proteins to form a transactivation complex that binds to antioxidant response elements (AREs) (3). _Nrf2_-deficient mice are highly susceptible to chemical-induced toxicity, carcinogenesis and oxidative burden (4), particularly in the lung, and this likely to be due to the loss of expression of detoxification, xenobiotic metabolism and redox genes (1,2,5–9). Of growing importance, other gene classes including those involved in protein transport, ubiquitination, phosphorylation, cell cycle, growth and apoptosis have been identified as potentially _Nrf2_-dependent and are altered in studies with phenolic antioxidant and isothiocyanate treatments (10–14).
To identify genes that are directly regulated by NRF2 requires evidence of binding at AREs and transactivation of transcription. Recent studies have used bioinformatics-based motif recognition (3,15–17), sequence conservation and gene expression information to identify genome-wide targets for NRF2 (18,19). Presumption of NRF2 occupancy of AREs has frequently been determined using electrophoretic mobility shift assays (EMSAs), luciferase constructs and immobilized oligonucleotide-binding assays (20), but chromatin immunoprecipitation (ChIP) is more definitive. Recently, Malhotra et al. (21) used deep sequencing of ChIP deoxyribonucleic acid (DNA) (ChIP-seq) to identify several hundred new Nrf2 targets in mouse embryonic fibroblasts using a genetic knockout strategy. Although there are more than 30 human genes demonstrated to be NRF2 dependent, less than half of the binding sites (AREs) have been validated by NRF2 ChIP. Thus, our goals for this work are to: (i) use ChIP-seq in human cells to localize and validate NRF2 binding at both known and ‘putative’ binding sites (i.e. targets lacking conclusive binding evidence in the human genome); (ii) discover novel candidate NRF2-mediated genes; and (iii) link these new candidates to biological processes in new ways.
Lymphoblastoid cell lines (LCLs) have been used to model exposure in human populations because of the rich genetic information available (22–24), and we have used them to examine NRF2 activation after treatment with the dietary isothiocyanate, sulforaphane (SFN). Dietary antioxidant supplementation with SFN in humans induces antioxidant genes in vivo (25), alters HDAC activity in peripheral blood mononuclear cells (26) and protects cells from diverse types of toxic environmental exposures (27). For example, dietary antioxidants seem to increase resistance to oxidative stress and mutagenicity in human lymphocytes (28,29), and this is likely due to activation of the NRF2 antioxidant pathway. Thus, activation of the NRF2-mediated pathway is increasingly proposed as a way to prevent or treat disease, including cancer (30), cardiovascular (31), obesity (32,33) and neurodegenerative disease (34). In addition, a Phase II therapeutic trial of an NRF2 activator has proven to be effective in chronic kidney disease (35).
To identify novel genes regulated by NRF2, we activated NRF2 to induce binding to DNA and compared the results of genome-wide techniques for detection of NRF2 occupancy [ChIP-on-DNA microarray (ChIP-on-chip) and ChIP-seq] with expression microarray data. This information was integrated with bioinformatics and independent functional analyses [ChIP-polymerase chain reaction (PCR) and short interfering RNA (siRNA) knockdown] of candidate gene expression in multiple cell types to support the selection of biologically plausible NRF2-dependent candidate genes for follow-up. In addition to validating numerous previously known and putative ARE sites in human cells, we have characterized novel NRF2-binding sites near genes that are involved in processes other than oxidant neutralization and xenobiotic metabolism, such as heme metabolism, apoptosis and cell death, immune response and retinoid signaling by the nuclear retinoid X receptor alpha (RXRA). Two identified NRF2 targets, RXRA and PPARGC1B, play central roles in lipid metabolism and adipocyte differentiation, and we further examined these genes in the context of the mouse 3T3-L1 adipocyte differentiation system. SFN treatment and Nrf2 knockdown alter expression of Rxra in differentiating adipocytes, and this was associated with impaired adipogenesis.
MATERIALS AND METHODS
Cell culture
We grew human lymphoblastoid cells (Coriell) as recommended by Coriell. BEAS-2B, A549 and 3T3-L1 cells were grown as recommended by ATCC. Details of cell culture experiments and sample preparation are provided in the Supplementary Materials and Methods section. 3T3-L1 cells were grown until confluent (designated as Day 0), and differentiation process was initiated by replacing growth medium with differentiation medium containing dexamethasone, 3-isobutyl-1-methylxanthine and insulin (DMI) cocktail as per published methods and stained with Oil Red O (32,36,37). 3T3-L1 cells with stable shRNA knockdown for Nrf2, Keap1 and scramble shRNA were generated as described in Pi et al. (32). Insulin solution (human, I9278), 3-isobutyl-1-methylxanthine (IBMX, I7018) and dexamethasone (D1756) were obtained from Sigma (St Louis, MO, USA) and d,l-SFN from Calbiochem.
ChIP and ChIP-Seq
For each tested LCL (total 6), we performed ChIP in biological duplicate or triplicate and combined replicates after validating SFN-induced NRF2 binding at known ARE loci for every experiment (Figure 1B and Supplementary Figure S2). Details are in the Supplementary Methods. ChIP DNA was quantified using Picogreen (Invitrogen), and sequencing libraries were prepared using the standard Illumina ChIP-seq protocol. The National Center for Genome Resources (Santa Fe, NM, USA) created the libraries and sequenced the immunoprecipitated samples on the Illumina Genome Analyzer II. Reads were mapped to NCBI human reference genome (build 36.3, March 2008) using Burrows-Wheeler Alignment (BWA) Tool (38). Sequencing data has been deposited to the National Center for Biotechnology Information GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE37589.
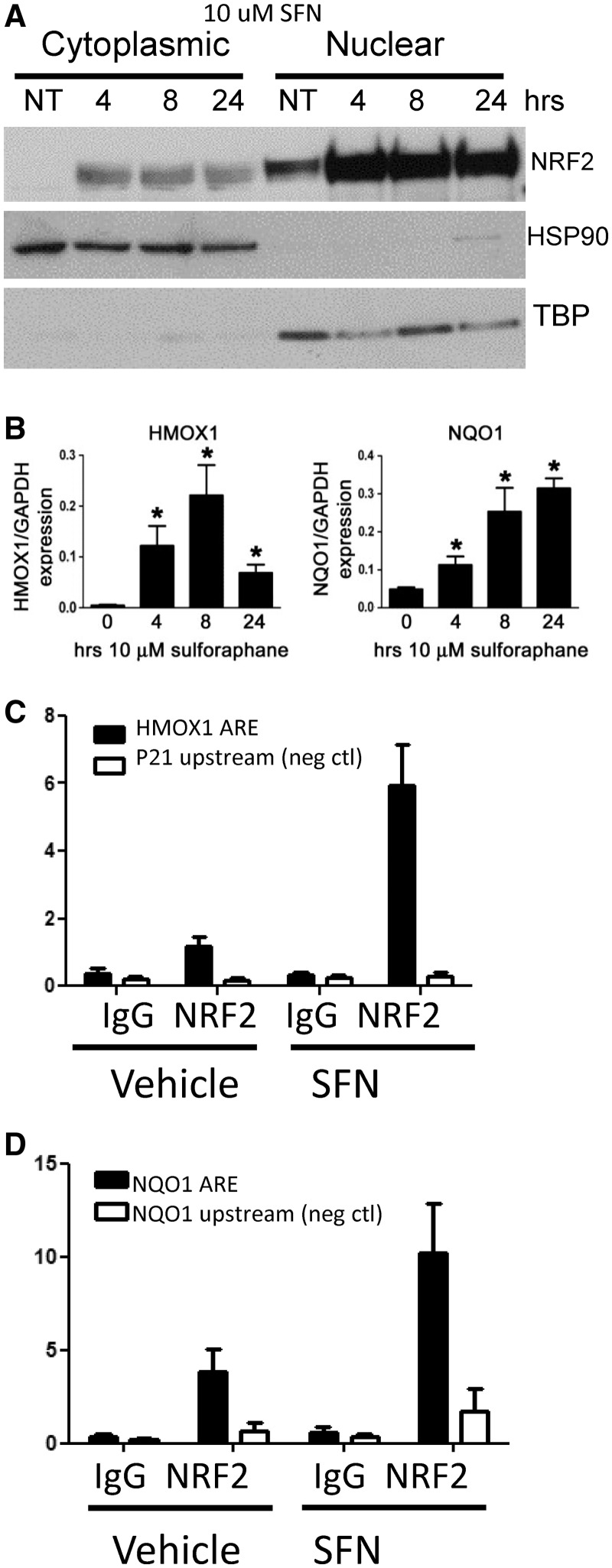
Figure 1.
SFN activates NRF2 nuclear translocation, downstream ARE gene expression and binding to ARE genes in lymphoblastoid cell lines. (A) NRF2 protein levels increase in nucleus after 4-h 10 -µM SFN treatment, and these levels persist through 24 h. TBP, TATA-binding protein, HSP90. (B) Gene expression of known NRF2-regulated genes, HMOX1 and NQO1, significantly increase after 4-, 8- and 24-h 10 -µM SFN exposures. Gene expression values are normalized to GAPDH. Bars represent average of six independent experiments ± SEM; *P < 0.05. (C) ChIP-PCR analysis demonstrates that 5-h 10 -µM SFN treatment increases NRF2 binding at known ARE loci in the promoter regions of HMOX1 and (D) NQO1. A small, but significant, amount of binding was also seen in vehicle control-treated cells. Bars represent amplified target loci normalized to input DNA, as described in ‘Materials and Methods’ section. Controls: immunoprecipitation (IP) with non-specific, species-matched IgG antibody and amplification of loci lacking a functional ARE motif (NQO1 promoter 2-kb upstream of functional AREs and p21 promoter region). Data in (C) are means ± SEM of seven independent experiments in a single cell line; (D) displays mean ± SEM of six independent ChIP experiments in six different LCLs. Enrichment graphs for several individual experiments are shown in Supplementary File 1, Supplementary Figure S2. Each ChIP experiment was tested in this way before sequencing.
ChIP-seq data analysis
The uniquely mapped short reads were used to identify regions of the genome with significant enrichment in NRF2-associated DNA sequences, hereafter referred to as ‘peak regions’ because of their appearance in genome-wide density plots. The peak detection was performed by QuEST 2.4 software (39) using the ‘Transcription factor binding site’ setting (bandwidth of 30 bp, region size of 300 bp) and the ‘stringent peak calling’ parameters (corresponding to 50-fold ChIP to input enrichment for seeding the regions and 3-fold ChIP enrichment for extending the regions). The three sequenced lanes were examined individually for reproducibility, and then unique reads were combined and reanalyzed to produce composite binding regions. ENCODE data (40) were downloaded from the UCSC Browser.
Gene expression microarray
To measure whole-genome expression in SFN-treated LCLs, we used Illumina HumanRef-8 v.3.0.Expression BeadChips.
