TARGETING STAT3 ABROGATES EGFR INHIBITOR RESISTANCE IN CANCER (original) (raw)
. Author manuscript; available in PMC: 2013 Sep 15.
Abstract
PURPOSE
EGFR is upregulated in most epithelial cancers where signaling through EGFR contributes to cancer cell proliferation and survival. The limited clinical efficacy of EGFR inhibitors suggests that identification of resistance mechanisms may identify new pathways for therapeutic targeting. Signal Transducer and Activator of Transcription-3 (STAT3) is upregulated in many cancers and activated via both EGFR-dependent and EGFR-independent pathways. In the present study, we tested the consequences of STAT3 inhibition in EGFR inhibitor resistant head and neck squamous cell carcinoma (HNSCC) and bladder cancer models to determine if STAT3 blockade can enhance responses to EGFR targeting.
EXPERIMENTAL DESIGN
pSTAT3 expression was assessed in human HNSCC tumors that recurred following cetuximab treatment. Cetuximab sensitive and cetuximab resistant cell lines were treated with a STAT3 decoy to determine EC50 concentrations and the effects on STAT3 target gene expression by western blotting. In vivo assays included evaluation of anti-tumor efficacy of STAT3 decoy in cetuximab sensitive and cetuximab resistant models followed by immunoblotting for STAT3 target protein expression.
RESULTS
Targeting STAT3 with a STAT3 decoy reduced cellular viability and the expression of STAT3 target genes in EGFR inhibitor resistance models. The addition of a STAT3 inhibitor to EGFR blocking strategies significantly enhanced anti-tumor effects in vivo. Biopsies from HNSCC tumors that recurred following cetuximab treatment demonstrated increased STAT3 activation compared with pretreatment biopsies.
CONCLUSIONS
These results suggest that STAT3 activation contributes to EGFR inhibitor resistance both in HNSCC and bladder cancer where concomitant targeting of STAT3 may represent an effective treatment strategy.
Keywords: cancer, epidermal growth factor receptor, Signal Transducers and Activators of Transcription, STAT3 decoy, cetuximab
Introduction
Epidermal growth factor receptor (EGFR) is hyperactivated in multiple cancers and has emerged as a validated therapeutic target in several solid tumors (1). EGFR monoclonal antibodies such as cetuximab and panitumumab (Erbitux, Vectibix), are FDA (Food and Drug Administration)-approved for the treatment of advanced head and neck squamous cell carcinoma (HNSCC) and/or colorectal cancer in combination with either radiotherapy or chemotherapy. The EGFR-selective tyrosine kinase inhibitor (TKI) erlotinib (Tarceva) is approved for the treatment of non-small cell lung cancer (NSCLC) and pancreatic cancer, although profound efficacy is generally limited to NSCLC tumors harboring EGFR activating mutations (2–8). The paucity of EGFR inhibitor resistance models and the limited availability of tumor biopsies in the setting of EGFR inhibitor resistance have contributed to an incomplete understanding of the mechanisms that contribute to intrinsic or acquired resistance to EGFR targeting in some cancers. Elucidation of EGFR inhibitor resistance mechanisms may identify pathways that can be targeted to enhance treatment responses.
Overactivation of multiple signaling pathways contribute to EGFR inhibitor resistance as cancers of different origins employ different mechanisms to escape EGFR thereapy. In erlotinib resistant lung cancer cells, increased expression of Interleukin-6 (IL-6) has been shown to be responsible for the EGFR-independent Signal Transducer and Activator of Transcription-3 (STAT3) phosphorylation (9). Overactivation of vascular endothelial growth factor (VEGF) has been shown to play a role in resistance to anti-EGFR therapy and combined blockade of VEGF and EGFR pathways with DC101, an anti-VEGF receptor monoclonal antibody, and cetuximab, respectively have shown greater inhibition of tumor growth than single agent in both gastric and colon cancer (10). Overexpression of HER-2, the second member of the erbB family, contributes to EGFR inhibitor resistance and targeting both EGFR and HER-2 using a dual tyrosine kinase inhibitor such as lapatinib showed activity in breast cancer cell lines overexpressing HER-2 (11).
STAT3, a member of the STAT family of transcription factors, is activated in several cancers (12). STAT3 tyrosine phosphorylation can be induced by stimulation of upstream receptor and/or nonreceptor kinases including EGFR(13), IL-6/gp130 and Janus kinases (JAKs) (14), and Src family kinases (15). STAT3 activation has been identified in the setting of resistance to EGFR tyrosine kinases inhibitors in preclinical models of glioma and HNSCC (12, 16) and resistance to neoadjuvant EGFR TKI treatment of NSCLC patients was associated with elevated STAT3 activity in patient tumors (17). These cumulative results suggest that STAT3 may be activated in the setting of resistance to EGFR inhibitor therapy where targeting STAT3 may overcome either de novo or acquired resistance.
In the absence of a small molecule with STAT3-selective activity, we developed a transcription factor decoy oligonucleotide, which has been shown to block STAT3-mediated DNA binding and inhibit tumor cell proliferation in vitro and xenograft growth in vivo in a wide variety of preclinical cancer models including xenografts and transgenic models (18–25). Combined treatment of HNSCC cell lines with the STAT3 decoy and EGFR TKI was associated with enhanced anti-tumor effects (26). In the present study, we tested the anti-tumor effects of STAT3 inhibition using the STAT3 decoy in preclinical cancer models of intrinsic or acquired resistance to EGFR TKI or cetuximab in tumor models not characterized by activating EGFR mutations. Furthermore, assessment of pSTAT3 in human HNSCC tumors that recurred following cetuximab treatment demonstrated increased pSTAT3 staining compared with levels in pretreatment biopsies. These findings suggest that targeting STAT3 may enhance the anti-tumor effects of EGFR inhibitors.
Materials and Methods
Cell line validation
The HNSCC cell lines Cal33, 686LN, HN5, OSC19 and the bladder cancer cell line T24 were validated using the AmpFlSTR® Profiler Plus™ kit from PE Biosystems (Foster City, CA) according to the manufacturer’s instructions.
Cell culture
Head and neck squamous cell carcinoma cell lines Cal33 (a kind gift from Jean Louis Fischel, Centre Antoine Lacassagne, Nice, France), HN5 and OSC19 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Mediatech, Inc., Herndon, VA) containing 10% heat-inactivated fetal bovine serum (FBS) at 37°C with 5% CO2. 686 LN (a kind gift from Georgia Chen, University of Emory, Atlanta, GA) was maintained in DMEM/F12 media (1:1) from GIBCO (Carlsbad, CA) containing 10% heat-inactivated fetal bovine serum ISC BioExpress (Kaysville, UT). The T24 bladder cancer cell line was obtained from American type culture collection (ATCC). The cetuximab resistant cell lines, T24 PR1, T24 PR2, and T24 PR3 were generated in vivo by exposing tumor-bearing athymic nude mice generated from the parental cell line T24, to increasing concentrations of cetuximab over a 3 month period, as described previously (27). T24 cells were cultured in DMEM (Mediatech, Inc., Herndon, VA) containing 10% heat-inactivated fetal bovine serum. The cetuximab resistant cell lines, T24 PR1, T24 PR2 and T24 PR3 were maintained in presence of cetuximab at a concentration of 100 nM in DMEM containing 10% heat-inactivated fetal bovine serum.
