Gene Fusions Associated with Recurrent Amplicons Represent a Class of Passenger Aberrations in Breast Cancer (original) (raw)
Abstract
Application of high-throughput transcriptome sequencing has spurred highly sensitive detection and discovery of gene fusions in cancer, but distinguishing potentially oncogenic fusions from random, “passenger” aberrations has proven challenging. Here we examine a distinctive group of gene fusions that involve genes present in the loci of chromosomal amplifications—a class of oncogenic aberrations that are widely prevalent in breast cancers. Integrative analysis of a panel of 14 breast cancer cell lines comparing gene fusions discovered by high-throughput transcriptome sequencing and genome-wide copy number aberrations assessed by array comparative genomic hybridization, led to the identification of 77 gene fusions, of which more than 60% were localized to amplicons including 17q12, 17q23, 20q13, chr8q, and others. Many of these fusions appeared to be recurrent or involved highly expressed oncogenic drivers, frequently fused with multiple different partners, but sometimes displaying loss of functional domains. As illustrative examples of the “amplicon-associated” gene fusions, we examined here a recurrent gene fusion involving the mediator of mammalian target of rapamycin signaling, RPS6KB1 kinase in BT-474, and the therapeutically important receptor tyrosine kinase EGFR in MDA-MB-468 breast cancer cell line. These gene fusions comprise a minor allelic fraction relative to the highly expressed full-length transcripts and encode chimera lacking the kinase domains, which do not impart dependence on the respective cells. Our study suggests that amplicon-associated gene fusions in breast cancer primarily represent a by-product of chromosomal amplifications, which constitutes a subset of passenger aberrations and should be factored accordingly during prioritization of gene fusion candidates.
Introduction
Chromosomal amplifications and translocations are among the most common somatic aberrations in cancers [1,2]. Gene amplification is an important mechanism for oncogene overexpression and activation. Numerous recurrent loci of chromosomal amplifications have been characterized in breast cancer, which result in gain of copy number and overexpression of oncogenes such as ERBB2 on 17q12 (the definitive molecular aberration in 20%–30% of all breast cancers) [3,4], as well as many other oncogenic drivers including Myc [5], EGFR [6], FGFR1 [7], CyclinD1 [8], RPS6KB1 [9], and others [10]. Chromosomal translocations leading to generation of gene fusions represent another prevalent mechanism for the expression of oncogenes in epithelial cancers [11]. Recently, we described the discovery and characterization of recurrent gene fusions in breast cancer involving MAST family serine threonine kinases and Notch family of transcription factors [12]. Interestingly, we also observed a large number of gene fusions, including some recurrent fusions involving known oncogenes localized at loci of chromosomal amplifications.
Here we carried out a systematic analysis of the association between gene fusions and genomic amplification by integrating RNA-Seq data with array comparative genomic hybridization (aCGH)-based whole-genome copy number profiling from a panel of breast cancer cell lines. We examined a set of “amplicon-associated gene fusions” that refer to all the fusions where one or both gene partners are localized to a site of chromosomal amplification. Specifically, we assessed the functional relevance of two amplicon-associated fusion genes involving oncogenic kinases EGFR and RPS6KB1 in the context of prioritizing fusion candidates important in tumorigenesis. Our results suggest that recurrent gene fusions localized to recurrent amplicons, displaying allelic imbalance between the fusion partners, may represent an epiphenomenon of genomic amplification cycles not essential for cancer development.
Materials and Methods
Gene Fusion Data Set
Chimeric transcript candidates were primarily obtained from paired-end transcriptome sequencing of breast cancer from a total of more than 49 cell lines and 40 tissue samples described previously [12]. aCGH data were generated using Agilent Human Genome 244A CGH Microarrays (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions, and data were analyzed using CGH Analytics (Agilent Technologies). Copy number alterations were assessed using ADM-2, with the threshold a setting of 6.0 and a bin size of 10.
RNA Isolation and Complementary DNA Synthesis
Total RNA was isolated using TRIzol and RNeasy Kit (Invitrogen, Carlsbad, CA) with DNase I digestion according to the manufacturer's instructions. RNA integrity was verified on an Agilent Bioanalyzer 2100 (Agilent Technologies). Complementary DNA was synthesized from total RNA using Superscript III (Invitrogen) and random primers (Invitrogen).