De novo motif discovery and identification of putative NRF2_-_binding sites
To determine whether our ChIP-seq experiment allows unambiguous recovery of the DNA motif responsible for sequence-specific NRF2 protein binding, we applied de novo motif discovery to the ChIP-seq peak regions. Using the Gibbs motif sampler (41,42), we searched all regions for enriched sequence patterns without any assumption. To identify putative NRF2-binding sites, we used a position weight matrix method based on a curated list of known AREs, as described previously (19).
Real-time quantitative PCR
For ChIP-PCR, we isolated ChIP DNA, as described earlier and tested for enrichment of ARE loci in HMOX1, NQO1 and other loci using the Power SYBR PCR Master Mix (ABI). For gene-specific expression analysis, we isolated total RNA isolated using the RNeasy kit (Qiagen) and treated with DNase. We reverse transcribed complementary DNA using SuperScript First-Strand Synthesis System (Invitrogen) with poly-T primers following manufacturer’s instructions and used TaqMan assays or exon spanning primers (Supplementary Table S6).
NRF2 gene silencing
We silenced NRF2 in BEAS-2B and A549 cells using a reverse transfection protocol using Ambion siRNAs. Nrf2 and Keap1 were silenced in 3T3-L1 cells using short hairpin RNA (shRNA) as described in Pi et al. (32). Additional details are available in Supplementary Materials and Methods section.
RESULTS
Activating NRF2 in LCLs
After exposure to oxidative or electrophilic stress, NRF2 dissociates from the cytosolic inhibitor KEAP1. It subsequently translocates to the nucleus (Figure 1A), forming a heterodimer with small MAF proteins (i.e. MAFG) and targeting ARE-binding sequences to activate transcription. To identify NRF2 occupancy at novel sites, we used a non-cytotoxic dose of the dietary isothiocyanate SFN to robustly activate NRF2, induce DNA binding and the transcription of downstream target genes in human LCLs from the HapMap CEU collection (Figure 1A–C; Supplementary Figure S1) (6,9). Figure 1A displays NRF2 protein translocation to the nuclear cellular fraction after 10 µM SFN treatment at 4, 8 and 24 h, and these levels were sustained for at least 24 h. Using real-time PCR, we verified NRF2-mediated gene transcription of well-known ARE genes, HMOX1 (43) and NQO1 (15) at 4, 8 and 24 h (Figure 1B). Importantly, using ChIP real-time PCR, we verified NRF2 binding at known NRF2-binding loci in the promoters of either HMOX1 (Figure 1C) or NQO1 (Figure 1D and Supplementary Figure S2) after SFN treatment in each of the independent ChIP experiments carried out in replicate in multiple LCLs. We observed that in untreated cells, low levels of NRF2 protein were found in the nucleus consistent with NRF2 occupancy at the HMOX1 and NQO1 promoters (Figure 1A, C and D). Both of these phenomena may be due to background oxidative stress in cells cultured at ambient oxygen level (44). These experiments established NRF2 activation, promoter occupancy and inducible downstream gene expression from 4 to 24 h SFN treatment in our cell culture model system.
Genomic distribution of NRF2 binding
We selected six unrelated HapMap CEU LCLs to treat with SFN, and following verification of NRF2 occupancy by ChIP-PCR (Figure 1D), genome-wide NRF2 binding was determined with sequencing. We isolated immunoprecipitated chromatin using a monoclonal antibody specific for human NRF2 from replicate cultures for each cell line treated with vehicle only (0.1% dimethylsulphoxide (also referred to as No Treatment, NT)) or 10 µM SFN for 5 h. We sequenced ChIP DNA with an Illumina Genome Analyzer II and aligned the 36-base sequence reads to NCBI human genome build 36.3 using the BWA program (Supplementary Table S1) (45). In addition, we sequenced input DNA and DNA immunoprecipitated with immunoglobulin G (IgG) antibody. To detect ChIP-seq peaks, we used the QuEST program (39) with its ‘stringent peak calling’ parameters and compared peak calling using a variety of reference sequences from input and IgG ChIP (Supplementary Table S2). Supplementary Figure S3 displays an example of how uniquely mapped sequence reads upstream of KEAP1 were distributed on both forward and reverse strands and then converted to density profiles for peak calling. We examined significant peaks in individual sequencing runs and only considered further those peak regions appearing in at least two sequencing runs from independent ChIP experiments (Supplementary Figure S4 and Supplementary Data set 1). To increase the sequencing depth and to further assess peak quality, we pooled the uniquely mapped reads from the three SFN and NT sequencing runs (Supplementary Figure S4). We identified 849 NRF2-occupied regions after SFN treatment (mean reads/peak = 22.7; total reads within peaks = 19 345) and for untreated, vehicle-only samples, 205 peak regions (mean reads/peak = 8.9; total reads within peaks = 1843). Of the 205 peak regions in the vehicle-only treated samples, 39 (19%) overlapped with peak regions in the SFN-treated samples. The 4.1-fold increase in the number of peak regions and 10.4-fold increase in uniquely mapped reads within peaks for the SFN-treated versus untreated cells were consistent with both the increase of NRF2 in the nucleus and the increased expression of known NRF2-mediated genes in SFN-treated cells. Additionally, many peak regions in both samples were just above background levels, therefore we also assessed the number and frequency distribution of ChIP-seq reads per peak in the SFN-treated sample and considered peaks with more than 15 unique reads (upper 95% of the mean) as high confidence (HC) peaks (242 of 849 peak regions). Only 19 of 205 peak regions in the untreated sample had more than 15 unique reads.
We assessed the location of sequence reads and ChIP-seq regions in relationship to the nearest gene transcriptional start site (TSS) based on the NCBI human RefSeq gene database (26 733 genes), Supplementary Figure S5A–S5C. NRF2-binding sites have been reported as _cis_-acting elements located at an average distance of ∼1800 bp from the gene TSS, based on 39 currently known functional human AREs (Supplementary Table S3). Among the SFN-treated ChIP-seq peaks, we observed that 716 of the 849 (84%) peak regions were within 100 kb of a gene TSS (Supplementary Figure S5B). Of these regions, 37% (268) of these regions were within 5 kb of gene TSS, and this represents a ∼10-fold enrichment of peaks near TSS over background (37% versus 3.6% of the genome maps to within 5 kb of a TSS). Similarly, when we consider the HC peak regions, we observed 205 of 242 (85%) were within 100 kb of TSS, and 28% were within 5 kb of the TSS. When we mapped NRF2 ChIP-seq peaks relative to gene structural features, most peaks (64%) were located within 10-kb upstream to 10-kb downstream of genes (Supplementary Figure S5C). Overall, these data suggest that most NRF2-binding sites, including the known AREs, are indeed cis acting, but there are certainly many HC-binding sites (37/242, 15%) that are quite distant from a TSS (>100 kb).
Binding motif analysis and overlap with NF-E2 binding
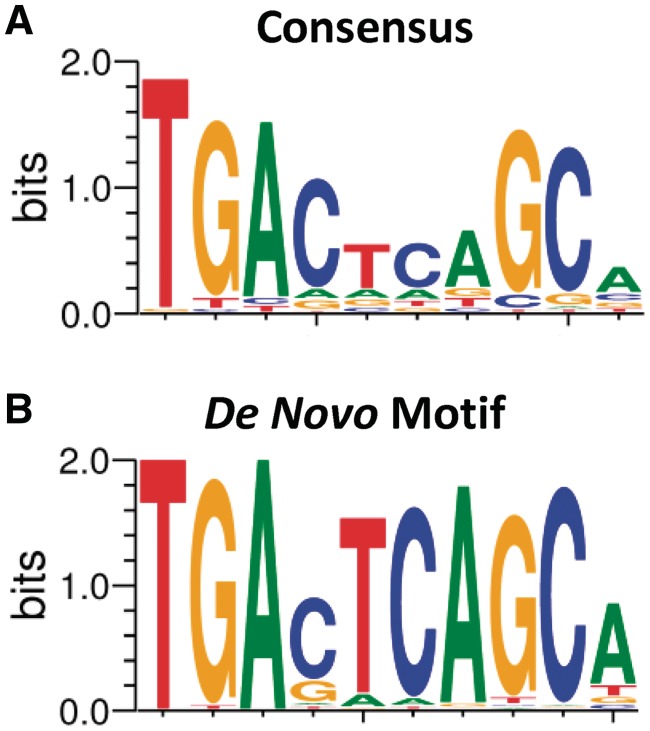
To determine whether our ChIP-seq experiment recovered the DNA motif assumed to be responsible for sequence-specific NRF2 protein–DNA binding (Figure 2A), we applied de novo motif discovery to the ChIP-seq peak regions. Using the Gibbs motif sampler (41,42), we searched all regions for enriched sequence patterns without any assumption. The top enriched de novo motif (Figure 2B) closely matches the core NRF2 consensus binding motif (Figure 2A), described previously in many studies (3,16,19). Using a position weight matrix based on known AREs (19), we determined that 643 of 849 (76%) peak regions obtained from the SFN-treated samples had at least one ARE. Of the 242 HC NRF2-binding regions, we observed that 231 peak regions (95%) contained one or more AREs. The lower ARE frequency (76%) in the lower confidence peak regions (composed of 15 reads or less) may be due to a higher number of false-positive signals among the lower confidence peaks. The locations of >90% of AREs identified were within 100 bp of the peak maximum (Supplementary Figure S5D).
Figure 2.
ChIP-seq peaks contain NRF2-binding motif. (A) NRF2-binding motif based on known AREs and (B) most enriched sequence pattern in the 849 ChIP-seq peak regions based on a de novo analysis using Gibbs motif sampler.
NRF2, encoded by the NFE2L2 gene, is closely related to the erythroid-specific regulator NF-E2, which has a similar AP1-like binding motif (TGA(G/C)TCA). Given the similarity of their binding motifs, we reasoned that these two DNA-binding proteins should share ChIP-seq-binding regions. We examined the overlap between NRF2-binding regions in lymphoid cells and NF-E2-binding regions recently determined in the erythroid cell line K562 by the Snyder lab in the ENCODE project (40) and found that 112 of 242 HC peaks (46%) were shared between these related proteins (5.6-fold enrichment over the mean overlap [8%, 20/242] between NRF2-binding sites and the 14 ENCODE TFs determined by ChIP-seq; Fisher’s exact test, P < 0.0001), further supporting the functionality of the identified NRF2-binding regions. Although NF-E2 is exclusive to erythroid cells, NRF2 is widely expressed among tissues. Notably, NRF2 has recently been identified as a key regulator of oxidative stress response in erythroid cells in sickle cell disease (46).