Immunohistochemical analysis and construction of tissue microarrays
Tumor biopsies were obtained from 7 HNSCC patients prior to cetuximab treatment and 15 patients following cetuximab treatment under a protocol approved by the Institutional Review Board at the University of Pittsburgh (IRB#991206). Informed consent was obtained from all subjects. The average composite score (intensity of staining x the percentage of tumor cells that stained positively) of pre- and post-cetuximab treated tumors are represented. Using a manual tissue array instrument (MTA-1; Beecher Instruments), a paraffin core of 1.0-mm was taken from a representative region of the donor block and arrayed into a blank recipient paraffin block in duplicate. The newly constructed array block was then warmed to 37°C for 10 minutes to allow annealing of donor cores to the paraffin wax of the recipient block while minimizing core loss. Donor cores ranged from 2 to 4 mm in length. Immunohistochemistry was carried out on formalin-fixed paraffin-embedded tissue microarray (TMA) sections by using antibodies against pSTAT3 (1:75 dilution, 1:75 overnight 4°C incubation, Santa Cruz Biotechnology). Tissue microarray sections were subjected to antigen retrieval for 15 minutes in 0.01 mol/L citrate buffer. TMAs were blocked and stained with primary antibodies. Following three 5-minute washes, TMAs were incubated with biotinylated anti-rabbit secondary antibody followed by treatment with avidin biotin complex. Signal was developed with 3,30-diaminobenzidine (DAB) substrate, modestly counterstained with hematoxylin, and slides were analyzed microscopically. Immunohistochemical staining was assessed semiquantitatively for each core. The percentage of immunoreactive cells was recorded and rounded to the nearest 10 percentile. Cytoplasmic staining was graded for intensity (0-negative, 1-weak, 2-moderate and 3-strong) A composite score was obtained by multiplying the intensity and the percentage staining score. The scores across replicate cores on slides were averaged. The average composite score for both antibodies were graphed by GraphPad Prism Software.
Reagents
Cetuximab (Erbitux) was purchased from the research pharmacy at the University of Pittsburgh Cancer Institute. Erlotinib (OSI-774; Tarceva) was purchased from NorthWest Pharmacy, Langley, BC, Canada.
STAT3 decoy design and synthesis
The STAT3 decoy sequence was 5′-CATTTCCCGTAAATC-3′, 3′-GTAAAGGGCATTTAG-5′ and the mutant control decoy sequence, which differed by one nucleotide at position 9 (G to T), was 5′-CATTTCCCTTAAATC-3′, 3′-GTAAAGGGAATTTAG-5′, as described previously (18). The single-stranded sense and antisense oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) with phosphorothioate modifications of the residues on 3′ and 5′ ends, as previously described (18).
Dose-response experiments
HN5, OSC19, T24 and the TKI and cetuximab resistant cells (686LN, Cal33, T24 PR1, T24 PR2 and T24 PR3) were seeded in 24-well plates in DMEM containing FBS. After 24 h, cells were transfected with increasing concentrations of STAT3 decoy (26). After 4 h of transfection, media was replaced with DMEM containing 10% FBS. After 72 h, MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assays were performed to determine percent cell viability. To determine the dose response of erlotinib in the parental (T24) and cetuximab resistant bladder cancer cell lines (T24 PR1, T24 PR2 and T24 PR3) as well as the TKI sensitive (HN5 and OSC19) and resistant (686LN and Cal33) HNSCC cell lines, the cells were plated in 24-well plates in DMEM containing FBS. After 24 h, cells were treated with varying concentrations of erlotinib in DMEM containing FBS. DMSO served as a vehicle control. After 72 h, MTT assays were performed to determine percent cell viability.
ELISA
Cutiximab resistant (T24 PR1, T24 PR2 and T24 PR3) and sentitive (T24) bladder cancer cells were plated and after twenty-four hours, cells were cultured in serum-free medium, and 24 h later, cells were collected and analyzed in duplicate with a human IL6 ELISA Kit (R&D Systems). This experiment was repeated three times. Results are reported as means ± standard deviation.
Immunoblotting
Cells were plated at a density of 8×105 cells/10 cm plate and after 24 h transfected with the EC50 concentrations of STAT3 decoy for the respective cell lines. After 4 h, the transfection media was replaced with DMEM + 10% FBS. After 24 h, the cells were harvested and protein content was determined using Bradford’s reagent (BIO-RAD, Hercules, CA) (26). Proteins (40 μg/lane) were separated by 10 % sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), probed with rabbit anti-human cyclin D1 polyclonal antibody or mouse anti-human Bcl-XL monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA), and developed using the enhanced chemiluminescence (ECL) detection system (Amersham Life Sciences Inc., Arlington Heights, IL). The membranes were stripped and re-probed with rabbit anti-human _β_-tubulin polyclonal antibody (Abcam Inc, Cambridge, MA), as a loading control. Densitometric analyses were performed using Image J software.
In vivo tumor xenograft studies
(A) Female athymic nude mice nu/nu (4–6 weeks old; 20 g; Harlan Sprague–Dawley) were inoculated with T24 cells (2×106 cells) into the right and left flanks resulting in two tumors per mouse (9 mice/group). Similarly, another group of mice (8 mice/group) were inoculated with the C225 resistant cells T24 PR3 (2×106 cells) in both the flanks. Once the tumors were palpable, mice were treated with STAT3 decoy/STAT3 mutant and cetuximab. Intratumoral injection of the STAT3 decoy/STAT3 mutant (50 μg) was delivered daily. Cetuximab was injected intraperitoneally (IP) at a dose of 1 mg/mouse, three times a week. Mice were sacrificed after 20 days and tumors were harvested for analysis. Animal care was in strict compliance with institutional guidelines established by the University of Pittsburgh, the Guide for the Care and Use of Laboratory Animals (National Academy of Sciences, 1996) and the Association for Assessment and Accreditation of Laboratory Animal Care International. (B) Female athymic nude mice (10 mice) nu/nu (4–6 weeks old; 20 g; Harlan Sprague–Dawley) were inoculated with 686LN cells (1×106 cells) into the right and left flanks resulting in two tumors per mouse. Once the tumors were palpable, mice were treated with STAT3 decoy/STAT3 mutant and cetuximab. Intratumoral injection of the STAT3 decoy/STAT3 mutant (50 μg) was delivered daily. Cetuximab was injected intraperitoneally (IP) at a dose of 0.2 mg/mouse, two times a week and the treatment was continued until day 20. Animal care was in strict compliance with institutional guidelines established by the University of Pittsburgh, the Guide for the Care and Use of Laboratory Animals (National Academy of Sciences, 1996) and the Association for Assessment and Accreditation of Laboratory Animal Care International.
