Mutations in the mechanotransduction protein PIEZO1 are associated with hereditary xerocytosis (original) (raw)
Abstract
Hereditary xerocytosis (HX, MIM 194380) is an autosomal dominant hemolytic anemia characterized by primary erythrocyte dehydration. Copy number analyses, linkage studies, and exome sequencing were used to identify novel mutations affecting PIEZO1, encoded by the FAM38A gene, in 2 multigenerational HX kindreds. Segregation analyses confirmed transmission of the PIEZO1 mutations and cosegregation with the disease phenotype in all affected persons in both kindreds. All patients were heterozygous for FAM38A mutations, except for 3 patients predicted to be homozygous by clinical and physiologic studies who were also homozygous at the DNA level. The FAM38A mutations were both in residues highly conserved across species and within members of the Piezo family of proteins. PIEZO proteins are the recently identified pore-forming subunits of channels that mediate mechanotransduction in mammalian cells. FAM38A transcripts were identified in human erythroid cell mRNA, and discovery proteomics identified PIEZO1 peptides in human erythrocyte membranes. These findings, the first report of mutation in a mammalian mechanosensory transduction channel-associated with genetic disease, suggest that PIEZO proteins play an important role in maintaining erythrocyte volume homeostasis.
Introduction
Hereditary xerocytosis (also known as HX or dehydrated stomatocytosis, DHSt; OMIM 194380) is an autosomal dominant hemolytic anemia characterized by primary erythrocyte dehydration.1 HX erythrocytes exhibit decreased total cation and potassium content that are not accompanied by a proportional net gain of sodium and water. HX patients typically exhibit mild to moderate, compensated hemolytic anemia. Erythrocyte mean corpuscular hemoglobin concentration is increased and erythrocyte osmotic fragility is decreased, both reflecting cellular dehydration.
A locus for HX on chromosome 16 (16q23-q24) was first identified in a large, 3-generation Irish family.2 This locus was refined to _D16S511_-16qter via study of 10 kindreds with variants of HX, pseudohyperkalemia, or nonimmune hydrops fetalis.3,4 Recent studies of one of the original HX families from Rochester, NY,5 and of a large HX family from Manitoba, Canada6 confirmed the linkage of the disease phenotype to chromosome 16q, and refined the candidate region to 16q24.2-16qter, a 2.4 million-bp interval containing 51 known or predicted genes.6 To identify the HX-associated genetic locus, we performed high-resolution single nucleotide polymorphism (SNP) typing and whole-exome sequencing on selected persons from both the New York and Canadian HX kindreds.
In the refined candidate region, no regions of copy number variation were detected at 16q24.2-16qter. A large region of homozygosity was detected in this region in DNA from a presumed homozygote from the New York kindred. Exome sequencing identified novel mutations affecting PIEZO1 (encoded by the FAM38A gene) in both HX kindreds. Segregation analyses confirmed transmission of the PIEZO1 mutations and cosegregation with the disease phenotype in all affected persons in both the New York and Canadian kindreds. All patients were heterozygous for the mutations, except for 3 New York patients predicted to be homozygous for HX by clinical and physiologic studies who were also homozygous at the DNA level. The FAM38A mutations were both in residues highly conserved across species and within members of the Piezo family of proteins.
PIEZO1 and PIEZO2 have recently been identified as proteins involved in mechanosensation and stretch-activated cation channel activation.7 The ability of a cell to sense mechanical signals is a fundamental characteristic of all or almost all cells, and this process has been linked to ion channels, specifically stretch-activated cation channels.8 Although the molecular identify of these channels has been well characterized in bacteria for many years, in higher organisms, the molecular identity of mechanosensory ion channels has until recently been unknown.9
The identification of PIEZO1 mutations associated with HX is the first report of human disease associated with mutation in a mammalian mechanosensory transduction ion channel. PIEZO proteins may play an important role in maintaining erythrocyte volume homeostasis.
Methods
Patients
Family A: New York pedigree.
Miller et al from Rochester, NY described a Swiss-German pedigree with dominantly inherited congenital hemolytic anemia associated with stomatocytosis, reticulocytosis, and shortened erythrocyte survival.5 Intracellular potassium and total monovalent cations were mildly decreased, and erythrocyte osmotic fragility was decreased. The pedigree's lineage was traced to the mid-18th century. In one loop of the pedigree, paternal and maternal great-great-grandmothers were sisters. This consanguineous mating produced 3 children predicted to be HX homozygotes, based on clinical and laboratory data. This research was approved by the Institutional Review Board at Yale University School of Medicine.
Family B: Canadian pedigree.
Houston et al from Winnipeg, MB described a large Canadian kindred with typical features of HX, including well-compensated hemolysis associated with elevated mean corpuscular hemoglobin concentration and decreased erythrocyte osmotic fragility.6
Copy number analyses
DNA from 1 New York heterozygote (M1-24), 1 predicted New York homozygote (M1-44), and 1 Canadian heterozygote (HA128) was analyzed using the Illumina Human660W-Quad SNP genotyping array. Genotyping was performed at the Yale Center for Genome Analysis using standard Illumina protocols. The HG18 version of the Log R Ratio and B Allele Frequency values for each sample was exported from the Illumina GenomeStudio Version 2010.1 software. The PennCNV software (Version 2011-05-03) was used to detect copy number changes using the B Allele Frequency and Log R Ratio data.10 Copy number change regions were intersected with RefSeq exonic regions using the BEDtools 2.11.2 software to identify genes that had copy number changes.11
Targeted exon capture
Targeted regions of genomic DNA were captured using a NimbleGen SeqCap EZ Exome Version 2.0 solution-based capture system according to the manufacturer's protocol. The design targets 329 028 coding exons from the CCDS database (September 2009) and the RefSeq database (January 2010), as well as 710 microRNAs from miRBase (Version 14, September 2009). Probes were designed against the February 2009 hg19 human genome sequence with 2.1 million probes capturing approximately 44.1 MB of targeted sequence.
