Small Molecule Inhibitors of MicroRNA miR-21 Function (original) (raw)
. Author manuscript; available in PMC: 2012 Aug 28.
Published in final edited form as: Angew Chem Int Ed Engl. 2008;47(39):7482–7484. doi: 10.1002/anie.200801555
MicroRNAs (miRNAs) have recently emerged as an important class of gene regulators, and their misregulation has been linked to a variety of cancers. Small molecule inhibitors of miRNAs would be important tools to elucidate the detailed mechanisms of miRNA function and provide lead structures for the development of new therapeutics. We are reporting a cellular screen for miRNA pathway inhibitors and the first small molecule modifiers of miRNA function.
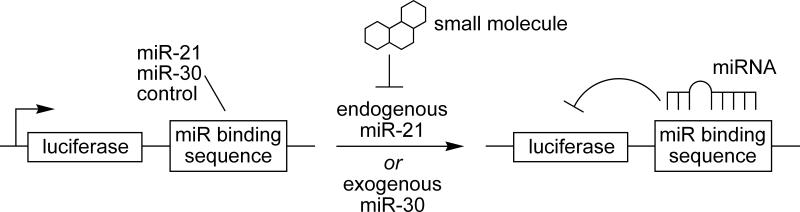
miRNAs are single-stranded noncoding RNAs of 21-23 nucleotides. They are a novel class of gene regulators that function by binding to the 3’ untranslated region of target messenger RNAs leading to either suppression of their translation or acceleration of their degradation.[1] The majority of miRNAs are initially transcribed as primary miRNAs (primiRNAs),[2] which are further processed in the nucleus by the enzyme Drosha, thus transforming pri-miRNAs into shorter stem-loop-structured, double-stranded RNAs called precursor miRNAs (pre-miRNAs).[3] Pre-miRNAs are then transported from the nucleus to the cytoplasm and are processed by Dicer into mature miRNAs.[4] Mature miRNAs enter the effector complex, called the RNA-induced silencing complex (RISC), to then target single-stranded complementary mRNAs (Supporting Figure 1).[5] It is estimated that miRNAs are involved in the regulation of about 30% of all genes and almost every genetic pathway.[6] Moreover, recent evidence suggests that they can function as oncogenes and tumor suppressors.[7, 8] Thus, small molecule regulation of misregulated miRNAs has the potential to provide a new area of therapeutics. So far, specific miRNA inhibition has been only achieved by antisense nucleic acids.[9] We developed an assay for small molecule inhibitors of miRNA function and discovered potentially specific miRNA pathway inhibitors. Although inhibitors of the siRNA pathway have been identified,[10] to our knowledge no small molecule inhibitors of the miRNA pathway have been reported. We selected miR-21 as a target miRNA due to its documented function as an anti-apoptotic factor in cancer cells and its elevated levels in various cancers such as breast, ovarian, and lung cancer as well as glioblastomas.[7, 11] Lentiviral reporter constructs for miRNA activity were assembled by introducing the complementary sequences of mature miR-21, the specificity control miR-30, or a negative control linker sequence (a site with no detectable recognition by natural miRNAs) downstream of a luciferase reporter gene (Supporting Figure 2). These plasmids serve as sensors to detect the presence of specific mature miRNAs (Scheme 1).
Scheme 1.
Luciferase expression under control of a miRNA binding sequence in the 3’ untranslated region (3’ UTR) provides an efficient miRNA assay. Endogenous miR-21 (HeLa cells) or exogenous miR-30 downregulate luciferase activity when paired with their specific binding sequence.
The reporter constructs were stably introduced into HeLa cells which express high levels of miR-21, but relatively low levels of miR-30.[12] In order to test the miRNA specificity of the reporter system, cells that contained both the Luc-miR-30A reporter construct and a construct expressing exogenous primary miR-30 were assayed. These cells displayed a greatly diminished luciferase signal compared to cells with a mismatched Luc-miR-30A reporter/miR-21 combination (Supporting Figure 3), demonstrating that the Luc-miR-21 and Luc-miR-30A reporters are specific and react only to miR-21 and miR-30, respectively. The ability to detect endogenous miR-21 was proven by the fact that the Luc-miR-21 reporter, when introduced into HeLa cells, led to a 90% decreased luciferase signal in comparison to the control luciferase-linker construct, visualizing the high level of endogenous miR-21 expression in HeLa cells (Supporting Figure 4). As expected, the miR-30A reporter displayed only a modest decrease since HeLa cells express relatively low levels of endogenous miR-30.
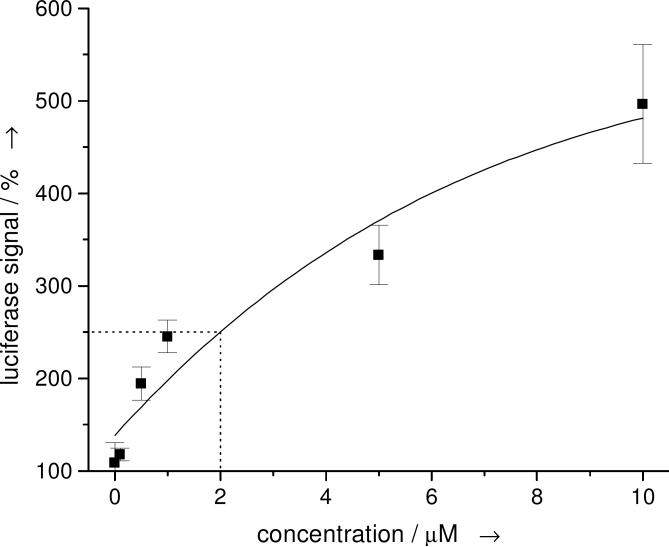
Subsequently, a primary screen of >1000 compounds from our own compound collection and the Library of Pharmacologically Active Compounds (Sigma-Aldrich) was conducted at a 10 μM compound concentration and an initial hit compound 1 was discovered. This diazobenzene led to an increase of the luciferase signal by 251% compared to untreated cells (the DMSO control had no effect on the luciferase signal; Supporting Figure 5). Through several rounds of screening and structural modification a preliminary structure-activity relationship was developed (Supporting Figure 6). Acylation and alkylation of the amino group in 1 led to diminished activities. However, the screening of a wide range of molecules structurally related to the azobenzene core delivered the highly active compound 2 (5-fold increase of the luciferase signal at 10 μM, Figure 1 and 2A). Other molecules derived from 2 through introduction of an amino or nitro group in the 4’ position led to 12% or 64% reduced activity, respectively. Other amide substituents displayed a substantial loss of activity (24-53%), with the exception of allyl and propyl groups, which showed 11% and 16% lower activity, respectively. An exchange of the amide for a sulfonamide delivered compounds with no activity and, interestingly, the styrene analog of 2 had a 40% lower activity. Thus far, 2 is the most effective small molecule inhibitor of microRNA miR-21 function inducing a 485% increase in the luciferase reporter signal at 10 μM. The diazobenzene 2 does not display cytotoxic effects at this concentration as determined by an MTT assay (data not shown). Dose response studies from 0-10 μM revealed a concentration dependence of the luciferase signal with an EC50 of 2 μM (Figure 1).
Figure 1.
Dose-response curve of 2, revealing an EC50 of 2 μM and a luciferase signal increase of ~500% at 10 μM. The error bars represent the standard deviation from three independent measurements.
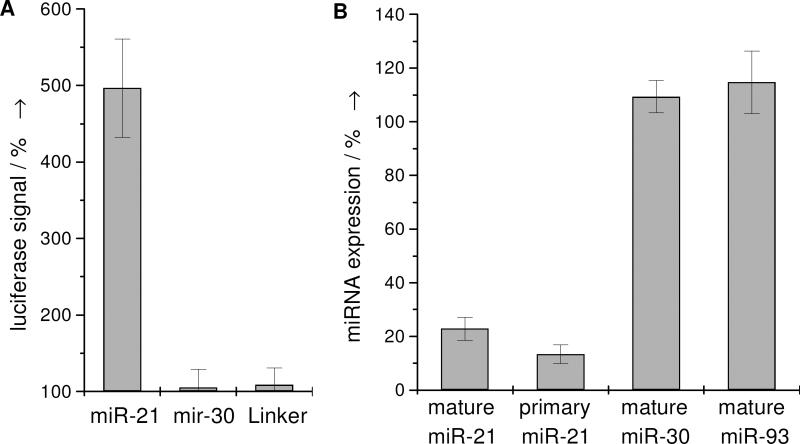
Figure 2.
A) Change in luciferase signal of cells treated with 2 (10 μM) relative to a DMSO control. B) Mature or primary miRNA levels in cells treated with 2 (10 μM) relative to a DMSO control, as determined by quantitative RT-PCR. The error bars represent the standard deviation from three independent measurements.
Several experiments were conducted in order to investigate the mode of action of the inhibitor 2. The compound does not affect the luciferase signal in HeLa cells expressing the Luc-linker control sequence (Figure 2A), thus indicating that it does not increase the luciferase signal through means other than inhibiting the miRNA pathway. Furthermore, HeLa cells expressing both the miR-30 luciferase reporter construct and miR-30 were treated with 2. In this case, no increase of the luciferase signal was detected (Figure 2A), demonstrating that 2 is potentially specific towards miR-21 and does not have a general effect on the common miRNA pathway.
The specificity of 2 for the inhibition of miR-21 function was further validated by measuring intracellular miRNA levels via quantitative RT-PCR (Figure 2B). We found that levels of the stably expressed, exogenous mature miR-30, endogenous mature miR-93 and endogenous non-miRNA genes such as E-chaderin, ID1, RAP1A, Fibronectin are not reduced by treatment with 2 (Figure 2B, Supporting Figure 7). Gratifyingly, miR-21 expression is reduced by 78% compared to the DMSO control in HeLa cells. Quantitative RT-PCR experiments with primers specific for the primary miR-21 (pri-miR-21) sequence but not mature or precursor miR-21 revealed that the pri-miR-21 levels in cells treated with 2 were reduced by 87% (Figure 2B). Similar observations were also made in three other cell lines, MCF-7, A172, and MDA-MB-231, which endogenously express miR-21 (Supporting Figure 7 & 8). These results strongly suggest that compound 2 is an inhibitor that targets the transcription of the miR-21 gene into pri-miR-21, but not downstream processes of the common miRNA pathway.
In summary, we developed a method to identify inhibitors of the miRNA pathway in live cells, specifically of miR-21 an important anti-apoptotic factor in several cancers. A screening of >1000 small organic molecules followed by a structure activity relationship analysis of an initial hit delivered the azobenzene 2 as a specific and efficient inhibitor of miR-21 expression. Research on miRNAs is still in its infancy and the biogenesis of many miRNAs (including miR-21) is not fully understood, thus specific inhibitors of the miRNA pathway (like 2) will be unique tools for the investigation of miRNAs and their involvement in various types of diseases.
experimental section
Screening protocol
HeLa cells were cultured in DMEM and 10% FBS. 2500 HeLa cells containing the Luc-miR-21 construct were plated into each well of a 384-well plate and incubated overnight at 37 °C with 5% CO2. Plates were centrifuged for 90 min at 2,500 × g. Medium was changed 24 h postinfection and compounds were added to a final concentration of 10 μM. Luciferase signals were determined 48 hours after compound treatment using a Bright-Glo luciferase assay kit (Promega) according to the manufacturer's protocol. The data of three independent experiments was averaged.
Supplementary Material
Supporting Information
Footnotes
**
This research has been supported in part by the National Institutes of Health (NS059478-01) and Accelerate Brain Cancer Cure. DDY acknowledges a graduate research fellowship from the ACS Medicinal Chemistry Division. AD is a Beckman Young Investigator and a Cottrell Scholar. QH acknowledges support from the Elsa Pardee Foundation, Breast Cancer Alliance, and the V Foundation.
Contributor Information
Kiranmai Gumireddy, The Wistar Institute, 3601 Spruce Street Philadelphia, PA 19104, USA.
Douglas D. Young, Department of Chemistry North Carolina State University Raleigh, NC 27695-8204, USA
Xin Xiong, Department of Chemistry North Carolina State University Raleigh, NC 27695-8204, USA.
John B. Hogenesch, Department of Pharmacology Institute for Translational Medicine and Therapeutics University of Pennsylvania School of Medicine Philadelphia, PA 19104-6160, USA
Qihong Huang, The Wistar Institute, 3601 Spruce Street Philadelphia, PA 19104, USA Fax: (+) 1-215-898-7952 qhuang@wistar.orghttp://www.wistar.org/research\_facilities/huang/research.htm.
Alexander Deiters, Department of Chemistry North Carolina State University Raleigh, NC 27695-8204, USA Fax: (+) 1-919-515-5079 alex_deiters@ncsu.eduhttp://www4.ncsu.edu/~adeiter/.
references
- 1.a Carthew RW. Curr. Opin. Genet. Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]; b He L, Hannon GJ. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]; c Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Cullen BR. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 3.a Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]; b Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]; c Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.a Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]; b Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]; c Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]; d Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Genes. Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Yi R, Qin Y, Macara IG, Cullen BR. Genes. Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Hammond SM. Curr. Opin. Genet. Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]; b Hammond SM, Bernstein E, Beach D, Hannon GJ. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]; c Hutvagner G, Zamore PD. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- 6.Hwang HW, Mendell JT. Br. J. Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan JA, Krichevsky AM, Kosik KS. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 8.a Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. Proc. Natl. Acad. Sci. U.S.A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Chiu YL, Dinesh CU, Chu CY, Ali A, Brown KM, Cao H, Rana TM. Chem. Biol. 2005;12:643–648. doi: 10.1016/j.chembiol.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 11.a Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. Jama. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]; c Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 12.Schmittgen TD, Jiang J, Liu Q, Yang L. Nucleic Acids Res. 2004;32:e43. doi: 10.1093/nar/gnh040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information