Statistical Analyses
The relationship between pSTAT3 expression and EC50 for erlotinib in the four HNSCC cell lines were analyzed using nonparametric correlation (Spearman) and two-tailed test for comparing two groups. The in vitro comparison of pSTAT3 expression in the sensitive (T24) and the cetuximab resistant bladder cancer cell lines (T24 PR1, T24 PR2 and T24 PR3) were analyzed using the Mann-Whitney test for comparing two groups. All tests were exact and two-tailed. The in vitro comparison of IL-6 production in the sensitive (T24) and the cetuximab resistant bladder cancer cell lines (T24 PR1, T24 PR2 and T24 PR3) were analyzed using the Mann-Whitney test for comparing two groups. All tests were approximate and two-tailed. In vitro comparisons of Bcl-XL and cyclin D1 expression levels and the differences in STAT3 in HNSCC patient biopsies between treatment groups were conducted using the Mann-Whitney test for comparing two groups. All tests were exact and two-tailed. For the statistical analyses of the in vivo experiment involving the cetuximab resistant bladder cancer cell line T24 PR3 and the isogenic parental cell line T24 treated with STAT3 decoy and cetuximab, all comparisons were conducted on tumor volume measurements on day 20, the last day of the xenograft experiment. The two cell lines (parental and resistant) were compared by testing the difference between the two groups of mice, each bearing one cell line, with a two-tailed Wilcoxon test. The effect of STAT3 decoy was evaluated by testing whether the inter-flank tumor volumes (STAT3 decoy on one flank, mutant control decoy on the other) differed from zero by a two-tailed signed rank test. For the statistical analyses of the in vivo experiment involving the TKI resistant HNSCC cell line treated with STAT3 decoy and cetuximab, tumor volumes were natural log transformed. A mixed effects polynomial regression model was fit to the tumor growth curves for each treatment group. Structured covariance matrices were estimated for with-mouse variation across time. A restricted maximum likelihood test was constructed to determine the appropriate choice for model parameters. A two degree of freedom test was constructed to simultaneously test the equality of the linear and quadratic regression parameters. Residuals were inspected to evaluate the adequacy of fit.
Results
Phospho-STAT3 levels and erlotinib sensitivity in HNSCC cell lines
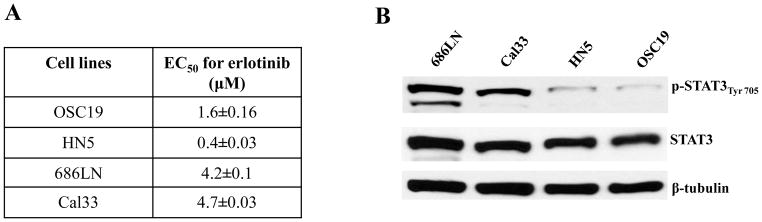
With the goal of comparing STAT3 activation in cells that are sensitive or resistant to EGFR TKI, we treated HNSCC cell lines with varying doses of erlotinib. Two HNSCC cell lines demonstrated relatively high EC50 concentrations (4.6 μM and 4.3 μM for 686LN and Cal33, respectively) and two HNSCC cell lines with lower EC50 concentrations (1.4 μM and 0.364 μM for OSC19 and HN5, respectively) (Figure 1A). We next assessed the levels of total STAT3 and tyrosine705 phosphorylated STAT3 (pSTAT3) in the four cell lines. The cells with greater EC50 for erlotinib (686LN and Cal33) exhibited increased pSTAT3 expression relative to the cells which were more sensitive to erlotinib and demonstrated less pSTAT3 expression (OSC19 and HN5), with Spearman correlation coefficient (r) of 1, p=0.08 (Figure 1B). Although, these results do not suggest a significant correlation but the trend indicated that STAT3 activation may be associated with intrinsic EGFR TKI resistance in HNSCC cell lines.
Figure 1.
pSTAT3 expression levels correlate with erlotinib sensitivity in HNSCC cell lines. (A) Representative HNSCC cell lines (OSC19, HN5, 686LN and Cal33) were treated with varying concentrations of erlotinib. After 72h, MTT assays were performed and EC50 values were calculated. The experiment was performed two times with similar results. (B) Representative HNSCC cell lines (OSC19, HN5, 686LN and Cal33) were seeded in 10 cm plates (1×106 cells) and after 24h, cells were harvested to obtain cell lysates. Forty micrograms of protein/lane were subjected to electrophoresis and immunoblotted for pSTAT3Tyr705 and total STAT3. β-tubulin was used as a loading control. The HNSCC cell lines with higher EC50s for erlotinib (Cal33 and 686LN) demonstrated higher pSTAT3 levels compared with the HNSCC cell lines exhibiting lower EC50s for erlotinib (OSC19 and HN5). Pearson correlation analysis showed pSTAT3 expression and EC50 for erlotinib were positively correlated (r=0.094) with a statistically significant difference (p = 0.026) for the HNSCC cell lines. The experiment was repeated 2 times with similar results.
Increased expression of phosphotyrosine STAT3 (pSTAT3) in cetuximab resistant cell lines
We next sought to determine whether pSTAT3 levels are increased in the setting of acquired resistance to the monoclonal antibody cetuximab. Since HNSCC cell lines are not reproducibly growth inhibited by cetuximab in vitro, while HNSCC cell line xenografts are uniformly sensitive to cetuximab in vivo, it was not possible to determine the association between STAT3 activation and intrinsic resistance to cetuximab. Treatment of cetuximab sensitive (T24) and cetuximab resistant bladder cancer cell lines (T24 PR1, T24 PR2 and T24 PR3) with erlotinib demonstrated comparable EC50 values (see Supplementary Figure S1). We recently developed an in vivo model of acquired resistance to cetuximab using T24 bladder carcinoma cells (27). As shown in Figure 2A–B, expression of pSTAT3 was increased from 3.2–3.9 fold in the cetuximab resistant cell lines (T24 PR1, T24 PR2 and T24 PR3) compared with the isogenic parental cell line, which retains sensitivity to cetuximab. Although levels of total and/or phosorylated EGFR and JAKS 1–3 did not differ between cetuximab sensitive and cetuximab resistant cells, a significant increase in IL-6 secretion was found in the cetuximab resistant bladder cancer cell lines (T24 PR1, T24 PR2 and T24 PR3) compared to the sensitive T24 cells (Figure 2C; p ~0.005).
Figure 2.
(A) pSTAT3 protein expression is increased in cetuximab resistant cancer cells. Cetuximab sensitive parental T24 cells and isogenic clones demonstrating acquired resistance to cetuximab (T24 PR1, T24 PR2 and T24 PR3) were subjected to immunoblotting for pSTAT3Tyr705 and STAT3. β-tubulin was used as a loading control. (B) pSTAT3 expression was increased by 3.2–3.9-fold in the cetuximab resistant T24 PR1, T24 PR2 and T24 PR3 cells compared to the isogenic cetuximab sensitive T24 cells (p=0.03). The experiment was repeated four times with similar results. (C) IL6-secreted protein levels increase in cetuximab resistant bladder cancer cells (T24 PR1, T24 PR2 and T24 PR3, p~0.005) compared to the sensitive counterpart (T24) as confirmed by ELISA.
