Abrogation of disease development in plants expressing animal antiapoptotic genes (original) (raw)
Abstract
An emerging topic in plant biology is whether plants display analogous elements of mammalian programmed cell death during development and defense against pathogen attack. In many plant–pathogen interactions, plant cell death occurs in both susceptible and resistant host responses. For example, specific recognition responses in plants trigger formation of the hypersensitive response and activation of host defense mechanisms, resulting in restriction of pathogen growth and disease development. Several studies indicate that cell death during hypersensitive response involves activation of a plant-encoded pathway for cell death. Many susceptible interactions also result in host cell death, although it is not clear how or if the host participates in this response. We have generated transgenic tobacco plants to express animal genes that negatively regulate apoptosis. Plants expressing human Bcl-2 and Bcl-xl, nematode CED-9, or baculovirus Op-IAP transgenes conferred heritable resistance to several necrotrophic fungal pathogens, suggesting that disease development required host–cell death pathways. In addition, the transgenic tobacco plants displayed resistance to a necrogenic virus. Transgenic tobacco harboring Bcl-xl with a loss-of-function mutation did not protect against pathogen challenge. We also show that discrete DNA fragmentation (laddering) occurred in susceptible tobacco during fungal infection, but does not occur in transgenic-resistant plants. Our data indicate that in compatible plant–pathogen interactions apoptosis-like programmed cell death occurs. Further, these animal antiapoptotic genes function in plants and should be useful to delineate resistance pathways. These genes also have the potential to generate effective disease resistance in economically important crops.
Keywords: programmed cell death, resistance
Regulation of cell death often is crucial to the outcome of plant–pathogen interactions. Host cell death can occur in both resistant (incompatible) and susceptible (compatible) plant–pathogen interactions, although the mechanism(s) and relevant pathways are poorly understood. In either case, pathogen challenge can trigger groups of plant cells to die. The cellular characteristics of the death process strongly implicates specific signals and autonomous cellular biochemical processes that execute individual cells. Recent findings show similarities in microbial infection mechanisms of animals, humans, and plants (1–3). An emerging question concerns whether common features exist between programmed cell death pathways in plants and those in other eukaryotes. In animals, programmed cell death (PCD), or its morphological equivalent, apoptosis, is genetically controlled cellular suicide (4). It is a complex process essential for development, maintenance of cellular homeostasis, and defense against environmental insults including pathogen attack. Apoptosis induces characteristic morphological and biochemical alterations in cells and results in the orchestrated disassembly of the cell in a noninflammatory manner (5, 6).
Many incompatible plant–pathogen interactions induce hypersensitive lesions and activate host defense mechanisms (7). The hypersensitive response has been defined as rapid, localized death of plant cells associated with resistance to pathogen attack. This response is invoked by pathogenic fungi, viruses, bacteria, and nematodes (8). Visually, this reaction appears as a region of dead cells at the point of pathogen recognition by the plant. Although this scenario resembles PCD in mammalian systems, specific illustrations of a functionally conserved program in plants exhibiting the hallmark characteristics of PCD are limited and molecular details of this process in plants remain unclear (9). One of the few examples of apoptosis in plant disease resistance is described by Ryerson and Heath (10), where hypersensitive resistance in cowpea to the cowpea rust fungus (Uromyces vignae) is accompanied by degradation of host DNA into oligonucleosomal fragments as well as terminal deoxynucleotidyltransferase-mediated UTP end labeling-positive cells, both common events in PCD. Similar indicators of PCD can appear in the diseased state (susceptible or compatible reaction). For example, in plants that are sensitive to toxin-producing fungi, e.g.,Fusarium moniliforme, Alternaria alternata, and_Cochliobolus victoriae_, certain features of apoptosis are observed (11, 12). In addition, cucumber mosaic virus D satellite RNA has been shown to induce morphological features of PCD in infected tomato (13). Thus, PCD in plants can accompany both susceptible and resistant reactions, suggesting that overlapping biochemical pathways are operative in these two contrasting outcomes.
Apoptosis in animals is controlled through functionally conserved genes, initially identified in_Caenorhabditis elegans_ (14, 15). Both proapoptotic (CED-3, CED-4) and antiapoptotic (CED-9) regulators have been identified in C. elegans, as have the analogous mammalian counterparts, namely, caspases, APAF-1, and Bcl-2, respectively (reviewed in ref. 4). A key finding is the complementation of nematode CED-9 by human Bcl-2 (16), indicating functional conservation between nematode and human cell death pathways. Genes of the Bcl-2 family have been identified in mammalian viruses,Drosophila, and mice, among others. Moreover, two classes of baculovirus antiapoptotic proteins [p35 and inhibitor of apoptosis (IAP)] inhibit apoptosis in humans, and certain human IAPs are functional in flies with IAP defects (17). Thus, regulators of cell death are functionally conserved across broad taxonomic distances within animals.