Quantitative Real-time Polymerase Chain Reaction
Primers for validation of candidate gene fusions were designed using the National Center for Biotechnology Information Primer Blast (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), with primer pairs spanning exon junctions amplifying 70- to 110-bp products for every chimera tested. Quantitative polymerase chain reaction (QPCR) was performed using SYBR Green MasterMix (Applied Biosystems, Carlsbad, CA) on an Applied Biosystems StepOne Plus Real-Time PCR System. All oligonucleotide primers were obtained from Integrated DNA Technologies and are listed in Table W1. GAPDH wasusedasendogenous control. All assays were performed twice, and results were plotted as average fold change relative to GAPDH.
Cell Proliferation Assays
Cells were transfected with small interfering RNAs (siRNAs) using Oligofectamine reagent (Life Sciences, Carlsbad, CA), and 3 days after transfection, the cells were plated for proliferation assays. At the indicated times, cell numbers were counted using Coulter Counter (Indianapolis, IN).
Western Blot
Cell pellets were sonicated in NP-40 lysis buffer (50 mM Tris-HCl, 1% NP-40, pH 7.4; Sigma, St. Louis, MO), complete protease inhibitor mixture (Roche, Indianapolis, IN), and phosphatase inhibitor (EMD Bioscience, San Diego, CA). Immunoblot analysis was carried out using antibodies for ERBB2 (MS-730-PABX; Thermo Scientific, Fremont, CA) and RPS6KB1 (2708S; Cell Signaling, Danvers, MA). Human β-actin antibody (Sigma, St. Louis, MO) was used as a loading control.
Knockdown Assays
Short hairpin RNAs (shRNAs; Table W1) were transduced in presence of 1 µg/ml polybrene. All siRNA transfections were performed using Oligofectamine reagent (Life Sciences). For siRNA knockdown experiments, multiple custom siRNA sequences targeting the ARID1A-MAST2 fusion (Thermo, Lafayette, CO) were used [12].
Results
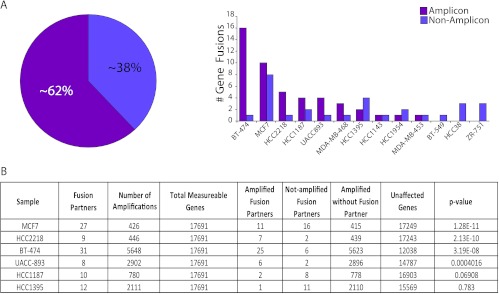
Paired-end transcriptome sequencing of breast cancer cell lines and tissues led to the identification of an average of more than four gene fusions per breast cancer sample [12]. Interestingly, we observed that some of the cell lines with the largest number of gene fusions also harbored many well-known chromosomal amplifications, prompting us to examine a likely association between genomic amplifications and gene fusions. To assess copy number alterations at the chromosomal coordinates of the fusion genes, we analyzed aCGH (244K Agilent array) data in a set of 14 cell lines (Table W2) and observed that as many as 62% of the total number of fusions were associated with regions of amplifications (Figure 1_A_). The genes involved in fusions were found to be significantly associated with their genomic amplification status based on Fisher exact t test (P < .0004), in four of six cell lines with the maximum number of fusions, including BT-474, MCF7, HCC2218, and UACC893 (Figure 1_B_).
Figure 1.
Distribution of gene fusions across breast cancer cell lines. (A) Pie chart representation of the relative proportion of gene fusions associated with loci of genomic amplifications compared to unamplified loci (left) and bar graph representation of the relative distribution of gene fusions across different breast cancer cell lines (right). (B) Table summarizing the statistical significance of association between gene fusions and chromosomal amplifications in breast cancer cell lines with the highest number of gene fusions in A (using Fisher exact t test, sorted by P value).
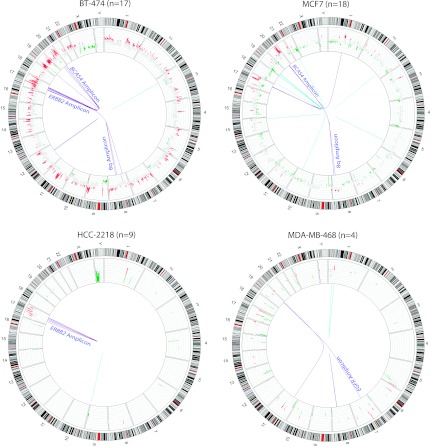
Examining the distribution of fusion genes in individual samples revealed that a majority of the gene fusions were associated with 17q12 amplicon harboring ERBB2 and 17q23 amplicon that includes genes such as BCAS3, RPS6KB1, and TMEM49, 20q13 amplicon with BCAS4 and the chr8q amplicon commonly found amplified in breast cancer (Table W2 and Figures 2 and W1). Interestingly, the breast cancer cell line BT-474 that harbors both the chr17 amplicons and the chr20 amplicon and MCF7 with prominent amplifications in chr17, chr20, and chr8q showed the maximum number of gene fusions observed in a sample, accounting for as many as 26 gene fusions associated with amplicons compared against only 9 in unamplified loci (Figures 1 and 2 and Table W2).
Figure 2.
Graphical representation of integrative analysis of gene fusions with copy number analysis. Circos plots of the genome-wide distribution of gene fusions along with status of copy number alterations. Red and green peaks represent amplifications and deletions; purple and cyan lines represent the fusions associated with amplicons and nonamplicons, respectively. “_n_” refers to the total number of fusions identified.
In the backdrop of a large number of somatic aberrations seen in cancers, any “recurrent” events observed across samples are generally regarded as potentially “driving” tumorigenesis. Interestingly, among the more than 380 gene fusions reported in our compendium of breast cancer fusions [12], as many as 62 genes were found to be recurrent partners (appear at least twice). Among these, whereas the MAST and Notch fusions were shown to be functionally recurrent and potentially driving aberrations in up to 5% to 7% of breast cancers, 33 of other recurrent gene fusions were found to be associated with known frequent amplicons, including ERBB2, BCAS3/4, and chr8q. Among these, three fusions each involved the ikaros family zinc finger protein 3 transcription factor (IKZF3 on chr17q12 amplicon) and breast carcinoma amplified sequence 3 (BCAS3 on chr17q23 amplicon) as 3′ partners—all with different 5′ partners. Similarly, tripartite motif containing 37 (TRIM37 on chr17q23) was a common 5′ partner in three distinct gene fusions with different 3′ partners (Table W2). To further expand our integrative analysis of copy number aberrations and gene fusions, next we used the breast cancer aCGH data [13,14] and observed gene fusion-associated amplicons in MCF7, BT-474, and MDA-MB-468, HCC-1187 as seen in our data as well as in an additional panel of cell lines, including ZR-75-30, SUM190, MDA-MB-361, HCC-1428, and HCC-1569 (Figure W2). Clearly, apart from triggering overexpression of constituent genes, our observations strongly suggest that the loci of chromosomal amplifications also serve as “hot-spots” for the generation of recurrent gene fusions.
Next, to assess whether amplicon-associated gene fusions impart oncogenic phenotypes on the cells, we examined the open reading frames (ORFs), functional domains/motifs, and conservation of fusion architecture across different samples. Among recurrent fusion candidates within amplicons, we focused on known cancer-associated partner genes such as kinases, oncogenes, tumor suppressors, or known fusion partners in the Mitelman Database of chromosomal aberrations in cancer [15] and observed several functionally plausible gene fusions. Here we describe our observations with two specific examples of gene fusions involving oncogenic kinases.
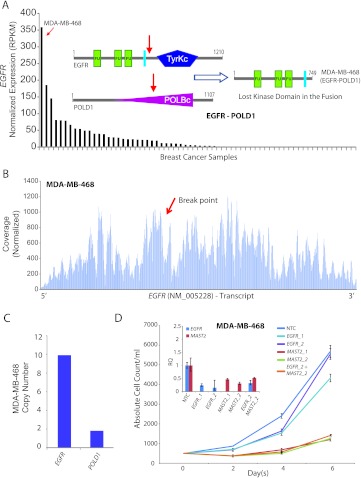
The triple-negative breast cancer cell line MDA-MB-468 is known to show an overexpression of epidermal growth factor receptor (EGFR) [16]. In our transcriptome sequencing compendium of 89 breast cancer cell lines and tissues, the highest expression of EGFR is observed in MDA-MB-468 (Figure 3_A_), potentially resulting from a focal amplification at chr7p12 (Figure 2). In addition, we detected an EGFR fusion transcript (EGFR-POLD1) in this cell line, encoding the N-terminal portion of EGFR, completely devoid of the tyrosine kinase domain (Figure 3_A_, inset). However, the uniform read-coverage observed across the full length of the EGFR transcript in this sample (Figure 3_B_), precluded the existence of any exon imbalance, suggesting that even as the kinase domain is lost in the fusion, the full-length EGFR protein is expressed in this cell line. Further, we observed a remarkable mismatch between the copy numbers of EGFR and its fusion partner POLD1 (Figure 3_C_) that supports a predominant expression of full-length EGFR compared with the EGFR-POLD1 chimera. This is unlike the observation in case of MAST kinase fusions in breast cancer characterized in our previous study [12], in which case a marked exon imbalance in coverage was observed (Figure W3). Considering that the MDA-MB-468 harbors both MAST2 and EGFR fusions, we were intrigued to assess its relative “dependence” on both the kinases. Surprisingly, a profound reduction in cell proliferation was observed on siRNA knockdown of MAST2, whereas EGFR knockdown showed little effect (Figure 3_D_). Next, testing the possibility of EGFR amplicon potentially cooperating with MAST2, we found that the effect of combined knockdown of EGFR and MAST2 was comparable with that of MAST2 knockdown alone (Figure 3_D_), further suggesting that EGFR amplification does not signify a driver aberration. In this context, the EGFR fusion transcript that represents a miniscule fraction of overall EGFR expression and encodes only the N-terminal portion lacking the kinase domain was reckoned to be inconsequential.
Figure 3.
(A) Normalized expression (RPKM) of EGFR in descending order of expression in a panel of breast cancer samples obtained from RNA-Seq. Schematic representation of wild-type EGFR and POLD1 proteins with putative breakpoints indicated by red arrows and the domain structure of the putative fusion protein (inset). (B) Plot of normalized coverage of EGFR transcript in MDA-MB-468 cell line showing the location of the breakpoint (indicated by red arrow). (C) Bar graph representing the copy number of EGFR and POLD1 in MDA-MB-468. (D) Proliferation assay showing absolute cell count (y axis) over a time course (x axis) after knockdown with EGFR and/or MAST2 siRNAs in MDA-MB-468. QPCR assessment of knockdown efficiencies relative to nontargeted control (NTC; inset).
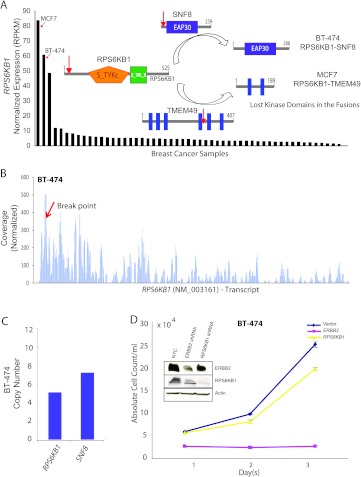
Next, we looked at recurrent gene fusions involving oncogenic serine threonine kinase ribosomal protein S6 kinase on chr17q23 frequently amplified in breast cancers [17–20] identified in BT-474 (RPS6KB1-SNF8) and MCF7 (RPS6KB1-VMP1). Both of these cell lines harbor amplifications at the RPS6KB1 locus and express the highest levels of RPS6KB1 among all the samples examined (Figure 4_A_). Both the chimeric transcripts retain only the first exon of RPS6KB1 and the respective open reading frames show a complete loss of the kinase domain (Figure 4_A_, inset). We also observed an even read coverage across the RPS6KB1 transcript in both fusion-positive cell lines, similar to a representative benign mammary epithelial cell line, albeit at a much higher level, indicating that full-length RPS6KB1 protein is encoded in these samples (Figures 4_B_ and W4_A_). Further, the difference between the copy number observed between the fusion partners in both the RPS6KB1 fusions (Figures 4_C_ and W4_B_) indicates an allelic imbalance between the full-length and the putative fusion genes. Next, considering that BT-474 is an _ERBB2_-positive cell line, we tested potential dependence of these cells on the RPS6KB1 protein. Surprisingly, similar to our observations with EGFR knockdown in MDA-MB-468 cells, here we observed only a small effect on cell proliferation after shRNA knockdown of RPS6KB1, in dramatic contrast to the effect of ERBB2 knockdown (Figure 4_D_). Notably, the shRNA knockdown of RPS6KB1 led to a significant depletion of the full-length protein yet it did not affect cell proliferation compared with ERBB2 protein depletion (Figure 4_D_, inset). Therefore, BT-474 cells do not display a dependence on RPS6KB1 protein, and considering that the RPS6KB1 fusion product is completely devoid of all functional domains of RPS6KB1, including the kinase domain, this fusion also likely represents a passenger event.
Figure 4.
(A) Normalized expression (RPKM) of RPS6KB1 in descending order of expression in a panel of breast cancer samples obtained from RNA-Seq. Schematic representation of wild-type RPS6KB1, TMEM49, and SNF8 proteins with putative breakpoints indicated by red arrows and the domain structure of the putative fusion proteins in BT-474 and MCF7 (inset). (B) Plot of normalized coverage of RPS6KB1 transcript in BT-474 cell line showing the location of the breakpoint (indicated by red arrow). (C) Bar graph representing the copy number of RPS6KB1 and SNF8 in BT-474 (D) Proliferation assay showing absolute cell count (y axis) over a time course (x axis) after knockdown with RPS6KB1 and/or ERBB2 shRNAs in BT-474. Western blot assessment of the knockdown efficiency relative to nontargeted control (NTC). Actin was used as a loading control (inset).
Discussion
In our systematic search for gene fusions in breast cancer using high-throughput transcriptome sequencing, we observed a notably large number of fusion genes associated with many well characterized recurrent amplicons, including 17q12, 17q23, 20q13, and 8q, among others. Amplicon-associated gene fusions were found to involve complex and cryptic rearrangements, involving one or both partners within the amplicon site, with the chimeric transcript expression apparently concealed in the backdrop of highly expressed wild-type genes. The gene fusions considered here include only “expressed” chimeric transcripts derived from known/annotated fusion partners. Chromosomal rearrangements that do not express chimeric transcripts or that involve unannotated fusion partners are excluded from this analysis. This likely accounts for the variability observed in the number of gene fusions scored across multiple samples with known amplicons. Because many of the fusions at the amplicons appeared to be recurrent, although frequently fused with multiple different partners, it led us to examine whether the recurrence was incidentally associated with recurrent amplicons or signified functionally important aberrations.
MDA-MB-468 represents a prototype triple-negative breast cancer cell line with a “basal-like” gene expression profile that shows an overexpression of the oncogenic kinase EGFR due to a focal amplification at chr7p12. Here we discovered a chimeric transcript involving EGFR. However, careful examination of this transcript revealed that the fusion encodes N-terminal EGFR protein, without the kinase domain. Transcriptome sequencing did not show evidence of fusion-associated exon imbalance in EGFR expression, suggesting that full-length EGFR is expressed in this cell line. In addition, the significantly higher genomic copy number of EGFR compared to its fusion partner POLD1 suggests that a minor allelic fraction of the EGFR is involved in fusion with POLD1, whereas other amplified copies of the gene express the full-length molecule. Technically, the detection and monitoring of the EGFR fusion transcript in the backdrop of extremely high levels of wild-type EGFR transcript is challenging; therefore, we chose to assess the dependency imparted by full-length EGFR. Interestingly, the knockdown of EGFR had only a slight effect on the proliferation of MDA-MB-468 cells, whereas a profound reduction in cell proliferation was observed on the knockdown the fusion gene MAST2. Combined knockdown of MAST2 and EGFR produced the same effect as that by MAST2 alone, further calling into question the credentials of EGFR as a driver aberration in MDA-MB-468 cells. Interestingly, MDA-MB-468 is known to be insensitive to EGFR inhibitors like erlotinib [21] and gefitinib [22].
Similarly, the recurrent gene fusions involving RPS6KB1 retain only the first exon, and the chimeric ORFs show a complete loss of the kinase domain in breast cancer cell lines BT-474 and MCF7. Similar to the EGFR fusion, DNA copy number analysis and RNA-Seq data provided the evidence that full-length RPS6KB1 protein is encoded in both these cell lines. Notably, both BT-474 and MCF7 have been shown to express high levels of full-length RPS6KB1 protein [23], suggesting that these cells exhibit elevated activity of RPS6KB1 as a result of amplification, independent of the fusion. Again, similar to EGFR knockdown in MDA-MB-468, RPS6KB1 knockdown in BT-474 (an ERBB2-positive cell line) showed an insignificant effect on cell proliferation compared to ERBB2 knockdown. Interestingly, in a previous study, knockdown of RPS6KB1 was found to have no effect on apoptosis in both BT-474 and MCF7 breast cancer cells [24].
In the light of our observations, we surmise that repeated breaks and rejoining of chromosomes during chromosomal amplifications led to the generation of amplicon-associated gene fusions. Loci of recurrent genomic amplifications thus engender “pseudo” recurrent gene fusions that may largely represent passenger aberrations involving random breakpoints. The two cell lines with established drivers—ERBB2 in BT-474 and MAST2 in MDA-MB-468—made it possible for us to assess the relative importance of amplicon fusions involving RPS6KB1 and EGFR, respectively. In cases where a driver is not clearly apparent, a more careful examination of all plausible fusion candidates will be required. Importantly, even as our study primarily pertains to breast cancers based on available data and a well-documented preponderance of copy number aberrations in breast cancers [10], we expect the association between amplicons and gene fusions to be consistent in other cancers as well. We argue here for a measure of caution in considering the functional implications of recurrent gene fusions associated with amplifications because these may be simply a result of massive chromosomal upheaval at the amplicons, not representing clonally selected oncogenic events.
Supplementary Material
Supplementary Figures and Tables
Acknowledgments
The authors thank Vishal Kothari and Nikita Consul for technical help.
Footnotes
1
This project was supported in part by Department of Defense Breast Cancer Research Program (W81XWH-08-0110), American Association for Cancer Research Stand Up to Cancer (SU2C) award, the National Functional Genomics Center (W81XWH-11-1-0520) from Department of Defense to A.M.C., and, in part, by the US National Institutes of Health through the University of Michigan's Cancer Center Support grant 5 P30 CA46592. A.M.C. is supported by the US National Cancer Institute's Early Detection Research Network (U01 CA111275). A.M.C. is supported by the Doris Duke Charitable Foundation Clinical Scientist Award and the Burroughs Welcome Foundation Award in Clinical Translational Research. A.M.C. is an American Cancer Society Research professor and Taubman Scholar. S.D. is supported by Howard Hughes Medical Institute Medical Student Fellowship.
2
This article refers to supplementary materials, which are designated by Tables W1 and W2 and Figures W1 to W4 and are available online at www.neoplasia.com.
References
- 1.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 3.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 4.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 5.Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688–1695. doi: 10.1054/bjoc.2000.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhargava R, Gerald WL, Li AR, Pan Q, Lal P, Ladanyi M, Chen B. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol. 2005;18:1027–1033. doi: 10.1038/modpathol.3800438. [DOI] [PubMed] [Google Scholar]
- 7.Elbauomy Elsheikh S, Green AR, Lambros MB, Turner NC, Grainge MJ, Powe D, Ellis IO, Reis-Filho JS. FGFR1 amplification in breast carcinomas: achromogenic in situ hybridisation analysis. Breast Cancer Res. 2007;9:R23. doi: 10.1186/bcr1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elsheikh S, Green AR, Aleskandarany MA, Grainge M, Paish CE, Lambros MB, Reis-Filho JS, Ellis IO. CCND1 amplification and cyclin D1 expression in breast cancer and their relation with proteomic subgroups and patient outcome. Breast Cancer Res Treat. 2008;109:325–335. doi: 10.1007/s10549-007-9659-8. [DOI] [PubMed] [Google Scholar]
- 9.Inaki K, Hillmer AM, Ukil L, Yao F, Woo XY, Vardy LA, Zawack KF, Lee CW, Ariyaratne PN, Chan YS, et al. Transcriptional consequences of genomic structural aberrations in breast cancer. Genome Res. 2011;21:676–687. doi: 10.1101/gr.113225.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chinnaiyan AM, Palanisamy N. Chromosomal aberrations in solid tumors. Prog Mol Biol Transl Sci. 2010;95:55–94. doi: 10.1016/B978-0-12-385071-3.00004-6. [DOI] [PubMed] [Google Scholar]
- 12.Robinson DR, Kalyana-Sundaram S, Wu YM, Shankar S, Cao X, Ateeq B, Asangani IA, Iyer M, Maher CA, Grasso CS, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollack JR, Sorlie T, Perou CM, Rees CA, Jeffrey SS, Lonning PE, Tibshirani R, Botstein D, Borresen-Dale AL, Brown PO. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci USA. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greshock J, Naylor TL, Margolin A, Diskin S, Cleaver SH, Futreal PA, deJong PJ, Zhao S, Liebman M, Weber BL. 1-Mb resolution array-based comparative genomic hybridization using a BAC clone set optimized for cancer gene analysis. Genome Res. 2004;14:179–187. doi: 10.1101/gr.1847304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Cancer Genome Anatomy Project. 2010. [March 2012]. Available at: http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- 16.Hyatt DC, Ceresa BP. Cellular localization of the activated EGFR determines its effect on cell growth in MDA-MB-468 cells. Exp Cell Res. 2008;314:3415–3425. doi: 10.1016/j.yexcr.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barlund M, Monni O, Kononen J, Cornelison R, Torhorst J, Sauter G, Kallioniemi O-P, Kallioniemi A. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res. 2000;60:5340–5344. [PubMed] [Google Scholar]
- 18.Couch FJ, Wang XY, Wu GJ, Qian J, Jenkins RB, James CD. Localization of PS6K to chromosomal region 17q23 and determination of its amplification in breast cancer. Cancer Res. 1999;59:1408–1411. [PubMed] [Google Scholar]
- 19.Monni O, Barlund M, Mousses S, Kononen J, Sauter G, Heiskanen M, Paavola P, Avela K, Chen Y, Bittner ML, et al. Comprehensive copy number and gene expression profiling of the 17q23 amplicon in human breast cancer. Proc Natl Acad Sci USA. 2001;98:5711–5716. doi: 10.1073/pnas.091582298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–322. doi: 10.1023/a:1023081624133. [DOI] [PubMed] [Google Scholar]
- 21.Bartholomeusz C, Yamasaki F, Saso H, Kurisu K, Hortobagyi GN, Ueno NT. Gemcitabine overcomes erlotinib resistance in EGFR-overexpressing cancer cells through downregulation of Akt. J Cancer. 2011;2:435–442. doi: 10.7150/jca.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiello MR, D'Alessio A, De Luca A, Carotenuto A, Rachiglio AM, Napolitano M, Cito L, Guzzo A, Normanno N. AZD3409 inhibits the growth of breast cancer cells with intrinsic resistance to the EGFR tyrosine kinase inhibitor gefitinib. Breast Cancer Res Treat. 2007;102:275–282. doi: 10.1007/s10549-006-9340-7. [DOI] [PubMed] [Google Scholar]
- 23.Yamnik RL, Digilova A, Davis DC, Brodt ZN, Murphy CJ, Holz MK. S6 kinase 1 regulates estrogen receptor alpha in control of breast cancer cell proliferation. J Biol Chem. 2009;284:6361–6369. doi: 10.1074/jbc.M807532200. [DOI] [PubMed] [Google Scholar]
- 24.Heinonen H, Nieminen A, Saarela M, Kallioniemi A, Klefstrom J, Hautaniemi S, Monni O. Deciphering downstream gene targets of PI3K/mTOR/70S6K pathway in breast cancer. BMC Genomics. 2008;9:348. doi: 10.1186/1471-2164-9-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures and Tables