Localization of binding sites in known and novel NRF2-dependent genes
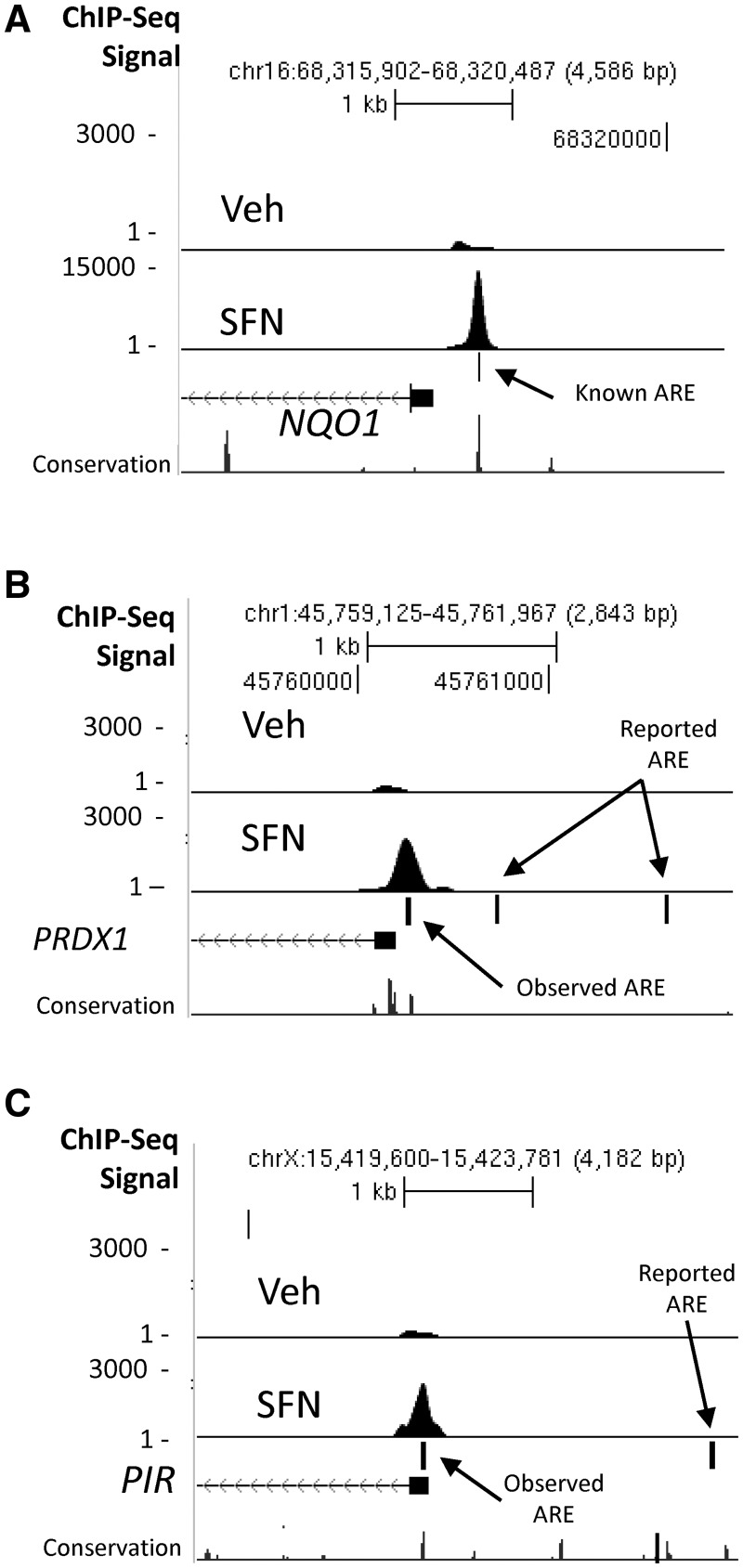
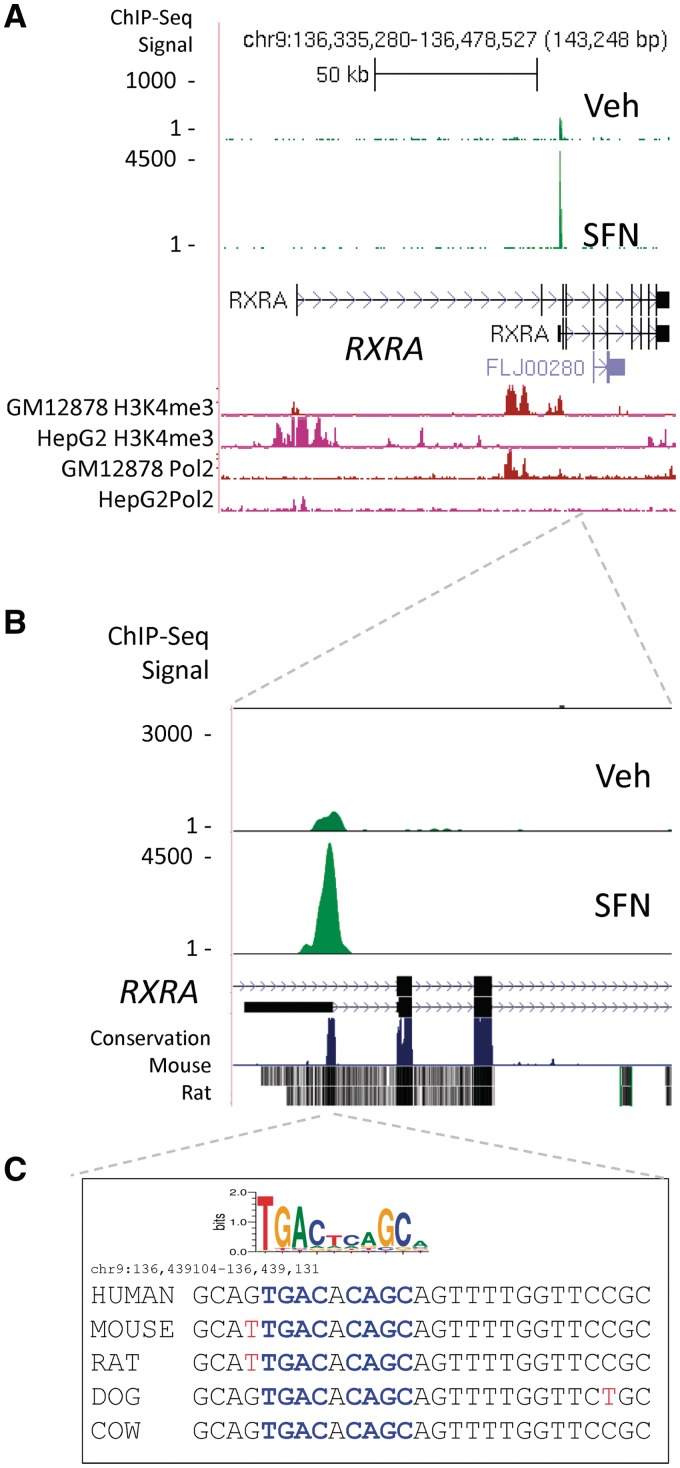
Figure 3A SFN track displays an example of SFN-induced NRF2 ChIP-seq peak region localized over a known functional ARE in the NQO1 promoter (47). Supplementary Data set 1 lists genes with significant ChIP-seq peaks in the SFN and NT sample. Seventeen of the top 25 genes ranked as the most significant binding peaks have been previously described as bone fide or putative NRF2-dependent genes, and these ChIP-seq data provide further evidence of NRF2-binding-mediated regulation for these genes (e.g. FTL, FTH1, HMOX1, NQO1, TXN, GCLC, TXNRD1, ME1 and PRDX1). In addition, we identified binding locations in several putative ARE genes previously reported as part of an NRF2-regulated gene expression signature in airway cells (PIR, TKT, ABCB6, SQSTM1, GSR and TALDO1) (48). Among human genes considered to have functional ARE sequences, we observed that some (e.g. PRDX1 and PIR) (Figure 3B and C) displayed NRF2-binding peaks in locations different from those described in the literature. PRDX1 displayed a prominent peak over a consensus matching, evolutionarily conserved ARE sequence located in the proximal promoter (PRDX1, chr1:45 760 275) and PIR displayed a strong peak in the proximal promoter (Figure 3C, chrX: 15 421 342). In both cases, we did not detect binding at the locations reported by other investigators (48,49). Importantly, among the most significant HC peak regions (Supplementary Data set 1, SFN), there are many regions that have never been functionally linked with NRF2. These novel peak regions are nearby the TSS of genes, which include RXRA, AKIRIN2, GPI, KIFC3, ANKRD11, SH3TC1, CPEB2, CPEB3, PPARGC1B, UNKL and BMP10. Interestingly, the location of the peak region in the RXRA (RXRA) gene suggests that an alternative TSS may be used in LCLs. RXRA has several predicted alternative start sites and several reported alternatively spliced messenger ribonucleic acids (mRNAs). We observed a large NRF2-binding peak located over a conserved ARE sequence (Figure 4A–C) located in intron 2 near the TSS of one of the shorter transcripts. In Figure 4A and in the lower tracks on Supplementary Figure S6A–S6D, we display ENCODE ChIP-seq data for histone H3K4me3 (active promoter) and RNAPol2 (transcription) for GM12878 (LCL) and HepG2 (hepatocarcinoma). The relative locations of these signals, near 5′ TSS for HepG2 and near alternate TSS in GM12878, suggest that transcription of RXRA may start at different TSS in these two lines.
Figure 3.
Examples of NRF2 ChIP-seq peaks at AREs in the gene promoter regions of (A) NQO1, (B) PIR and (C) PRDX1. Control reads from IgG IP are shown in the first track (IgG), vehicle-treated samples in the second track (Veh) and SFN-treated samples in the third track (SFN). Ticks represent reported ARE sites, based on previous literature (Supplementary File 1 and Supplementary Table S4) or observed AREs.
Figure 4.
ChIP-seq peak region near an alternative TSS for the RXRA gene. (A) ChIP-seq peak region is mapped ∼80-kb downstream of the RXRA TSS as defined by NCBI genome build 36.3. Tracks displaying ENCODE H3K4me3 and RNAPol2 ChIPseq data indicate open chromatin and active transcription near the alternate start site (Supplementary Figure S6). The peak region is within 1 kb of an alternative TSS of RXRA, seen in more detail in (B). (C) Sequence under the peak region contains a highly conserved ARE sequence, which closely matches the consensus sequence (blue) as shown by the sequence logo. Mouse and rat nucleotides not matching the human sequence are displayed in red.