In light of these observations, we undertook a transgenic, “comparative pathobiology” approach to evaluate effects of known regulators of animal apoptosis on plant–pathogen interactions. We focused these studies on several negative regulators of apoptosis. Specifically, we generated transgenic tobacco plants expressing: (i) human bcl-2, (ii) human bcl-xl, (iii) nematod_e ced-9_, and (iv) baculovirus op-iap. All of the products of these genes suppress cell death in appropriate cellular contexts (18–21). Given the broad taxonomic occurrence of regulators of PCD, coupled with structural and functional overlap among these proteins, we examined the phenotypical consequences of expressing this selected subset of genes in plants exposed to pathogen challenge. We demonstrate that expression of bcl-2, bcl-xl,ced-9, or op-iap in tobacco confers heritable resistance to three important fungal phytopathogens. In addition, these same transgenic plants show resistance to tomato spotted wilt virus (TSWV). We further show that DNA laddering occurs in susceptible but not resistant transgenic tobacco tissue during the normal course of infection by Sclerotinia sclerotiorum, a broad host range, necrotrophic fungus.
Materials and Methods
Plasmid Constructions and Plant Transformation.
The following antiapoptotic cDNA genes were used: (i) human bcl-2 [obtained from S. Korsmeyer, Dana–Farber Cancer Institute, Boston (19)], (ii)bcl-xl [obtained from C. Thompson, University of Chicago (20)], (iii) C. elegans ced-9 [obtained from R. Horvitz, Massachusetts Institute of Technology, Cambridge (15)], and (iv) Baculovirus op-iap [obtained from L. Miller, University of Georgia, Athens (21)]. Separate binary vectors were constructed that contained these genes under the control of the cauliflower mosaic virus 35S promoter and the Agrobacterium nopaline synthase terminator (22).
For bcl-2 and ced-9 constructs, based on available sequence information, PCR primers were designed that introduced an _Nco_I site at the 5′ end of the ORF and an_Xba_I site at the 3′ end of the ORF. For bcl-2, the following primers were used: 5′-TTTTTCCTCTGGGAGGGCCATGGCGCACGCTGG-3′ and 5′-TTTTTTCTAGATGCTCTTCGGGCGTGG-3′.
For ced-9, the following primers were used: 5′-GAATTCCGGTTTGAGCCATGGCGACACGCTGCACGGCG-3′ and 5′-TTTTTTCTAGAAATACGTTACTTCAAGCTG-3′. PCR was performed and the products were digested with _Nco_I and _Xba_I and subcloned into the plant expression cassette pRTL2 (23). The expression cassettes containing bcl-2 and ced-9 were subcloned into the binary vector pPZP212 (24) as _Hin_dIII fragments. The resulting constructs are referred to as pPTN161 and pPTN147, respectively. A similar strategy was used for Op-IAP, except that _Nco_I and _Bam_HI sites were engineered by PCR by using the following primer set: 5′-GTTGCAGACCATGGCCAGCTCCCGAGCAATTGGC-3′ and 5′-TTTTTGGATCCTTTTATTGTTACACTTGG-3′. The op-iap construct is referred to as pPTN148.
Bcl-xl wt was digested with _Xba_I and_Bsp_HI and subcloned into pRTL2. The resulting plasmid was digested with _Pst_I and ligated into pPZP211. To generate a point mutation in bcl-xl, the gene was amplified with primers harboring a G-to-A substitution at codon 138 (G138A). This site-directed mutation was inserted into pZP211 in an identical manner.
Agrobacterium tumefaciens strain C58C1 (25) was used in all tobacco transformations. The binary vectors pPTN147 (ced-9), pPTN148 (op-iap), pPTN161 (bcl-2), pPTN250 (bcl-xl), and pPTN251 (_bcl-xl_-G138A) were mobilized into C58C1 by triparental mating (26).Agrobacterium transconjugants were selected on AB minimal medium supplemented with streptomycin (100 mg/liter), spectinomycin (100 mg/liter), rifampicin (50 mg/liter), gentamycin (50 mg/liter), and sucrose at 1.5% (wt/vol). Tobacco leaf tissue (cv. Glurk, genotype NN; Turkish, genotype nn) was transformed and plants were regenerated as described (27).
DNA/RNA Isolation and Electrophoresis.
DNA and RNA from tobacco leaves were isolated as described (28, 29). RNA blot hybridization and membrane washing were performed under stringent conditions (30). Immunoblots were done as described (31). Anti-human Bcl-2 antiserum was provided by J. Reed (The Burnham Institute, La Jolla, CA) (32), and antibody to Bcl-xl was from Chemicon. IAP antiserum was not available. The CED-9 antibody was kindly provided by H. R. Horvitz and Brad Hersh (Massachusetts Institute of Technology, Cambridge). ELISA detection of TSWV was done with a Pathoscreen kit (Agdia, Elkhart, IN).
Plant Inoculation and Disease-Resistance Assays.
The following fungi were evaluated: Sclerotinia sclerotiorum isolate 1980 (33), Botrytis cinerea (ATCC 11542), and_Cercospora nicotianae_ (kindly provided by G. Upchurch, North Carolina State University, Raleigh). In addition, transgenic tobacco plants were tested for reaction to a necrogenic virus, TSWV. Transgenic and control tobacco were grown at 25°C with 16-h light periods in the greenhouse. Leaves of 5-week-old tobacco plants were used for all experiments. Detached-leaf assays were experimentally convenient, and they responded similarly to inoculated whole plants under greenhouse conditions.