Exome sequencing
Cluster generation was performed on the Illumina Cluster Station. The captured, purified, and clonally amplified libraries targeting the exomes from the HX family members were sequenced on a HiSeq 2000 with paired-end sequencing at a read length of 75 bp. The Illumina Version 1.7.48 Pipeline was used for image analysis and base calling.
Exome sequence analysis
Fastq sequence reads were aligned to the human genome (hg19build37) using BWA 0.5.9 software.12 Variant analysis was performed using GATK analysis Version 1.0.5336 pipeline13,14 as follows: Duplicate reads were identified using the Picard Version 1.41 MarkDuplicates software (http://picard.sourceforge.net) and were not used for variant identification.12 Sequencing reads in the region of insertion/deletions and clustered SNPs in dbSNP (build 130) and the 1000 genomes (August 4, 2010 release) databases were realigned using the GATK Version 1.1-35 implementation of the Smith-Waterman alignment algorithm to reduce false positives.13
Base alignment quality scores were determined and used to further reduce false-positive calls.15 Individual base quality scores were recalibrated by the GATK software using dbSNP (build 130) and 629 complete human genome sequences from the 1000 genomes project (August 2010 release)16,17 and machine cycle and dinucleotide covariate. SNPs were called using the GATK Bayesian algorithm for variant discovery and genotyping, which uses base qualities and allele counts to determine probabilities for called variants. Indels were called using the GATK Unified Genotyper implementation of Dindel.18 Indels and novel SNPs were annotated using the SeattleSeq database (http://snp.gs.washington.edu/SeattleSeqAnnotation131/). The SeattleSeq analysis determines whether an SNP or indel changes a protein sequence or splice site, whether the nucleotide is conserved,19,20 and provides a Grantham score and polyphen classification to assess the likely effect of a mutation on protein function.21,22
mRNA expression analyses
Human CD34-selected stem and progenitor cells were cultured and selected to yield a population of erythroid cells (R3/R4) as previously described.23 RNA was prepared using the RNeasy kit (QIAGEN) according to the manufacturer's instructions. RNA was treated with DNase I and reverse-transcribed with oligo(dT) primer using a SuperScript first-strand synthesis system (Invitrogen). Reverse transcription products were amplified by real-time PCR with primer pairs tiled across the FAM38A locus (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Sanger sequencing
Sanger sequencing was performed on an Applied Biosystems 3130XL capillary sequencer. Primers for amplification of genomic DNA are listed in supplemental Table 2.
Erythrocyte membrane isolation and digestion
Erythrocyte membrane ghosts were prepared as described.24 Protein from 100 μL enriched RBC membrane fraction was precipitated with the methanol/chloroform method as described by Wessel and Flugge.25 The protein pellet was dissolved in 30 μL 8M urea, 0.4M Tris-HCl buffer, pH 8.0 (23°C) by vortex, and an aliquot was used for protein quantitation by UV-VIS spectrophotometry at 280 nm on a NanoDrop (Thermo Fisher Scientific). Cysteines were reduced with 4.1mM DTT (10 minutes at 60°C), and the reaction was quenched on ice. Carbamidomethylation of cysteines was carried out for 30 minutes at room temperature in the dark using 8.3mM indole-3-acetic acid IAA. Excess IAA was quenched with DTT and the reaction diluted to ∼ 1M urea with 200 μL 90mM Tris-HCl buffer, pH 8.0 (23°C) with 2mM CaCl2. Digestion with trypsin was carried out at 37°C for 15 hours using a trypsin/protein ratio of 1:15. The reaction was quenched with 12 μL 20% trifluoroacetic acid. Peptides were desalted by reversed-phase chromatography using C18 UltraMicroSpin columns. Peptides were dried in a vacuum centrifuge, dissolved in 20 μL 3:8 volume/volume 70% FA/0.1% trifluoroacetic acid and quantified by UV-Vis on the NanoDrop. The peptide concentration was adjusted to 0.6 μg/μL for liquid chromatography-mass spectrometry (LC-MS).
Mass spectrometry
LC-MS was performed on a triple TOF 5600 (AB Sciex) with nanospray source connected to a nanoAcquity UPLC (Waters). The mobile phases for LC separation consisted of water (A) and acetonitrile (B) containing 0.1% of FA, respectively. A total of 3 μg of peptides was injected onto a Waters Symmetry C18, 5-μm particle size, 180 μm ID × 20 mm nano Acquity UPLC trap column, which was connected to a Waters BEH 30 C18, 1.7-μm particle size, 75-μm ID × 150 mm capillary column operated at 45°C. Peptides were trapped for 3 minutes at 1% B with a flow rate of 5 μL/min. Gradient elution was carried out using a flow rate of 0.5 μL/min with a 2-step gradient from 5% to 40% B in 160 minutes and 40% to 85% B in 3.3 minutes, respectively.