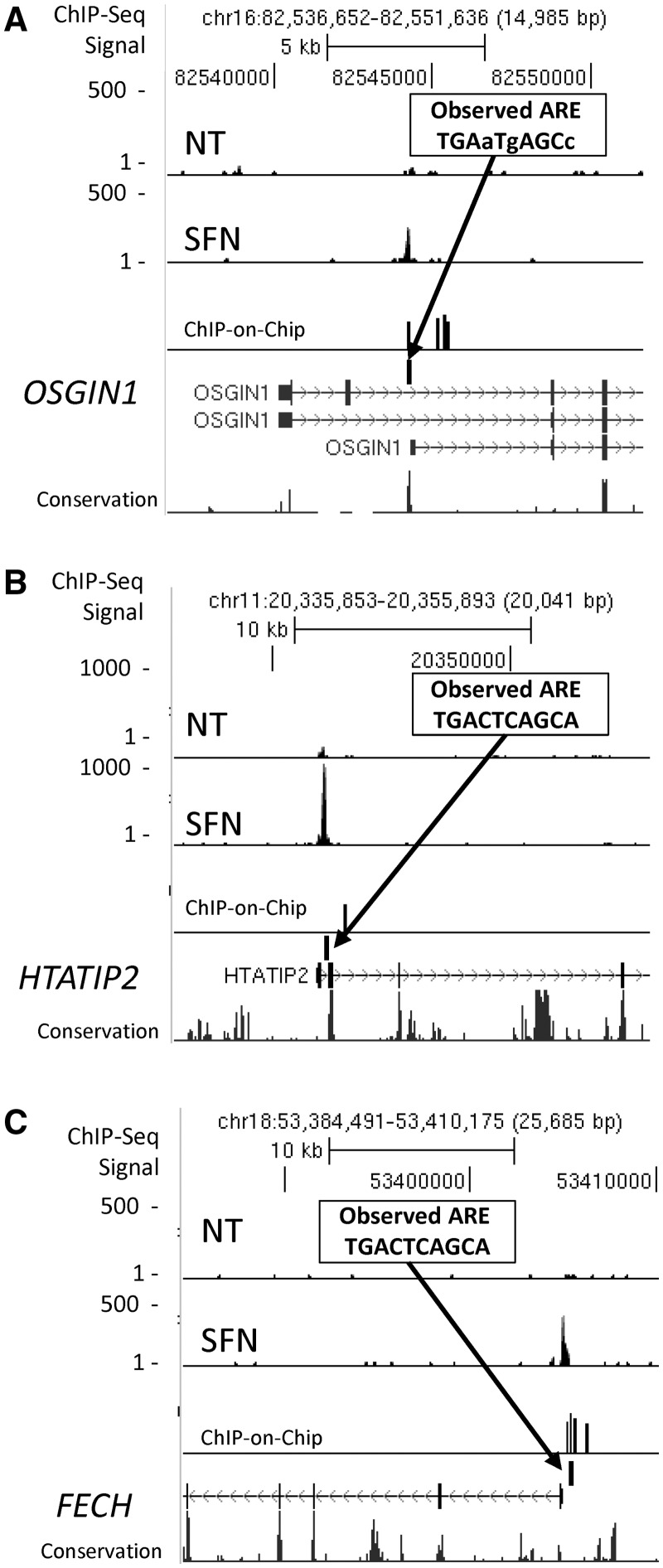
We did additional ChIP experiments to prioritize candidates for further analysis, including a ChIP-on-chip experiment using the Agilent Human Promoter array, and determined the overlap with ChIP-seq peaks (Supplementary Data set 1). Three examples of candidate NRF2 genes (OSGIN1, HTATIP2 and FECH) with highly significant ChIP-seq peaks, ChIP-on-chip signals and good ARE sequence motifs within the ChIP-seq peaks are shown in Figure 5A–C. For each of the binding peaks associated with the eight genes shown in Figures 3–5 and Supplementary Figure S3, as well as eight additional genes listed in Supplementary Table S6, we further validated binding by ChIP-PCR in independent experiments. We detected NRF2 occupancy at 10 × 10 HC peaks and 5 × 5 lower ranked peaks (S6). Due to the importance of RXRA in numerous physiological processes, we carried out experiments to validate NRF2 binding and inducible expression in additional cell types. Significant NRF2 binding in RXRA was detected in untreated LCLs (Figure 4A and B; Supplementary Figure S6E), in SFN-exposed BEAS-2B bronchial epithelial cells (Supplementary Figure S6E) and mouse 3T3-L1 cells (Supplementary Figure S6H). In addition, using ENCODE data for the GM12878 cell line, we detected nucleosome displacement indicated by collocation of H3K4me1 displacement and DNase hypersensitivity at the NRF2-binding site (Supplementary Figure S6F).
Figure 5.
Peak regions that matched in ChIP-seq and ChIP-on-chip. Three examples of genes, OSGIN1 (A), HTATIP2 (B) and FECH (C), which have not been previously described as regulated by NRF2. Insets display ARE motif under the ChIP-peak regions. Lower case nucleotides represent mismatch with ARE consensus.
SFN-mediated gene expression
We used Illumina human Ref-8 microarrays to assess NRF2-mediated gene expression in the six sequenced lines and in the 60 unrelated CEU HapMap LCLs after 24 h exposure to 10 µM SFN as part of a related, ongoing genetic study in our laboratory (50). Comparing mean log 2 expression values using _t_-tests and fold-change values for treated versus untreated across all cell lines, we found 508 genes changed by 1.3-fold or greater with false discovery rate (FDR) _q_-value < 0.05 (Supplementary Data set 1). Similar results were obtained when analysis was carried out using only the six cell lines that were sequenced but with less statistical confidence (e.g. n = 6 versus n = 60). Among genes with both ChIP-seq peaks and gene expression changes, there were significantly more ChIP-seq peak regions near up-regulated genes (20.6%; 60/291) than down-regulated genes (4.6%; 10/217; P < 0.0001, Fisher’s exact test). None of the down-regulated genes displayed HC ChIP-seq peaks. These data support findings in the existing literature suggesting that NRF2 is primarily a positive regulator of gene transcription rather than a suppressor (51). Considering the 242 HC peak regions, expression of 97 of the nearby genes was near or below the array detection limit in the LCLs (log 2 expression < 6.0), including some with highly significant NRF2 binding (e.g. HBE1, KIFC3, BMP10 and ANKRD11).
A number of known NRF2-regulated genes (FTL, PRDX1, GSTP1, PSMA3 and TKT) were highly expressed but showed minimal change following treatment (<30%). Based on the presence of strong H3K4me3 marks (enrichment at enhancer/promoter elements, ENCODE data not shown), expression of these genes may already be at maximal levels under NT conditions. They were among the top 2% highest expression values in the untreated samples, and after SFN treatment, despite the very large increase in NRF2 binding at their AREs (5- to 20-fold increase), only a small increase in expression signal was observed. Presumably, there are other determinants, such as tissue-specific cofactors, negative feedback loops, epigenetic or signaling mechanisms, which affect both basal expression and NRF2-mediated transcriptional regulation of these highly expressed genes in lymphoblastoid cells. Among all genes with expression and ChIP-seq peaks, 67 showed both NRF2 occupancy and SFN-altered expression >30%, and we carried out further analysis of this group. RXRA induction by SFN was validated in a time course experiment in LCLs (Supplementary Figure S6I) and in several additional cell lines (HepG2, Supplementary Figure S6I; 3T3-L1; Figure 6D and F).
Figure 6.
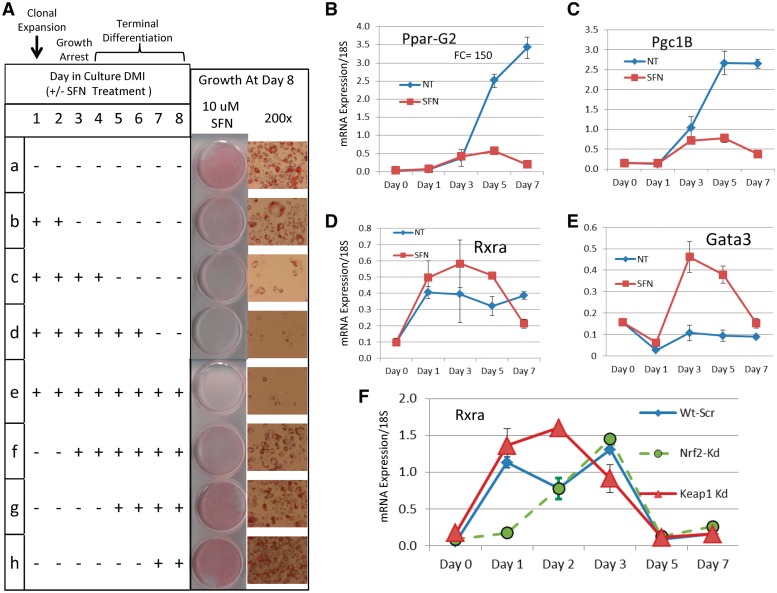
Effects of SFN treatment on 3T3-L1 adipocytes during differentiation. (A) Table displays eight treatment conditions (SFN ± starting and ending on different days). Growth at day 8 is shown in photographs at two magnifications. Treatment for 2, 4, 6 or 8 days inhibits adipogenesis. Treatments starting at day 2 or later have less effect. (B) PparG2 expression by real-time PCR increases rapidly after day 3 under standard differentiation conditions indicating commitment to terminal adipocyte state. Continuous SFN treatment prevents differentiation. (C) Rxra expression is higher under SFN treatment conditions. (D) Expression of Pgc1B, the PPAR coactivator (homolog of human PPARGC1B), is similar to Pparg2 both untreated and treated. (E) Gata3, a negative regulator of adipogenesis, dips initially, then increases dramatically by Day 3 with SFN treatment. (F) Rxra expression was tested in 3T3-L1 adipocytes with shRNA knockdown of Nrf2 (Nrf2-Kd, green) and Keap1 (Keap1-Kd, red). Keap1 knockdown genetically activates Nrf2. The Rxra expression profile under Keap1 knockdown (red triangles) is very similar to that observed for SFN treatment (C, red squares). Nrf2 knockdown (green) delays the rise in Rxra expression. This is accompanied by impaired adipogenesis (32).
MicroRNAs
We identified HC ChIP-Seq peak regions in the vicinity of several microRNAs (miR-365-1/miR-193B, miR-617, miR-592, miR-29B-1, miR-1207, miR-32, miR-181C, miR-200C and miR-550-1), the most significant peak occurring near the mir365-1/mir193B cluster (Supplementary Figure S7A–S7C). Among these, only miR-29B-1 has been linked with oxidative stress (52), although miR-365-1 was observed to be induced in bronchial epithelial cells of smokers compared with nonsmokers (53). Many of these microRNAs have been detected in tumors and decreased levels of miR-365/193B (54), miR-617 (55), miR-200C (56) and miR-29B (57) have been linked with tumor aggressiveness in a variety of cancers. Using real-time PCR assays, we tested expression of miR-365-1/miR-193B, miR-617, miR-592, miR-29B-1, miR-1207, mir32, miR-181C, miR-200C and miR-550-1. Of these, only miR-365-1, miR-193B, miR-181C and miR-29B-1 showed appreciable expression in lymphoblast cells. MiR-365-1 displayed variability after exposure (trending up and then significantly down), but only miR-29B-1 showed significant SFN-induced change relative to no treatment (Supplementary Figures S7D–S8C). The reduced expression of miR-29B-1 in response to SFN was consistent with Luna et al. (52) who observed oxidant exposure suppressed miR-29B-1.