For fungal detached-leaf assays, a minimum of two leaves per plant from a minimum of five independently transformed plant lines per transgene of both tobacco cultivars were inoculated by placing 5-mm-diameter agar plugs containing actively growing hyphal tips from 3-day-old colonies of either S. sclerotiorum or B. cinerea grown on potato dextrose agar. For inoculation with C. nicotianae, spore suspensions were sprayed directly on leaves. Spores were obtained by following standard procedures (34) and sprayed as a conidial suspension (104 conidia per ml) on tobacco plants until run-off. For viral assays, detached tobacco leaves were dusted with carborundum and rub-inoculated with TSWV-infected plant sap diluted 1:25 (wt/vol) in 10 mM sodium phosphate buffer, pH 7/10 mM sodium sulfite. After 5 min, leaves were washed thoroughly with water, placed in Petri dishes, and held at 25°C under constant illumination.
Leaves were placed on moistened sterile filter paper in glass Petri dishes and incubated for 3–7 days at 25°C and high humidity. All treatments were repeated on whole plants with similar results. Inoculated plants were placed in a mist chamber at 100% relative humidity at 25°C. A minimum of five independently transformed plant lines per transgene were used. All experiments were repeated at least three times.
Results
Establishment of Transgenic Plants and Analysis of Gene Expression.
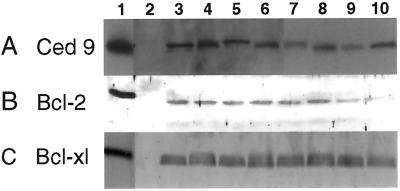
Single or low-copy-number, kanamycin-resistant transgenic tobacco plants harboring the various antiapoptotic genes were selected after Southern blot analysis (not shown). Nontransformed tobacco and tobacco containing vector alone served as controls. Glurk and Turkish lines were obtained with each of the transgenes (ced-9,bcl-2, bcl-xl, and op-iap). Northern blot analysis confirmed the expression of the transgenes, and protein expression levels in these plants were quantified by Western blot analysis (Fig. 1 and data not shown).
Figure 1.
Western blot assays of expression of Bcl-2-related proteins in vitro and in transformed tobacco lines. (A) CED-9 expression. Lanes: 1, His-tagged CED-9; 2, wild-type tobacco; 3–10, separate ced-9 transgenic plants. (B) Bcl-2 expression. Lanes: 1, monkey kidney cells expressing His-tagged Bcl-2; 2, wild-type tobacco; 3–10, separate_bcl-2_-transgenic plants. (C) Bcl-xl expression. Lanes: 1, monkey kidney cells expressing His-tagged Bcl-xl; 2, wild-type tobacco; 3–6, separate _bcl-xl_-transgenic plants; 7–10, transgenic (susceptible) plants with_bcl-xl_-G138A.
Bcl-2 Family of Proteins and Op-IAP Confer Resistance to Fungal Pathogens.
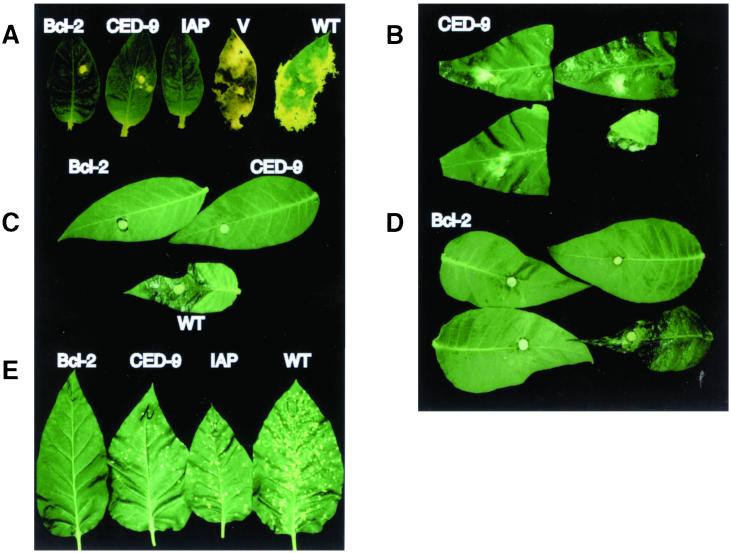
When transgenic tobacco harboring Bcl-2, CED-9, or Op-IAP was inoculated with S. sclerotiorum, plants were highly tolerant to this pathogen and, in most cases, completely resistant to infection. In contrast, plants transformed with vector alone or nontransformed tobacco were highly and typically susceptible (Fig.2A). S. sclerotiorum requires inoculation with a nutritional source, unlike most other fungi that can incite disease when delivered in water or buffer alone. As shown in Fig. 2A, the fungus, when inoculated with a rich nutritional source, grew vegetatively along the leaf surfaces of plants expressing the antiapoptotic genes, without infecting host tissue. The fungus eventually ceased growth, presumably because the nutritional source became depleted; even with extended incubation, the fungus still failed to colonize and infect plant tissue. In contrast, the fungus on control plants completely colonized and macerated leaf tissue during the same time period (Fig.2A). B. cinerea, also a broad-host-range necrotrophic fungal pathogen, showed a similar response when inoculated to the same transgenic tobacco lines (Fig. 2C and data not shown). C. nicotianae produces a non-host-specific toxin, cercosporin, which is required for pathogenicity. Tobacco harboring Bcl-2 was strikingly resistant to this pathogen (Fig.2E), whereas wild-type tobacco was extremely sensitive; intact plants were dead within 2 weeks. Transgenic tobacco with CED-9 and Op-IAP expressed symptoms, albeit to a reduced extent relative to control plants (Fig. 2E). Although_C. nicotianae_-infected tobacco containing CED-9 and Op-IAP showed symptoms of toxin-induced disease, these plants as well as the Bcl-2 tobacco all survived the inoculation and set seed, in contrast to the wild-type plants that were killed before they could set seed.