Growth of EGFR inhibitor-resistant cells is abrogated by STAT3 blockade
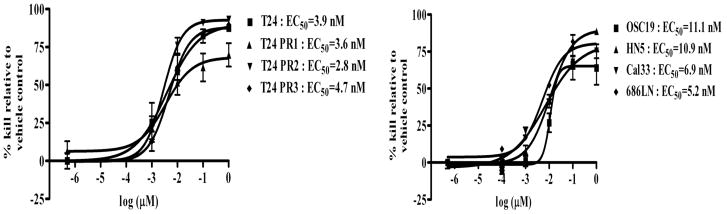
To determine if targeting STAT3 can reduce the growth rate of cells demonstrating intrinsic or acquired resistance to EGFR inhibitors, we first tested the effect of the STAT3 decoy on cell proliferation in the models of intrinsic erlotinib resistance and acquired cetuximab resistance. Cetuximab sensitive parental T24 cells and cetuximab resistant clones T24 PR1, T24 PR2 and T24 PR3, as well as the HNSCC cell lines demonstrating intrinsic erlotinib resistance (or sensitivity) were treated with increasing concentrations of STAT3 decoy or a mutant control decoy that differs only by a single base-pair and fails to interfere with STAT3-mediated DNA binding (18). In 72 h treatment assays, the STAT3 decoy exhibited highly similar EC50 values in both the parental and cetuximab resistant cell lines (T24:EC50=3.9 nM, T24 PR1:EC50=3.6 nM, T24 PR2:EC50=2.8 nM and T24 PR3:EC50=4.7 nM) (Figure 3). In the erlotinib sensitive and resistant HNSCC cell lines, the STAT3 decoy exhibited comparable EC50 values, ranging between 5–11 nM. Although these cells expressed different levels of pSTAT3, they exhibited similar sensitivity to the STAT3 decoy. Similar findings were observed with the preclinical JAK/STAT inhibitor JSI-124 (data not shown). These findings indicate that STAT3 targeting decreases cell proliferation in cancer cells that exhibit either intrinsic or acquired resistance to EGFR inhibitors.
Figure 3.
Targeting STAT3 inhibits proliferation of cancer cell lines with intrinsic or acquired resistance to EGFR inhibitors. Erlotinib sensitive (OSC19 and HN5) and erlotinib resistant (686LN and Cal33 cells) HNSCC cell lines and cetuximab sensitive (T24) and cetuximab resistant cancer cells (T24 PR1, T24 PR2 and T24 PR3) were treated with a range of concentrations of the STAT3 decoy. After 72h, MTT assays were performed and EC50 values were calculated. The experiment was performed three times with similar results. Each data point in the curve represents Mean ± Standard deviation.
STAT3 decoy downmodulates STAT3 target gene expression in EGFR inhibitor resistant cells
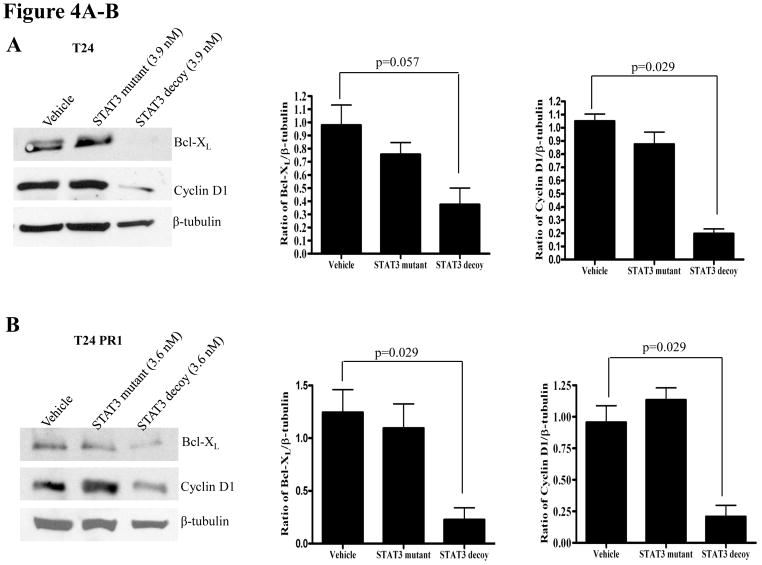
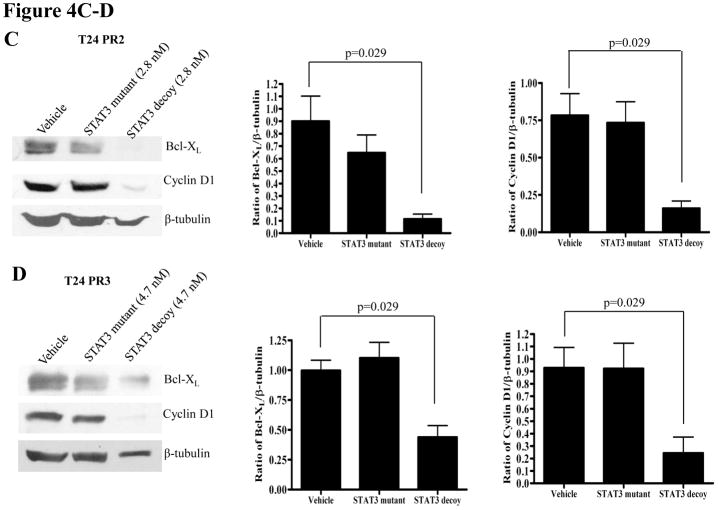
Abrogation of target gene expression is a biochemical indication that the STAT3 decoy is blocking STAT3-mediated signaling. To determine the effect of the STAT3 decoy on the expression of STAT3 target genes in EGFR inhibitor resistant models, cells were treated with EC50 concentrations of STAT3 decoy, followed by immunoblotting for the STAT3 target genes, cyclin D1 and Bcl-XL. _β_-tubulin was assessed as a loading control. In the parental cetuximab sensitive and the cetuximab resistant cells, treatment with STAT3 decoy resulted in a significant decrease in expression of Bcl-XL and cyclin D1 in T24 (p=0.057, 0.029 respectively), T24PR1 (p=0.029, 0.029 respectively), T24PR2 (p=0.029, 0.029 respectively) and T24PR3 (p=0.029, 0.029 respectively) cells, when compared to treatment with vehicle (Figure 4A–D, respectively). Similarly, treatment with STAT3 decoy led to downmodulation of cyclin D1 and Bcl-XL in both the erlotinib sensitive (Supplementary Figure S2 A–B) and resistant HNSCC cell lines (Supplementary Figure S2 C–D). In the erlotinib sensitive and resistant HNSCC cells, treatment with the STAT3 decoy led to a significant decrease in expression of Bcl-XL and cyclin D1 in OSC19 (p=0.029, 0.029 respectively), HN5 (p=0.057, p=0.029 respectively), Cal33 (p=0.057, 0.029 respectively) and 686LN (p=0.029, 0.029 respectively) cells, in comparison with treatment with vehicle (Supplementary Figure S2 A–D, respectively)
Figure 4.
