Impaired Nipple Development and Parturition in LGR7 Knockout Mice (original) (raw)
Abstract
LGR7 is a G-protein coupled receptor with structural homology to the gonadotrophin and thyrotrophin receptors. Recently, LGR7 was deorphanized, and it was shown that relaxin is the ligand for LGR7. To further study the function of this receptor, mice deficient for LGR7 were generated by replacing part of the transmembrane-encoding region with a LacZ reporter cassette. Here we show that LGR7 is expressed in various tissues, including the uterus, heart, brain, and testis. Fertility studies using female LGR7−/− mice showed normal fertility and litter size. However, some females were incapable of delivering their pups, and several pups were found dead. Moreover, all offspring died within 24 to 48 h after delivery because female LGR7−/− mice were unable to feed their offspring due to impaired nipple development. In some male LGR7−/− mice, spermatogenesis was impaired, leading to azoospermia and a reduction in fertility. Interestingly, these phenomena were absent in mutant mice at older ages or in later generations. Taken together, results from LGR7 knockout mice indicate an essential role for the LGR7 receptor in nipple development during pregnancy. Moreover, a defect in parturition was observed, suggesting a role for LGR7 in the process of cervical ripening.
LGR7 is a leucine-rich repeat-containing G-protein coupled receptor that was identified on the basis of its structural homology to the follicle-stimulating hormone, luteinizing hormone, and thyrotropin receptor family (4, 5). An interesting feature of these receptors are their large extracellular domains with leucine-rich repeats, which are commonly found in proteins involved in protein-protein interactions (8). Recently, LGR7 has been deorphanized, and it was shown that relaxin is the high-affinity ligand (6). The relaxin hormone has been studied in detail over the last few decades and has been implicated in many physiological processes related to the female reproductive system, including the induction of collagen remodeling and softening of the tissues of the birth canal prior to delivery. Also, the inhibition of uterine contractile activity and development of growth and differentiation of the mammary gland have been reported (2). In addition, relaxin has been implicated to trigger blood vessel dilatation in several organs and tissues, such as rat and mouse uterine endometrium and myometrium and mammary glands. Moreover, it has been shown that relaxin increases coronary flow, an effect which was paralleled by an increase of the production of nitric oxide, a potent vasodilatory agent (15).
The observation that mice deficient for relaxin are unable to deliver milk to their pups due to impaired mammary gland development and the absence of nipple enlargement at the end of pregnancy has provided the first evidence for the function of relaxin. Moreover, it was reported that some of the mice were unable to deliver, most probably due to the absence of cervical softening or contractile activity (20). Phenotypic data related to the role of relaxin cardiac function indicate a gender-specific phenotype in which males were characterized by a defect in left-ventricular filling, which is likely caused by an increase in collagen content in the left ventricle (3).
Expression analysis using reverse transcriptase PCR or multiple-tissue Northern blot analysis has revealed that LGR7 is expressed in a variety of tissues, including the uterus, brain, kidney, ovary, placenta, heart, skin, and prostate. Using LGR7-specific antibodies, the presence of the receptor in some of these tissues was confirmed (6).
In addition to a role as LGR7 ligand, relaxin has also been shown to be capable of activating the LGR8 receptor, albeit with much lower affinity. The LGR8 receptor has previously been shown to be essential for testicular descent, a function that was also described for its ligand Insl3 (14, 16). On the basis of these data, a physiological relationship between relaxin and LGR8 was postulated. However, based on knockout data that show identical phenotypes for LGR8 and Insl3 but no overlap with the phenotype of relaxin-deficient mice, it has been unequivocally demonstrated that Insl3 is the cognate ligand for LGR8 and that relaxin is not able to compensate for a lack of Insl3 (11).
To obtain further insight into the function of the LGR7 receptor, particularly in terms of the male and female reproductive system, mice deficient for the LGR7 receptor were generated. Here we report that female LGR7−/− mice show normal fertility but severe impairment of nipple development during pregnancy, resulting in an inability of the animals to feed their offspring. Moreover, defective parturition reflecting impaired cervical ripening was observed. A minority of male LGR7−/− mice exhibited a transitory disrupted spermatogenesis and reduced fertility; the transitory aspect may reflect redundancy and/or may be caused by differences in genetic background.
MATERIALS AND METHODS
Generation of LGR-7-deficient mice.
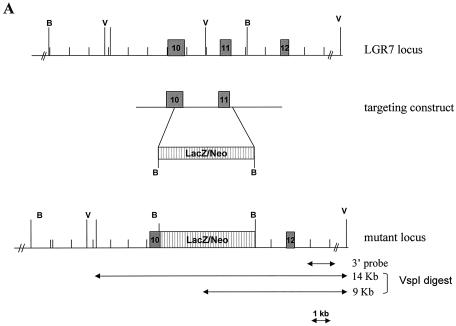
Homologous recombination in embryonic stem (ES) cells was used to disrupt the murine LGR7 gene. A replacement vector was designed to delete part of exon 10 and exon 11, the sequences encoding part of the transmembrane region of the gene. The deleted sequence was replaced with a LacZ/MC1-Neo selection cassette (Fig. 1A) such that the LacZ reporter gene was in frame with the LGR7 coding sequence. The targeting vector was electroporated into 129 Sv/Evbrd cells, after which colonies were selected in the presence of G418 and 1-(2-deoxy-2-fluoro-β-d-arabinofuranosyl)-5-iodouracil (FIAU). G418-FIAU-resistant colonies were isolated and analyzed for homologous recombination by Southern blot analysis (Fig. 1B). ES cell clones characterized by correct homologous recombination at both sides of the vector and a single integration event were selected for blastocyst injections. Chimeras obtained were mated to C57BL/6 (albino) females to generate heterozygous mice. The heterozygous (LGR7+/−) mice were then interbred to produce homozygous (LGR−/−) knockout mice.
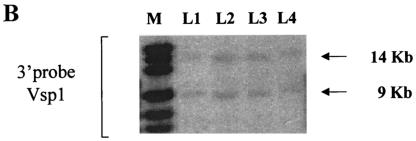
FIG. 1.
Targeted disruption of the LGR7 gene. (A) Schematic representation of the LGR7 locus, targeting construct, and mutant locus. Dark boxes represent exons of LGR7. The expected fragment lengths after _Vsp_I digestion are 9 kb for the wild type and 14 kb for the mutant allele. B, _Bam_HI; V, _Vsp_I. (B) Targeted ES cells: Southern blot analysis of _Vsp_I-restricted DNA from clones L1, L2, L3, and L4; analysis was performed with a 3′ probe. M, marker.