Figure 2.
Antiapoptotic genes confer resistance to necrotrophic fungi in transgenic plants. Detached Glurk tobacco leaves were inoculated with three species of fungi. (A) Leaves inoculated with_S. sclerotiorum_ from plants transformed with_bcl-2_, ced-9, bcl-xl, or_op-iap_, as indicated. Leaf V is from a plant transformed with binary vector pZP212 alone. (B) Leaves inoculated with S. sclerotiorum from F1 progeny of plants transformed with ced-9. (C) Leaves inoculated with B. cinerea from plants transformed with_bcl-2_ or ced-9, as indicated. (D) Leaves inoculated with B. cinerea from F1 progeny of plants transformed with_bcl-2_. (E) Leaves inoculated with_C. nicotianae_ from plants transformed with_bcl-2_, ced-9, or op-iap, as indicated. In A, C, and_E_, WT indicates a leaf from an untransformed control plant. In B and D, the leaf shown at_Lower Right_ is from a kanamycin-sensitive F1 plant.
The tobacco plants were selfed for genetic analysis. A total of 15 kanamycin-sensitive progeny plants lacking transgene expression were susceptible to all tested fungi, whereas plants selected for kanamycin resistance and transgene expression were resistant and generally identical in phenotype to the parental, primary transformant (e.g., Fig. 2 B and D, S. sclerotiorum and_B. cinerea_ inoculations, respectively). Progeny leaves in Fig. 2B were similar in size when inoculated. The susceptible leaf (Fig. 2B Lower Right) became extensively macerated. In all cases, the fungal resistance phenotypes mediated by bcl-2, ced-9, or_op_-iap transgenes appeared to be dominant and cosegregated with the selectable marker gene. Both Turkish and Glurk cultivars responded identically after fungal challenge, indicating no tobacco genotype-specific effects in the transgenic plants after pathogen challenge.
Bcl-2 Family Proteins and Op-IAP Confer Resistance to a Viral Pathogen.
Plants expressing antiapoptotic genes also were challenged with TSWV. Certain plant host–virus combinations produce necrotic lesions as a disease symptom, regardless of host genotype. In such combinations the plant is called a local lesion host of the virus. TSWV causes large, spreading necrotic lesions on inoculated leaves (35), followed by systemic infection and necrosis. Typical lesions were observed on control plants for TSWV (Fig. 3). Necrotic symptoms were greatly reduced or absent on the tobacco leaves expressing Bcl-2 (not shown) as well as CED-9 and Op-IAP after inoculation with TSWV (Fig. 3). Like the fungal pathogens, this effect was independent of the tobacco genetic background because transgenic Turkish and Glurk cultivars gave identical results. However, we have preliminary evidence that a cultivar-specific HR is not inhibited by these genes because tobacco mosaic virus induced local lesions on all four types of transgenic Glurk tobacco, which has the N gene for resistance (unpublished data).
Figure 3.
Transgenic plants expressing antiapoptotic genes have altered response to TSWV. TSWV lesions on detached Glurk tobacco leaves. (A) Three leaves from plants expressing CED-9. (B) Three leaves from Op-IAP-expressing plants. In both_A_ and B, leaf 4 is from an untransformed control plant.
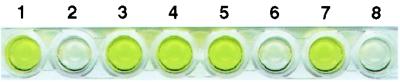
ELISA assays demonstrated that both necrotic lesions and surrounding areas of inoculated leaves of the control plants contained TSWV proteins (Fig. 4). On the Bcl-2, CED-9, and Op-IAP plants, however, TSWV proteins were present in the inoculated areas of the leaves but were not detectable in the surrounding, uninoculated regions of the leaves immediately adjacent to the inoculated region (Fig. 4). This suggests that virus spread from the site of inoculation was impaired in Bcl-2, CED-9, and Op-IAP plants. The resistance phenotypes also cosegregated with the transgenes in the progeny of selfed plants.
Figure 4.
ELISA of TSWV accumulation in detached leaves from transformed tobacco lines. Wells 1 and 2, positive and negative controls, respectively. Wells 3, 5, and 7 are samples from inoculated half-leaves; wells 4, 6, and 8 are from the uninoculated half of the same leaves. Plants are transformed with vector only (wells 3 and 4), with ced-9 (wells 5 and 6), or with op-iap (wells 7 and 8).