Pathway analysis
SFN is a potent NRF2 activator, but it can also regulate gene expression through a number of other mechanisms including histone modification and alteration of MAPK, NF-κB or AP-1 pathways (58). We did pathway analysis to assess whether there were gene sets (or pathways) enriched in the differentially expressed genes and/or genes with NRF2 ChIP-seq peaks. We used Ingenuity Pathway Analysis (IPA) to analyze gene enrichment in toxicity response pathways for all genes with SFN-altered gene expression (508 genes) and the smaller set of genes with both expression changes and ChIP-seq peak regions within 100 kb of the TSS (Supplementary Table S4). Five pathways were significantly enriched when examining all SFN-responsive genes (Supplementary Figure S9. Open bars. Significance threshold shown represents Fisher’s exact test with FDR correction, P < 0.05). These were oxidative stress response mediated by NRF2; cell cycle: G2/M DNA damage checkpoint regulation; cholesterol biosynthesis; LXR/RXR activation and aryl hydrocarbon receptor signaling. These data clearly indicate that the NRF2 pathway is involved in SFN-mediated gene responsiveness, but based on expression, several other processes are altered including cholesterol biosynthesis and RXR activation. When we limited analysis to the 67 SFN-responsive genes with ChIP-seq peak regions (dark bars), only the two pathways related to oxidative stress were significantly enriched in the IPA analysis. Genes with significant expression changes but no NRF2 binding at AREs may represent downstream targets of NRF2-regulated genes. To examine the 67 SFN-responsive genes with ChIP-seq peaks in more detail, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID) functional annotation clustering tool (59,60). Using this tool to integrate information from Gene Ontology, BioCarta, KEGG and more than 40 other annotation categories, we identified broader functional categories (Table 1 and Supplementary Data set 1 DAVID). The most enriched functional categories were those involved in redox homeostasis and heme metabolism, vitamin metabolic process, oxidation reduction and regulation of apoptosis and cell death.
Table 1.
DAVID functional annotation clustering of 67 NRF2-bound genes with gene expression changesa
| Clustered category | Enrichment score |
|---|---|
| Cellular redox homeostasis | 2.59 |
| Heme metabolism | 2.39 |
| Vitamin metabolic process | 2.37 |
| Oxidoreductase activity | 2.34 |
| Apoptosis and cell death | 2.33 |
| Iron binding (oxidoreductase activity) | 1.78 |
| Glutathione metabolism | 1.62 |
Selection and validation of human NRF2 candidate genes
To select promising novel NRF2-regulated gene candidates for follow-up under NRF2 knockdown conditions, we identified genes with matched peak regions in both ChIP-seq and ChIP-on-chip data sets that were within 2.5 kb of the TSS and possessing an ARE within the peak. This subset included 29 gene candidates, 10 of which had known or putative functional AREs (three known NRF2 targets with peaks ∼4.5 kb from the TSS were also included, HMOX1, FTH1 and TFE3). An example of one of these candidates was FECH, the gene that codes for the enzyme ferrochelatase that carries out the final step in heme biosynthesis. FECH displayed both ChIP-seq and ChIP-on-chip peaks, was induced 1.3-fold and had an ARE in its proximal promoter (Figure 5C). Given the coverage limitations of ChIP-on-chip, we also selected 14 genes that had a ChIP-seq peak region within 2.5 kb of the TSS, contained an ARE motif and exhibited transcriptional regulation by SFN but lacked a ChIP-on-chip result. Among these was RXRA, which had a highly significant ChIP-seq peak region, contained a highly conserved ARE and displayed up-regulation of gene expression (1.5-fold). The ChIP-seq peak was not close to the primary NCBI-defined TSS but was within 1 kb of an alternative TSS (Figure 4). We also chose to examine a pair of genes, NCAPD2 and GAPDH, with a binding region between them containing a highly conserved ARE, to test whether they ‘shared’ regulation by NRF2. These 14 additional tested genes included two well-known NRF2 pathway genes, MAFG and KEAP1 (KEAP1, Supplementary Figure S3), that had binding sites identified in the mouse (61,62) but not in the human genome. This set of genes is not intended to be comprehensive but represents a sampling of new candidates.
Due to the importance of the NRF2 pathway in the airway response to oxidants (e.g. hyperoxia), we tested the impact of NRF2 silencing on expression of candidate genes using two airway cell lines (A549 and BEAS-2B) that are commonly used for NRF2-related oxidative stress studies. Effective silencing of NRF2 in LCLs was not achieved with siRNA or shRNA. However, after transient siRNA silencing of NRF2 transcripts, A549 showed 94% reduction and BEAS-2B showed 86% reduction of expression compared with non-specific siRNA-transfected controls. NRF2 silencing led to significantly reduced expression of six of the seven known and five of the six putative NRF2-regulated genes in both A549 and BEAS-2B cells (P < 0.05, _t_-test; Supplementary Table S5). For many genes, we observed mRNA reduction with NRF2 silencing at both baseline and after SFN treatment consistent with high baseline NRF2 activity in these cell lines (63). NRF2 silencing significantly altered expression of 20 of 30 of the candidate genes in both A549 and BEAS-2B cells including RXRA (Supplementary Table S5). Three genes (GPD2, NDUFAF4 and HIST1H4H) had reduced expression with NRF2 silencing in one of the lines only; four did not change and three were not expressed. Interestingly, we observed strongly increased MT2A (metallothionein 2 A) expression in NRF2-silenced A549 cells at baseline and a larger increase under SFN treatment. This suggests the possibility of direct negative regulation of MT2A by NRF2 in A549 cells. This validation set is not comprehensive, and additional genes remain to be validated by silencing and ChIP-PCR, but these experiments reveal a number of potentially important new NRF2 target genes, particularly RXRA. Further analysis will be needed to establish their functional importance within the NRF2-regulated pathway.
NRF2 activation and RXRA expression in adipocyte differentiation
We identified strong NRF2 binding within RXRA and observed SFN-induced RXRA gene expression in multiple human cell lines following SFN treatment. In addition to RXRA, the PPAR coactivator PPARGC1B also displayed NRF2 binding, suggesting a novel role for NRF2 in pathways regulated by retinoids and involving RXRA. RXRA function is not well characterized in the cell types we initially investigated, but NRF2-binding sites in RXRA displayed 100% human/rodent homology (Figure 4). Thus, to explore the functional regulation of Rxra by Nrf2 activation, we chose to use the well-established mouse 3T3-L1 adipocyte differentiation model that is regulated by the Pparg/Rxra heterodimer. This model system has been used to implicate NRF2 in adipocyte biology (32,33,37), although the mechanism has remained somewhat inconclusive. To investigate whether Nrf2-dependent perturbation of Rxra could impact adipogenesis, we induced 3T3-L1 cells to differentiate and treated the cells with SFN for varying times, as shown in Figure 6A. Treatment of 3T3-L1 cells with 10 µm SFN for 2, 4, 6 and 8 days produced increasing levels of inhibition of differentiation (Figure 6A, tracks b–e). A single SFN dose on Day 1 produced detectable inhibition (Figure 6A, track b), whereas repeat doses (every 2 days) produced complete inhibition (tracks c–e). Notably, treatments starting at Day 3, or after, had much less effect, indicating that the inhibitory effect of SFN activation of Nrf2 occurs early in the cascade of signals driving adipogenesis. A similar pattern was observed with 5 µm SFN but with reduced effect (data not shown); toxicity was not evident. Nrf2 ChIP-PCR of the Nrf2-binding site in the Rxra gene in SFN-treated 3T3-L1 cells displayed strong enrichment relative to NT (Supplementary Figure S6H).
Figure 6B displays the real-time PCR profile of the master regulator of adipocyte differentiation, Pparg2, in differentiating adipocytes over 7 days under standard DMI conditions with and without SFN (NT vs. SFN; these are shown as conditions ‘a’ and ‘e’ in the Figure 6A table). Under standard differentiation conditions (NT), Pparg2 expression rapidly increases after Day 1 to a maximum of ∼150-fold as the cells become committed to terminal differentiation. However, under SFN treatment, Pparg2 remains low, consistent with failing to progress to terminal differentiation. A similar pattern was observed for Cebpa (Supplementary Figure S10A). Oxidative stress genes known to be regulated by Nrf2 (Nqo1 and Gsta1) showed modest but significant mRNA changes under differentiation conditions but displayed large, rapid increases during the first 3 days with SFN treatment and then declined to baseline levels (Supplementary Figure S10B–S10D). Expression of the PPARG coactivator, Pgc1b (Figure 6E; mouse homolog of PPARGC1B) mirrored Pparg2, suggesting its regulation is tied to differentiation status rather than Nrf2 activation.
Rxra expression, heterodimerization with Pparg, binding to DR1 sites and transactivation of adipocyte-specific genes are critical events in adipogenesis. Rxra expression was increased under SFN treatment relative to NT (standard DMI conditions) between Days 1 and 5 (Figure 6C, red squares and Supplementary Figure S10D). Interestingly, transcription factor Gata3, a negative regulator of adipocyte differentiation (64,65), was dramatically induced by SFN between Days 1 and 3 of treatment, and such levels of Gata3 have been demonstrated to suppress adipogenesis through direct binding to the Pparg promoter and interaction with Cebpa protein. We also used shRNA knockdown of Nrf2 and Keap1 (the negative regulator of Nrf2) and examined the real-time PCR profiles of Rxra in differentiating adipocytes under standard DMI conditions (Figure 6F). Relative to Wt-Scr, Rxra expression is reduced in Nrf2-Kd (green) and increased in Keap1-Kd (Keap1 knockdown produces constitutive activation of Nrf2). The delayed induction of Rxra expression in Nrf2-Kd cells may be related to the impaired adipogenesis observed in Nrf2-Kd cells demonstrated by Pi et al. (32). Importantly, the expression pattern observed for genetic activation of Rxra in the Keap1 knockdown cells (Figure 6F, red triangles) was consistent with that observed in SFN-treated cells (Figure 6D).
DISCUSSION
We present the first genome-wide binding data for human NRF2, the transcription factor that regulates anti-oxidative response. Following activation of NRF2 with the dietary antioxidant SFN, we used chromatin immunoprecipitation, massively parallel sequencing, microarray technologies, siRNA and bioinformatics to identify novel NRF2 human gene targets and to verify known or putative AREs. ChIP-seq identified 849 total binding regions and 242 HC regions in SFN-treated lymphoblastoid cells; most are novel targets in humans, and 110 regions overlap with ChIP-seq identified Nrf2 target genes in mouse embryonic fibroblasts (21). Within these binding regions, a de novo motif analysis identified the most enriched motif, and it perfectly matches the core ARE motif. Within the HC NRF2-bound peak regions, 96% contained one or more ARE sequences, and many, such as PRDX, PIR, RXRA, KEAP1, mir-29B and mir-365-1, were highly conserved between humans and rodents (Figures 3B, C and 4C, Supplementary Figures S3, S7C and S8B). The high frequency of AREs in these peaks and their position near the peak maximum support functionality for these binding sites. In addition, 50% of these peaks colocate with ChIP-seq regions determined for the structurally related erythroid-specific factor, NF-E2. The large number of genes that display NRF2 binding but no change in gene expression suggests that additional factors, perhaps coactivators and chromatin state, are involved in transcription regulation of this pathway. In addition, there are many genes that display expression changes but lack NRF2 binding, and these may be regulated indirectly by changes in the redox environment.