(A–D) Decreased STAT3 target gene expression in cetuximab sensitive (or resistant) cells following treatment with the STAT3 decoy. Cetuximab sensitive (T24) and cetuximab resistant (T24PR1, T24PR2 and T24PR3) bladder cancer cells were treated with STAT3 decoy at EC50 concentrations. As controls, cells were treated with vehicle alone, or the mutant control STAT3 decoy. After 72 h, cells were harvested and proteins (40 μg/lane) were subjected to electrophoresis and immunoblotted for cyclin D1 or Bcl-XL. β-tubulin was used as a loading control. The experiment was repeated 4 times with similar results.
Targeting STAT3 augments the in vivo anti-tumor effects of cetuximab in both cetuximab sensitive and cetuximab resistant models
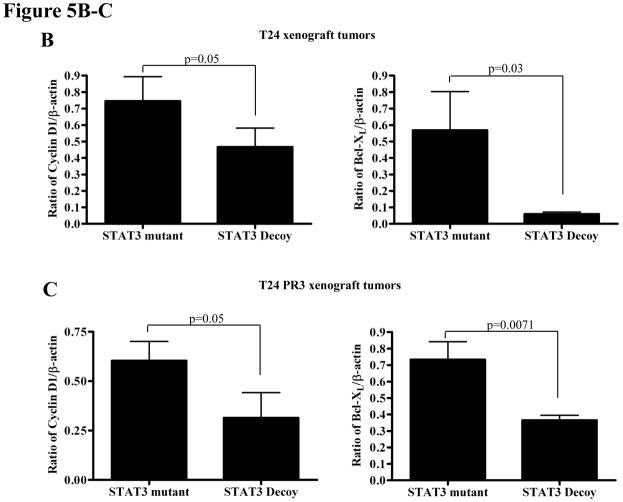
Despite widespread EGFR expression in most epithelial malignancies, cetuximab is only effective in a subset of cancer patients. Since pSTAT3 levels are elevated in HNSCC cells [Fig. 2 was done in vitro, not with xenograft tumors] demonstrating acquired cetuximab resistance, we assessed the anti-tumor effects of the STAT3 decoy in combination with cetuximab in cetuximab sensitive (T24) and cetuximab resistant xenografts (T24 PR3). Mice bearing xenograft tumors (9 mice/group) were treated with daily intratumoral injections (50 μg) of the STAT3 decoy (or the mutant control decoy) in combination with intraperitoneal cetuximab (1 mg), three times a week. As shown in Figure 5A, treatment with the STAT3 decoy plus cetuximab demonstrated a small, but significant growth inhibition of tumors generated from the cetuximab sensitive parental cell line (T24) when compared to treatment with STAT3 mutant control decoy plus cetuximab (p=0.0078). Profoundly, treatment of cetuximab resistant tumors with STAT3 decoy in combination with cetuximab resulted in substantial and significant tumor growth inhibition in comparison to treatment with STAT3 mutant control decoy and cetuximab (p=0.0078). At the end of treatment, tumors were harvested and assessed for expression of proteins encoded by STAT3 target genes. Cetuximab-sensitive tumors treated with STAT3 decoy plus cetuximab demonstrated a decrease in Bcl-XL (p=0.03) and cyclin D1 (p=0.05) expression when compared to treatment with STAT3 mutant control decoy plus cetuximab. Similarly, cetuximab-resistant tumors treated with STAT3 decoy plus cetuximab demonstrated in a decrease in Bcl-XL (p=0.0071) and cyclin D1 (p=0.05) expression when compared to treatment with STAT3 mutant control decoy plus cetuximab (Figure 5B–C). These results indicate that inhibition of STAT3 may be an effective therapeutic strategy in the setting of cetuximab resistance.
Figure 5.
STAT3 decoy inhibits in vivo growth of cetuximab sensitive and resistant xenograft tumors, while downregulating target gene expression. (A) T24 and T24 PR3 cells (2 × 106 cells) were inoculated subcutaneously in 9 athymic nude mice per cell line in the right and left flanks. After 4 days, when the tumors were clearly palpable, the tumor on the right flank was treated with daily injections of the STAT3 decoy (50 μg) and the tumor on the left flank was treated with the STAT3 mutant control decoy (50 μg). Mice were also given cetuximab at a dose of 1 mg/mouse, three times a week intraperitoneally. Tumors were measured three times per week. At the end of day 20, the cetuximab sensitive parental xenografts (T24) treated with STAT3 decoy + cetuximab showed a significant decrease in tumor volume compared to the T24 xenografts treated with STAT3 mutant control decoy + cetuximab (p=0.0078). In addition, the cetuximab resistant T24 PR3 xenografts treated with STAT3 decoy + cetuximab showed a significant decrease in tumor volume compared to the T24 PR3 xenografts treated with STAT3 mutant control decoy + cetuximab (p=0.0078). (B) STAT3 decoy downmodulates STAT3 target gene expression in cetuximab resistant xenograft tumors. Mice from the experiment described in panel A were sacrificed after 20 days and tumors were harvested for analysis of STAT3 target gene expression. In the cetuximab sensitive parental xenografts (T24) treated with STAT3 decoy + cetuximab, decreases in Bcl-XL (p=0.03) and cyclin D1 (p=0.05) were observed when compared to tumors treated with STAT3 mutant control decoy + cetuximab (upper panel). Similarly, decreases in Bcl-XL (p=0.0071) and cyclin D1 (p=0.05) expressions were observed in the cetuximab resistant tumors (T24 PR3) treated with STAT3 decoy + cetuximab compared to the tumors treated with STAT3 mutant control decoy + cetuximab (lower panel).
STAT3 decoy inhibits tumor growth in vivo of TKI resistant HNSCC xenograft
The effect of STAT3 decoy was also assessed in combination with cetuximab in TKI resistant xenograft tumors generated from the HNSCC cell line 686LN. 686LN (1 × 106 cells) were inoculated subcutaneously in 10 athymic nude mice into the right and left flank. After 8 days, when the tumors were clearly palpable, mice were treated with daily intratumoral injections of the STAT3 decoy/STAT3 mutant (50 μg). Cetuximab was administered at a dose of 0.2 mg/mouse, two times a week intraperitoneally. Tumors were measured three times per week. At the end of day 20, TKI resistant HNSCC xenografts treated with STAT3 decoy + cetuximab demonstrated a significant decrease in tumor volume when compared to the xenografts treated with STAT3 mutant control + cetuximab (p<0.0001) (Supplementary Figure S3).
Human HNSCC tumors that recur following cetuximab treatment demonstrate increased p-STAT3
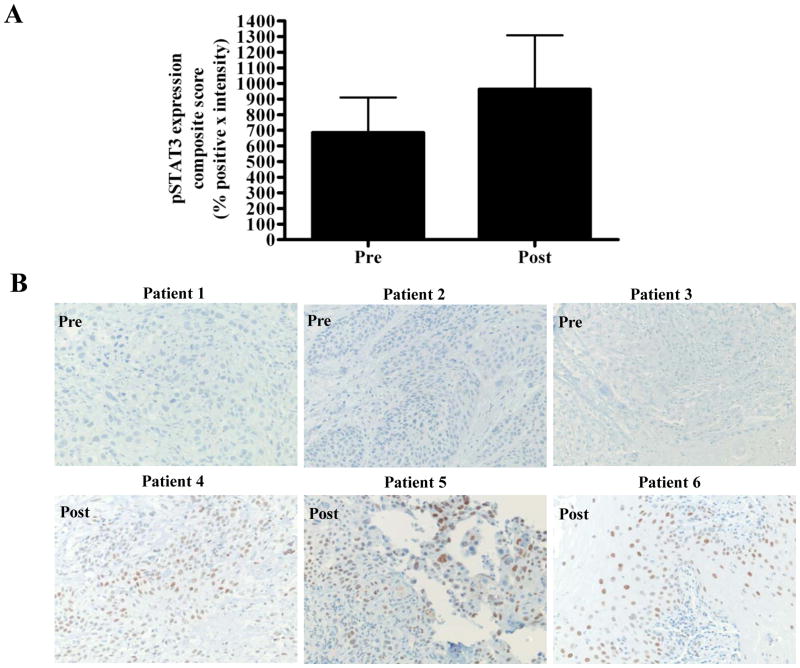
Cetuximab was FDA-approved for the treatment of HNSCC in 2006. To date, no study has systematically characterized HNSCC tumors that persist or develop following treatment with cetuximab. To begin to understand the mechanisms of clinical resistance to cetuximab, we created a tissue microarray containing HNSCC tumor samples before and/or following treatment with cetuximab-containing regimens at the University of Pittsburgh, as described previously (28). Immunohistochemical (IHC) staining of the TMA with anti-pSTAT3 was carried out. Expression of pSTAT3 expression in tumor samples following disease progression after cetuximab treatment was elevated compared with levels in tumors biopsies obtained prior to cetuximab administration (Figure 6A). Figure 6B demonstrates representative pSTAT3 IHC from 3 tumors prior to cetuximab treatment (upper panel) and 3 tumors following cetuximab treatment (lower panel). These findings from a small and heterogeneous HNSCC patient cohort suggest that pSTAT3 may represent a potential therapeutic target to improve responses to EGFR targeted therapy.
Figure 6.
Human HNSCC tumors from cetuximab-treated patients exhibit increased pSTAT3 expression compared with pretreatment tumors. (A) Cumulative results from 7 tumors prior to cetuximab treatment and 15 tumors follow cetuximab treatment from TMA stained with pSTAT3 antibody. The average composite score (intensity of staining x the percentage of tumor cells that stained positively) of pre- and post-cetuximab treated tumors are represented. (B) Representative pSTAT3 IHC from 3 tumors prior to cetuximab treatment (upper panel) and 3 tumors following cetuximab treatment (lower panel).
Discussion
Persistent activation of STAT3 has been implicated in conferring resistance to conventional therapies in some malignancies. Elevated levels of STAT3 have been reported in several drug resistant cancer cells where inactivation of STAT3 reversed the multi-drug resistant phenotype (29).
Several strategies have been developed to target the STAT3 pathway in settings of intrinsic resistance to chemotherapy and radiation in preclinical cancer models. In an in vivo model of non small cell lung cancer (NSCLC), inhibition of STAT3 using the STAT3 pathway inhibitor cucurbitacin I (JSI-124), a triterpenoid derivative, was able to overcome resistance to chemotherapy or radiotherapy (30). Inhibition of STAT3 reportedly sensitized glioma cells to temozolomide, an alkylator-based chemotherapy (31). In bladder cancer, aberrant STAT3 activation has been associated with chemoresistance, where subsequent inhibition of STAT3 activation by STAT3 siRNA or treatment with the JAK2 inhibitor AG-490, increased the sensitivity of the cells to chemotherapeutic agents (32). Others have reported that AG-490 treatment restores chemosensitivity in drug-resistant hematopoietic tumor cells (33). Using in vivo HNSCC models, cucurbitacin I treatment enhanced the inhibitory effects of ionizing radiation and abrogated radioresistance (34).
Several studies have demonstrated that acquired therapeutic resistance is associated with enhanced activation of the STAT3 where inhibiting the STAT3 pathway can restore drug sensitivity (35, 36). In gastric cancer, the STAT3 pathway has been shown to be involved in acquired drug resistance and inhibition of STAT3 by the STAT3-SH2 antagonist, 5,15-diphenylporphyrin (DPP), sensitized resistant cells to chemotherapy (37). In ovarian cancer models, acquisition of drug resistance was correlated with STAT3 activation both in vitro where resistance to paclitaxel has been associated with increased expression of pSTAT3 as well as in a human tumor tissue array, where recurrent or metastatic lesions demonstrated increased pSTAT3 expression compared with levels in the primary tumor (38). They further demonstrated that in vitro inhibition of the STAT3 pathway using the triterpenoid CDDO-Me reverses paclitaxel resistance in ovarian cancer cell lines (39). In breast cancer, tamoxifen resistance was associated with activation of STAT3, supporting the use of STAT3 inhibitors to overcome acquired tamoxifen resistance (40). Both constitutive and inducible STAT3 activation has been reported in acute myeloid leukemias (AML) and AML cell lines where the use of the small molecule probe C188 that targets the phosphotyrosine (pY) peptide binding site within the STAT3 SH2 domain can sensitize the drug-resistant tumor cells (41).
Several studies have implicated STAT3 activation in EGFR resistance. In hepatocellular carcinoma, cetuximab resistance was mediated via STAT3 activation and combination therapy using both inhibitors of EGFR and STAT3 in vitro, enhanced growth inhibition (42). In NSCLC STAT3 activation conferred resistance to NSCLC against gefitinib, which was restored upon suppression of STAT3 activity, suggesting that in NSCLC patients that are insensitive to EGFR inhibitors, STAT3 targeting maybe considered as an alternative therapy (43). In a pilot study where activated signaling molecules were evaluated in NSCLC patients who received gefitinib prior to surgical resection of tumor, pSTAT3 levels were elevated in the surgically resected tumor tissue implicating STAT3 as a potential candidate in mediating primary resistance (17). In high grade glioma, elevated levels of pSTAT3 has been linked to chemoresistance and blockade of STAT3 signaling sensitized the glioma cells to chemotherapy, thus providing a rationale for use of targeted therapies against STAT3 (16). In the present study, we identified STAT3 activation and increased IL-6 production in preclinical cancer models of intrinsic and acquired resistance to EGFR inhibitors. Our results demonstrate that targeting STAT3 using the STAT3 decoy in cetuximab or TKI resistant cells sensitizes the cells to EGFR inhibitor treatment in vitro and in vivo. The STAT3 decoy has also been tested in leukemia where inhibition of hyperactivated STAT3 in adriamycin resistant K562/A02 leukemia cells by the STAT3 decoy increased the sensitivity of the cells to adriamycin (44). Further investigation demonstrated activation of pSTAT3 in human HNSCC tumors that recurred following cetuximab treatment, suggesting that STAT3 activation is associated with cetuximab resistance. These cumulative findings suggest that strategies that inhibit STAT3 may abrogate therapeutic resistance to EGFR inhibitors.
Supplementary Material
Figure 1
Figure 2
Figure 3
Figure Legend
Statement of Translational Relevance.
Cumulative evidence suggests that Signal Transducer and Activator of Transcription-3 (STAT3) may contribute to therapeutic resistance and targeting STAT3 represents a potential strategy to improve treatment responses. Although compounds that decrease STAT3 activation have been used to reduce therapeutic resistance, none of the agents selectively or specifically inhibit STAT3. In this study, human head and neck squamous cell carcinoma (HNSCC) that recurred following cetuximab treatment demonstrated increased pSTAT3 expression. Treatment with a transcription factor decoy oligonucleotide specifically targeting STAT3 inhibited the growth of preclinical cancer models that were resistant to EGFR inhibitors. These findings suggest that targeting STAT3 may be effective in the setting of EGFR inhibitor resistance to augment treatment responses.
Acknowledgments
Financial support: This work was supported by NIH grants R01CA098372 and P50CA097190 as well as the American Cancer Society to JRG, and NIH grant R01CA137260 to DEJ.
We are grateful to Kelly M. Quesnelle, University of Pittsburgh, for providing the cetuximab resistant bladder cancer cell models.
References
- 1.Camp ER, Summy J, Bauer TW, Liu W, Gallick GE, Ellis LM. Molecular mechanisms of resistance to therapies targeting the epidermal growth factor receptor. Clin Cancer Res. 2005;11:397–405. [PubMed] [Google Scholar]
- 2.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358:1160–74. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 3.Harandi A, Zaidi AS, Stocker AM, Laber DA. Clinical Efficacy and Toxicity of Anti-EGFR Therapy in Common Cancers. J Oncol. 2009;2009:567486. doi: 10.1155/2009/567486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamatodani T, Ekblad L, Kjellen E, Johnsson A, Mineta H, Wennerberg J. Epidermal growth factor receptor status and persistent activation of Akt and p44/42 MAPK pathways correlate with the effect of cetuximab in head and neck and colon cancer cell lines. J Cancer Res Clin Oncol. 2009;135:395–402. doi: 10.1007/s00432-008-0475-2. [DOI] [PubMed] [Google Scholar]
- 5.Schilder RJ, Sill MW, Chen X, Darcy KM, Decesare SL, Lewandowski G, et al. Phase II study of gefitinib in patients with relapsed or persistent ovarian or primary peritoneal carcinoma and evaluation of epidermal growth factor receptor mutations and immunohistochemical expression: a Gynecologic Oncology Group Study. Clin Cancer Res. 2005;11:5539–48. doi: 10.1158/1078-0432.CCR-05-0462. [DOI] [PubMed] [Google Scholar]
- 6.Small EJ, Fontana J, Tannir N, DiPaola RS, Wilding G, Rubin M, et al. A phase II trial of gefitinib in patients with non-metastatic hormone-refractory prostate cancer. BJU Int. 2007;100:765–9. doi: 10.1111/j.1464-410X.2007.07121.x. [DOI] [PubMed] [Google Scholar]
- 7.Thatcher N, Chang A, Parikh P, Rodrigues Pereira J, Ciuleanu T, von Pawel J, et al. Gefitinib plus best supportive care in previously treated patients with refractory advanced non-small-cell lung cancer: results from a randomised, placebo-controlled, multicentre study (Iressa Survival Evaluation in Lung Cancer) Lancet. 2005;366:1527–37. doi: 10.1016/S0140-6736(05)67625-8. [DOI] [PubMed] [Google Scholar]
- 8.Goodin S. Erlotinib: optimizing therapy with predictors of response? Clin Cancer Res. 2006;12:2961–3. doi: 10.1158/1078-0432.CCR-06-0426. [DOI] [PubMed] [Google Scholar]
- 9.Yao Z, Fenoglio S, Gao DC, Camiolo M, Stiles B, Lindsted T, et al. TGF-beta IL-6 axis mediates selective and adaptive mechanisms of resistance to molecular targeted therapy in lung cancer. Proc Natl Acad Sci U S A. 2010;107:15535–40. doi: 10.1073/pnas.1009472107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabernero J. The role of VEGF and EGFR inhibition: implications for combining anti-VEGF and anti-EGFR agents. Mol Cancer Res. 2007;5:203–20. doi: 10.1158/1541-7786.MCR-06-0404. [DOI] [PubMed] [Google Scholar]
- 11.Morgillo F, Cantile F, Fasano M, Troiani T, Martinelli E, Ciardiello F. Resistance mechanisms of tumour cells to EGFR inhibitors. Clin Transl Oncol. 2009;11:270–5. doi: 10.1007/s12094-009-0354-6. [DOI] [PubMed] [Google Scholar]
- 12.Kijima T, Niwa H, Steinman RA, Drenning SD, Gooding WE, Wentzel AL, et al. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13:355–62. [PubMed] [Google Scholar]
- 13.Kim HS, Park YH, Lee J, Ahn JS, Kim J, Shim YM, et al. Clinical impact of phosphorylated signal transducer and activator of transcription 3, epidermal growth factor receptor, p53, and vascular endothelial growth factor receptor 1 expression in resected adenocarcinoma of lung by using tissue microarray. Cancer. 2010;116:676–85. doi: 10.1002/cncr.24748. [DOI] [PubMed] [Google Scholar]
- 14.Schindler C, Darnell JE., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–51. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 15.Cao X, Tay A, Guy GR, Tan YH. Activation and association of Stat3 with Src in v-Src-transformed cell lines. Mol Cell Biol. 1996;16:1595–603. doi: 10.1128/mcb.16.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14:6042–54. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haura EB, Sommers E, Song L, Chiappori A, Becker A. A pilot study of preoperative gefitinib for early-stage lung cancer to assess intratumor drug concentration and pathways mediating primary resistance. J Thorac Oncol. 2010;5:1806–14. doi: 10.1097/JTO.0b013e3181f38f70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A. 2003;100:4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 20.Zhang XZJ, Wei H, Tian Z. STAT3-decoy oligodeoxynucleotide inhibits the growth of human lung cancer via down-regulating its target genes. Oncol Rep. 2007;17:1377–82. [PubMed] [Google Scholar]
- 21.Sun Z, Yao Z, Liu S, Tang H, Yan X. An oligonucleotide decoy for Stat3 activates the immune response of macrophages to breast cancer. Immunobiology. 2006;211:199–209. doi: 10.1016/j.imbio.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Shen J, Li R, Li G. Inhibitory effects of decoy-ODN targeting activated STAT3 on human glioma growth in vivo. In Vivo. 2009;23:237–43. [PubMed] [Google Scholar]
- 23.Pedranzini L, Leitch A, Bromberg J. Stat3 is required for the development of skin cancer. J Clin Invest. 2004;114:619–22. doi: 10.1172/JCI22800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun X, Zhang J, Wang L, Tian Z. Growth inhibition of human hepatocellular carcinoma cells by blocking STAT3 activation with decoy-ODN. Cancer Lett. 2008;262:201–13. doi: 10.1016/j.canlet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 25.Souissi I, Najjar I, Ah-Koon L, Schischmanoff PO, Lesage D, Le Coquil S, et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-kappaB. BMC Cell Biol. 2011;12:14. doi: 10.1186/1471-2121-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boehm AL, Sen M, Seethala R, Gooding WE, Freilino M, Wong SM, et al. Combined targeting of epidermal growth factor receptor, signal transducer and activator of transcription-3, and Bcl-X(L) enhances antitumor effects in squamous cell carcinoma of the head and neck. Mol Pharmacol. 2008;73:1632–42. doi: 10.1124/mol.107.044636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quesnelle KM, Grandis JR. Dual kinase inhibition of EGFR and HER2 overcomes resistance to cetuximab in a novel in vivo model of acquired cetuximab resistance. Clin Cancer Res. 17:5935–44. doi: 10.1158/1078-0432.CCR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhola NE, Thomas SM, Freilino M, Joyce S, Sahu A, Maxwell J, et al. Targeting GPCR-mediated p70S6K activity may improve head and neck cancer response to cetuximab. Clin Cancer Res. 17:4996–5004. doi: 10.1158/1078-0432.CCR-10-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Xiao W, Wang L, Tian Z, Zhang J. Deactivation of signal transducer and activator of transcription 3 reverses chemotherapeutics resistance of leukemia cells via down-regulating P-gp. PLoS One. 2011;6:e20965. doi: 10.1371/journal.pone.0020965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsu HS, Huang PI, Chang YL, Tzao C, Chen YW, Shih HC, et al. Cucurbitacin I inhibits tumorigenic ability and enhances radiochemosensitivity in nonsmall cell lung cancer-derived CD133-positive cells. Cancer. 117:2970–85. doi: 10.1002/cncr.25869. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Chen L, Bao Z, Li S, You G, Yan W, et al. Inhibition of STAT3 reverses alkylator resistance through modulation of the AKT and beta-catenin signaling pathways. Oncol Rep. 26:1173–80. doi: 10.3892/or.2011.1396. [DOI] [PubMed] [Google Scholar]
- 32.Chen RJ, Ho YS, Guo HR, Wang YJ. Long-term nicotine exposure-induced chemoresistance is mediated by activation of Stat3 and downregulation of ERK1/2 via nAChR and beta-adrenoceptors in human bladder cancer cells. Toxicol Sci. 115:118–30. doi: 10.1093/toxsci/kfq028. [DOI] [PubMed] [Google Scholar]
- 33.Alas S, Bonavida B. Inhibition of constitutive STAT3 activity sensitizes resistant non-Hodgkin’s lymphoma and multiple myeloma to chemotherapeutic drug-mediated apoptosis. Clin Cancer Res. 2003;9:316–26. [PubMed] [Google Scholar]
- 34.Chen YW, Chen KH, Huang PI, Chen YC, Chiou GY, Lo WL, et al. Cucurbitacin I suppressed stem-like property and enhanced radiation-induced apoptosis in head and neck squamous carcinoma--derived CD44(+)ALDH1(+) cells. Mol Cancer Ther. 9:2879–92. doi: 10.1158/1535-7163.MCT-10-0504. [DOI] [PubMed] [Google Scholar]
- 35.Bewry NN, Nair RR, Emmons MF, Boulware D, Pinilla-Ibarz J, Hazlehurst LA. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Mol Cancer Ther. 2008;7:3169–75. doi: 10.1158/1535-7163.MCT-08-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J, Bi C, Janakakumara JV, Liu SC, Chng WJ, Tay KG, et al. Enhanced activation of STAT pathways and overexpression of survivin confer resistance to FLT3 inhibitors and could be therapeutic targets in AML. Blood. 2009;113:4052–62. doi: 10.1182/blood-2008-05-156422. [DOI] [PubMed] [Google Scholar]
- 37.Yang J, Huang J, Dasgupta M, Sears N, Miyagi M, Wang B, et al. Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci U S A. 107:21499–504. doi: 10.1073/pnas.1016147107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duan Z, Foster R, Bell DA, Mahoney J, Wolak K, Vaidya A, et al. Signal transducers and activators of transcription 3 pathway activation in drug-resistant ovarian cancer. Clin Cancer Res. 2006;12:5055–63. doi: 10.1158/1078-0432.CCR-06-0861. [DOI] [PubMed] [Google Scholar]
- 39.Duan Z, Ames RY, Ryan M, Hornicek FJ, Mankin H, Seiden MV. CDDO-Me, a synthetic triterpenoid, inhibits expression of IL-6 and Stat3 phosphorylation in multi-drug resistant ovarian cancer cells. Cancer Chemother Pharmacol. 2009;63:681–9. doi: 10.1007/s00280-008-0785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishii Y, Waxman S, Germain D. Tamoxifen stimulates the growth of cyclin D1-overexpressing breast cancer cells by promoting the activation of signal transducer and activator of transcription 3. Cancer Res. 2008;68:852–60. doi: 10.1158/0008-5472.CAN-07-2879. [DOI] [PubMed] [Google Scholar]
- 41.Redell MS, Ruiz MJ, Alonzo TA, Gerbing RB, Tweardy DJ. Stat3 signaling in acute myeloid leukemia: ligand-dependent and -independent activation and induction of apoptosis by a novel small-molecule Stat3 inhibitor. Blood. 117:5701–9. doi: 10.1182/blood-2010-04-280123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W, Shen X, Xia X, Xu G, Ma T, Bai X, et al. NSC 74859-mediated inhibition of STAT3 enhances the anti-proliferative activity of cetuximab in hepatocellular carcinoma. Liver Int. 32:70–7. doi: 10.1111/j.1478-3231.2011.02631.x. [DOI] [PubMed] [Google Scholar]
- 43.Chiu HC, Chou DL, Huang CT, Lin WH, Lien TW, Yen KJ, et al. Suppression of Stat3 activity sensitizes gefitinib-resistant non small cell lung cancer cells. Biochem Pharmacol. 81:1263–70. doi: 10.1016/j.bcp.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Zhang X, Xiao W, Wang L, Tian Z, Zhang J. Deactivation of signal transducer and activator of transcription 3 reverses chemotherapeutics resistance of leukemia cells via down-regulating P-gp. PLoS One. 6:e20965. doi: 10.1371/journal.pone.0020965. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1
Figure 2
Figure 3
Figure Legend