Pathogen Resistance Is Reversed by Loss-of-Function Mutation.
An important consideration in these observations is the question: Are these antiapoptotic gene products directly affecting cell death/resistance pathways in the plant or are they causing a general, stress response in the plant that accounts for resistance? To address this question, we generated an additional set of transgenic tobacco expressing the Bcl-2 family member Bcl-xl and Bcl-xl containing a G138A mutation (Fig. 1B, lanes 6–9). Glycine 138 is located in the conserved BH-1 domain of Bcl-xl, which is essential for its antiapoptotic activity (36). Results from 10 independent, transformed tobacco lines indicate that Bcl-xl protects tobacco from_S. sclerotiorum_ infection, as was the case for Bcl-2 (Fig.5). Significantly, the mutant Bcl-xl (G138A) expressing tobacco responded as untransformed control plants although in some lines lesions were slower to develop (Fig. 5). Selfed Bcl-xl plants also showed segregation of resistance and transgene expression (data not shown). None of the selfed progeny containing the G138A mutation showed resistance. Western blot analysis of these tobacco lines showed equivalent steady-state protein levels in both Bcl-xl (wt) and Bcl-xl (G138A) lines (Fig. 1B). Thus, our data indicate that resistance to S. sclerotiorum requires a functional Bcl-xl protein.
Figure 5.
Plants expressing wild-type Bcl-xl, but not mutant Bcl-xl, are resistant to S. sclerotiorum. Shown are detached Glurk tobacco leaves inoculated with S. sclerotiorum. Leaves A and B are from plants transformed with bcl-xl, and leaves C and D are from plants transformed with mutant_bcl-xl_ (G138A).
Disease Development and Apoptosis.
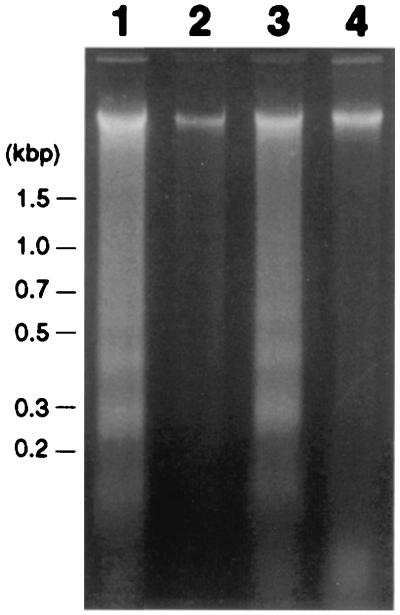
One classical biochemical method for demonstrating apoptosis is the presence of oligonucleosome-sized fragments of DNA that, when electrophoresed on agarose gels, produce a characteristic “ladder.” Genomic DNA was isolated from tobacco leaves after inoculation with S. sclerotiorum. As shown in Fig.6, lanes 1 and 3, the isolated DNA was fragmented and formed characteristic ladders. Lane 1 shows DNA isolated 4 h after inoculation and lane 3 shows it 8 h after inoculation. There were no macroscopic symptoms at either time point, but, by 8 h, fungal growth was evident. By 18 h postinoculation, symptoms of disease were clearly visible (Fig.2A). Thus, before discernible disease development occurred, the fungus was inducing DNA fragmentation in the host plant. Of note, transgenic tobacco harboring Bcl-xl not only was resistant to the fungus, but DNA laddering did not occur during the same time period (Fig. 6, lanes 2 and 4) or even later time points (not shown). Similar results occurred with tobacco harboring CED-9 and Bcl-2 (not shown). These observations suggest that S. sclerotiorum induces PCD during successful infection. Further, this pathogen-induced plant cell death has some similarity to PCD in animals.
Figure 6.
DNA laddering in susceptible tobacco after inoculation with S. sclerotiorum. Ethidium bromide-stained agarose gel of DNA extracted from nontransgenic tobacco (lanes 1 and 3) and the transgenic line expressing Bcl-2 (lanes 2 and 4) after challenge with S. sclerotiorum. DNA was extracted 4 (lanes 1 and 2) and 8 h (lanes 3 and 4) postinoculation.
Effects of Transgene Expression on Plant Physiology.
Transgenic tobacco constitutively expressing IAP grew, flowered, and set seed in a manner similar to wild-type tobacco, with no detectable morphological or physiological alterations. However, plant growth and morphology were affected in some lines expressing either Bcl-2, Bcl-xl, or CED-9. Lines with high Bcl-2 expression were male sterile. High-Bcl-xl-expressing lines also were male sterile and stunted, and they exhibited stem bleaching, flower deformation, and slightly altered leaf pigmentation (data not shown). CED-9 transgenic plants had variegated leaf pigmentation. These highly expressing transgenic plant lines were also extremely resistant to the pathogens tested. In contrast, moderately expressing transgenic lines (e.g., Figs. 2 and 3) showed none of these altered growth patterns but retained pathogen resistance.