RXRα, with the sixth most significant binding peak, is a notable addition to the NRF2-regulated pathway. It is an important nuclear retinoid receptor that heterodimerizes with RARα, RARβ, RXRβ, LXR, VDR and PPARs to mediate the transcriptional regulation of genes involved in diverse cellular processes including cell differentiation, cell cycle, growth and response to steroids, fatty acids and vitamins. Although we present the first evidence that NRF2 regulates a retinoid receptor, our finding is not the first report of an interaction between the retinoid pathway and NRF2. Recent evidence suggests that oral retinoic acid may inhibit NRF2-mediated protective responses to carcinogen exposure (66). In addition, we and others (32,67) have recently identified that Pparg, the RXRA binding partner, is a Nrf2-regulated gene in mice. In this work, we observe highly significant NRF2-binding peaks near malic enzyme 1 (ME1), a PPAR signaling pathway gene involved in lipogenesis and the PPAR gamma coactivator 1B (PGC1B and PPARGC1B). In mouse studies that focus on dietary fat, activation of the NRF2 pathway prevents high-fat diet-induced obesity in mice, and although the mechanism is not well understood (33), a number of reports suggest that the effect is through the PPAR signaling pathway (32,68). In addition, NRF2 seems to be essential for normal adipocyte differentiation in mice (32). We observed that activation of Nrf2 in adipocytes, either by SFN or genetically (Keap1-shRNA), induced Rxra relative to standard conditions (Figure 6B and D). In SFN-treated cells, after Day 1, this was accompanied by a rapid increase in Gata3, the negative regulator of adipogenesis. Considering our evidence that NRF2 activation and binding mediate upregulation of RXRA in lymphocytes and adipocytes, and the clear evidence that PPARG/RXRA heterodimers are critical regulators of adipocyte differentiation (36,69), we hypothesize that NRF2-mediated regulation of RXRA expression in adipocytes may affect PPARG-regulated lipid synthesis and fat accumulation. If altered RXRA expression affected RXRA occupancy (36), this may explain how NRF2 activation inhibits fat accumulation (33). Recent evidence indicates that the ratio of RXRA to RXRB in adipocytes is important for PPARG function (70), and perturbation of this ratio could be the mechanism. Thus, altered expression of RXRA after exposure to NRF2-activating stimuli could impact a number of outcomes including lipid synthesis and the cellular response to endogenous or therapeutic retinoids. A more detailed investigation of NRF2-mediated effects on RXRA expression during adipocyte differentiation is warranted.
Our ChIP-seq analysis also identified peak regions near the TSS of many known or putatively identified NRF2-regulated genes. However, we observed the location of these peak regions did not always match the previously published positions (Supplementary File 1, Supplementary Table S4 and Figure 3A–C). For the PRDX1 and PIR ChIP-seq sites, comparative genomics analysis reveals consensus matching, evolutionary conserved AREs under the peaks (Figure 3B and C), whereas the reported NRF2-binding sites (48,49) have a poor match with the consensus and displayed little or no conservation. While not every human ARE displays evolutionary conservation, a preponderance of established loci do (19,71), strongly supporting functionality for the locations identified in this report.
Our analysis has revealed a number of additional novel gene candidates that seem to expand the role of NRF2 in regulating oxidoreductases (AIFM2, GPD2, HTATIP2 and NDUFAF4) and metabolism genes (GNPDA1), including those involved in heme metabolism (AMBP, FECH). It is well known that NRF2 mediates the regulation of several genes involved in iron metabolism (FTL, FTH1) and the heme degradation pathway (HMOX1). However, NRF2-dependent regulation of the gene AMBP, a protein that binds heme, degrades it and is induced by reactive oxygen species (ROS) in erythroid cell lines (72), is a novel finding. In addition, we demonstrate for the first time that expression of FECH (ferrochelatase), the enzyme catalyzing the final metabolic step of heme biosynthesis IX, is regulated by NRF2. The presence of NRF2 binding in the globin locus, near HBE1 and HBG1 is also notable, given the potential role of NRF2 in modulating sickle cell disease (46).
Numerous genes identified by our ChIP-seq experiments suggest a role for NRF2 in cell cycle regulation and tumor growth. Recent demonstration that genetic activation of NRF2 in Keap1−/− MEFs can enhance cell proliferation (21) and the identification of tumor mutations that activate the NRF2 transcriptional pathway (73–75) are consistent with these findings. DAVID analysis of SFN-regulated genes that possessed ChIP-seq peak regions clearly indicated enrichment of functional annotation categories involved in cellular growth and death processes (Table 1). Many of the genes in this category, such as HTATIP2, OSGIN1 (76) and MAFG (77) have dual roles in oxidative stress response and cell regulation. Among these, HTATIP2 is an oxidoreductase that induces apoptosis under oxidative stress conditions by stabilization of p53 mRNA (78). This interconnection between antioxidant response, redox signaling and cell cycle regulation underscores the complexity and context dependence of NRF2-modulated responses. NRF2 activation is protective in countless exposure scenarios, and NRF2 activators inhibit growth of some tumor cell lines (14,73,74); however, constitutive activation in tumors by mutation drives proliferation and resistance to treatments. Genes, such as SLC3A2, implicated in teratocarcinoma formation in mice (79) and adenocarcinoma (80) and TFE3, regulator of the growth factor transforming growth factor beta, are further examples of proliferation and differentiation-related genes that are involved in tumorigenesis. Translocations in the TFE3 promoter region have been associated with sporadic cases of renal cancer (81). We also identified NRF2 binding near a number of microRNAs, some of which are implicated in tumor aggressiveness, and we observed NRF2-mediated suppression of miR29B following SFN treatment. Although follow-up of these findings is beyond the scope of this study, further characterization of the impact of NRF2 activation on genes involved in tumor growth is essential.
Finally, we demonstrated NRF2 occupancy of genes involved in the activity and homeostasis of NRF2 itself, such as KEAP1, MAFG and SQSTM1. Induction of MAFG, the heterodimeric partner of NRF2, is mediated by NRF2, but binding had not been validated (17,82). The autophagy receptor, SQSTM1/p62, is both a target for NRF2 and a positive regulator of NRF2 (83). Of particular interest is the observation that NRF2 binds an ARE motif in the KEAP1 promoter. Sustained NRF2 activity levels likely lead to NRF2-dependent production of KEAP1, which in turn leads to suppressed NRF2-mediated gene expression through a negative feedback mechanism.
Although NRF2 is the master regulator of oxidative stress response, including during human erythroblast differentiation (46), it is increasingly evident that NRF2 mediates the expression of genes involved in other diverse processes (21,68,84). For example, Nrf2 has been identified as a positive regulator of Notch1 (85), a negative regulator of osteoblast differentiation in mice (86) and necessary for adipocyte differentiation (32). Our experiments have further extended the role of NRF2 signaling in adipogenesis. NRF2 regulation of RXRA in adipogenesis suggests the potential for NRF2 activators to therapeutically manipulate RXRA in numerous retinoid-mediated pathways. Further characterization of how these new targets fit into an expanding NRF2-regulated network should reveal how NRF2 activation and suppression can impact processes such as cell homeostasis, adipogenesis, proliferation, environmental response and disease etiology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–6, Supplementary Figures 1–10, Supplementary Methods, Supplementary Data set and Supplementary References [87–112].
FUNDING
The Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health [ZO1-ES-100475-M-0001 and ES065079-15 to D.A.B. and ES016005 to J.P.]. Funding for open access charge: Laboratory of Molecular Genetics National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
The authors thank Dr. Shuangshuang Dai and John Grovenstein at NIEHS-EITS for excellent computational and database support; Drs Paul Wade, David Fargo, Hye-Youn Cho and Mr Gary Pittman of NIEHS for helpful discussions and review and the NIEHS Microarray Core for generating gene expression and ChIP-on-chip profiles.