PCR genotyping.
PCR analysis on isolated genomic DNA obtained from tail biopsy samples was used to screen genotypes. The primers used for the wild-type allele were 5′-ACTGAGCAGTTCCTCCGGGTAA-3′ and 5′-AGGTGGTTCTCGCCAAACGC-3′. Primers used to detect the mutated allele were 5′-CTTGGGTGGAGAGGCTATTC-3′ and 5′-AGGTGAGATGACAGGAGATC-3′. PCR conditions were 100 ng of each primer, 1 U of Taq polymerase (Amersham Pharmacia Biotech, Inc.), 200 μM deoxynucleoside triphosphates (Amersham Pharmacia Biotech, Inc.), and 1× PCR buffer (Amersham Pharmacia Biotech, Inc.) in a volume of 50 μl. Amplification was carried out by using the following protocol: denaturing for 5 min at 95°C followed by 30 cycles consisting of denaturing at 95°C for 30 s, annealing at 57°C (mutant allele) or 59°C (wild-type allele) for 30 s, and extension at 72°C for 1 min in a Perkin-Elmer DNA thermal cycler model 9700. Samples were subsequently analyzed on 2% agarose gels. A 269-bp fragment was generated from the wild-type allele, whereas the mutant allele generated a 280-bp fragment.
Animals and husbandry.
Animals were housed in macrolon cages (eight animals/cage) at 19 to 21°C; the relative humidity was 50 to 60%, and an artificial light cycle of 12 h alternating with 12 h of darkness was offered. Standard pelleted food (Special Diet Services, Witham, Essex, England) and water were provided ad libitum during the experiment. The animals were observed for morbidity and mortality daily. Additionally, the animals were regularly observed for any behavioral or physical abnormalities.
All animal experiments were approved by the Organon Animal Ethics Committee.
Staining for β-galactosidase (β-Gal) activity.
LGR7 expression profiling using the LacZ reporter gene was performed on heterozygous male and female LGR7 mutant mice. As controls, wild-type mice were chosen. About 40 organs and tissues were dissected from a single mouse and fixed for 30 to 90 min (depending on the organ size) at room temperature in 0.2% glutaraldehyde, 1.5% paraformaldehyde, 2 mM MgCl2, 5 mM EGTA, and 100 mM sodium phosphate (pH 7.3). Tissues were rinsed three times for 20 min in 0.2% NP-40, 0.1% sodium deoxycholate, 2 mM MgCl2, 100 mM sodium phosphate, and they were then stained for 48 h in 5 mM K3Fe(CN)6, 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (dissolved in dimethylformamide) per ml, 0.2% NP-40, 0.1% sodium deoxycholate, 2 mM MgCl2, and 100 mM sodium phosphate (pH 7.3). After three washes for 10 min in 0.2% NP-40, 0.1% sodium deoxycholate, 2 mM MgCl2, and 100 mM sodium phosphate, the tissues were fixed overnight in phosphate-buffered-4% formaldehyde and subsequently rinsed twice for 5 min in phosphate-buffered saline (PBS). Tissues were serially dehydrated in ethanol (50 to 100%, twice for 60 min each) and twice in 100% 2-propanol (60 min each, at room temperature and 60°C). The tissues were then infiltrated twice for 60 min with 1:1 mix of preheated 2-propanol and paraffin, infiltrated three times for 60 min in paraffin, and embedded. Sections (thickness, 3 to 4 μm) were made and counterstained with nuclear fast red.
Histology.
Phenotypic analysis was performed on 12-week-old males and females from the F2 generation (wild-type, heterozygous, and homozygous; six mice/group). Before autopsies were conducted, blood was obtained from each mouse. All internal organs were visually examined for signs of abnormalities. The following organs were dissected free of adjacent fat and other surrounding tissues, and their weights were recorded: heart; spleen; thymus; liver and gall bladder; kidneys; adrenal glands; brain; testes; epididymides; seminal vesicles, coagulating glands, and prostate gland; ovaries; and uterus. The organ weight relative to terminal body weight, determined prior to autopsy, was calculated. Samples of all organs and tissues from the animals were preserved in 4% buffered formaldehyde (except one testis that was preserved in Bouin), dehydrated, and embedded in paraffin wax. Sections (thickness, 3 to 4 μm) made from these blocks were stained with the hematoxylin and eosin (HE) staining method or periodic acid-Schiff reagents as described previously (1) and examined histopathologically.
Detection of apoptosis.
Formalin-fixed and paraffin-embedded sections from testes and epididymides of wild-type and homozygous males were used for the detection of DNA fragmentation by the terminal deoxynucleotidyltransferase (TdT)-mediated dUTP-biotin nick end labeling (TUNEL) method. Deparaffinized tissue sections were incubated for 15 min at room temperature with DNase-free proteinase K (20 μg/ml, pH 8.0), rinsed in demineralized water, blocked for endogenous peroxidase activity with 2% H2O2 in PBS for 15 min, rinsed in demineralized water, and preincubated with TdT buffer [equilibration buffer (pH 6.8); 200 mM sodium cacodylate, 2.5 mM cobalt chloride, 25 mM Tris-HCl, 0.2 mM 1,1,1-trichloro-2,2-bis(4-chlorophenyl)ethane (DDT), 0.25 mg of bovine serum albumin per ml] for 5 min followed by incubation with TdT buffer containing nucleotide mix (50 μM dUTP-biotin, 0.2 to 0.4 U of TdT enzyme) for 1 to 2 h at 37°C. At this point, the reaction was stopped with SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) twice for 15 min; sections were then incubated with 2% bovine serum albumin in PBS for 10 min, rinsed in PBS, incubated with an avidin-biotin peroxidase complex (1:200) by using a Vectastain Elite kit for 30 min, rinsed with PBS, incubated with metal-enhanced 1,4-diamino-2-butanone in stable peroxidase buffer (1:10) for 10 min, rinsed in demineralized water twice for 5 min, and counterstained with hematoxylin.
Blood analyses.
Blood was collected via the vena cava under inhalation anesthesia (isoflurane). Blood samples were collected with K3EDTA or evaporated lithium-heparinate used as the anticoagulant depending on the subsequent analysis and rotated for 10 min (only K3EDTA samples) on an RM 5 rotator. Analyses were carried out by the Biochemical Analysis Team, Toxicology and Drug Disposition, N.V. Organon. Hematology and blood clotting parameters were determined by using a Cell-Dyn 3500 hematology analyzer (Abbott, Santa Clara, Calif.). Hematological parameters determined were red blood cell count, plasma hemoglobin concentration, hematocrit, mean corpuscular volume, mean corpuscular hemoglobin, mean corpuscular hemoglobin concentration, and reticulocyte count. The following white blood cell subpopulations were quantified and expressed in absolute numbers and as the percentage of the white blood cell count: white blood cell count, neutrophilic granulocyte count, lymphocyte count, monocyte count, eosinophilic granulocyte count, and basophilic granulocyte count. For assessing blood clotting, the thrombocyte count was taken. Blood biochemistry parameters (aspartate aminotransferase, alanine aminotransferase, alkaline phosphatase, total bilirubin, glucose, urea, triglycerides, cholesterol, total protein, albumin, creatinine, lactate dehydrogenase, creatine kinase, gamma glutamyl transpeptidase, sodium, potassium, calcium, and chloride) were determined by using a Hitachi-911 analyzer (Roche, Mannheim, Germany).
Histopathology.
HE-stained sections of the following organs and tissues were examined microscopically in all wild-type and homozygous mutant animals: spleen, thymus, mesenteric lymph node, bone marrow, endocrine pancreas, thyroid gland, parathyroid glands, adrenal glands, pituitary gland, heart, aorta, exocrine pancreas, salivary glands, liver and gall bladder, esophagus, stomach, duodenum, jejunum, ileum, cecum, colon, rectum, lungs, trachea, kidneys, urinary bladder, testes, epididymides, seminal vesicles, coagulating glands, prostate gland, ovaries, uterus, vagina, skeletal muscle, diaphragm, sternum, femur, joint, vertebrae, peripheral nerve, brain, spinal cord, optic nerve, skin, mammary glands, eye, harderian gland, lacrimal gland, and tongue.
Statistical evaluation.
Hematological, blood clotting, biochemical, and body and organ weight parameters were expressed as means ± standard errors of the means of the individual data. Statistical analysis was performed by using the Student t test (two tailed). P values of <0.05 were considered significant.
Fertility studies.
Timed matings were carried out by housing one male with one or two females in a cage. The mating period lasted at least 5 days. Each day, females were evaluated for the presence of vaginal plugs. Gestation day 0.5 was defined by the presence of a vaginal plug.
RESULTS
Targeted disruption of LGR7.
The LGR7 gene was disrupted by replacing two exons encoding part of the seven-transmembrane region with the LacZ reporter cassette. This modification prevented expression of a functional receptor. Consequently, LGR7 was no longer present in the cell membrane, and cells would not be capable of mediating the action of relaxin and/or other ligands via the cyclic AMP signaling pathway. Correct homologous recombination was confirmed in ES cells by Southern blot analysis (Fig. 1B).
LacZ expression under control of the LGR7 promoter.
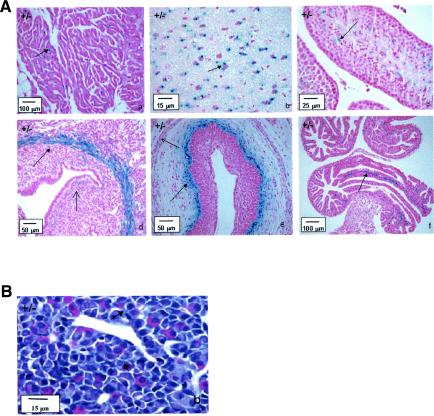
Due to the insertion of the LacZ reporter cassette into the LGR7 locus, tissues expressing LacZ can easily be studied on basis of their β-Gal activity. Analysis of more than 40 organs and tissues indicated the presence of β-Gal activity in a limited number of tissues, including the uterus, brain, heart, vagina, oviduct, and testis (Fig. 2A). Interestingly, high levels of β-Gal expression were also detected in the pituitary glands of pregnant mice (Fig. 2B), whereas β-Gal expression could not be detected in the pituitary gland of nonpregnant females or in the males (data not shown). In the salivary glands, epididymides, prostate gland, coagulating glands, preputial glands, and the gastrointestinal tract, positive staining was found in both wild-type and heterozygous mice, indicating the presence of endogenous β-Gal.
FIG. 2.
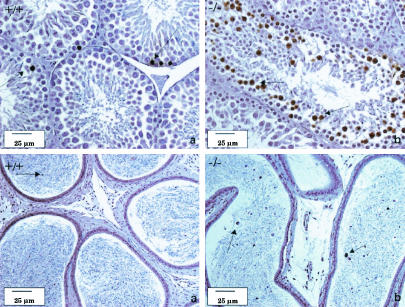
(A) β-Gal activity in tissues from 8- to 10-week-old male and female LGR7+/− mice. (a) Heart (atrium). Shown is local accumulation of staining (arrow) in the myocytes, not associated with the nucleus. (b) Brain (frontal cortex). Shown is strong staining (arrow) in the neurons, adjacent to the nuclei. (c) Testis. Shown is positive staining (arrow) in the late stages of spermatogenesis, near the spermatids; weak staining in the Leydig cells is also shown. (d) Uterus. Shown is strong staining in the circular layer of the myometrium (closed arrow); weak staining in the lamina propria under the luminal epithelium (open arrow) and the longitudinal layer of the myometrium is also shown. (e) Vagina. Shown is strong staining below the basal layer of the vaginal epithelium (closed arrow); weak staining in smooth muscle layer (open arrow) is also shown. (f) Oviduct. Shown is positive staining (arrow) in the vascular supporting tissue between the lining epithelium; weak staining in the endothelium of the blood vessels is also shown. (B) β-Gal activity in the pituitary gland of a pregnant LGR7+/− mouse. Positive staining (arrows) in several hormone-producing cells in the pars distalis is shown.
LGR7-knockout phenotype.
No obvious gross anatomical or behavioral differences or relevant changes in hematological, blood clotting (Table 1), or biochemical parameters (Table 2) were observed when comparing 8- to 10-week-old homozygous LGR7 knockout mice from the F2 generation with wild-type and heterozygous littermates. Organ weights did not differ among the different female groups (Table 3), and histopathological abnormalities were not observed for any of the organs and tissues studied (data not shown).
TABLE 1.
Overview of hematological and blood clotting parameters in control and LGR7 knockout micea
| Parameter and unitsb | Data for males of genotype: | Data for females of genotype: | ||||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | +/+ | +/− | −/− | |
| Red blood cells (1012/liter) | 9.29 ± 0.21 | 9.34 ± 0.13 | 9.35 ± 0.11 (5) | 9.39 ± 0.17 | 8.97 ± 0.19 | 9.44 ± 0.06 (4) |
| Hemoglobin (mmol/liter) | 8.97 ± 0.17 | 8.90 ± 0.07 | 8.74 ± 0.12 (5) | 9.25 ± 0.12 | 8.73 ± 0.10d | 9.19 ± 0.13 (4) |
| Hematocrit (liter/liter) | 0.461 ± 0.01 | 0.446 ± 0.01 | 0.449 ± 0.01 (5) | 0.460 ± 0.01 | 0.439 ± 0.00 | 0.461 ± 0.01 (4) |
| MCV (fl) | 49.7 ± 0.69 | 47.8 ± 0.39c | 48.0 ± 0.45 (5) | 49.0 ± 0.48 | 48.9 ± 0.58 | 48.8 ± 1.09 (4) |
| MCH (fmol) | 0.97 ± 0.01 | 0.95 ± 0.01 | 0.94 ± 0.01c (5) | 0.99 ± 0.01 | 0.97 ± 0.01 | 0.97 ± 0.02 (4) |
| MCHC (mmol/liter) | 19.5 ± 0.13 | 20.0 ± 0.18c | 19.5 ± 0.10 (5) | 20.1 ± 0.12 | 19.9 ± 0.14 | 20.0 ± 0.10 (4) |
| Reticulocytes | ||||||
| 109/liter | 378 ± 49 | 347 ± 41 | 331 ± 46 (5) | 355 ± 62 | 416 ± 48 | 273 ± 54 (4) |
| % RBC | 4.1 ± 0.6 | 3.7 ± 0.4 | 3.6 ± 0.5 | 3.8 ± 0.7 | 4.7 ± 0.6 | 2.9 ± 0.6 |
| White blood cells (109/liter) | 6.87 ± 0.68 | 7.04 ± 0.74 | 6.68 ± 0.41 (5) | 6.44 ± 0.60 | 5.00 ± 0.54 | 6.09 ± 0.93 (4) |
| Neutrophils | ||||||
| 109/liter | 1.04 ± 0.32 | 1.71 ± 0.67 | 0.83 ± 0.16 (5) | 0.51 ± 0.06 | 0.45 ± 0.04 | 0.55 ± 0.04 (4) |
| % WBC | 14.4 ± 4.1 | 21.9 ± 6.1 | 12.4 ± 2.0 | 8.4 ± 1.3 | 9.3 ± 0.9 | 9.5 ± 1.5 |
| Lymphocytes | ||||||
| 109/liter | 5.25 ± 0.57 | 4.65 ± 0.40 | 5.37 ± 0.42 (5) | 5.52 ± 0.60 | 4.24 ± 0.55 | 5.03 ± 0.81 (4) |
| % WBC | 77.3 ± 5.2 | 68.5 ± 7.1 | 80.1 ± 2.3 | 85.0 ± 1.6 | 83.8 ± 2.2 | 82.4 ± 1.8 |
| Monocytes | ||||||
| 109/liter | 0.49 ± 0.11 | 0.56 ± 0.12 | 0.37 ± 0.06 (5) | 0.27 ± 0.03 | 0.22 ± 0.08 | 0.34 ± 0.08 (4) |
| % WBC | 7.0 ± 1.4 | 7.8 ± 1.4 | 5.8 ± 1.2 | 4.5 ± 0.9 | 5.1 ± 2.0 | 5.7 ± 1.3 |
| Eosinophils | ||||||
| 109/liter | 0.09 ± 0.02 | 0.12 ± 0.03 | 0.11 ± 0.03 (5) | 0.14 ± 0.03 | 0.09 ± 0.02 | 0.16 ± 0.07 (4) |
| % WBC | 1.4 ± 0.3 | 1.9 ± 0.5 | 1.7 ± 0.4 | 2.1 ± 0.3 | 1.8 ± 0.3 | 2.4 ± 0.7 |
| Basophils (109/liter) | 0.00 | 0.00 | 0.00 (5) | 0.00 | 0.00 | 0.00 (4) |
| Thrombocytes (109/liter) | 893 ± 21 (5) | 930 ± 31 | 947 ± 50 (4) | 796 ± 50 (3) | 776 ± 60 | 732 ± 31 (3) |
TABLE 2.
Overview of biochemical parameters in plasma from control and LGR7 knockout micea
| Parameter and unitsb | Data for males of genotype: | Data for females of genotype: | ||||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | +/+ | +/− | −/− | |
| ASAT (U/liter) | 59 ± 4.2 | 57 ± 3.4 | 43 ± 3.1c | 49 ± 3.7 (5) | 43 ± 0.7 | 45 ± 2.9 (5) |
| ALAT (U/liter) | 51 ± 4.2 | 42 ± 2.8 | 34 ± 3.0d | 33 ± 3.7 (5) | 27 ± 1.9 | 30 ± 2.1 (5) |
| Alk. Phos. (U/liter) | 89 ± 4.2 | 89 ± 3.4 | 110 ± 7.1c | 130 ± 7.6 (5) | 121 ± 13.2 | 146 ± 9.6 (5) |
| Bilirubin (μmol/liter) | 1.9 ± 0.3 | 1.6 ± 0.1 | 2.1 ± 0.1 | 2.1 ± 0.2 (5) | 2.1 ± 0.2 | 2.1 ± 0.3 (5) |
| Glucose (mmol/liter) | 8.7 ± 0.5 | 9.1 ± 0.4 | 8.9 ± 0.5 | 8.8 ± 0.3 (4) | 8.6 ± 0.8 | 7.7 ± 0.5 (5) |
| Urea (mmol/liter) | 11.1 ± 0.8 | 9.4 ± 1.0 | 9.7 ± 0.3 | 8.7 ± 0.6 (5) | 8.5 ± 1.1 | 9.4 ± 0.8 (5) |
| Triglycerides (mmol/liter) | 1.25 ± 0.14 | 1.11 ± 0.19 | 1.12 ± 0.10 | 0.72 ± 0.08 (5) | 0.73 ± 0.09 | 0.75 ± 0.15 (5) |
| Cholesterol (mmol/liter) | 2.60 ± 0.26 | 2.39 ± 0.12 | 2.32 ± 0.10 | 1.91 ± 0.16 (5) | 2.23 ± 0.26 | 1.91 ± 0.14 (5) |
| Total protein (g/liter) | 50.6 ± 1.4 | 52.4 ± 1.6 | 53.2 ± 1.4 | 50 ± 1.2 (4) | 51 ± 0.9 | 52 ± 0.9 (5) |
| Albumin (g/liter) | 29.7 ± 0.5 | 28.4 ± 0.6 | 30.1 ± 1.0 | 33.9 ± 0.8 (4) | 33.8 ± 0.3 | 34.4 ± 0.9 (5) |
| Creatinine (μmol/liter) | 12.0 ± 0.5 | 11.5 ± 0.8 | 10.8 ± 0.9 | 11.0 ± 0.7 (5) | 11.8 ± 1.0 | 11.0 ± 0.8 (5) |
| LDH (U/liter) | 263 ± 31 | 239 ± 18 | 200 ± 22 | 264 ± 61 (5) | 195 ± 27 | 206 ± 25 (5) |
| Creatinine kinase (U/liter) | 99 ± 13 | 108 ± 16 | 95 ± 12 | 122 ± 42 (5) | 106 ± 34 | 86 ± 13 (5) |
| Gamma GT (U/liter) | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 (5) | 0 ± 0 | 0 ± 0 (4) |
| Sodium (mmol/liter) | 156 ± 0.4 | 154 ± 0.8 | 155 ± 0.5 | 152 ± 0.4 (5) | 153 ± 0.5 | 153 ± 0.2d (5) |
| Potassium (mmol/liter) | 4.4 ± 0.2 | 4.6 ± 0.3 | 4.4 ± 0.1 | 4.2 ± 0.2 (5) | 4.0 ± 0.2 | 4.3 ± 0.1 (5) |
| Calcium (mmol/liter) | 2.37 ± 0.05 | 2.37 ± 0.04 | 2.43 ± 0.04 | 2.38 ± 0.02 (4) | 2.41 ± 0.02 | 2.37 ± 0.02 (5) |
| Choride (mmol/liter) | 112 ± 1.4 | 114 ± 0.8 | 111 ± 0.8 | 113 ± 0.6 (5) | 112 ± 1.0 | 113 ± 0.6 (5) |
TABLE 3.
Overview of absolute organ weights and organ weight/body weight ratios in control and LGR7 knockout micea
| Organ and units | Data for males of genotype: | Data for females of genotype: | ||||
|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | +/+ | +/− | −/− | |
| Body wt (g) | 30.3 ± 1.8 | 29.6 ± 1.2 | 30.2 ± 1.6 | 23.8 ± 1.2 | 25.5 ± 0.9 | 24.4 ± 1.3 |
| Heart | ||||||
| mg | 153 ± 10 | 156 ± 11 | 151 ± 7 | 126 ± 6 | 141 ± 8 | 122 ± 6 |
| % | 0.50 ± 0.01 | 0.52 ± 0.03 | 0.50 ± 0.01 | 0.53 ± 0.02 | 0.55 ± 0.03 | 0.50 ± 0.02 |
| Spleen | ||||||
| mg | 100 ± 6 | 109 ± 4 | 99 ± 11 | 106 ± 9 | 119 ± 4 | 94 ± 8 |
| % | 0.33 ± 0.02 | 0.37 ± 0.02 | 0.33 ± 0.02 | 0.45 ± 0.04 | 0.47 ± 0.01 | 0.38 ± 0.02 |
| Thymus | ||||||
| mg | 58.7 ± 3.3 | 66.5 ± 6.5 | 68.8 ± 4.4 | 69.1 ± 7.6 | 55.8 ± 10.3 | 71.2 ± 6 |
| % | 0.20 ± 0.01 | 0.23 ± 0.02 | 0.23 ± 0.02 | 0.29 ± 0.03 | 0.22 ± 0.03 | 0.29 ± 0.02 |
| Liver | ||||||
| g | 1.73 ± 0.09 | 1.55 ± 0.07 | 1.68 ± 0.09 | 1.21 ± 0.07 | 1.43 ± 0.05 | 1.23 ± 0.07 |
| % | 5.75 ± 0.27 | 5.26 ± 0.19 | 5.55 ± 0.06 | 5.11 ± 0.21 | 5.62 ± 0.12 | 5.05 ± 0.12 |
| Kidneys | ||||||
| mg | 409 ± 32 | 381 ± 28 | 439 ± 23 | 283 ± 23 | 311 ± 13b | 314 ± 19 |
| % | 1.34 ± 0.04 | 1.28 ± 0.05 | 1.46 ± 0.04 | 1.18 ± 0.05 | 1.22 ± 0.02 | 1.29 ± 0.06 |
| Adrenal glands | ||||||
| mg | 9.9 ± 1.2 | 8.1 ± 0.8 | 8.2 ± 0.9 | 10.5 ± 1.3 | 11.4 ± 0.8 | 11.3 ± 0.6 |
| % | 0.034 ± 0.004 | 0.028 ± 0.003 | 0.028 ± 0.004 | 0.044 ± 0.005 | 0.045 ± 0.004 | 0.047 ± 0.004 |
| Brain | ||||||
| mg | 479 ± 9 | 475 ± 12 | 505 ± 9 | 497 ± 8 | 499 ± 8 | 496 ± 5 |
| % | 1.60 ± 0.09 | 1.61 ± 0.03 | 1.70 ± 0.09 | 2.11 ± 0.10 | 1.96 ± 0.06 | 2.06 ± 0.11 |
| Testes | ||||||
| mg | 205 ± 17 | 206 ± 21 | 117 ± 15c | |||
| % | 0.68 ± 0.04 | 0.69 ± 0.05 | 0.39 ± 0.05d | |||
| Epididymides | ||||||
| mg | 84.0 ± 6.2 | 79.0 ± 7.5 | 62.3 ± 4.3b | |||
| % | 0.28 ± 0.02 | 0.27 ± 0.02 | 0.21 ± 0.02 | |||
| Seminal vesicles | ||||||
| mg | 326 ± 19 (5) | 335 ± 32 | 280 ± 18 | |||
| % | 1.08 ± 0.08 | 1.12 ± 0.08 | 0.93 ± 0.04 | |||
| Ovaries | ||||||
| mg | 13.5 ± 0.9 | 13.1 ± 0.7 | 14.1 ± 0.9 | |||
| % | 0.057 ± 0.003 | 0.052 ± 0.003 | 0.059 ± 0.005 | |||
| Uterus | ||||||
| mg | 101 ± 12 | 104 ± 20 | 94 ± 14 | |||
| % | 0.43 ± 0.05 | 0.41 ± 0.08 | 0.38 ± 0.04 |
In the males, no relevant changes in organ weight changes were found for the homozygous mutant males compared to the organ weight changes of their heterozygous and wild-type littermates (except for the testes and the epididymides [Table 3]). Testis weight in 12-week-old homozygous LGR7 males from the F2 generation was significantly reduced compared to that of wild-type and heterozygous littermates. Epididymis weight in the homozygous LGR7 males was slightly reduced compared with that in the wild-type and heterozygous littermates.
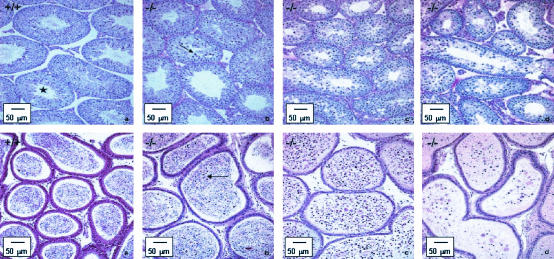
Histopathological analysis of the testes of mutant animals revealed defects in spermatogenesis, designated mild (3 of 6), moderate (2 of 6) or marked (1 of 6) (Fig. 3). Both moderate and marked phenotypes resulted in azoospermia.
FIG. 3.
Bouin-fixed and periodic acid-Schiff-stained sections of the testes (top) and epididymides (bottom) of LGR7 males. (a) Wild-type male. Shown is normal histological appearance of the testis with seminiferous tubules in all stages of spermatogenesis including stage 12 (asterisk) with normal meiotic cells; only some apoptotic spermatogenic cells were present at the basal layer (not indicated). The sperm count in the acini of the epididymis was normal. Panels b, c, and d represent photomicrographs of tissue from LGR7−/− males. (b) Shown are mild disrupted spermatogenesis in the testis with apoptosis of meiotic cells (arrow) and morphology of the epididymis comparable to that of the wild type but with several apoptotic cells in the lumina (arrow). Shown are moderate (c) and marked (d) disrupted spermatogenesis in the testes. No sperm, but only cell debris and necrotic material within the lumina in the epididymides (azoospermia), were present.
LGR7−/− males from the F1 generation (10 weeks and 6 months old) showed the same effects in terms of weight and histological appearance (mild disrupted spermatogenesis). Interestingly, however, in 7-month-old males from the F2 generation and in mice from the F3 generation, spermatogenesis defects were not observed.
Increased apoptosis in testis.
Regular apoptosis of spermatogenic cells in the testes is required to maintain proper testicular homeostasis. While in wild-type males TUNEL-positive nuclei in the spermatogenic cells were found with a normal frequency mainly in the basal layer of the testes, the testes of LGR7−/− males with mild disrupted spermatogenesis were characterized by apoptosis of stage 12 meiotic cells (17). However, spermatogenesis was not completely blocked in these animals, as spermatids and mature spermatozoa could still be observed in the seminiferous tubules. Apoptosis of the spermatogenic and meiotic cells in the testes and epididymides was confirmed by using the TUNEL method (Fig. 4).
FIG. 4.
Formalin-fixed sections of the testes (top) and epididymides (bottom) of LGR7 males (tissues stained with the TUNEL method). (a) Wild-type male. Shown is normal regular apoptosis of spermatogenic cells in the seminiferous tubules of the testis (arrows); some apoptotic spermatogenic cells in the acini of the epididymis (arrow) are also shown. (b) LGR7−/− male. Shown are apoptotic stage 12 meiotic cells in the testes (arrows) and increased numbers of apoptotic cells in the lumina of the epididymis (arrows).
Fertility studies.
Breeding of the first three LGR7−/− female mice from the F1 generation with wild-type males resulted in only one pregnant female that was incapable of delivering pups.
Breeding of LGR7−/− female mice from the F2 and F3 generations with wild-type males revealed that the females were fertile and produced normal-sized litters (Table 1). The pregnancy rate (number of pregnant females/total number of females) was 100%. In addition, gestation length (about 19.5 days, data not shown) was similar for all groups.
In the litters obtained from the knockout females, 25 of 162 pups were found dead (distributed among 9 of 21 litters). Macroscopically, no malformations were detected. Moreover, one female appeared to be incapable of delivering all the pups. More than 24 h after the start of delivery, three of eight pups were still in utero. Finally, all live offspring died within 24 to 48 h after delivery. Upon analysis, it was noted that none of the pups appeared to have milk in their stomachs, obviously caused by the inability of the mother to deliver milk.
Breeding of LGR7−/− male mice with wild-type females indicated reduced fertility in mice from the F2 generation (Table 4). The pregnancy rate was 33% in matings that used 11-week-old knockout males from the F2 generation versus about 65% in matings with 4.5- to 7-month-old LGR7−/− males from the F2 or F3 generation.
TABLE 4.
Fertility data of wild-type and homozygous male and female LGR7 mice
| Mating cross and ratioa | Generation/age | Pregnancy rate | Litter size | No. of pups found dead/live |
|---|---|---|---|---|
| WT M × WT F (1:1) | F3/11 wk | 6/6 | 9 ± 1 | 0/54 |
| WT M × WT F (1:1) | F3/4.5 mo | 6/6 | 7 ± 2 | 0/41 |
| KO M × WT F (1:2) | F2/14 wk | 8/24 | 8 ± 2 | 0/63 |
| KO M × WT F (1:1) | F2/7 mo | 7/11 | 9 ± 3 | 0/62 |
| KO M × WT F (1:1) | F3/4.5 mo | 4/6 | 10 ± 1 | 0/38 |
| KO M × KO F (1:1) | F3/11 wk | 5/8 | 9 ± 1 | 11/41b |
| WT M × KO F (1:1) | F2/14 wk | 8/8 | 10 ± 2 | 6/76b |
| WT M × KO F (1:1) | F3/11 wk | 5/5 | 9 ± 1 | 7/45b |
Histology of the nipple and mammary gland.
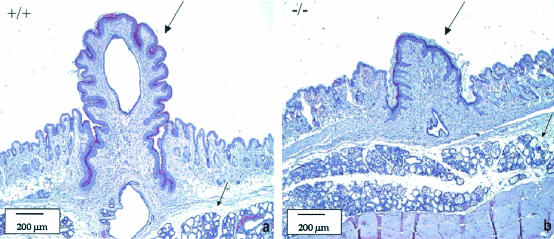
To establish the cause of the inability of the LGR7−/− females to feed their young, we investigated whether there were any differences between nipple and/or mammary gland development in pregnant wild-type versus LGR7−/− females. Indeed, in pregnant homozygous null mutants, nipple size was significantly reduced compared to that of their wild-type littermates. The mammary glands from homozygous females after delivery were lactating and appeared histologically similar to those from the wild-type mice (Fig. 5).
FIG. 5.
Histology of nipples and mammary glands from LGR7 mothers by using HE staining. Panel a represents results from the wild-type female; panel b represents those from the LGR7−/− female. The lactating nipple (large arrow) of the wild-type mother (a) was much longer than the nipple (large arrow) of the LGR7−/− mother (b) that was not able to support suckling. Lactating mammary glands (lobular hyperplasia with prominent secretion, small arrows) in both wild-type and LGR7−/− mice are also shown.
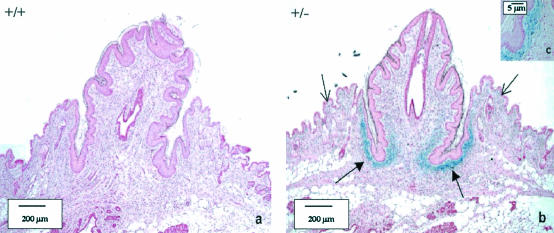
The nipples from the LGR7−/− mothers were much smaller than those from the wild-type mothers. To establish whether LGR7 is expressed during nipple development, a β-Gal staining was performed on the nipples of pregnant wild-type and heterozygous LGR7 females. As shown in Fig. 6, strong positive staining was found in the connective tissue at the base of the nipple in heterozygous LGR7 mice and not in wild types. Staining was also detected in some connective tissue cells just below the epithelium.
FIG. 6.
LGR-7 expression in the nipples of pregnant LGR-7 females on day 18.5 of gestation. (a) Wild-type female; (b) LGR7+/− female. In the pregnant LGR7+/− female (panel b), strong positive LacZ staining at the base of the nipple (closed arrows) is shown; some staining was also found in single cells just below the epithelium (open arrows). (c) Detail of panel b.
DISCUSSION
The LGR7 receptor was identified based on its homology to the other members of the glycoprotein hormone receptor subfamily and shares the highest homology (60% sequence identity) to the recently identified LGR8 receptor (16). However, while LGR8 is mainly expressed during embryogenesis and has been shown to be involved in testicular descent (16), LGR7 expression was shown to be restricted to a few organs and tissues. Using the LGR7 LacZ knockin construct, we found that our data confirmed the highly specific expression of LGR7 in the uterus, brain, oviduct, vagina, and heart. Moreover, these data are in line with relaxin binding studies that indicated the presence of a relaxin receptor in these organs and tissues (9, 13). Interestingly, β-Gal activity was also observed in testis—more specifically, in spermatids—representing the final stages of spermatogenesis.
The data we have obtained regarding the phenotype of female LGR7−/− mice show a remarkable similarity with the phenotype described for the relaxin-deficient female mice. Both knockouts showed no gross abnormalities, female fertility was not affected, litter sizes were normal and, most strikingly, nipple enlargement during pregnancy was impaired. For rats, relaxin has been shown to directly stimulate the growth of nipples during pregnancy (10). In relaxin-deficient mice, loosening of the underlying connective tissue was absent in the lactating nipples when compared to those of their wild-type littermates (20). This correlates well with the expression of LGR7 in the connective tissue of the nipples of pregnant LGR+/− females. As a consequence of the impairment of nipple development, all offspring died within 24 to 48 h after delivery due to the inability of the mothers to feed their young. Our data, however, do not confirm a role for LGR7 in mammary gland development during late pregnancy, as no abnormalities in glandular tissue or fat composition were observed. On the basis of our data, we concluded that LGR7 is a crucial factor for nipple development during pregnancy, most likely with relaxin as the sole ligand involved. Although relaxin was also found to activate LGR8 in vitro (3), our knockout data and data for the LGR8, Insl3, and relaxin knockout models demonstrate that relaxin and Insl3 are the cognate ligands for LGR7 and LGR8, respectively.
The timed expression of LGR7 during pregnancy in the nipple and pituitary gland suggests that pregnancy hormones such as estrogen or progesterone might be involved in regulating LGR7 gene expression in these tissues. Indeed, it was found that the LGR7 promoter contains a progesterone (but not an estrogen)-responsive element (data not shown). Moreover, it has been shown that the progesterone receptor is expressed in the pituitary gland upon induction with estrogen in sexually mature females but not in prepubescent 3-week-old females (7). It is therefore tempting to speculate that LGR7 expression during pregnancy in the pituitary gland, and most likely also in the nipple, is mediated by the progesterone receptor.
The observation that some pregnant females were incapable of delivering their pups and that several pups were found dead may be indicative of a role for LGR7 in parturition. Parturition involves the orderly and carefully timed events in both the uterus (onset of rhythmic contractions of the myometrium) and the cervix (tissue remodeling resulting in increased tissue softening) which allow delivery of the fetus. The presence of LGR7 in both the myometrium and cervix was confirmed by in situ hybridization (data not shown), by β-Gal staining, and by immunohistochemistry (3). However, data obtained regarding the average delivery time indicated no abnormalities (∼19.5 days). Moreover, histopathological abnormalities of the myometrium were not observed, suggesting that a delay or defect in uterine contractions is not the cause of defects in parturition. Instead, LGR7 deficiency may result in impaired cervical ripening. This phenotype has also been described in mice deficient for 5α-reductase type 1 (12). In these mice, the observed parturition defect could be corrected by the administration of relaxin. While alterations in relaxin expression could not account for the defective cervical ripening, it was hypothesized that the phenotype observed was caused by impaired expression of the relaxin receptor. Additional supporting evidence comes from studies using the relaxin knockout; these studies showed that collagen levels increase in the cervix during late pregnancy, a factor which may contribute to the partial parturition defect observed in these mice (19). Our data support this hypothesis, but additional histopathological studies of the cervix during the various stages of pregnancy to confirm these findings will be required.
Although initial reports concluded normal male fertility in relaxin knockouts (20), more recent data indicate a key role for relaxin in prostate growth, male reproductive tract development, and maintenance of male fertility. Moreover, it was concluded that relaxin may mediate its effect by serving as an antiapoptotic factor (18). Although our findings do not confirm a role of LGR7 in prostate development, impaired fertility was indeed observed in male LGR7−/− mice—a finding that correlated with increased apoptosis in testis. However, the reduced male fertility phenotype observed was absent in later generations and raises the question of whether genetic background and/or redundancy may be involved. Redundancy due to expression of a single homologous gene such as LGR8 in the same cell types of the testis, however, seems unlikely in view of the high interanimal variation observed. A more complex compensation mechanism involving larger numbers of genes, as can be anticipated in a variable genetic background, therefore seems more likely. The latter is supported by the observation that fertility defects were no longer observed in mice from the F3 generation that had been backcrossed with C57BL/6, suggesting that subtle differences in the genetic background may be responsible for the variation in the male fertility phenotype. Backcrossing to either the 129Sv or C57BL/6 background may result in the identification of other genetic factors involved in the function of LGR7 during the final stages of spermatogenesis.
Acknowledgments
The LGR7 knockout mice were generated at and contracted from Lexicon Genetics. We thank P. W. J. de Leeuw and J. H. M. I. Brands for performing the blood analyses.
REFERENCES
- 1.Bancroft, J. D., and A. Stevens. 1996. Theory and practice of histological techniques, 4th ed. Churchill Livingstone, Ltd., Edinburgh, Scotland.
- 2.Bani, D. 1997. Relaxin: a pleiotropic hormone. Gen. Pharmacol. 28**:**13-22. [DOI] [PubMed] [Google Scholar]
- 3.Du, X. J., C. S. Samuel, X. M. Gao, L. Zhao, L. J. Parry, and G. W. Tregear. 2003. Increased myocardial collagen and ventricular diastolic dysfunction in relaxin-deficient mice: a gender-specific phenotype. Cardiovasc. Res. 57**:**395-404. [DOI] [PubMed] [Google Scholar]
- 4.Hsu, S. Y., S. G. Liang, and A. J. Hsueh. 1998. Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol. Endocrinol. 12**:**1830-1845. [DOI] [PubMed] [Google Scholar]
- 5.Hsu, S. Y., M. Kudo, T. Chen, K. Nakabayashi, A. Bhalla, P. J. van der Spek, M. van Duin, and A. J. Hsueh. 2000. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol. Endocrinol. 14**:**1257-1271. [DOI] [PubMed] [Google Scholar]
- 6.Hsu, S. Y., K. Nakabayashi, S. Nishi, J. Kumagai, M. Kudo, O. D. Sherwood, and A. J. Hsueh. 2002. Activation of orphan receptors by the hormone relaxin. Science 295**:**671-674. [DOI] [PubMed] [Google Scholar]
- 7.Ismail, P. M., J. Li, F. J. DeMayo, B. W. O'Malley, and J. P. Lydon. 2002. A novel LacZ reporter mouse reveals complex regulation of the progesterone receptor promoter during mammary gland development. Mol. Endocrinol. 16**:**2475-2489. [DOI] [PubMed] [Google Scholar]
- 8.Kobe, B., and J. Deisenhofer. 1995. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374**:**183-186. [DOI] [PubMed] [Google Scholar]
- 9.Kohsaka, T., G. Min, G. Lukas, S. Trupin, E. T. Campbell, and O. D. Sherwood. 1998. Identification of specific relaxin-binding cells in the human female. Biol. Reprod. 59**:**991-999. [DOI] [PubMed] [Google Scholar]
- 10.Kuenzi, M. J., B. A. Connolly, and O. D. Sherwood. 1995. Relaxin acts directly on rat mammary nipples to stimulate their growth. Endocrinology 136**:**2943-2947. [DOI] [PubMed] [Google Scholar]
- 11.Kumagai, J., S. Y. Hsu, H. Matsumi, J. S. Roh, P. Fu, J. D. Wade, R. A. Bathgate, and A. J. Hsueh. 2002. INSL3/Leydig insulin-like peptide activates the LGR8 receptor important in testis descent. J. Biol. Chem. 277**:**31283-31286. [DOI] [PubMed] [Google Scholar]
- 12.Mahendroo, M. S., A. Porter, D. W. Russell, and R. A. Word. 1999. The parturition defect in steroid 5α-reductase type 1 knockout mice is due to impaired cervical ripening. Mol. Endocrinol. 13**:**981-992. [DOI] [PubMed] [Google Scholar]
- 13.Min, G., and O. D. Sherwood. 1996. Identification of specific relaxin-binding cells in the cervix, mammary glands, nipples, small intestine, and skin of pregnant pigs. Biol. Reprod. 55**:**1243-1252. [DOI] [PubMed] [Google Scholar]
- 14.Nef, S., and L. F. Parada. 1999. Cryptorchidism in mice mutant for Insl3. Nat. Genet. 22**:**295-299. [DOI] [PubMed] [Google Scholar]
- 15.Novak, J., L. A. Danielson, L. J. Kerchner, O. D. Sherwood, R. J. Ramirez, P. A. Moalli, and K. P. Conrad. 2001. Relaxin is essential for renal vasodilation during pregnancy in conscious rats. J. Clin. Investig. 107**:**1469-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Overbeek, P. A., I. P. Gorlov, R. W. Sutherland, J. B. Houston, W. R. Harrison, H. L. Boettger-Tong, C. E. Bishop, and A. Agoulnik. 2001. A transgenic insertion causing cryptorchidism in mice. Genesis 1**:**26-35. [DOI] [PubMed] [Google Scholar]
- 17.Russell, L. D., A. P. S. Hikim, R. A. Ettlin, and E. D. Clegg. 1990. Histological and histopathological evaluation of the testis, 1st ed. Cache River Press, St. Louis, Mo.
- 18.Samuel, C. S., H. Tian, L. Zhao, and E. P. Amento. 2003. Relaxin is a key mediator of prostate growth and male reproductive tract development. Lab. Investig. 83**:**1055-1067. [DOI] [PubMed] [Google Scholar]
- 19.Zhao, L., C. S. Samuel, G. W. Tregear, F. Beck, and E. M. Wintour. 2000. Collagen studies in late pregnant relaxin null mice. Biol. Reprod. 63**:**697-703. [DOI] [PubMed] [Google Scholar]
- 20.Zhao, L., P. J. Roche, J. M. Gunnersen, V. E. Hammond, G. W. Tregear, E. M. Wintour, and F. Beck. 1999. Mice without a functional relaxin gene are unable to deliver milk to their pups. Endocrinology 140**:**445-453. [DOI] [PubMed] [Google Scholar]