Discussion
The indispensable role of PCD in higher eukaryotic development and its response to environmental stress and pathogen attack is well established. Here, we show that negative regulators of apoptosis, when expressed in tobacco, conferred resistance phenotypes to several fungal pathogens and TSWV under conditions that normally result in necrotic disease. Moreover, we show that DNA ladders were induced specifically during disease development by a fungal pathogen but not in plants expressing cytoprotective antiapoptotic genes. Although any single criterion is not conclusive, these results are consistent with the model that S. sclerotiorum induces disease in tobacco by activating a plant PCD pathway. This pathway was inhibited by animal antiapoptotic genes, thereby preventing fungal colonization, DNA fragmentation, and subsequent disease development.
Coupled with recent reports, these data considerably strengthen the parallels between the hypersensitive response in plants and PCD in animals. For example, Bcl-xl and CED-9 have been reported to protect tobacco plants from cell death induced by either UV irradiation or paraquat treatment (37). In addition, in temperature-shift experiments after tobacco mosaic virus inoculation with Bcl-xl transgenic plants, expression of pathogenesis-related proteins PR1 and PR3 was reduced and expression of tobacco mosaic virus coat protein was increased. Lesion size was reported to be slightly larger in the transgenic plants (37), although the overall role of these transgenes with respect to plant disease remained an open question. Previous studies showed that tomato protoplasts treated with the mycotoxin fumonisin B1 underwent PCD-like response as indicated by DNA ladders and terminal deoxynucleotidyltransferase-mediated UTP end labeling-positive reacting cells (11). Recently, Ausubel and colleagues (38, 39) have shown that fumonisin B1 treatment of Arabidopsis protoplasts results in terminal deoxynucleotidyltransferase-mediated UTP end labeling-positive reacting cells and also that toxin infiltration of intact plants resulted in cell death, generation of reactive oxygen intermediates, and other biochemical markers associated with plant defense responses. Those authors also suggested that toxin-mediated PCD may be important during successful plant infection by pathogens. Bax, a death-promoting member of the Bcl-2 family known to kill yeast cells (40, 41), recently was introduced and expressed in tobacco cells. Cell death of these tobacco cells occurred in a manner similar to hypersensitive response (42). However, no distinctive morphological changes occurred, although Bax did localize to mitochondria. To date, no plant homologues of caspases or Bcl-2-like proteins have been identified, although caspase-like activity has been detected (43). Further, cysteine proteases are important in a number of plant PCD situations, including senescence (44, 45), xylogenesis (46), and oxidative stress (47). It is possible that functional analogs may be obscure at the sequence level, which is consistent with the divergence of plant and animal species. However, there is growing circumstantial evidence that similar pathways exist in plants and animals. Thus, although the extent to which animal and plant apoptosis pathways overlap is not yet resolved, there are several examples plant cell death accompanied by features of apoptosis.
A unifying aspect of our results with the fungi tested is the fact that all three pathogens are necrotrophs, i.e., they require host plant cell death to grow, colonize, and reproduce in the plant. In the case of_S. sclerotiorum_, it has been assumed that this aggressive, indiscriminate pathogen, with an impressive arsenal of degradative enzymes and toxins, simply ramifies through defenseless plant tissue. Our data suggest that this interaction is more sophisticated—that the pathogen specifically interacts with the plant by triggering host-cell-death pathways. Inhibition of this pathway presumably by antiapoptotic gene products prevents fungal infection despite the fact that the fungus has its full complement of virulence determinants. Necrotrophic pathogens may need to co-opt plant host apoptotic pathways for successful colonization and subsequent disease development. Suppression of plant cell death may improve resistance to necrotrophs.
The effect of Bcl-2 expression on C. nicotianae infection is of interest. This fungus synthesizes a light-activated polyketide toxin, cercosporin (48). CED-9- and IAP-containing transgenic tobacco plants were symptomatic but highly tolerant. Bcl-2 transgenic plants, on the other hand, were extremely resistant (Fig.2E). Cercosporin generates reactive oxygen species such as singlet oxygen, which is required for induction of host cell death. Bcl-2 reportedly interferes with cell death mediated by reactive oxygen possibly by promoting the scavenging of free radicals (49). Consistent with Bcl-2 protecting plants from reactive oxygen species is the recent report of paraquat resistance in transgenic Bcl-xl- and CED-9-containing tobacco (37). Paraquat is a contact herbicide that kills plant cells by causing the production of active oxygen species. Moreover, Bcl-2 expression reduced paraquat-induced apoptosis in mouse cells (50). It will be of interest to determine whether Bcl-2 functions similarly in plants, particularly because pathogen attack is known to generate reactive oxygen species (51).
The mechanisms leading to necrosis in response to virus infection are poorly understood. Clearly, in the case of TSWV, lesion formation at the site of inoculation does not prevent systemic invasion of the plant. However, the transgenes not only prevented the typical local necrotic response to virus inoculations, but limited virus movement or reproduction as well. Perhaps, TSWV causes physiological changes in the host before productive infection, and necrosis is one consequence of this elicitation. Molecular details of virus-induced symptomatology are unknown, precluding speculation on the effects of Bcl-2, CED-9, or Op-IAP. Because these genes also block fungal-induced necrosis, presumably a common pathway is involved in both cases.