The mass spectrometer was operated in information-dependent aquisition mode with a single 250-ms high-resolution time-of-flight MS survey scan (m/z = 400-1250) followed by up to 20 MS/MS scans (m/z = 100-1500) at more than 15 000 resolution and 50-ms accumulation time. Precursor ions exceeding 125 counts were fragmented with collision energy of 37 eV ± 15 eV using N2 as the collision gas. Fragmented peptides were set on an exclusion list for 10 seconds.
Protein database searching
All tandem MS spectra were searched in-house using MASCOT Version 2.2.0 against the human SwissProt database release February 2011. Search criteria were: Enzyme:Trypsin, allowing 1 missed cleavage. The precursor ion mass tolerance was set to 20 ppm for the precursor and 0.2 Da for fragment ions, respectively. Fixed modifications were carbamidomethyl at cysteine, and the variable modification was oxidation of M. The overall false discovery rate estimated by MASCOT was better than 3.5%.
Results
Copy number analyses
To interrogate the 16q24.2-16qter candidate region for variations in copy number, high-resolution SNP typing with genomic DNA from affected persons of both the New York and Canadian HX kindreds was performed using Illumina Human660W-Quad BeadChip arrays. This array contains 657 366 markers and includes 3512 markers in the 16q24-16qter HX locus. No regions of copy number variation were detected in the 16q24.2-16qter candidate region (data not shown).
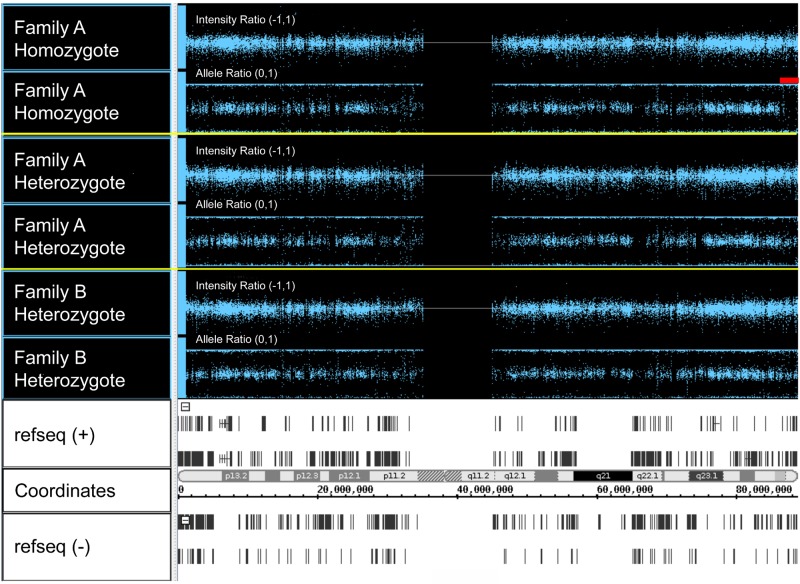
In one loop of the New York pedigree, the parents were unknowingly distantly related, and clinical and laboratory data predicted that 3 of their children were HX homozygotes (see “Mutation detection”). We analyzed the genotyping data in the 16q24.2-16qter candidate region in one of these children. This patient (III-15; UZ1-34) has an extended block of homozygosity on chromosome 16 starting after 87445363 (last heterozygous SNP), before 87445839 (first homozygous SNP) and extending to 90109711, the last called SNP at the telomeric end of chromosome 16 (Figure 1). This region contains 100 homozygous SNPs. There are 3 possible heterozygous SNPs at the very end of 16q that could be miscalls (2 SNPs are in 1000 genomes) or not a variant at all. No other extended regions of homozygosity are present on chromosome 16.
Figure 1.
Copy number analyses via single nucleotide polymorphism microarray. Whole-genome SNP typing was performed using DNA from the patients indicated and the Illumina HumanHap 550 Bead Chip array. The figure shows SNPs from 3 patients on chromosome 16. For each patient, the top panel represents the intensity ratio, and the bottom panel represents the allele ratio. Intensity ratio is a measure of patient SNP log signal intensity compared with reference sample SNP intensity; 2 copies = intensity ratio of 0; 1 copy = intensity ratio of −1. Most values are centered around zero, indicating diploid copy number. Allele ratio is the ratio of the signal for one of 2 genotypes over the total signal. Homozygous SNPs localize to 0 or 1; heterozygous SNPs localize to 0.5. In Family A homozygote III-15, genotyping data in the 16q24.2-16qter candidate region demonstrate an extended block of homozygosity extending to the telomeric end of chromosome 16 (red bar). Note that the intensity ratio in this region is unchanged, indicating the homozygosity is not caused by a large deletion. The bottom panel shows, from top to bottom, (+) strand genes, cytogenetic bands, chromosome position, and (−) strand genes.
Exome sequencing and novel variant identification
Whole-exome sequencing was performed using Nimblegen solution-based exon capture and high throughput sequencing. Paired-end DNA sequencing data were aligned to the February 2009 GRCh37 hg19 version of the human genome sequence. High mean coverage of more than 65 was achieved and more than 92.1% of all targeted bases were read more than 10 times for all the samples, respectively, more than sufficient to determine genotypes in the 16q24-16qter region and to identify novel variants in HX family members with high specificity (supplemental Table 3).
Genotypes for single nucleotide and indel variants in the exome targeted regions were called for HX and normal samples using the GATK Unified Genotyper. Variants with quality score less than 50 and that were in dbSNP were filtered out, and the remaining variants were submitted to the SeattleSeq annotation pipeline. Any variant that did not change protein sequences or mRNA splicing was removed.
Candidate gene identification
Assuming a model with dominant inheritance, we searched the exome data for variants in the 16q24.2-16qter candidate region present in the New York HX patients, but not in control unaffected samples. Two genes (FAM38A and TPSAB1) were identified in the candidate region.
We repeated the same analyses using a subset of patients from the Canadian HX family. Five genes (FAM38A, ACSM2A, E4F1 CENPBD1, and C16orf55) were identified in the candidate region. Thus, novel, nonsynonymous variants in the FAM38A gene, which encodes the protein PIEZO1, were present in both kindreds.
Mutation detection
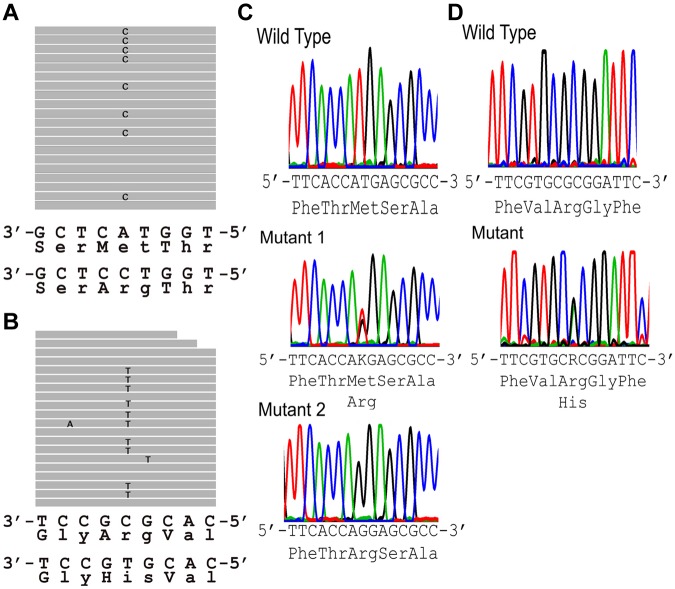
Sequence analysis of the FAM38A gene in DNA from the New York HX patients revealed abnormal DNA sequences in the alignment of multiple short sequencing reads in exon 46 (Figure 2A). The corresponding DNA sequence revealed a heterozygous T to G substitution (nucleotide position chromosome 16:88 783 293, hg19) changing a Met (ATG) to Arg (AGG) at amino acid 2225. Sequence analysis of the FAM38A gene in DNA from the Canadian HX patients revealed abnormal DNA sequences in the alignment of multiple short sequencing reads in exon 51 (Figure 2A). The corresponding DNA sequence revealed a heterozygous G to A substitution (nucleotide position chromosome 16:88 782 212, hg19) changing an Arg (CGC) to His (CAC) at amino acid 2456. These 2 variants were not found in dbSNP Version 135, which contains phase 1 of the 1000 genomes project, and they were not found in approximately 3200 alleles of the National Heart, Lung, and Blood Institute exome sequencing project.
Figure 2.
Exome sequencing. (A-B) Sequence tags. (A) New York HX patient II-10 (exon 46). Eight of 19 full-length reads have a C instead of an A (G replacing T on opposite strand), leading to a missense mutation, Met to Arg at amino acid 2225. (B) Canadian HX patient IV-4 (exon 51). Ten of 20 full-length reads have a T instead of a C (A replacing G on opposite strand), leading to a missense mutation, Arg to His at amino acid 2456. (C-D) Sanger sequencing confirmation of FAM38A gene mutations. (C) New York kindred. Top: Partial exon 46 wild-type sequence. Middle: corresponding sequence from a New York family HX heterozygote. Bottom: corresponding from a New York family HX homozygote. (D) Canadian kindred. Top: partial exon 51 wild-type sequence. Bottom: corresponding sequence from a Canadian HX heterozygote. The .bam file yielding the sequence traces from exome sequencing in panels A and B places the FAM38A gene in reverse complement orientation. The letters K and R are designations in the IUPAC code where K represents the nucleotides G or T and R represents the nucleotides A or G.
No other deleterious variants were detected in the coding regions of the FAM38A gene. The mutations in exons 46 and 51 of the FAM38A gene identified by exome sequencing were validated by conventional Sanger sequencing (Figure 2B).
No mutations were identified in the SLC4A1 gene locus, previously associated with a subset of HX patients.
Inheritance and mutation analyses
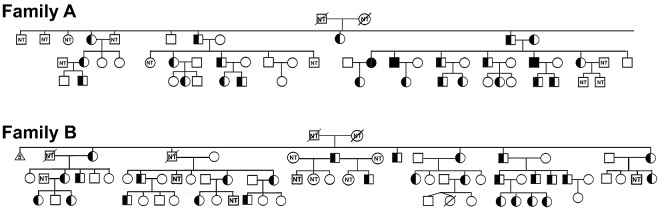
Segregation analyses confirmed transmission of the FAM38A mutations and cosegregation with the disease phenotype in affected persons in both the New York and Canadian kindreds (Figure 3). The mutations were absent in non-HX family members. Heterozygous persons exhibit well-compensated hemolysis, elevated mean corpuscular hemoglobin concentration, and decreased osmotic fragility.5,6 In one loop of the New York family, the paternal and maternal great-great-grandmothers were sisters, and 3 offspring of this consanguineous mating were identified as homozygotes based on clinical and laboratory data, including severe hemolytic anemia with reticulocytosis, shortened red blood cell survival, stomatocytosis, hyperbilirubinemia, and markedly altered erythrocyte sodium and potassium levels.5 DNA analyses confirmed all 3 patients were indeed homozygous for the M2225R mutation.
Figure 3.
Cosegregation of FAM38A gene mutations with HX phenotype. The FAM38A gene mutations detected by exome sequencing in affected HX persons cosegregated with disease phenotype in all affected persons in both kindreds. Family A indicates New York pedigree. As predicted by clinical, genetic, and biochemical studies, the parents and affected HX persons are heterozygous for the FAM38A mutation and persons III-14, III-15, and III-21 are homozygous for the FAM38A mutation. Family B indicates Canadian pedigree. Inheritance of the FAM38A mutation is heterozygous. “NT” in the symbols indicates persons who were “not tested” and clinical, laboratory, biochemical, and genetic data are not available.
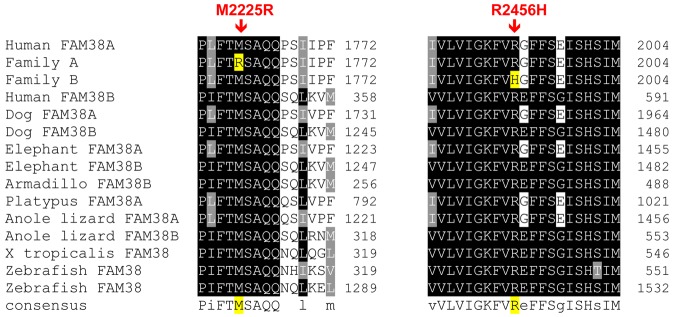
The missense mutations were both predicted to be pathogenic by PolyPhen, a tool that predicts possible impact of an amino acid substitution on the structure and function of human proteins using physical and comparative considerations. The mutations were both in conserved resides, with highly significant phastCons scores, M2225R 1.0 and R2456H 0.812, and highly significant conservation scores, M2225R 4.34 and R2456H 3.43. These amino acids are highly conserved across vertebrate species, including the clades of placental mammals, the extant Eutherians, and within members of the PIEZO family of proteins (Figure 4).
Figure 4.
Conservation of mutations across vertebrate species. The mutant amino acid residues identified in HX patients are conserved across vertebrate species, including the clades of placental mammals, the extant Eutherians, and within members of the Piezo family of proteins.
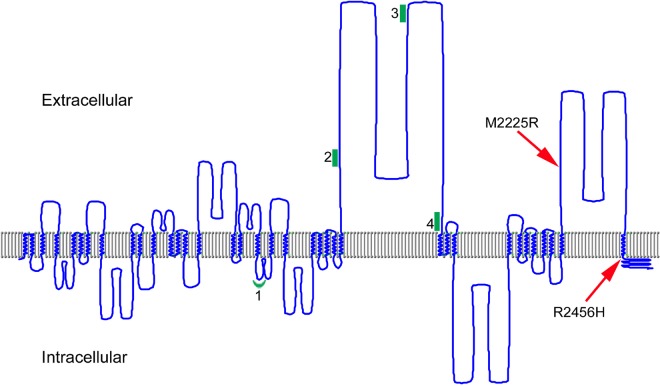
The amino acids affected by the FAM38A missense mutations identified in the HX kindreds are both located in the COOH-terminal region of PIEZO1 (Figure 5).
Figure 5.
A model of human PIEZO1. The predicted transmembrane regions of PIEZO1 (UniProt accession Q92508) were displayed using TMRPres2D software.26 UniProt predictions are based on the regions that have α-helical transmembrane potential in at least 2 of the predictive tools, TMHMM, Memsat, Phobius, and the hydrophobic moment plot method of Eisenberg et al.28 Arrows indicate the location of HX-associated mutations; red line, the location of PIEZO1 peptides identified in erythrocyte membranes; and green, the locations of peptides identified in discovery proteomics of erythrocyte membrane ghosts.
PIEZO1 mRNA expression in erythroid cells
There are limited data regarding expression of PIEZO1 in erythroid cells. We sought to ensure that both the FAM38A gene and its protein product were expressed in erythroid cells. mRNA was prepared from cultured, primary human erythroid progenitor cells, cDNA prepared by reverse transcription, and then amplified with 4 sets of primers tiled across the FAM38A locus (supplemental Figure 1A).
These cDNA transcripts were amplified (supplemental Figure 1B), subcloned, and sequenced. These overlapping cDNA transcripts corresponded to the human full-length FAM38A cDNA (GI: 257196141), which is homologous to the murine Fam38A transcript of Piezo1 associated with induction of mechanically activated cellular currents.7 Microarray analyses performed by Keller et al demonstrated moderate levels of FAM38A mRNA (GEO accession no. GDS2431)27 in cultured human erythroid progenitor cells differentiated in vitro, confirming our observations.
Unbiased discovery proteomics
Western blot analyses of wild-type R3/R4 cell proteins and human erythrocyte ghosts using 3 different commercial antibodies gave inconsistent and irreproducible results. Thus, we performed a series of mass spectrometry-based, unbiased proteomic analyses of human erythrocyte ghosts to search for PIEZO1-derived peptides. This discovery proteomics strategy yielded 4 peptides corresponding to PIEZO1: GFALYNAANLK (peptide 1), QHQLAPLPAQAVFASGTR (peptide 2), YLLTQELLQGGEVHR (peptide 3), and IPELEEAELFAEGQGR (peptide 4). These peptides were aligned to the predicted human PIEZO1 amino acid sequence. These peptides are spread across the PIEZO1 protein (Figure 5; supplemental Figure 2). The identification of peptide 1 indicates that the long isoform of PIEZO1, similar to murine Piezo1 associated with a unitary conductance approximately 25 pS current in transfected HEK293 cells on membrane deformation,7 is present in human erythrocyte membranes.
Discussion
Mechanotransduction, the conversion of mechanical force into biologic signals, is a fundamental physiologic process of mammalian cells. It influences many critical processes, including embryonic development, tactile, pain, and auditory sensation, regulation of vascular tone, flow sensing in the kidney, and muscle and tendon stretch.8 The primary mechanism for mechanosensation is linked to calcium permeable, stretch-activated cation channels. In bacteria, the molecular identity of mechanosensitive ion channels has been understood at the molecular level for many years. However, in higher organisms, the molecular identity of mechanosensory ion channels has been unknown.9
Recently, PIEZO proteins have been identified as mechanically activated ion channels in eukaryotes.7 Originally identified in the murine neuroblastoma cell line N2A, PIEZO proteins induce mechanically activated cationic currents in various cell types.29 PIEZO-induced currents are blocked by gadolinium, ruthenium red, and the peptide GsMTx4 (Grammostola spatulata mechanotoxin 4),30,31 commonly used to inhibit mechanosensitive currents. Murine Piezo1 assembles into a homo-multimeric complex that is fully active without other proteins. Similar to murine Piezo protein, the single Piezo protein in Drosophila induces mechanically activated cation currents.32
Protein structure prediction programs indicate that PIEZO proteins are very large integral membrane proteins with 24 to 40 transmembrane domains located across the protein (Figure 5).7 What is the role of PIEZO1 in erythroid cells? Does it regulate a stretch-activated calcium pathway? On deformation, calcium enters the erythrocyte through an as yet unidentified pathway.33,34 Recent studies indicate that local deformations in the erythrocyte membrane can transiently activate a calcium-permeability pathway leading to increased intracellular calcium, activation of potassium currents through the calcium-sensitive Gardos channel, and secondary anion currents through 5-nitro-2-(3-phenylpropylamino)benzoic acid and diisothiocyanostilbene-2,2′-disulfonate-inhibitable channels.35 However, this and other previous work provides no evidence for mechanically induced PIEZO-associated currents through a 10- to 25-pS channel in wild-type mature erythrocytes either in whole cell configuration or in single-channel recordings after deformation.35
Identification of this stretch-induced calcium-activated cation pathway and determination of whether PIEZO channels participate is of great physiologic relevance to erythrocyte biology. Circulatory shear stress has been suggested to cause reversible increases in erythrocyte calcium permeability.34,36 Erythrocyte aging has been associated with alterations in membrane permeability for calcium.37–39 Localized membrane deformation has been associated with increased calcium in the process of apical alignment in the initial steps of malaria invasion.38 Finally, in sickle cell disease, deoxygenated, circulating erythrocytes exhibit reversible increases in membrane permeability to cations through an as yet unidentified pathway known as Psickle,40,41 and membrane deformation is necessary for this increase in sickling-induced cation permeability.42 Increased calcium levels mediated by Psickle lead to activation of the Gardos channel, a critical step in the dehydration of sickle erythrocytes.43–46 A recent study shows similarities between erythrocyte nonspecific cation conductance pathways and Psickle and demonstrate sensitivity of the pathway to GsMTx4, indicating properties of a mechanosensitive ion channel.47
PIEZO1 may serve as a sensor for cell swelling, as GsMTx4 blockade inhibits both mechanically activated currents and whole-cell regulatory volume decrease in NRK-49 cells.31,48 Perhaps PIEZO proteins play a previously unrecognized role in erythrocyte volume regulation, with PIEZO1-mutant erythrocytes gradually becoming dehydrated during their repeated cycles of travel through the microcirculation, associated with changes in oxygenation/deoxygenation.
Unraveling the effects of these HX-associated mutations will provide insight into the structure and function of PIEZO1 and will shed additional insight into the pathogenesis of primary and secondary disorders of erythrocyte hydration. The mutations may disrupt critical protein-protein interactions, may influence PIEZO1 function in erythroid cells, or may impair PIEZO1 trafficking to the erythrocyte membrane. Juxtamembrane mutations of highly conserved arginine residues have been shown to disrupt transporter membrane trafficking, such as occurs with anion exchanger 1 in hereditary spherocytosis and distal renal tubular acidosis,49,50 perhaps because of misfolding or mutation of arginine-based endoplasmic reticulum retention/retrieval signals.51
Another major unsolved puzzle in PIEZO1-associated HX is the great phenotypic variability exhibited between patients. Associated clinical syndromes include familial pseudohyperkalemia with or without hematologic abnormalities. Some patients exhibit in utero hydrops fetalis, not associated with fetal anemia that typically resolves spontaneously before or shortly after birth without any subsequent sequelae. Many patients exhibit marked iron overload, suggesting either a pattern of abnormal iron homeostasis or marked dyserythropoiesis.
In conclusion, PIEZO proteins are the first mammalian mechanosensory signal transduction proteins identified to date. When mutated, PIEZO1 is associated with the phenotype of hereditary xerocytosis. Determination of the structure and function of PIEZO proteins will shed light on the erythrocyte hydration pathways mediated by PIEZO proteins in mature erythrocytes.
Supplementary Material
Supplemental Methods, Figures, and Tables
Acknowledgments
The authors thank the patients for their participation and Dr Joseph Hoffman for critical manuscript review.
This work was supported in part by the Doris Duke Foundation (P.G.G.), and the Manitoba Medical Services Foundation (R.Z.). B.L.H. was a recipient of an American Society of Hematology Trainee Award.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.Z., B.L.H., D.S.H., Y.M., J.R., V.P.S., and P.G.G. participated in experimental design, execution, and interpretation; B.S. participated in patient phenotyping; and all authors wrote and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick G. Gallagher, Department of Pediatrics, Yale University School of Medicine, 333 Cedar St, PO Box 208064, New Haven, CT 06520-8064; e-mail: patrick.gallagher@yale.edu.
References
- 1.Bruce LJ. Hereditary stomatocytosis and cation-leaky red cells: recent developments. Blood Cells Mol Dis. 2009;42(3):216–222. doi: 10.1016/j.bcmd.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Carella M, Stewart G, Ajetunmobi JF, et al. Genomewide search for dehydrated hereditary stomatocytosis (hereditary xerocytosis): mapping of locus to chromosome 16 (16q23-qter). Am J Hum Genet. 1998;63(3):810–816. doi: 10.1086/302024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grootenboer S, Schischmanoff PO, Laurendeau I, et al. Pleiotropic syndrome of dehydrated hereditary stomatocytosis, pseudohyperkalemia, and perinatal edema maps to 16q23-q24. Blood. 2000;96(7):2599–2605. [PubMed] [Google Scholar]
- 4.Iolascon A, Stewart GW, Ajetunmobi JF, et al. Familial pseudohyperkalemia maps to the same locus as dehydrated hereditary stomatocytosis (hereditary xerocytosis). Blood. 1999;93(9):3120–3123. [PubMed] [Google Scholar]
- 5.Miller DR, Rickles FR, Lichtman MA, La Celle PL, Bates J, Weed RI. A new variant of hereditary hemolytic anemia with stomatocytosis and erythrocyte cation abnormality. Blood. 1971;38(2):184–204. [PubMed] [Google Scholar]
- 6.Houston BL, Zelinski T, Israels SJ, et al. Refinement of the hereditary xerocytosis locus on chromosome 16q in a large Canadian kindred. Blood Cells Mol Dis. 2011;47(4):226–231. doi: 10.1016/j.bcmd.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Coste B, Mathur J, Schmidt M, et al. Piezo1 and Piezo2 are essential components of distinct mechanically activated cation channels. Science. 2010;330(6000):55–60. doi: 10.1126/science.1193270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nilius B. Pressing and squeezing with Piezos. EMBO Rep. 2010;11(12):902–903. doi: 10.1038/embor.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsunozaki M, Bautista DM. Mammalian somatosensory mechanotransduction. Curr Opin Neurobiol. 2009;19(4):362–369. doi: 10.1016/j.conb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K, Li M, Hadley D, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17(11):1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Depristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H. Improving SNP discovery by base alignment quality. Bioinformatics. 2011;27(8):1157–1158. doi: 10.1093/bioinformatics/btr076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albers CA, Lunter G, Macarthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21(6):961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185(4154):862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- 23.Steiner LA, Maksimova Y, Schulz V, et al. Chromatin architecture and transcription factor binding regulate expression of erythrocyte membrane protein genes. Mol Cell Biol. 2009;29(20):5399–5412. doi: 10.1128/MCB.00777-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesi SL, Knowles WJ, Morrow JS, Bologna M, Marchesi VT. Abnormal spectrin in hereditary elliptocytosis. Blood. 1986;67(1):141–151. [PubMed] [Google Scholar]
- 25.Wessel D, Flugge UI. A method for the quantitative recovery of protein in dilute-solution in the presence of detergents and lipids. Anal Biochem. 1984;138(1):141–143. doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 26.Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics. 2004;20(17):3258–3260. doi: 10.1093/bioinformatics/bth358. [DOI] [PubMed] [Google Scholar]
- 27.Keller MA, Addya S, Vadigepalli R, et al. Transcriptional regulatory network analysis of developing human erythroid progenitors reveals patterns of coregulation and potential transcriptional regulators. Physiol Genom. 2006;28(1):114–128. doi: 10.1152/physiolgenomics.00055.2006. [DOI] [PubMed] [Google Scholar]
- 28.Eisenberg D, Luthy R, Bowie JU. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- 29.Coste B, Xiao B, Santos JS, et al. Piezo proteins are pore-forming subunits of mechanically activated channels. Nature. 2012;483(7388):176–181. doi: 10.1038/nature10812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae C, Sachs F, Gottlieb PA. The mechanosensitive ion channel Piezo1 is inhibited by the peptide GsMTx4. Biochemistry. 2011;50(29):6295–6300. doi: 10.1021/bi200770q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delmas P, Hao J, Rodat-Despoix L. Molecular mechanisms of mechanotransduction in mammalian sensory neurons. Nat Rev. 2011;12(3):139–153. doi: 10.1038/nrn2993. [DOI] [PubMed] [Google Scholar]
- 32.Kim SE, Coste B, Chadha A, Cook B, Patapoutian A. The role of Drosophila Piezo in mechanical nociception. Nature. 2012;483(7388):209–212. doi: 10.1038/nature10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brain MC, Pihl C, Robertson L, Brown CB. Evidence for a mechanosensitive calcium influx into red cells. Blood Cells Mol Dis. 2004;32(3):349–352. doi: 10.1016/j.bcmd.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Johnson RM. Membrane stress increases cation permeability in red cells. Biophys J. 1994;67(5):1876–1881. doi: 10.1016/S0006-3495(94)80669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyrda A, Cytlak U, Ciuraszkiewicz A, et al. Local membrane deformations activate Ca2+-dependent K+ and anionic currents in intact human red blood cells. PLoS One. 2010;5(2):e9447. doi: 10.1371/journal.pone.0009447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larsen FL, Katz S, Roufogalis BD, Brooks DE. Physiological shear stresses enhance the Ca2+ permeability of human erythrocytes. Nature. 1981;294(5842):667–668. doi: 10.1038/294667a0. [DOI] [PubMed] [Google Scholar]
- 37.Lew VL, Daw N, Perdomo D, Etzion Z, Bookchin RM, Tiffert T. Distribution of plasma membrane Ca2+ pump activity in normal human red blood cells. Blood. 2003;102(12):4206–4213. doi: 10.1182/blood-2003-06-1787. [DOI] [PubMed] [Google Scholar]
- 38.Lew VL, Tiffert T. Is invasion efficiency in malaria controlled by pre-invasion events? Trends Parasitol. 2007;23(10):481–484. doi: 10.1016/j.pt.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Romero PJ, Romero EA. Differences in Ca2+ pumping activity between sub-populations of human red cells. Cell Calcium. 1997;21(5):353–358. doi: 10.1016/s0143-4160(97)90028-2. [DOI] [PubMed] [Google Scholar]
- 40.Lew VL, Bookchin RM. Ion transport pathology in the mechanism of sickle cell dehydration. Physiol Rev. 2005;85(1):179–200. doi: 10.1152/physrev.00052.2003. [DOI] [PubMed] [Google Scholar]
- 41.Lew VL, Ortiz OE, Bookchin RM. Stochastic nature and red cell population distribution of the sickling-induced Ca2+ permeability. J Clin Invest. 1997;99(11):2727–2735. doi: 10.1172/JCI119462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohandas N, Rossi ME, Clark MR. Association between morphologic distortion of sickle cells and deoxygenation-induced cation permeability increase. Blood. 1986;68(2):450–454. [PubMed] [Google Scholar]
- 43.Browning JA, Ellory JC, Gibson JS. Pathophysiology of red cell volume. Contrib Nephrol. 2006;152:241–268. doi: 10.1159/000096327. [DOI] [PubMed] [Google Scholar]
- 44.Browning JA, Staines HM, Robinson HC, Powell T, Ellory JC, Gibson JS. The effect of deoxygenation on whole-cell conductance of red blood cells from normal individuals and sickle cell patients. Blood. 2007;109(6):2622–2629. doi: 10.1182/blood-2006-03-001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brugnara C. Sickle cell disease: from membrane pathophysiology to novel therapies for prevention of erythrocyte dehydration. J Pediatr Hematol Oncol. 2003;25(12):927–933. doi: 10.1097/00043426-200312000-00004. [DOI] [PubMed] [Google Scholar]
- 46.Joiner CH. Gardos pathway to sickle cell therapies? Blood. 2008;111(8):3918–3919. doi: 10.1182/blood-2008-01-135350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma YL, Rees DC, Gibson JS, Ellory JC. The conductance of red blood cells from sickle cell patients: ion selectivity and inhibitors. J Physiol. 2012;590(Pt 9):2095–2105. doi: 10.1113/jphysiol.2012.229609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua SZ, Gottlieb PA, Heo J, Sachs F. A mechanosensitive ion channel regulating cell volume. Am J Physiol Cell Physiol. 2010;298(6):C1424–C1430. doi: 10.1152/ajpcell.00503.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quilty JA, Li J, Reithmeier RA. Impaired trafficking of distal renal tubular acidosis mutants of the human kidney anion exchanger kAE1. Am J Physiol Renal Physiol. 2002;282(5):F810–F820. doi: 10.1152/ajprenal.00216.2001. [DOI] [PubMed] [Google Scholar]
- 50.Quilty JA, Reithmeier RA. Trafficking and folding defects in hereditary spherocytosis mutants of the human red cell anion exchanger. Traffic. 2000;1(12):987–998. doi: 10.1034/j.1600-0854.2000.011208.x. [DOI] [PubMed] [Google Scholar]
- 51.Michelsen K, Yuan H, Schwappach B. Hide and run: arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 2005;6(8):717–722. doi: 10.1038/sj.embor.7400480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Methods, Figures, and Tables