REFERENCES
- 1.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 2.Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer. Lett. 2001;174:103–113. doi: 10.1016/s0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 3.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991;266:11632–11639. [PubMed] [Google Scholar]
- 4.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 5.Lee JM, Calkins MJ, Chan K, Kan YW, Johnson JA. Identification of the NF-E2-related factor-2-dependent genes conferring protection against oxidative stress in primary cortical astrocytes using oligonucleotide microarray analysis. J. Biol. Chem. 2003;278:12029–12038. doi: 10.1074/jbc.M211558200. [DOI] [PubMed] [Google Scholar]
- 6.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 7.Yates MS, Kwak MK, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, Baumgartner KJ, Roebuck BD, Liby KT, Yore MM, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66:2488–2494. doi: 10.1158/0008-5472.CAN-05-3823. [DOI] [PubMed] [Google Scholar]
- 8.Cho HY, Reddy SP, Debiase A, Yamamoto M, Kleeberger SR. Gene expression profiling of NRF2-mediated protection against oxidative injury. Free Radic. Biol. Med. 2005;38:325–343. doi: 10.1016/j.freeradbiomed.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 9.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61:3299–3307. [PubMed] [Google Scholar]
- 10.Barve A, Khor TO, Nair S, Lin W, Yu S, Jain MR, Chan JY, Kong AN. Pharmacogenomic profile of soy isoflavone concentrate in the prostate of Nrf2 deficient and wild-type mice. J. Pharm. Sci. 2008;97:4528–4545. doi: 10.1002/jps.21311. [DOI] [PubMed] [Google Scholar]
- 11.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Cancer Lett. 2006;243:170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- 12.Nair S, Xu C, Shen G, Hebbar V, Gopalakrishnan A, Hu R, Jain MR, Lin W, Keum YS, Liew C, et al. Pharmacogenomics of phenolic antioxidant butylated hydroxyanisole (BHA) in the small intestine and liver of Nrf2 knockout and C57BL/6J mice. Pharm. Res. 2006;23:2621–2637. doi: 10.1007/s11095-006-9099-x. [DOI] [PubMed] [Google Scholar]
- 13.Shen G, Xu C, Hu R, Jain MR, Nair S, Lin W, Yang CS, Chan JY, Kong AN. Comparison of (-)-epigallocatechin-3-gallate elicited liver and small intestine gene expression profiles between C57BL/6J mice and C57BL/6J/Nrf2 (-/-) mice. Pharm. Res. 2005;22:1805–1820. doi: 10.1007/s11095-005-7546-8. [DOI] [PubMed] [Google Scholar]
- 14.Kensler TW, Wakabayashi N, Slocum SL, Skoko JJ, Shin S. When Nrf2 talks, who's listening? Antioxid. Redox Signal. 2010;13:1649–1663. doi: 10.1089/ars.2010.3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nioi P, McMahon M, Itoh K, Yamamoto M, Hayes JD. Identification of a novel Nrf2-regulated antioxidant response element (ARE) in the mouse NAD(P)H:quinone oxidoreductase 1 gene: reassessment of the ARE consensus sequence. Biochem. J. 2003;374(Pt 2):337–348. doi: 10.1042/BJ20030754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc. Natl Acad. Sci. USA. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamamoto T, Kyo M, Kamiya T, Tanaka T, Engel JD, Motohashi H, Yamamoto M. Predictive base substitution rules that determine the binding and transcriptional specificity of Maf recognition elements. Genes Cells. 2006;11:575–591. doi: 10.1111/j.1365-2443.2006.00965.x. [DOI] [PubMed] [Google Scholar]
- 18.Taylor RC, Acquaah-Mensah G, Singhal M, Malhotra D, Biswal S. Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLoS Comput. Biol. 2008;4:e1000166. doi: 10.1371/journal.pcbi.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Tomso DJ, Chorley BN, Cho HY, Cheung VG, Kleeberger SR, Bell DA. Identification of polymorphic antioxidant response elements in the human genome. Hum. Mol. Genet. 2007;16:1188–1200. doi: 10.1093/hmg/ddm066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chorley BN, Wang X, Campbell MR, Pittman GS, Noureddine MA, Bell DA. Discovery and verification of functional single nucleotide polymorphisms in regulatory genomic regions: current and developing technologies. Mutat. Res. 2008;659:147–157. doi: 10.1016/j.mrrev.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res. 2010;38:5718–5734. doi: 10.1093/nar/gkq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan S, Bleibel WK, Huang RS, Shukla SJ, Wu X, Badner JA, Dolan ME. Mapping genes that contribute to daunorubicin-induced cytotoxicity. Cancer Res. 2007;67:5425–5433. doi: 10.1158/0008-5472.CAN-06-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459:587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choy E, Yelensky R, Bonakdar S, Plenge RM, Saxena R, De Jager PL, Shaw SY, Wolfish CS, Slavik JM, Cotsapas C, et al. Genetic analysis of human traits in vitro: drug response and gene expression in lymphoblastoid cell lines. PLoS Genet. 2008;4:e1000287. doi: 10.1371/journal.pgen.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riedl MA, Saxon A, Diaz-Sanchez D. Oral sulforaphane increases phase II antioxidant enzymes in the human upper airway. Clin. Immunol. 2009;130:244–251. doi: 10.1016/j.clim.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp. Biol. Med. 2007;232:227–234. [PMC free article] [PubMed] [Google Scholar]
- 27.Cho HY, Kleeberger SR. Nrf2 protects against airway disorders. Toxicol. Appl. Pharm. 2010;244:43–56. doi: 10.1016/j.taap.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 28.Porrini M, Riso P, Oriani G. Spinach and tomato consumption increases lymphocyte DNA resistance to oxidative stress but this is not related to cell carotenoid concentrations. Eur. J. Nutr. 2002;41:95–100. doi: 10.1007/s003940200014. [DOI] [PubMed] [Google Scholar]
- 29.Tope AM, Rogers PF. Evaluation of protective effects of sulforaphane on DNA damage caused by exposure to low levels of pesticide mixture using comet assay. J. Environ. Sci. Health B. 2009;44:657–662. doi: 10.1080/03601230903163624. [DOI] [PubMed] [Google Scholar]
- 30.Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol. Appl. Pharm. 2010;244:66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mann GE, Bonacasa B, Ishii T, Siow RCM. Targeting the redox sensitive Nrf2-Keap1 defense pathway in cardiovascular disease: protection afforded by dietary isoflavones. Curr. Opin. Pharmacol. 2009;9:139–145. doi: 10.1016/j.coph.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Pi JB, Leung L, Xue P, Wang WP, Hou YY, Liu DX, Yehuda-Shnaidman E, Lee C, Lau J, Kurtz TW, et al. Deficiency in the nuclear factor E2-related factor-2 transcription factor results in impaired adipogenesis and protects against diet-induced obesity. J. Biol. Chem. 2010;285:9292–9300. doi: 10.1074/jbc.M109.093955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin S, Wakabayashi J, Yates MS, Wakabayashi N, Dolan PM, Aja S, Liby KT, Sporn MB, Yamamoto M, Kensler TW. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-Imidazolide. Eur. J. Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calkins MJ, Johnson DA, Townsend JA, Vargas MR, Dowell JA, Williamson TP, Kraft AD, Lee JM, Li J, Johnson JA. The Nrf2/ARE pathway as a potential therapeutic target in neurodegenerative disease. Antioxid. Redox. Sign. 2009;11:497–508. doi: 10.1089/ars.2008.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pergola PE, Raskin P, Toto RD, Meyer CJ, Huff JW, Grossman EG, Krauth M, Ruiz S, Audhya P, Christ-schmidt H, et al. Bardoxolone methyl and kidney function in CKD with type 2 diabetes. N. Engl. J. Med. 2011;365:327–336. doi: 10.1056/NEJMoa1105351. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPAR gamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Gene Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, Yamamoto M, Kensler TW. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol. Cell. Biol. 2007;27:7188–7197. doi: 10.1128/MCB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. ENCODE Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence CE, Altschul SF, Boguski MS, Liu JS, Neuwald AF, Wootton JC. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science. 1993;262:208–214. doi: 10.1126/science.8211139. [DOI] [PubMed] [Google Scholar]
- 43.Alam J, Wicks C, Stewart D, Gong P, Touchard C, Otterbein S, Choi AM, Burow ME, Tou J. Mechanism of heme oxygenase-1 gene activation by cadmium in MCF-7 mammary epithelial cells. Role of p38 kinase and Nrf2 transcription factor. J. Biol. Chem. 2000;275:27694–27702. doi: 10.1074/jbc.M004729200. [DOI] [PubMed] [Google Scholar]
- 44.Lees SJ, Childs TE, Booth FW. p21(Cip1) expression is increased in ambient oxygen, compared to estimated physiological (5%) levels in rat muscle precursor cell culture. Cell Prolif. 2008;41:193–207. doi: 10.1111/j.1365-2184.2008.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sangokoya C, Telen MJ, Chi JT. microRNA miR-144 modulates oxidative stress tolerance and associates with anemia severity in sickle cell disease. Blood. 2010;116:4338–4348. doi: 10.1182/blood-2009-04-214817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaiswal AK. Regulation of genes encoding NAD(P)H:quinone oxidoreductases. Free Radic. Biol. Med. 2000;29:254–262. doi: 10.1016/s0891-5849(00)00306-3. [DOI] [PubMed] [Google Scholar]
- 48.Hubner RH, Schwartz JD, De Bishnu P, Ferris B, Omberg L, Mezey JG, Hackett NR, Crystal RG. Coordinate control of expression of Nrf2-modulated genes in the human small airway epithelium is highly responsive to cigarette smoking. Mol. Med. 2009;15:203–219. doi: 10.2119/molmed.2008.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YJ, Ahn JY, Liang P, Ip C, Zhang Y, Park YM. Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 2007;67:546–554. doi: 10.1158/0008-5472.CAN-06-2401. [DOI] [PubMed] [Google Scholar]
- 50.Chorley BN, Wang X, Campbell MR, Kleeberger SR, Bell DA. Identifying functional polymorphisms that alter sulforaphane-induced gene expression via the NRF2 pathway Cancer Prev. Res. 2010;3:B53. [Google Scholar]
- 51.Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79:1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 52.Luna C, Li GR, Qiu JM, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol. Vis. 2009;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- 53.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang XL, Ergun A, Lu JN, Liu G, Zhang XH, Bowers J, et al. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc. Natl Acad. Sci. USA. 2009;106:2319–2324. doi: 10.1073/pnas.0806383106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li ZY, Hassan MQ, Jafferji M, Aqeilan RI, Garzon R, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Biological Functions of miR-29b Contribute to Positive Regulation of Osteoblast Differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang H, Gu J, Wang KK, Zhang W, Xing J, Chen Z, Ajani JA, Wu X. MicroRNA expression signatures in Barrett's esophagus and esophageal adenocarcinoma. Clin. Cancer Res. 2009;15:5744–5752. doi: 10.1158/1078-0432.CCR-09-0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu J, Ohuchida K, Mizumoto K, Sato N, Kayashima T, Fujita H, Nakata K, Tanaka M. MicroRNA, hsa-miR-200c, is an independent prognostic factor in pancreatic cancer and its upregulation inhibits pancreatic cancer invasion but increases cell proliferation. Mol. Cancer. 2010;9:169. doi: 10.1186/1476-4598-9-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Garzon R, Heaphy CEA, Havelange V, Fabbri M, Volinia S, Tsao T, Zanesi N, Kornblau SM, Marcucci G, Calin GA, et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myzak MC, Dashwood RH. Chemoprotection by sulforaphane: keep one eye beyond Keap1. Cancer Lett. 2006;233:208–218. doi: 10.1016/j.canlet.2005.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 60.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 61.Lee OH, Jain AK, Papusha V, Jaiswal AK. An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 2007;282:36412–36420. doi: 10.1074/jbc.M706517200. [DOI] [PubMed] [Google Scholar]
- 62.Katsuoka F, Motohashi H, Engel JD, Yamamoto M. Nrf2 transcriptionally activates the mafG gene through an antioxidant response element. J. Biol. Chem. 2005;280:4483–4490. doi: 10.1074/jbc.M411451200. [DOI] [PubMed] [Google Scholar]
- 63.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tong Q, Dalgin G, Xu HY, Ting CN, Leiden JM, Hotamisligil GS. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290:134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 65.Tong Q, Tsai J, Tan G, Dalgin G, Hotamisligil GS. Interaction between GATA and the C/EBP family of transcription factors is critical in GATA-mediated suppression of adipocyte differentiation. Mol. Cell. Biol. 2005;25:706–715. doi: 10.1128/MCB.25.2.706-715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc. Natl Acad. Sci. USA. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho HY, Gladwell W, Wang XT, Chorley B, Be D, Reddy SP, Kleeberger SR. Nrf2-regulated PPAR gamma Expression Is Critical to Protection against Acute Lung Injury in Mice. Am. J. Resp. Crit. Care. 2010;182:170–182. doi: 10.1164/rccm.200907-1047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sykiotis GP, Habeos IG, Samuelson AV, Bohmann D. The role of the antioxidant and longevity-promoting Nrf2 pathway in metabolic regulation. Curr. Opin. Clin. Nutr. 2011;14:41–48. doi: 10.1097/MCO.0b013e32834136f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert CJ, Liu XS, et al. PPAR gamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Gene Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lefebvre P, Lefebvre B, Benomar Y, Guedin A, Langlois A, Hennuyer N, Dumont J, Bouchaert E, Dacquet C, Penicaud L, et al. Proteasomal degradation of retinoid X receptor alpha reprograms transcriptional activity of PPAR gamma in obese mice and humans. J. Clin. Invest. 2010;120:1454–1468. doi: 10.1172/JCI38606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horvath MM, Wang X, Resnick MA, Bell DA. Divergent evolution of human p53 binding sites: cell cycle versus apoptosis. PLoS Genet. 2007;3:e127. doi: 10.1371/journal.pgen.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Olsson MG, Allhorn M, Olofsson T, Akerstrom B. Up-regulation of alpha1-microglobulin by hemoglobin and reactive oxygen species in hepatoma and blood cell lines. Free Radic. Biol. Med. 2007;42:842–851. doi: 10.1016/j.freeradbiomed.2006.12.017. [DOI] [PubMed] [Google Scholar]
- 73.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 2009;34:176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 74.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008;58:262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh A, Bodas M, Wakabayashi N, Bunz F, Biswal S. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010;13:1627–1637. doi: 10.1089/ars.2010.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao H, Li P, Venters BJ, Zheng S, Thompson PR, Pugh BF, Wang Y. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J. Biol. Chem. 2008;283:20060–20068. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catani L, Vianelli N, Amabile M, Pattacini L, Valdre L, Fagioli ME, Poli M, Gugliotta L, Moi P, Marini MG, et al. Nuclear factor-erythroid 2 (NF-E2) expression in normal and malignant megakaryocytopoiesis. Leukemia. 2002;16:1773–1781. doi: 10.1038/sj.leu.2402597. [DOI] [PubMed] [Google Scholar]
- 78.Zhao J, Chen J, Lu B, Dong L, Wang H, Bi C, Wu G, Guo H, Wu M, Guo Y. TIP30 induces apoptosis under oxidative stress through stabilization of p53 messenger RNA in human hepatocellular carcinoma. Cancer Res. 2008;68:4133–4141. doi: 10.1158/0008-5472.CAN-08-0432. [DOI] [PubMed] [Google Scholar]
- 79.Feral CC, Nishiya N, Fenczik CA, Stuhlmann H, Slepak M, Ginsberg MH. CD98hc (SLC3A2) mediates integrin signaling. Proc. Natl Acad. Sci. USA. 2005;102:355–360. doi: 10.1073/pnas.0404852102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kaira K, Oriuchi N, Imai H, Shimizu K, Yanagitani N, Sunaga N, Hisada T, Kawashima O, Kamide Y, Ishizuka T, et al. CD98 expression is associated with poor prognosis in resected non-small-cell lung cancer with lymph node metastases. Ann. Surg. Oncol. 2009;16:3473–3481. doi: 10.1245/s10434-009-0685-0. [DOI] [PubMed] [Google Scholar]
- 81.Franzini A, Picozzi SC, Politi PL, Barana L, Bianchi F, Alfano G, Gatti G, Fanciullacci F, Gariboldi M, Strada M, et al. A case of renal cancer with TFE3 gene fusion in an elderly man. Clinical, radiological and surgical findings. Urol. Int. 2007;78:179–181. doi: 10.1159/000098080. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Chorley BN, Pittman GS, Kleeberger SR, Brothers J, Liu G, Spira A, Bell DA. Genetic variation and antioxidant response gene expression in the bronchial airway epithelium of smokers at risk for lung cancer. PLoS One. 2010;5:e11934. doi: 10.1371/journal.pone.0011934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jain A, Lamark T, Sjottem E, Larsen KB, Awuh JA, Overvatn A, McMahon M, Hayes JD, Johansen T. p62/SQSTM1 is a target gene for transcription factor nrf2 and creates a positive feedback loop by inducing antioxidant response element-driven gene transcription. J. Biol. Chem. 2010;285:22576–22591. doi: 10.1074/jbc.M110.118976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 2010;3:re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration. Sci. Signal. 2010;3:ra52. doi: 10.1126/scisignal.2000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J. Biol. Chem. 2006;281:18015–18024. doi: 10.1074/jbc.M600603200. [DOI] [PubMed] [Google Scholar]
- 87.Dunning MJ, Smith ML, Ritchie ME, Tavare S. Beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 88.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 89.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 90.Keum YS, Yu S, Chang PP, Yuan X, Kim JH, Xu C, Han J, Agarwal A, Kong AN. Mechanism of action of sulforaphane: inhibition of p38 mitogen-activated protein kinase isoforms contributing to the induction of antioxidant response element-mediated heme oxygenase-1 in human hepatoma HepG2 cells. Cancer Res. 2006;66:8804–8813. doi: 10.1158/0008-5472.CAN-05-3513. [DOI] [PubMed] [Google Scholar]
- 91.Dhakshinamoorthy S, Long DJ, 2nd, Jaiswal AK. Antioxidant regulation of genes encoding enzymes that detoxify xenobiotics and carcinogens. Curr. Top. Cell Regul. 2000;36:201–216. doi: 10.1016/s0070-2137(01)80009-1. [DOI] [PubMed] [Google Scholar]
- 92.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 93.Peirson SN, Butler JN, Foster RG. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 2003;31:e73. doi: 10.1093/nar/gng073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Erickson AM, Nevarea Z, Gipp JJ, Mulcahy RT. Identification of a variant antioxidant response element in the promoter of the human glutamate-cysteine ligase modifier subunit gene. Revision of the ARE consensus sequence. J. Biol. Chem. 2002;277:30730–30737. doi: 10.1074/jbc.M205225200. [DOI] [PubMed] [Google Scholar]
- 95.Arinze IJ, Kawai Y. Transcriptional activation of the human Galphai2 gene promoter through nuclear factor-kappaB and antioxidant response elements. J. Biol. Chem. 2005;280:9786–9795. doi: 10.1074/jbc.M414006200. [DOI] [PubMed] [Google Scholar]
- 96.Banning A, Deubel S, Kluth D, Zhou Z, Brigelius-Flohe R. The GI-GPx gene is a target for Nrf2. Mol. Cell Biol. 2005;25:4914–4923. doi: 10.1128/MCB.25.12.4914-4923.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 2005;280:16891–16900. doi: 10.1074/jbc.M500166200. [DOI] [PubMed] [Google Scholar]
- 98.Hintze KJ, Theil EC. DNA and mRNA elements with complementary responses to hemin, antioxidant inducers, and iron control ferritin-L expression. Proc. Natl Acad. Sci. USA. 2005;102:15048–15052. doi: 10.1073/pnas.0505148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim YC, Yamaguchi Y, Kondo N, Masutani H, Yodoi J. Thioredoxin-dependent redox regulation of the antioxidant responsive element (ARE) in electrophile response. Oncogene. 2003;22:1860–1865. doi: 10.1038/sj.onc.1206369. [DOI] [PubMed] [Google Scholar]
- 100.Kurz EU, Cole SP, Deeley RG. Identification of DNA-protein interactions in the 5' flanking and 5' untranslated regions of the human multidrug resistance protein (MRP1) gene: evaluation of a putative antioxidant response element/AP-1 binding site. Biochem. Biophys. Res. Commun. 2001;285:981–990. doi: 10.1006/bbrc.2001.5262. [DOI] [PubMed] [Google Scholar]
- 101.Lesniak W, Szczepanska A, Kuznicki J. Calcyclin (S100A6) expression is stimulated by agents evoking oxidative stress via the antioxidant response element. Biochim. Biophys. Acta. 2005;1744:29–37. doi: 10.1016/j.bbamcr.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 102.Li Y, Jaiswal AK. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of AP1 binding site contained within human antioxidant response element. J. Biol. Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 103.Moinova HR, Mulcahy RT. An electrophile responsive element (EpRE) regulates beta-naphthoflavone induction of the human gamma-glutamylcysteine synthetase regulatory subunit gene. Constitutive expression is mediated by an adjacent AP-1 site. J. Biol. Chem. 1998;273:14683–14689. doi: 10.1074/jbc.273.24.14683. [DOI] [PubMed] [Google Scholar]
- 104.Montano MM, Deng H, Liu M, Sun X, Singal R. Transcriptional regulation by the estrogen receptor of antioxidative stress enzymes and its functional implications. Oncogene. 2004;23:2442–2453. doi: 10.1038/sj.onc.1207358. [DOI] [PubMed] [Google Scholar]
- 105.Mulcahy RT, Wartman MA, Bailey HH, Gipp JJ. Constitutive and beta-naphthoflavone-induced expression of the human gamma-glutamylcysteine synthetase heavy subunit gene is regulated by a distal antioxidant response element/TRE sequence. J. Biol. Chem. 1997;272:7445–7454. doi: 10.1074/jbc.272.11.7445. [DOI] [PubMed] [Google Scholar]
- 106.Munzel PA, Schmohl S, Buckler F, Jaehrling J, Raschko FT, Kohle C, Bock KW. Contribution of the Ah receptor to the phenolic antioxidant-mediated expression of human and rat UDP-glucuronosyltransferase UGT1A6 in Caco-2 and rat hepatoma 5L cells. Biochem. Pharmacol. 2003;66:841–847. doi: 10.1016/s0006-2952(03)00389-7. [DOI] [PubMed] [Google Scholar]
- 107.Park EY, Rho HM. The transcriptional activation of the human copper/zinc superoxide dismutase gene by 2,3,7,8-tetrachlorodibenzo-p-dioxin through two different regulator sites, the antioxidant responsive element and xenobiotic responsive element. Mol. Cell Biochem. 2002;240:47–55. doi: 10.1023/a:1020600509965. [DOI] [PubMed] [Google Scholar]
- 108.Tsuji Y. JunD activates transcription of the human ferritin H gene through an antioxidant response element during oxidative stress. Oncogene. 2005;24:7567–7578. doi: 10.1038/sj.onc.1208901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, Xiao L, Thiagalingam A, Nelkin BD, Casero RA., Jr The identification of a cis-element and a trans-acting factor involved in the response to polyamines and polyamine analogues in the regulation of the human spermidine/spermine N1-acetyltransferase gene transcription. J. Biol. Chem. 1998;273:34623–34630. doi: 10.1074/jbc.273.51.34623. [DOI] [PubMed] [Google Scholar]
- 110.Wilson LA, Gemin A, Espiritu R, Singh G. ets-1 is transcriptionally up-regulated by H2O2 via an antioxidant response element. FASEB J. 2005;19:2085–2087. doi: 10.1096/fj.05-4401fje. [DOI] [PubMed] [Google Scholar]
- 111.Yueh MF, Tukey RH. Nrf2-keap1 signaling pathway regulates human UGT1A1 expression in vitro and in transgenic UGT1 mice. FASEB J. 2007;21:A1182–A1182. doi: 10.1074/jbc.M610790200. [DOI] [PubMed] [Google Scholar]
- 112.Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Nuclear factor erythroid 2-related factor 2 is a positive regulator of human bile salt export pump expression. Hepatology. 2009;50:1588–1596. doi: 10.1002/hep.23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data