An important consideration in these experiments concerns the specificity and mode of action of these transgenes. We believe these transgenes specifically confer resistance in our experiments for the following reasons: (i) only transgenic plants were resistant, (ii) inheritance of resistance was strictly correlated with expression of the transgene, and (iii) a loss-of-function mutant of Bcl-xl (G138A) failed to confer resistance under the same conditions, whereas wild-type Bcl-xl did, and this phenotype also was evident after selfing of the parent transgenic plants.
The mechanism of action of these antiapoptotic genes in plants, at present, is conjectural. In preliminary experiments, there was no consistent pattern of induction of defense genes (e.g., PR proteins; see Fig. 7, which is published as supplemental data on the PNAS web site, www.pnas.org) across independently generated transgenic lines, but we cannot rule out that these genes may, in some cases, trigger known defense-signaling pathways (e.g., salicylic acid, jasmonic acid). Even in mammalian systems, it is not entirely clear how, for example, Bcl-2 promotes cell survival. Several mechanisms have been proposed to explain how Bcl-2 prevents apoptosis (52), including prevention of cytochrome c release from mitochondria, prevention of reactive oxygen-induced cell death, dimerization with and inactivation of death-promoting apoptotic proteins such as Bax, and sequestration of caspase-activating proteins.
The IAP family of proteins likewise is important for the suppression of programmed cell death. Several IAPs are caspase inhibitors (16), but some IAPs clearly do not have any antiapoptotic function. In yeast and nematodes, IAP proteins are involved in cell division and cytokinesis, respectively (53, 54). Moreover, the molecular mechanism of the baculoviral IAP suppression of cell death is not entirely clear because no protease target is known. Given that no molecular markers involving plant cell-programmed cell death have been identified, it is premature to speculate on the mechanisms underlying our results. That characteristic DNA fragmentation was observed during successful fungal attack provides compelling evidence that there is at least some overlap between plant and animal cell death processes. It will be of interest to determine more precisely how these gene products interact with plant cells. These transgenes may serve as tools to dissect cell death pathways in plants. Moreover, these transgenes are viable candidates for engineering resistance into economically important crop plants.
Supplementary Material
Supplemental Figure
Acknowledgments
We thank John Reed, Dave Dunigan, Drake Stenger, Jim Alfano, and Les Lane for helpful discussions and comments on the manuscript. Robert Butchko is acknowledged for the supplemental data. John Reed, Bob Horvitz, and Brad Hersh are gratefully acknowledged for providing antibodies. This research was supported partially by the University of Nebraska Program in Comparative Pathobiology.
Abbreviations
PCD
programmed cell death
IAP
inhibitor of apoptosis
TSWV
tomato spotted wilt virus
References
- 1.Bergey D R, Howe G A, Ryan C A. Proc Natl Acad Sci USA. 1996;93:12053–12058. doi: 10.1073/pnas.93.22.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitham S, Dinesh-Kumar S D, Choi D, Hehl R, Corr C, Baker B. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- 3.Dwyer S C, Leoendre L, Low P S, Leto T L. Biochem Biophys Acta. 1996;1289:231–237. doi: 10.1016/0304-4165(95)00156-5. [DOI] [PubMed] [Google Scholar]
- 4.Lockshin R A, Zakeri Z, Tilly J L. When Cells Die. New York: Wiley–Liss; 1998. [Google Scholar]
- 5.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 6.Williams G T, Smith C A. Cell. 1993;74:777–779. doi: 10.1016/0092-8674(93)90457-2. [DOI] [PubMed] [Google Scholar]
- 7.Doke N, Chai H B, Kowaguchi A. In: Molecular Determinants of Plant Diseases. Nishimura S, Vance C P, Doke N, editors. Berlin: Springer; 1987. pp. 235–252. [Google Scholar]
- 8.Morel J B, Dangl J L. Cell Death Differ. 1997;4:671–683. doi: 10.1038/sj.cdd.4400309. [DOI] [PubMed] [Google Scholar]
- 9.Heath M. Eur J Plant Pathol. 1998;104:117–124. [Google Scholar]
- 10.Ryerson D E, Heath M C. Plant Cell. 1996;8:393–402. doi: 10.1105/tpc.8.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H, Li J, Bostock R, Gilchrist D G. Plant Cell. 1996;8:375–491. doi: 10.1105/tpc.8.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarre D A, Wolpert T J. Plant Cell. 1999;11:237–249. doi: 10.1105/tpc.11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu P, Roossinck M J. Plant Cell. 2000;12:1079–1092. doi: 10.1105/tpc.12.7.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis R E, Yuan J, Horvitz H R. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 15.Hengartner M O, Horvitz H R. Philos Trans R Soc London B. 1994;345:243–246. doi: 10.1098/rstb.1994.0100. [DOI] [PubMed] [Google Scholar]
- 16.Vaux D L, Weissman I L, Kim S K. Science. 1992;258:1955–1957. doi: 10.1126/science.1470921. [DOI] [PubMed] [Google Scholar]
- 17.Deveraux Q L, Reed J C. Genes Dev. 1999;13:239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 18.Reed J C. Oncogene. 1998;17:3225–3236. doi: 10.1038/sj.onc.1202591. [DOI] [PubMed] [Google Scholar]
- 19.Yang E, Korsmeyer S J. Blood. 1996;88:386–401. [PubMed] [Google Scholar]
- 20.Boise L H, Garcia M G, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 21.Crook N E, Clem R J, Miller L K. J Virol. 1993;67:2168–2174. doi: 10.1128/jvi.67.4.2168-2174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.An G. Plant Physiol. 1986;81:86–91. doi: 10.1104/pp.81.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carrington J C, Freed D D. J Virol. 1990;64:1590–1597. doi: 10.1128/jvi.64.4.1590-1597.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hajdukiewicz P, Suab Z, Maliga P. Plant Mol Biol. 1994;25:989–994. doi: 10.1007/BF00014672. [DOI] [PubMed] [Google Scholar]
- 25.Koncz C, Schell J. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- 26.Ditta G, Stanfield S, Corbin D, Helinski D R. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horsch R B, Fry J E, Hoffman N L, Eichholtz D, Rogers S G, Fraley R T. Science. 1985;227:1229–1231. [Google Scholar]
- 28.White J L, Kaper J M. J Virol Methods. 1989;23:83–94. doi: 10.1016/0166-0934(89)90122-5. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–162. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Krajewski S, Krajewski M, Shabaik A, Miyashita T, Wang H G, Reed J C. Am J Pathol. 1994;145:4701–4714. [PMC free article] [PubMed] [Google Scholar]
- 33.Dickman M B, Mitra A. Physiol Mol Plant Pathol. 1992;41:255–263. [Google Scholar]
- 34.Tuite J. Plant Pathological Methods. Minneapolis: Burgess; 1969. [Google Scholar]
- 35.Ie T S. C.M.I. Descriptions of Plant Viruses No. 39. Kew, U.K.: Commonwealth Mycological Institute; 1970. [Google Scholar]
- 36.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 37.Mitsuhara I, Malik K A, Miura M, Ohashi Y. Curr Biol. 1999;9:775–778. doi: 10.1016/s0960-9822(99)80341-8. [DOI] [PubMed] [Google Scholar]
- 38.Stone J M, Heard J E, Asai T, Ausubel F M. Plant Cell. 2000;12:1811–1822. doi: 10.1105/tpc.12.10.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asai T, Stone J M, Heard J E, Kovtin Y, Yorgey P, Sheem J, Ausubel F M. Plant Cell. 2000;12:1823–1835. doi: 10.1105/tpc.12.10.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zha H, Fisk H A, Haffe M P, Mahajan N, Herman B, Reed J C. Mol Cell Biol. 1996;16:6494–6508. doi: 10.1128/mcb.16.11.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jürgensmeier J M, Krajewski S, Armstrong R C, Wilson G M, Oltersdorf T, Fritz L C, Reed J C, Ottlie S. Mol Biol Cell. 1997;8:325–339. doi: 10.1091/mbc.8.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lacomme C, Santa Cruz S. Proc Natl Acad Sci USA. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Pozo O, Lam E. Curr Biol. 1998;8:1129–1132. doi: 10.1016/s0960-9822(98)70469-5. [DOI] [PubMed] [Google Scholar]
- 44.Drake R, John I, Farrell A, Cooper W, Schuch W, Grierson D. Plant Mol Biol. 1996;30:755–767. doi: 10.1007/BF00019009. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y, Hanson M R. Plant Physiol. 2000;122:1323–1333. doi: 10.1104/pp.122.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minami A, Fuuda H. Plant Cell Physiol. 1995;36:1599–1606. [PubMed] [Google Scholar]
- 47.Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A. Plant Cell. 1999;11:431–443. doi: 10.1105/tpc.11.3.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daub M E. Phytopathology. 1982;72:370–374. [Google Scholar]
- 49.Hockenbery D M, Oltvai Z N, Yin X M, Milliman C L, Korsmeyer S J. Cell. 1993;75:241–251. doi: 10.1016/0092-8674(93)80066-n. [DOI] [PubMed] [Google Scholar]
- 50.Fabisiak J P, Kagan V E, Ritou V B, Johnson D E, Lazo J S. Am J Physiol. 1997;272:675–684. doi: 10.1152/ajpcell.1997.272.2.C675. [DOI] [PubMed] [Google Scholar]
- 51.Bolwell G P. Curr Opin Plant Biol. 1999;2:293–294. doi: 10.1016/S1369-5266(99)80051-X. [DOI] [PubMed] [Google Scholar]
- 52.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 53.Uren A G, Beilharz T, O'Connell M J, Bugg S J, Van Driel R, Vaux D L, Lithgow T. Proc Natl Acad Sci USA. 1999;96:10170–10175. doi: 10.1073/pnas.96.18.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraser A G, James C, Evan G I, Hengartner M O. Curr Biol. 1999;9:292–301. doi: 10.1016/s0960-9822(99)80137-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure