PML promotes MHC class II gene expression by stabilizing the class II transactivator (original) (raw)
Promyelocytic leukemia (PML) protein binds to and stabilizes CIITA at PML nuclear bodies, which promotes expression of the MHC class II gene locus in response to interferon-γ exposure.
Abstract
Promyelocytic leukemia (PML) nuclear bodies selectively associate with transcriptionally active genomic regions, including the gene-rich major histocompatibility (MHC) locus. In this paper, we have explored potential links between PML and interferon (IFN)-γ–induced MHC class II expression. IFN-γ induced a substantial increase in the spatial proximity between PML bodies and the MHC class II gene cluster in different human cell types. Knockdown experiments show that PML is required for efficient IFN-γ–induced MHC II gene transcription through regulation of the class II transactivator (CIITA). PML mediates this function through protection of CIITA from proteasomal degradation. We also show that PML isoform II specifically forms a stable complex with CIITA at PML bodies. These observations establish PML as a coregulator of IFN-γ–induced MHC class II expression.
Introduction
Promyelocytic leukemia (PML) nuclear bodies, also known as the PML oncogenic domain or ND 10 (nuclear domain 10), are multiprotein subnuclear structures that were first observed 50 yr ago by electron microscopy as nuclear-dense granular bodies (de Thé et al., 1960). PML bodies are present in cell nuclei of almost all human tissues (Gambacorta et al., 1996; Cho et al., 1998). Usually, nuclei of cultured cells contain ∼5–30 PML bodies, ranging in size from ∼0.2 to 1 µm (Ascoli and Maul, 1991). It has been suggested that PML bodies act as dynamic nuclear organizing centers for the coordinated regulation of different cellular functions, including DNA damage response, apoptosis, senescence, and antiviral responses (Sternsdorf et al., 1997; Negorev and Maul, 2001; Bernardi and Pandolfi, 2007). This variety of functions may be attributable to regulated traffic at PML bodies of a selected number of nuclear proteins, which play key roles in these processes (Boisvert et al., 2001; Wiesmeijer et al., 2002; Weidtkamp-Peters et al., 2008; Brand et al., 2010). Six nuclear PML isoforms sharing the same N terminus but containing variable C termini generated by alternative splicing have been identified (Fagioli et al., 1992; Jensen et al., 2001) and are present at all PML bodies (Condemine et al., 2006).
Because components of the ubiquitin–proteasome and SUMO pathways accumulate in or adjacent to a subset of PML bodies, they have also been proposed as sites of specific protein turnover within the nucleus (Fabunmi et al., 2001; Lallemand-Breitenbach et al., 2001; Rockel et al., 2005; Saitoh et al., 2006; Scharf et al., 2007; Chen et al., 2008; Sharma et al., 2010). The expression of the PML gene and other genes encoding PML body components is strongly increased by interferons (IFNs) in vitro and in vivo. Therefore, PML isoforms and PML bodies have also been implicated in IFN response and immune surveillance pathways, including antiviral responses and inflammatory processes (Terris et al., 1995; Tavalai and Stamminger, 2008).
According to a recently curated PML body interactome, 166 different proteins can be physically and/or functionally linked to PML. Strikingly, one half of these factors are involved in transcriptional regulation (Van Damme et al., 2010). Many transcription factors transiently associate with PML bodies, in which their activity is modulated by posttranslational modifications (PTMs). This may result in either activation or repression of specific genes, depending on the context (Zhong et al., 2000). In addition, sites of nascent mRNA transcription and transcriptionally active specific genomic loci were found to be associated with the periphery of PML bodies (Grande et al., 1996; Boisvert et al., 2000; von Mikecz et al., 2000; Fuchsová et al., 2002; Kießlich et al., 2002, Wang et al., 2004; Xie and Pombo, 2006).
Most cell types do not express major histocompatibility (MHC) II genes constitutively, but their expression can be stimulated by IFN-γ (Collins et al., 1984). The MHC II expression pattern is controlled primarily by transcriptional regulation at the conserved MHC II gene promoters. The MHC II enhanceosome contains the constitutively expressed transcription factors regulatory factor X (RFX), nuclear factor-Y, and cAMP response element binding protein (Choi et al., 2011). MHC II gene expression is completely dependent on the expression of the MHC class II transactivator (CIITA), which is recruited in a coactivator-like fashion to the MHC II promoters through protein–protein interactions with RFX, cAMP response element binding protein, and nuclear factor-Y (Steimle et al., 1993; Reith et al., 2005). CIITA expression is induced by IFN-γ through the Janus kinase (JAK)–signal transducer and activator of transcription (STAT) pathway, and expression of exogenous CIITA is sufficient to induce MHC class II expression in many different cell types (Steimle et al., 1994). Shiels et al. (2001) and Wang et al. (2004) have previously demonstrated a highly nonrandom spatial association of the MHC locus with PML bodies, raising the hypothesis that PML (bodies) may be directly involved in transcriptional regulation of selectively associated genes. Subsequently, Gialitakis et al. (2010) have found that the MHC II gene DRA relocates to PML bodies upon induction with IFN-γ and that PML protein isoform IV may function to maintain a sustained transcription-permissive epigenetic state of the DRA gene. Here, we analyze potential functional relationships between PML (nuclear bodies) and CIITA-dependent MHC class II gene expression.
Results
IFN-γ increases the spatial proximity between PML bodies and the MHC II gene cluster
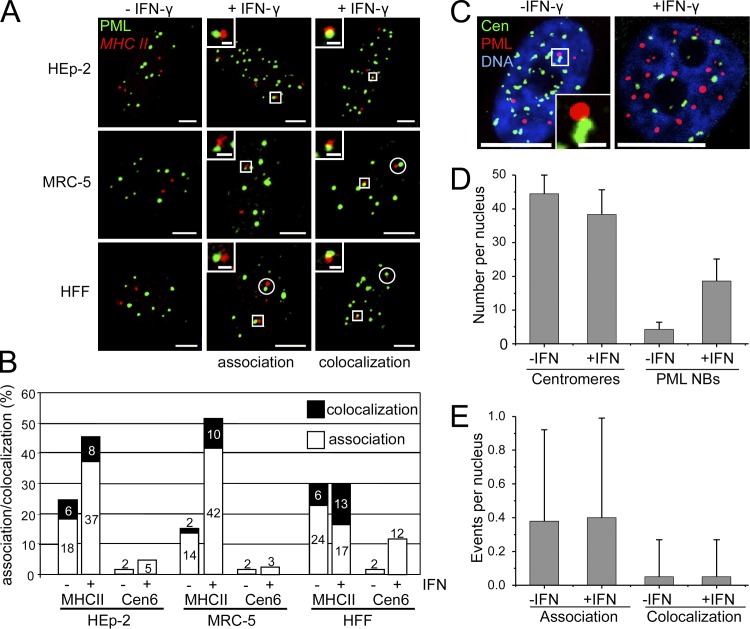
IFN-γ induces a stable relocation of the human leukocyte antigen (HLA) class II DRA gene to PML bodies in HeLa cells (Gialitakis et al., 2010). Here, we confirm and extend these observations for HEp-2 cells, the primary human lung fibroblast line MRC-5, and primary human foreskin fibroblasts (HFFs) using a MHC II–specific FISH probe (Fig. 1 A). To quantify the spatial proximity between PML bodies and the MHC II gene cluster, we assessed the frequency of lacking association (no contact), association (contact), and colocalization (considerable overlap) between the two signals in confocal images (Fig. 1, A and B). The definition colocalization included both considerable overlap and complete colocalization. This approach revealed an increase of association and colocalization events between PML bodies and the MHC II probe in HEp-2, MRC-5, and HFF after IFN-γ stimulation (Fig. 1 B). Quantification was based on the MHC II signal closest to the next PML body. However, the second MHC class II signal was also associated or colocalized with the PML body signal in >80% of MRC-5 cells or HFF in which the first signal was already associated or colocalized (Fig. 1 A, white circles; and not depicted). We also calculated the theoretical probability for random associations between PML bodies and the MHC class II signals by applying a self-developed algorithm that considers nuclear volume and number and size of objects in the nucleus. The probabilities for random signal overlap varied between 0.30 and 0.59% in MRC-5 and 4.48 and 8.77% in HEp-2 cells (Table S1) and were thus substantially lower than the measured values. Centromeric chromatin of chromosome 6 was not in spatial proximity to PML bodies, although IFN-γ induced a low degree of nonrandom signal overlap in HFFs (Fig. 1 B). Currently, we have no explanation for this phenomenon. It might be related to a dynamic, cell cycle–regulated connection between centromeres and PML bodies as described previously (Everett et al., 1999). To exclude that elevated numbers of PML bodies after IFN-γ treatment caused random proximity between PML bodies and genomic regions, we determined the association frequency between PML bodies and centromeres in MRC-5 fibroblasts (Fig. 1 C). Although the number of PML bodies increased three- to fourfold during IFN-γ treatment (Fig. 1 D), there was no increase of their signal overlap rate with the centromeres (Fig. 1 E). Signal overlap events between PML bodies and centromeres in IFN-γ–treated HEp-2 cells, or HFFs never exceeded 1 or 2%, respectively (unpublished data). The latter result appears to be contradictory to the data for HFF in Fig. 1 B, in which PML bodies were not found to be in close proximity to the centromere 6 region. Although we have no ready explanation for this phenomenon, the discrepancy might be explained by using HFFs from different population doublings. Despite this inconsistency, it is safe to conclude that IFN-γ specifically induces an increase of the spatial proximity between the MHC II gene cluster and PML bodies in HEp-2 cells as well as in primary human lung and skin fibroblasts.
Figure 1.
IFN-γ induces increased spatial proximity between PML bodies and the MHC II gene cluster. (A) Untreated (−IFN-γ) or IFN-γ–treated (+IFN-γ; 250 U/ml for 24 h) HEp-2 cells, MRC-5 cells, and primary human foreskin fibroblasts (HFF) were double labeled to detect the MHC class II gene locus and PML nuclear bodies by DNA-FISH and antibody staining, respectively. Fluorescence signals were acquired from the entire nuclear volume by confocal sectioning. Images display projections of the z stacks. Association was defined as touching or “kissing” of the red and the green signal, whereas colocalization displayed a considerable overlap indicated by yellow color. This definition was only applied to single confocal sections from the image stacks. Adjacent signals without touching were not scored. In the majority of primary fibroblasts (>80%) with association or colocalization events, the second MHC II signal was also found in the vicinity of a PML body (white circles). Insets show enlarged views of the areas indicated by white rectangles. Bars: (main images) 5 µm; (insets) 1 µm. (B) HEp-2, MRC-5, and HFFs were double labeled as described in A to detect either the MHC class II gene locus or the centromere 6 sequence. Association and colocalization events were scored as described in A. Bars show mean values in percentages from four independent experiments with n = 200 cells each. Standard deviation (not depicted) was in all cases <12% of the mean value. (C–E) Up-regulation of PML does not increase the association frequency between nuclear bodies and centromeres. Untreated and IFN-γ–treated MRC-5 cells were stained with anti-PML and anticentromere (Cen) antibodies. DNA was stained with DAPI. The image on the left side of C represents a maximum projection of a z stack through a nucleus of an untreated HEp-2 cell to visualize all centromeres. An association event between a PML body and a centromere (white box) is shown as a magnified view in the inset. The right panel of C shows a single midnucleus confocal section of an IFN-γ–treated HEp-2 cell. Bars: (main images) 10 µm; (inset) 1 µm. (D and E) The number of PML bodies and centromeres (D) as well as the number of association/colocalization events between the two structures (E) was determined from z stacks. Bars show mean values and standard deviations (n = 50). NB, nuclear bodies.
PML is required for efficient IFN-γ–induced MHC II expression
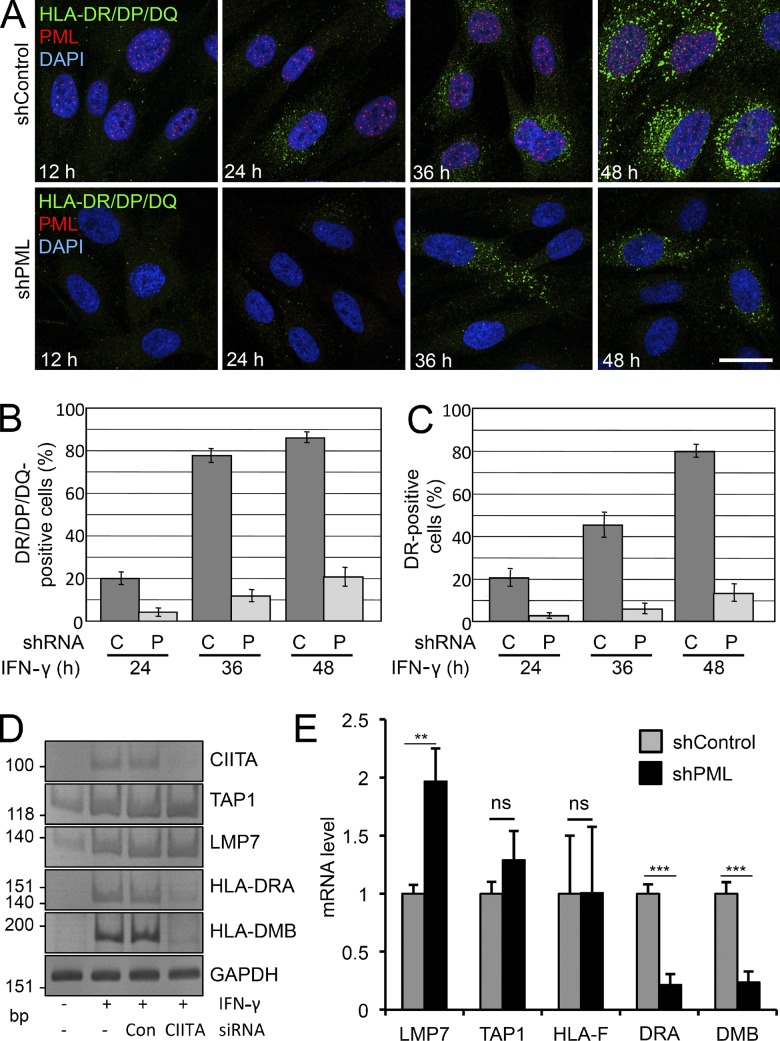
To investigate whether PML plays a role in MHC II expression, HFF stably expressing control or PML small hairpin RNA (shRNA) were treated with IFN-γ, and MHC II expression was monitored at different time points by indirect immunofluorescence microscopy using monoclonal antibodies against various class II proteins. In the presence of PML, IFN-γ–induced expression of HLA-DR/DP/DQ became visible after 24 h, and after 48 h, most cells were positive (Fig. 2 A). Coimmunostaining with anti-PML antibodies confirmed an IFN-γ–induced increase in the number of nuclear bodies. In contrast, cells expressing the PML-specific shRNA had no PML bodies and showed a strongly reduced induction of class II molecule expression (Fig. 2 A). A similar result was obtained in HEp-2 cells (Fig. S1). Quantification revealed that the number of MHC II–positive cells was diminished by a factor of ∼4 in HFF in the absence of PML (Fig. 2, B and C). To determine whether PML depletion interferes with MHC II expression at the transcript level, RT-PCR was performed. As expected, CIITA and MHC II mRNAs were undetectable in unstimulated cells. Transcription of IFN-γ–stimulated genes within the MHC II locus, including TAP1, LMP7, and MHC II antigens DRA and DMB, was clearly detected after 24 h of IFN-γ treatment (Fig. 2 D). TAP1 is activated by IFN-γ but not via CIITA (Wright, et al., 1998). LMP7 expression appears to be regulated by CIITA-dependent and -independent pathways (Londhe et al., 2012). Depletion of CIITA strongly diminished IFN-γ–induced transcription of the MHC II genes but not of TAP1 or LMP7. Importantly, IFN-γ–induced MHC II mRNA expression was also significantly reduced in PML-depleted HFFs, whereas transcription of TAP1 and the MHC class I gene HLA-F was not affected (Fig. 2 E). Interestingly, expression of LMP7 mRNA was up-regulated twofold in PML-depleted cells, which may be explained by PML-mediated selective gene regulation within the MHC class I cluster (Kumar et al., 2007). Collectively, these results demonstrate that down-regulation of PML leads to a decrease in IFN-γ–induced MHC II molecule expression at the transcriptional level.
Figure 2.
PML knockdown inhibits IFN-γ–induced MHC class II expression. (A) Primary HFFs stably transduced with control (shControl) or PML (shPML) shRNA vectors were grown on coverslips in the presence of IFN-γ. Samples were taken at different time points as indicated and processed for immunofluorescence staining to detect the MHC II antigens HLA-DR/DQ/DP, PML, and DNA. Images show midnucleus confocal sections. Bar, 20 µm. (B and C) Quantitation of HLA-DR/DP/DQ and HLA-DR–positive cells from experiments as shown in A. Bars represent mean values and standard deviations quantified from ≥500 cells each. C, control shRNA; P, PML shRNA. (D) CIITA depletion represses IFN-γ–induced expression of MHC class II genes but not the stimulation of TAP1 or LMP7 within the same genomic region (6p21.3; classical class II). Semiquantitative RT-PCR analysis of control and CIITA siRNA knockdown HEp-2 cells with and without IFN-γ treatment (24 h) as indicated. RT-PCR was performed with primers specific for the indicated mRNAs. (E) Quantitative RT-PCR of the indicated genes from HFFs stably transduced with control or PML shRNA vectors. Cells were incubated with 20 U/ml IFN-γ for 24 h. Error bars represent standard deviations from mean values obtained from four independent experiments. **, P < 10−2; ***, P < 10−3. Con, control.
PML regulates CIITA mainly at the posttranslational level
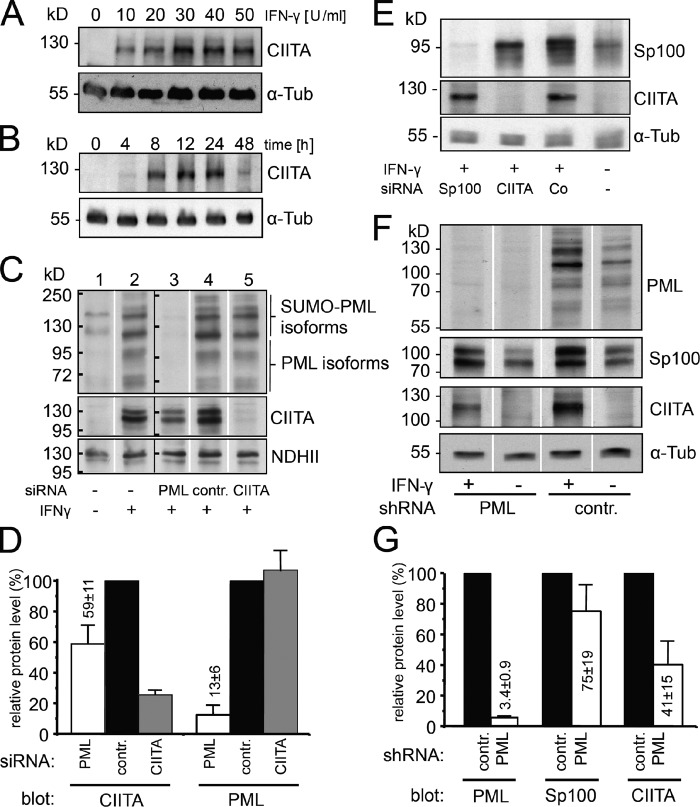
MHC class II mRNA expression is quantitatively dependent on the amount of CIITA (Otten et al., 1998). CIITA was up-regulated by IFN-γ at concentrations as low as 10 U/ml (Fig. 3 A), and induction peaked between 6 and 24 h (Figs. 3 B and 4 B). By Western blotting, various PML isoforms were detected (Fig. 3 C, lane 1). IFN-γ induced up-regulation of all PML-specific bands with similar efficiency, indicating no increase of particular PML protein products through IFN-γ (Fig. 3 C, lanes 2, 4, and 5). The IFN-γ–induced CIITA protein level was reduced to 59 ± 11% in HEp-2 cells, in which PML was down-regulated by siRNA to 13 ± 6% of control levels (Fig. 3 D). Down-regulation of CIITA had no effect on PML protein levels (Fig. 3, C and D). SP100, another constitutive IFN-γ–inducible PML body component, was also efficiently depleted in HEp-2 cells using specific siRNA, but this had no effect on the CIITA protein level (Fig. 3 E). In HFFs stably expressing shRNAs against PML, the amount of PML protein is reduced to a very low level (3.4 ± 0.9%; Fig. 3 F). Under these conditions, the CIITA protein level decreased to 41 ± 15% compared with control shRNA-treated fibroblasts (Fig. 3 G). Consistent with published data, we also found a reduction of the Sp100 level in PML knockdown fibroblasts (Fig. 3 G; Everett et al., 2006). We also analyzed the protein level of RFX5 and NFY-C, two of the constitutive components of the MHC II enhanceosome. This showed that PML depletion has no effect on the protein amount of these factors (Fig. S2), suggesting that PML knockdown does not result in reduced protein levels of all components of the MHC II enhanceosome.
Figure 3.
PML knockdown inhibits IFN-γ–induced CIITA expression. (A and B) IFN induced CIITA expression. CIITA protein levels were analyzed in Western blots of HEp-2 cells treated for 24 h with the indicated concentrations of IFN-γ. (B) Time course for CIITA induction in the presence of 100 U/ml IFN-γ. (C) RNAi-treated HEp-2 cells were cultured in the absence (−) or presence (+) of 10 U/ml IFN-γ for 24 h and then processed for immunoblotting to detect PML, CIITA, and as a loading control, nuclear DNA helicase II (NDHII). The allocation of PML isoforms and SUMOylated isoforms was performed according to Condemine et al. (2006). The data were obtained by loading samples on two large-well gels (one including the samples shown in lanes 1 and 2 and the other including the samples shown in lanes 3–5), transferring the gels to a membrane, cutting the membrane section for each sample into equal-sized strips, and probing of these strips in parallel for the indicated antibodies. The position of the molecular weight protein standard bands is indicated as black bars at the left side of the two blots originating from two independent gels. (D) Quantitation of protein levels after knockdown. Relative signal intensities of immunodetected bands on blots such as shown in C were determined by densitometry. Values were normalized to control siRNA-treated cells (contr.). Bars show mean values and standard deviations from at least three independent experiments. (E) Sp100 depletion does not impact on CIITA protein levels. RNAi-treated HEp-2 cells as indicated were cultured in the absence (−) or presence (+) of IFN-γ for 24 h and then processed for immunoblotting to detect Sp100, CIITA, and α-tubulin. (F) Inhibition of IFN-induced CIITA expression after PML depletion in primary HFFs. HHFs stably expressing control or PML-specific shRNAs and cultured in the absence or presence of 10 U/ml IFN-γ for 24 h were analyzed for PML, Sp100, CIITA, and α-tubulin expression levels. The data were obtained by loading samples on a large-well gel, transferring the gel to a membrane, cutting the membrane section for each sample into equal-sized strips, and probing these strips in parallel for the indicated antibodies. (G) Quantitation of protein levels after knockdown. Relative signal intensities of immunodetected bands on blots such as shown in F were determined by densitometry. Values were normalized to control siRNA treated cells (contr.). Bars show mean values and standard deviations from five independent experiments. White lines indicate that intervening lanes have been spliced out. α-Tub, α-tubulin.
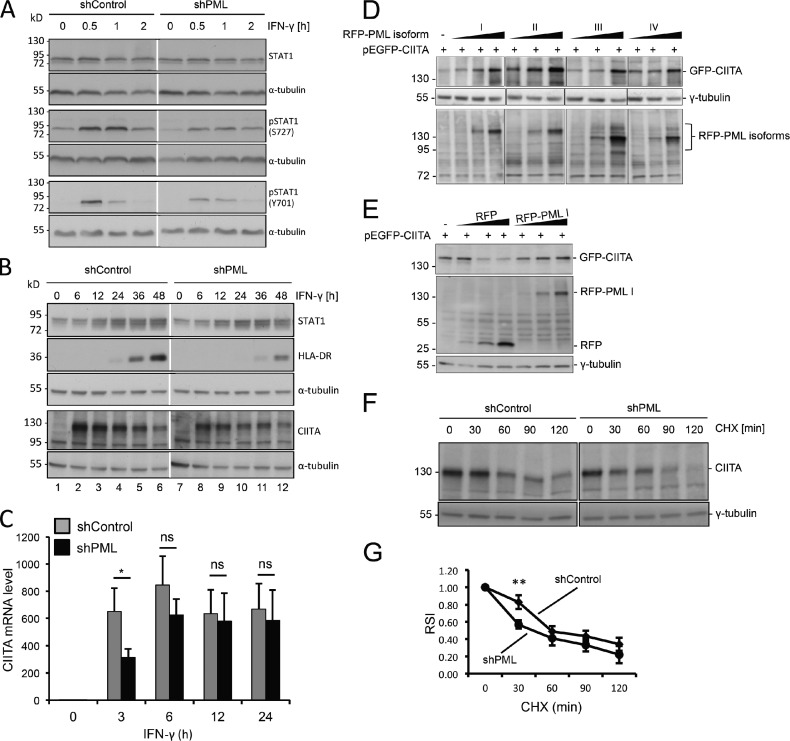
Figure 4.
PML regulates CIITA protein amount mainly through stabilization at the posttranslational level. (A and B) Primary HFFs stably expressing shControl or shPML vectors were treated with IFN-γ. Samples were taken at the indicated time points and analyzed by Western blotting for the expression of STAT1, phospho-STAT1, CIITA, HLA-DR, and α-tubulin. Numbers on the left indicate the positions of standard protein marker bands. (C) Primary HFFs stably expressing shControl or shPML vectors were treated with IFN-γ for 24 h. Samples were taken at the indicated time points, and CIITA mRNA levels were determined by quantitative real-time PCR. Error bars represent standard deviations from mean values obtained from three independent experiments. *, P < 0.05. (D–G) Stabilization of CIITA protein levels through PML. (D) PML isoform II is more efficient than other isoforms to stabilize CIITA at the protein level. HEp-2 cells were cotransfected with constant amounts of vector GFP-CIITA (300 ng) and increasing amounts (100, 300, and 600 ng) of vectors encoding RFP-tagged PML isoforms as indicated. Plasmid DNA concentration was kept constant by adding GFP control vector to a final amount of 1 µg. Western blots of whole-cell lysates from transfected cells were probed with anti-GFP, anti-RFP, and anti–γ-tubulin antibodies as indicated. Tubulin serves as a loading control for the GFP-CIITA data, whereas the band at 72 kD on the RFP-PML isoform blot is a cross-reacting band that serves as an internal control for equal loading. (E) Overexpressed RFP does not result in stabilization of GFP-CIITA at the protein level. HEp-2 cells were cotransfected with constant amounts of vector GFP-CIITA and increasing amounts of vectors encoding RFP alone or the RFP–PML I isoform as described for D. Western blots of whole cell lysates from transfected cells were probed with anti-GFP, anti-RFP, and anti–γ-tubulin antibodies as indicated. (F) PML depletion renders CIITA more sensitive to protein degradation. shControl- and shPML-expressing cells were treated with 20 µ/ml IFN-γ for 12 h to induce expression of endogenous CIITA. Protein synthesis in these cells was then inhibited by incubation with 20 µg/ml cycloheximide (CHX). Samples were removed at the indicated time points, and the amount of endogenous CIITA and γ-tubulin was analyzed in Western blots. (G) Quantitation of the effect of PML depletion on the CIITA protein amount at endogenous levels. CIITA band intensities as shown in F were quantitated from four independent experiments and are displayed as relative signal intensity (RSI) against the incubation time with cycloheximide. Data were normalized to the CIITA signal at 0 min. Data points show mean values and standard deviations (n = 4). **, P < 0.01. Black and white lines indicate that intervening lanes have been spliced out.
Next, we asked at which level PML might regulate the CIITA protein amount. IFN-γ induces transcription of the CIITA gene through the JAK–Stat pathway (Dong et al., 1999). Because it was demonstrated recently that PML depletion leads to reduced STAT1 phosphorylation (El Bougrini et al., 2011), we tested whether CIITA mRNA expression is affected through the JAK–STAT signaling pathway. Although overall STAT1 levels remained unchanged during the first 2 h of IFN-γ activation, phosphorylation of STAT1 at Serine 727 and Tyrosine 701 was reduced in PML-depleted cells (Fig. 4 A). These results confirm and extend previous observations (El Bougrini et al., 2011). At later time points (6–48 h), IFN-γ induced an up-regulation of STAT1, which was similar in shControl and shPML cells (Fig. 4 B). This observation suggests that the IFN-γ activation site/IFN-stimulated response element–dependent activation of the STAT1 gene is independent of PML. However, the CIITA protein amount was higher between 6 and 36 h of IFN-γ stimulation in cells proficient for PML (Fig. 4 B). The increase in CIITA level was accompanied by a subsequent induction of HLA-DR protein expression, which was clearly reduced in shPML cells compared with shControl cells (Fig. 4 B). Measuring the CIITA mRNA level showed that the PML status of cells is directly linked to the induction of CIITA mRNA in the early response to IFN-γ stimulation for ≤3 h (Fig. 4 C). This observation shows that PML regulates transcription of the CIITA gene by modulating the early STAT1 phosphorylation level. However, between 6 and 24 h after IFN-γ stimulation, the CIITA mRNA level was not significantly different in PML-proficient and -deficient cells (Fig. 4 C). It is during this period of time that the lack of PML leads to decreased protein level of CIITA (Fig. 4 B, blot CIITA, compare lanes 2–4 with lanes 8–10). This strongly suggested that PML stabilizes CIITA at the posttranscriptional level, in addition to its early regulatory effect on CIITA mRNA transcription via the JAK–STAT pathway.
To test whether PML can regulate the stability of CIITA, we performed cotransfection experiments. When constant amounts of the GFP-CIITA plasmid were cotransfected with increasing amounts of RFP-tagged PML isoforms, expression of GFP-CIITA was increased in a PML-dependent manner (Fig. 4 D). PML isoforms I to IV induced increased expression of GFP-CIITA, with PML isoform II yielding the strongest effect (Fig. 4 D). This result was obtained in three independent assays (unpublished data). Increased expression of RFP alone resulted in a decrease of the amount of GFP-CIITA indicating that PML and not RFP or increased plasmid concentration is responsible for the observed effects (Fig. 4 E). CIITA has a short half-life caused by rapid proteasomal degradation (Schnappauf et al., 2003). This allowed us to test whether PML also stabilizes CIITA at the endogenous level. The protein amount of CIITA was analyzed at various time points in cycloheximide-treated cells. This approach revealed rapid disappearance of the CIITA protein band within 2 h in control cells (Fig. 4 F). The decrease of the CIITA protein level after cycloheximide was stronger in PML-depleted fibroblasts (Fig. 4 F). Quantitation of signals confirmed that degradation of CIITA was significantly stronger in cells lacking PML (Fig. 4 G). These data demonstrate that also at the endogenous level, PML can regulate the amount of CIITA through protection from degradation.
CIITA forms a complex with PML isoform II at nuclear bodies
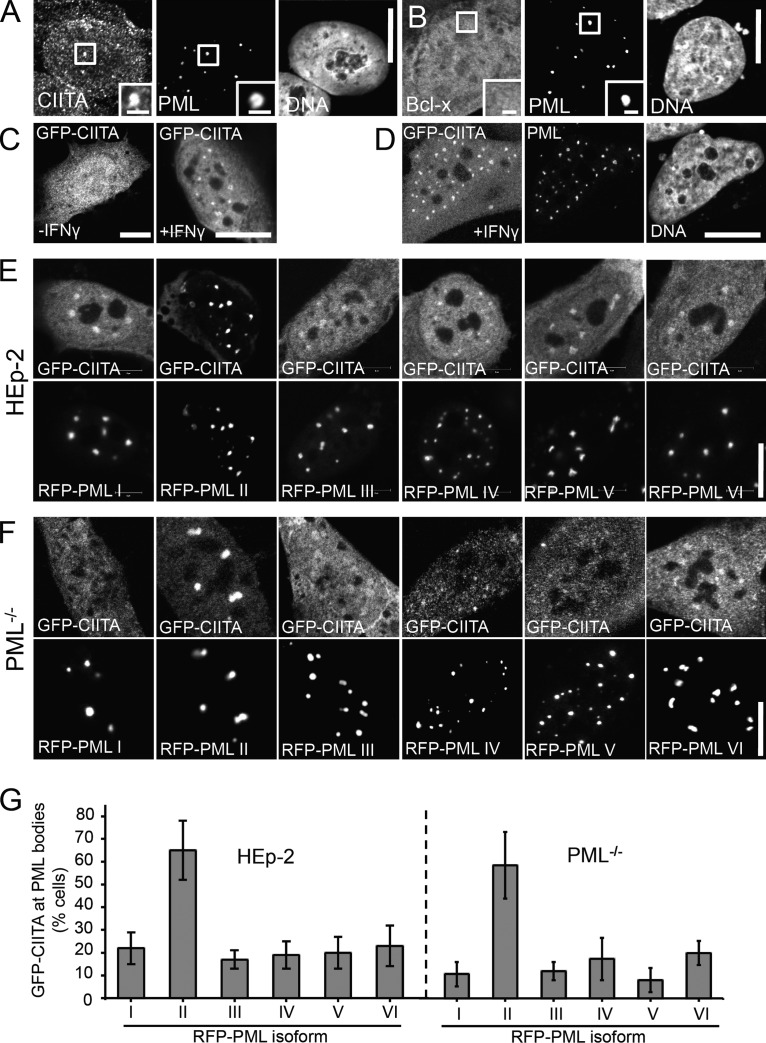
We then analyzed CIITA distribution in the nucleus in more detail (Fig. 5). All anti-CIITA antibodies tested in this study resulted in a cellular background immunofluorescence staining pattern even without IFN-γ treatment and IFN-γ induced only a slight increase of the anti-CIITA immunofluorescence signals (unpublished data). These observations indicated that the epitopes recognized by these antibodies are only partially accessible. We therefore tested several different established immunofluorescence protocols to find conditions, which result in more accessible epitopes (Guillot et al., 2004). This approach showed that only those protocols, which included a prepermeabilization step produced specific anti-CIITA immunofluorescence staining in IFN-γ–treated cells (Fig. 5 A and not depicted). When cells were permeabilized with Triton X-100 before fixation, endogenous CIITA was found in a dotlike pattern in the nucleus in addition to a microspeckled diffuse distribution (Fig. 5 A). The dotlike pattern of CIITA completely colocalized with PML bodies, showing that a subpopulation of endogenous CIITA resides at PML bodies (Fig. S3). We therefore conclude that epitopes recognized by the anti-CIITA antibody used here are inaccessible in normally fixed PML bodies but become accessible by detergent treatment before fixation. The same method did not lead to accumulation at PML nuclear bodies of the unrelated protein Bcl-x (Fig. 5 B).
Figure 5.
PML isoform II recruits CIITA into nuclear bodies. (A and B) CIITA is present at PML nuclear bodies at endogenous levels. HEp-2 cells grown on coverslips were treated or not treated with IFN-γ (10 U/ml for 24 h), permeabilized with 1.5% Triton X-100 for 10 min, and then fixed with 4% formaldehyde and processed for immunofluorescence detection of endogenous CIITA (A) or Bcl-x (B) and PML. Insets show magnified views of the areas indicated by rectangles. Images show midnucleus confocal sections. (C and D) GFP-CIITA accumulates at PML bodies in IFN-γ–treated cells. MRC-5 cells on coverslips were transfected with an expression plasmid encoding GFP-tagged CIITA and cultured in the absence (−IFN-γ) or presence (+IFN-γ) of 10 µ/ml IFN-γ for 24 h. The cells shown in C represent a maximum intensity 3D projection of the entire cellular GFP-CIITA distribution. All other images show midnucleus confocal sections. In D, the cells were coimmunostained with an antibody to detect endogenous PML. DNA was counterstained with DAPI. (E and F) HEp-2 cells (E) or PML-null NIH-3T3 cells (F) coexpressing GFP-CIITA and the indicated RFP-PML isoforms were grown on coverslips, fixed, and analyzed by fluorescence confocal microscopy. (G) The fluorescence intensity of GFP-CIITA was determined in PML nuclear bodies and in similar-sized regions in the nucleoplasm in cells as shown in E and F. The graphs show the percentage of double-transfected cells, in which the ratio of GFP-CIITA fluorescence in nuclear bodies was at least twofold higher than in the nucleoplasm. Bars represent mean values ± SD from n = 100 cells per transfection. Bars: (main images) 10 µm; (insets) 1 µm.
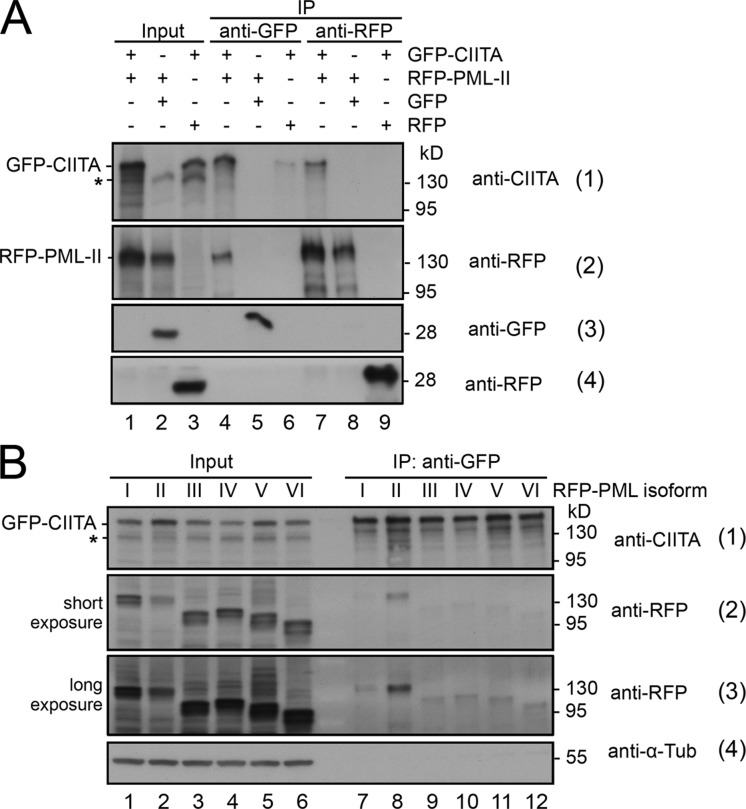
To circumvent the limitations of anti-CIITA antibody staining, we used GFP-tagged CIITA. Expression of GFP-CIITA induced a dose-dependent expression of MHC class II genes (Fig. S4 A). Thus, the GFP-CIITA fusion protein is functional, confirming our published data (Hake et al., 2000; Camacho-Carvajal et al., 2004). Within the nucleus, GFP-CIITA was distributed homogenously (Fig. 5 C, left). Upon IFN-γ treatment, GFP-CIITA also accumulated in a dotlike pattern in the nucleus of some but not all cells (Fig. 5 C, right; and not depicted). The dots represented PML bodies as revealed by anti-PML coimmunostaining (Fig. 5 D). Coexpression experiments revealed that all PML isoforms colocalized with GFP-CIITA in the nuclear bodies (Fig. 5 E). The accumulation of GFP-CIITA in PML bodies varied with overexpression of different PML isoforms but appeared most efficient in cells coexpressing RFP–PML II (Fig. 5 E). We obtained similar results in PML knockout mouse fibroblasts transfected with the different PML isoforms (Fig. 5 F). Quantification demonstrated that RFP–PML II most efficiently attracted GFP-CIITA into nuclear bodies of HEp-2 cells. Because the same result was obtained in PML knockout cells (Fig. 5 G), we conclude that the observed interaction is specific for PML isoform II. Coimmunoprecipitation experiments were performed to confirm complex formation. IFN-γ–treated HEp-2 cells were cotransfected with GFP-tagged CIITA and RFP-tagged PML isoform II. RFP–PML II was coimmunoprecipitated by GFP-CIITA (Fig. 6 A, blot 2, lane 4), and reciprocally, GFP-CIITA was coimmunoprecipitated by RFP–PML II (Fig. 6 A, blot 1, lane 7). As controls, immunoprecipitates of untagged GFP did not contain RFP–PML II (Fig. 6 A, blot 2, lane 5), and immunoprecipitates of untagged RFP did not contain GFP-CIITA (Fig. 6 A, blot 1, lane 8). Another set of coimmunoprecipitation experiments revealed that GFP-CIITA most efficiently formed a complex with GFP-tagged PML II (Fig. 6 B, blots 2 and 3, lane 8). Nevertheless, small amounts of the other RFP-tagged PML isoforms were also detected in GFP-CIITA immunoprecipitates (Fig. 6 B, blot 3, lanes 7 and 9–12).
Figure 6.
GFP-CIITA forms a complex with RFP-tagged PML isoform II. (A) HEp-2 cells were treated with 20 U/ml IFN-γ and transfected with the indicated expression vectors. After 24 h, GFP, RFP, and fusions thereof were immunoprecipitated from soluble cell lysates using GFP-Trap or RFP-Trap and subjected to Western blotting to detect CIITA, RFP-PML, GFP, and RFP. (B) CIITA specifically coimmunoprecipitates PML isoform II. HEp-2 cells expressing GFP-CIITA were treated with 20 U/ml IFN-γ and cotransfected with the six nuclear PML isoforms tagged with RFP. After 24 h, GFP-CIITA was immunoprecipitated using GFP-Trap, and precipitates were analyzed for the presence of CIITA and the RFP-tagged PML isoforms by Western blotting. The asterisks indicate endogenous CIITA. IP, immunoprecipitation. α-Tub, α-tubulin.
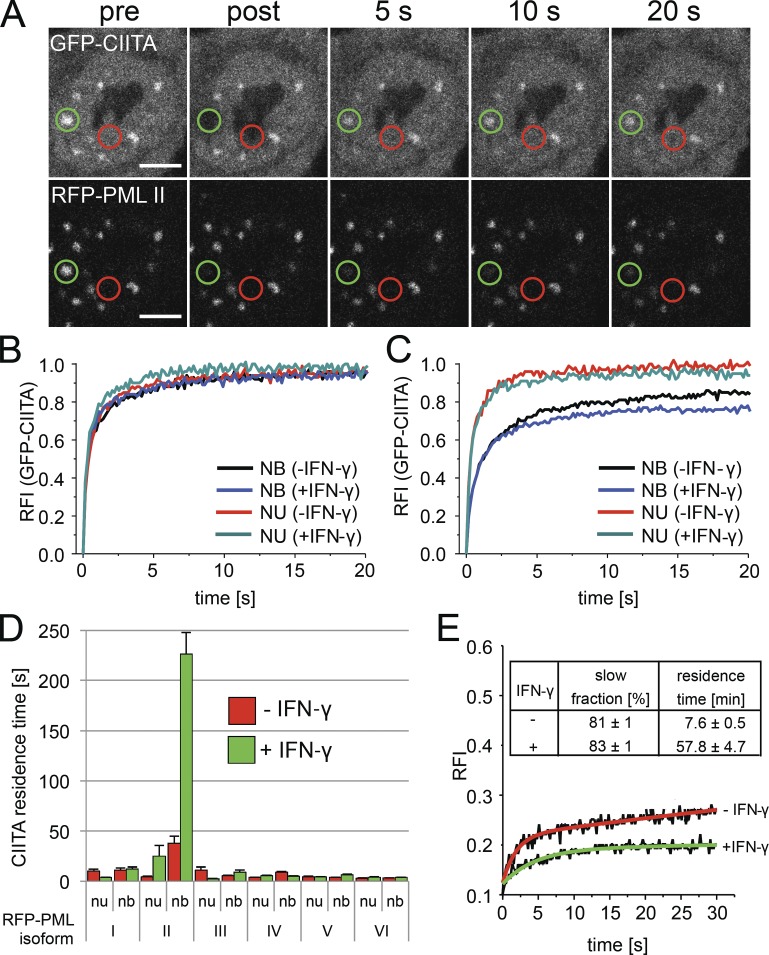
FRAP was used to determine protein complex formation at PML nuclear bodies in living cells. GFP-CIITA fluorescence was bleached at PML nuclear bodies or within the nucleoplasm (Fig. 7 A). Fluorescence recovery of GFP-CIITA after the bleach pulse was rapid within the nucleoplasm as well as at PML bodies (Fig. 7 A). Quantification of the FRAP data demonstrated that GFP-CIITA fluorescence recovered to ∼90% already after 5 s, and prebleach values were reached after 20 s in cells cotransfected with RFP-tagged PML isoform I (Fig. 7 B). This indicates that the complete pool of GFP-CIITA molecules exchanges rapidly at nuclear bodies and at chromatin. The latter is characteristic of typical transcription factor exchange at chromatin (Hager et al., 2009; Hemmerich et al., 2011). GFP-CIITA dynamics at PML bodies containing overexpressed RFP–PML I were very similar to those measured in the nucleoplasm, but the FRAP curve at PML bodies indicated a measurable decrease in GFP-CIITA mobility (Fig. 7 B). IFN-γ did not significantly change the mobility of GFP-CIITA in RFP–PML I-cotransfected cells (Fig. 7 B). Similar FRAP curves were observed in cells cotransfected with RFP-tagged PML isoforms III to VI (unpublished data).
Figure 7.
GFP-CIITA interacts with RFP-PML II at nuclear bodies. (A) FRAP was performed on HEp-2 cells coexpressing GFP-CIITA and RFP-PML isoform II. Fluorescence of both fluorophores was bleached in circular areas containing PML bodies (green circles) or in the nucleoplasm (red circles), and recovery was monitored over time. Bars, 5 µm. (B and C) FRAP of GFP-CIITA at PML bodies (nuclear bodies [NB]) and in the nucleoplasm (NU) was quantitated from experiments as shown in A in cells coexpressing RFP–PML I (B) or RFP–PML II (C) treated or not treated with IFN-γ for 24 h. (D) The residence time of the slow fraction of GFP-CIITA at PML nuclear bodies (nb) and in the nucleoplasm (nu) in cells coexpressing the indicated RFP-tagged PML isoforms was determined from FRAP experiments as described in the Materials and methods. Error bars represent standard deviations from mean values obtained from FRAP curves from three independent measurements with 10 cells each (n = 30). (E) IFN-induced retention of RFP–PML II at nuclear bodies. FRAP of RFP–PML II at nuclear bodies was monitored in the absence (−) or presence (+) of IFN-γ (24 h). The inset table shows the relative amounts and residence times of the slow fraction of RFP–PML II at nuclear bodies as determined from two-component exponential fit functions (red and green curves). RFI, relative fluorescence intensity.
In contrast, exchange of GFP-CIITA at PML bodies was substantially reduced in cells coexpressing RFP–PML II (Fig. 7 C). To quantify this difference, FRAP curves were fitted with exponential functions assuming two differently mobile GFP-CIITA populations as described previously (Hemmerich et al., 2008). The relative fractions and the recovery halftimes of GFP-CIITA in RFP-PML–coexpressing cells were determined from the two-component exponential fit functions (Table S2). The existence of at least two differently mobile fractions is consistent with previous analyses of transcription factor mobility in living cells. The initial fast recovery in FRAP experiments represents freely diffusing molecules, whereas the late phase of the FRAP curve is dominated by association/dissociation kinetics at chromatin. We determined the residence time of the slow exchanging population of GFP-CIITA from its exponential term of the fit function as previously described (Sprague et al., 2004). This approach revealed residence times of a few seconds for GFP-CIITA in chromatin as well as at nuclear bodies after cotransfection with RFP-tagged PML I and III to VI (Fig. 7 D, red bars). Similar results were obtained in IFN-γ–treated cells (Fig. 7 D, green bars). In contrast, the residence time of GFP-CIITA at nuclear bodies containing overexpressed RFP–PML II was 38 (±7) s, and this value increased to 226 (±22) s after IFN-γ treatment (Fig. 7 D). These data confirm that CIITA can directly or indirectly interact with PML isoform II at nuclear bodies and that this interaction is strongly enhanced by IFN-γ. All PML isoforms exchange at nuclear bodies with mean residence times of several minutes in living cells (Weidtkamp-Peters et al., 2008, Brand et al., 2010). We tested whether IFN-γ also had an effect on the exchange rate of PML II at nuclear bodies. Evaluation of the FRAP results indeed revealed that the exchange rate of GFP–PML II was decreased upon IFN-γ treatment compared with untreated cells (Fig. 7 E). The residence time of the slow exchanging fraction of GFP–PML II was decreased almost eightfold in the presence of IFN-γ (Fig. 7 E). Thus, the interaction between GFP-CIITA and RFP–PML II at nuclear bodies occurs on a hyperstable RFP–PML II scaffold to which GFP-CIITA can bind with residence times of several minutes.
Discussion
The MHC II gene cluster is subject to rapid and massive conformational and epigenetic changes upon IFN-γ treatment in cells without constitutive MHC class II molecule expression. IFN-γ treatment induces a rapid looping of the MHC gene cluster away from CT-6 followed by complete decondensation of the locus (Volpi et al., 2000; Müller et al., 2004; Branco and Pombo, 2006; Christova et al., 2007). Evidence for a direct role of PML bodies in transcriptional regulation of MHC II genes was recently established by the observation that the HLA class II DRA gene is specifically relocated to PML bodies in HeLa cells upon IFN-γ induction (Gialitakis et al., 2010). PML bodies were proposed in this study as coactivators of DRA transcription by maintaining H3K4me2 levels at the DRA gene promoter through complex formation between the K4 methyltransferase mixed lineage leukemia (MLL) protein and PML isoform IV (Gialitakis et al., 2010). Here, we confirm that IFN-γ specifically evokes an increase of the spatial proximity between PML bodies and the MHC II gene cluster in human cells lines and extend these observations to primary human fibroblasts. We show for the first time that PML regulates IFN-γ–induced MHC II mRNA expression mainly through stabilization of the CIITA at the protein level. We also demonstrate that PML II, but no other PML isoform, forms a complex with CIITA at PML bodies. Our quantitative FRAP data indicate that the (overexpressed) PML II–CIITA complex can be stabilized at PML bodies for several minutes by IFN-γ. Compared with other isoforms, PML II also exhibited the strongest effect with respect to CIITA stabilization. These observations suggest that PML bodies may act as coregulators of IFN-γ–induced MHC II expression through a functional interaction between PML II and CIITA.
There is accumulating evidence for transcriptional control of nuclear body–associated genes through PML: During programmed in vitro differentiation of mouse embryonic stem cells via neural precursor cells into postmitotic neurons, PML bodies are tightly associated with the Oct3/4 locus when this gene is active in embryonic stem cells. Shut down of Oct3/4 mRNA transcription in neural precursor cells and postmitotic neurons is directly correlated with the loss of contact between PML bodies and the OCT3/4 locus (Aoto et al., 2006). A high association rate (50%) was also observed between PML bodies and the p53 gene in interphase Jurkat cells, but the functional relevance of this interaction was not investigated (Sun et al., 2003). Specific associations of genomic regions with PML bodies has led to the idea that they act as anchoring points for specific chromatin regions, where they have a function analogous to that postulated for matrix attachment regions (Wang et al., 2004; Ching et al., 2005). This idea has been substantiated recently by the demonstration of the organization of the MHC class I locus into distinct higher-order chromatin loop structures based on the physical and functional interaction between PML and the matrix attachment region–binding protein SATB1. These studies indicated that a SATB1–PML body network intersects at the MHC I locus to coordinate expression of MHC I genes (Kumar et al., 2007). Recently, it was shown that a CTCF–CIITA–RFX network complex mediates an IFN-γ–inducible long-range interaction between HLA-DRB1, HLA-DQA1, and the XL9 enhancer positioned between these two MHC II genes. These interactions are believed to form 25- and 15-kb loop-type structures connecting the promoter regions of HLA-DRB1 and HLA-DQA1 (Majumder et al., 2008). Subsequently, the same group showed that CTCF is required for a structural reorganization of the complete MHC II locus into distinct loop structures in a CIITA-dependent manner (Majumder and Boss, 2010). However, endogenous CTCF (MacPherson et al., 2009) and GFP-tagged CTCF (our unpublished data) do not accumulate at PML bodies. The structural and functional relationship between CTCF-mediated loop formation and CIITA–PML-mediated transcriptional control of MHC II mRNA expression remains to be determined.
The nonrandom close proximity between PML bodies and the MHC II cluster is not restricted to genomic subregions of this locus because FISH probes from different regions of the MHC II cluster are in similarly close proximity to PML bodies (Shiels et al., 2001). We also observed that in >80% of IFN-γ–treated human fibroblasts showing a close proximity between a PML body with one MHC II allele, the second allele was also in close proximity to another PML body. MHC II genes are not subject to allelic exclusion (Majumder and Boss, 2010). Because of the stochastic nature of transcription initiation (Suter et al., 2011) and dynamic conformation changes of the activated MHC II genomic region (Müller et al., 2004; Christova et al., 2007), its close proximity to PML bodies may be detected in only a fraction of cells at a given time. In addition, PML body–associated genes within the MHC II locus are transcriptionally active as revealed by RNA FISH (Wang et al., 2004) and quantitative PCR (qPCR; Kumar et al., 2007), indicating that PML bodies have no default transcription-repressive effect on genes in close proximity. Collectively, it is safe to say that PML bodies are in very close proximity to transcriptionally active MHC II genes in a considerable fraction of IFN-γ–treated cells. However, it should also be pointed out that a physical association between PML bodies and the MHC II locus is not an absolute requirement for MHC II gene transcription because cells without PML nuclear bodies still express detectable amounts of MHC II mRNAs (Fig. 2 E) from which MHC II molecules are being translated (Fig. 4 B).
In contrast to our study, Wang et al. (2004) did not detect differences in the expression level of genes in the proximity of PML bodies after PML knockdown. In the current study, we have specifically analyzed the expression of CIITA-dependent genes, which have not been investigated in the Wang et al. (2004) study. Therefore, the results obtained by Wang et al. (2004) cannot be directly compared with our findings. Furthermore, the PML knockdown in the Wang study yielded ∼50% of the cells without PML bodies, and a considerable amount of PML protein was still detected by Western blotting after siRNA (Wang et al., 2004). It cannot be fully excluded that the PML knockdown was not efficient enough to reveal a PML-mediated influence on the expression of the genes analyzed in this study.
The transcription of MHC II genes is quantitatively controlled by the level of CIITA (Fig. S4 A; Otten et al., 1998). We uncovered here that PML is able to modulate CIITA expression at the transcriptional level through the JAK–STAT signaling pathway at an early time point (3 h) after IFN-γ induction (Fig. 4 A). Subsequently, there was no significant difference of the CIITA mRNA level between control and PML knockdown cells. Because the protein half-life of CIITA is only ∼30 min as a result of rapid proteasomal degradation (Schnappauf et al., 2003), one can assume that the reduced CIITA mRNA level after 3 h of IFN-γ induction has only a little effect on the CIITA protein amount produced from mRNA during the 6–24-h interval. Consistent with the idea of a PML-mediated stabilization of CIITA at the protein level, we observed substantially reduced CIITA protein amounts in PML-depleted cells between 6 and 24 h after IFN-γ addition and increased degradation of endogenous CIITA in the absence of PML. These observations indicate that PML functions to protect CIITA from proteasomal degradation. In a similar fashion, PML also protects other nuclear factors, such as the transcription factors p53 and p73, and the DNA damage response protein TopBP1 from proteasomal degradation by sequestration into PML nuclear bodies (Louria-Hayon et al., 2003; Xu et al., 2003; Bernardi et al., 2004; Bernassola et al., 2004). This is solid evidence to suggest that one major function of PML bodies is stabilization of specific nuclear factors at the protein level.
We have shown previously that GFP–PML II is stably bound to PML bodies with a mean residence time of 6.2 min (Weidtkamp-Peters et al., 2008; Brand et al., 2010). Because PML II is able to physically tether CIITA to PML bodies for several minutes (Fig. 5), they might serve as a structural hub or surface at which CIITA is stabilized through protection from degradation. Currently, we do not know whether the IFN-γ–induced relocation of the MHC II gene cluster to PML bodies is functionally connected to the stabilization of CIITA. PML bodies may facilitate efficient assembly of CIITA into MHC II enhanceosome complexes, and the transcriptional activity of these complexes may be increased by more spatial proximity between MHC II genes and the nuclear bodies. This model is consistent with our observation that GFP-tagged RFX-AP, RFX-B, and NFYa can also bind to PML bodies and that a small but significant amount of MHC II promoter chromatin coimmunoprecipitates with endogenous PML (our unpublished data). Interestingly, IFN-γ also induced tighter binding of GFP–PML II to PML bodies (Fig. 7 E), whereas exchange of GFP–PML V and GFP–PML VI was unaffected (not depicted). The IFN-γ–induced increased residence time of PML II at nuclear bodies may be one mechanism for prolonged retention and stabilization of CIITA at PML bodies. The activity of CIITA is regulated by PTMs, including phosphorylation, acetylation, and ubiquitination (Wu et al., 2009). PML bodies are potential hot spots for such PTMs (Bernardi and Pandolfi, 2007). Acetylation of CIITA by CBP at K141 and K144, which up-regulates CIITA activity at MHC II promoters (Spilianakis et al., 2000), could be mediated by the acetyltransferase CBP, which is also a PML body component (Boisvert et al., 2001). It will be interesting to reveal which PML body–localized PTMs contribute to CIITA stability.
The SATB1–PML, the MLL–PML IV, and the CIITA–PML II interaction networks (Kumar et al., 2007; Gialitakis et al., 2010; and this study, respectively) provide compelling evidence for a direct role of PML bodies in the transcriptional regulation of the human MHC locus (Shiels et al., 2001). In all of these studies, specific PML isoforms were shown to contribute to transcriptional regulation of MHC genes. The underlying mechanisms share common features such as close proximity between the PML body and MHC genes, but they are clearly different in nature, including regulated chromatin loop formation (SATB1–PML), epigenetic marking (MLL–PML IV), and regulation of CIITA protein levels (CIITA–PML II). Future studies will further unravel the molecular details of how PML or the PML nuclear bodies participate in these multilevel mechanisms underlying the control of MHC molecule expression.
Materials and methods
Cell culture and transfection
The following cell lines and primary cells were used: human HEp-2 cervix carcinoma epithelial cells (CCL23; American Type Culture Collection), human MRC-5 lung fibroblasts (CCL171; American Type Culture Collection), primary human dermal fibroblasts from foreskin or biopsies (provided by R.W. Kinne, Experimental Rheumatology Unit, University Hospital Jena, Jena, Germany), primary human dermal fibroblast lines stably transduced with control or PML shRNA vectors were established as described previously (Tavalai et al., 2006), and mouse 3T3-PML+/+ and 3T3-PML−/− cells (provided by T. Hofmann, University of Heidelberg, Heidelberg, Germany; Brand et al., 2010). Cells were cultured in DME supplemented with 10% fetal calf serum in a 9.5% CO2 atmosphere at 37°C. IFN-γ (I-3265; Sigma-Aldrich) was added to the cells as indicated. For live-cell imaging experiments, cells were seeded on 42-mm glass dishes (H. Saur Laborbedarf) and transfected with plasmid DNA 1–2 d before observation using transfection reagent (FuGENE HD; Roche) according to the manufacturer’s protocol.
Antibodies
The following primary antibodies were used: anti-PML rabbit antibody (ABD-030; Jena Biosciences), anti-PML mAb (PG-M3; Santa Cruz Biotechnology, Inc.), antitubulin mAb (T-9026; Sigma-Aldrich), anti-CIITA rabbit antibody (serum 21; Bontron et al., 1997), anti-Sp100 rabbit antibody (ABD-031; Jena Biosciences), anti-Sp100 rabbit antibody (GH3; provided by H. Will, Heinrich Pette Institute, Hamburg, Germany; Milovic-Holm et al., 2007), anti–HLA-DR mAb (G46-6; BD), anti–HLA-DR/DP/DQ mAb (TÜ-39; BD), anti–Bcl-x rabbit mAb (#1018-1; Epitomics; Enzo Life Sciences), anti–RFX-5 rabbit antibody (NB100-195; Novus Biologicals), and anti–NFY-C rabbit antibody (NBP10-19147; Novus Biologicals). The following secondary antibodies were used: Alexa Fluor 488-anti–mouse IgG (Invitrogen), Alexa Fluor 546-anti–sheep (Invitrogen), Cy2-, Cy3-, or Cy5-anti–rabbit IgG (highly cross-adsorbed; Dianova), and horseradish peroxidase–coupled anti–rabbit IgG and anti–mouse IgG (highly cross-adsorbed; Dianova).
Plasmids
The following plasmids were used for transient transfection assays: pEGFP–PML I to VI and pmRFP–PML I to VI were described in detail previously (Weidtkamp-Peters et al., 2008; Brand et al., 2010); pmRFP-C1/2/3 vectors were cloned by replacing the GFP cDNA in pEGFP-C1/2/3 with a cDNA encoding monomeric RFP (provided by R. Tsien, Stanford University, Stanford, CA) as a PCR fragment via the AgeI and BsrGI sites; pEGFP-CIITA is described in Schnappauf et al. (2003); plasmids pEGFP–RFX-5, pEGFP–RFX-AP, and pEGFP–RFX-B were provided by J. Boss (Emory University School of Medicine, Atlanta, GA). All pEGFP and pmRFP vectors contain full-length cDNAs of these proteins fused to the 3′ end of GFP or RFP, respectively, giving rise to N-terminally fluorescently labeled fusion proteins. NFY-expressing GFP plasmids were provided by D. Doenecke (University of Göttingen, Göttingen, Germany).
Western blots
Whole-cell extracts were electrophoresed on SDS-PAGE and transferred to nitrocellulose membrane (Protran; Schleicher & Schuell). The membrane was blocked in 5% skimmed milk/PBS-T (PBS + 0.1% [vol/vol] Tween 20 [Carl Roth]) and incubated with protein-specific primary and subsequently horseradish peroxidase–conjugated species-specific secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.). Signal was detected using the ECL reagent (GE Healthcare) on imaging film (BioMax; Kodak).
Immunocytochemistry and microscopy
Cells grown on 15-mm-diam coverslips were fixed with 4% formaldehyde for 10 min and permeabilized with 0.25% Triton X-100 for 3 min. Primary antibodies were incubated on cells for 45 min. After washing steps with PBS, secondary antibodies coupled to appropriate fluorophores were incubated on cells for 45 min followed by a DNA staining step using TO-PRO-3 (Invitrogen) for 10 min and mounting with antifade mounting medium (ProLong gold; Invitrogen). Other cell fixation methods included different protocols described in detail previously by Guillot et al. (2004). In brief, protocols were as follows: for protocol 1, cells on coverslips rinsed in PBS (10 mM Na2HPO4/NaH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4), fixed/permeabilized in ethanol (20 min at −20°C); for protocol 2, cells on coverslips were rinsed in PBS-Mg, fixed in 1.75% formaldehyde in PBS-Mg (10 mM Na2HPO4/NaH2PO4, pH 7.4, 150 mM NaCl, and 2 mM MgCl2; 20 min at 4°C), rinsed in PBS-Mg (15 min), and permeabilized with 0.5% Triton X-100 in PBS-Mg (15 min); for protocol 3, cells on coverslips were rinsed in PBS-Mg, permeabilized in 1.5% Triton X-100 in PBS-Mg (10 min at 4°C), rinsed, and fixed in 1.75% formaldehyde in PBS-Mg (20 min at 4°C); for protocol 4, cells on coverslips rinsed in PBS, fixed in 4% formaldehyde and 250 mM Hepes, pH 7.6 (10 min and 4°C), and refixed in 8% formaldehyde and 250 mM Hepes, pH 7.6 (50 min at 4°C) followed by permeabilization in 0.5% Triton X-100 in PBS (30 min with gentle rocking); for protocol 5, cells on coverslips were rinsed in PBS, fixed/permeabilized in 0.1% Triton X-100 in 4% formaldehyde and 250 mM Hepes, pH 7.6 (10 min at 4°C), refixed in 8% formaldehyde and 250 mM Hepes, pH 7.6 (50 min at 4°C), and permeabilized as in protocol 4; for protocol 6, cells on coverslips were rinsed in PBS, fixed in 4% formaldehyde and 125 mM Hepes, pH 7.6 (10 min at 4°C), and refixed in 8% formaldehyde and 125 mM Hepes, pH 7.6 (50 min at 4°C) followed by permeabilization in 0.5% Triton X-100 in PBS (30 min with gentle rocking). For microscopy of fixed cells, a laser-scanning confocal microscope (LSM 510 Meta or LSM 710/ConfoCor 3; Carl Zeiss) was used using a 63×-fold oil objective (numerical aperture of 1.3) and ZEN software (Carl Zeiss).
Immuno-FISH
For immuno-FISH analyses, cells were fixed in 4% and then in 8% formaldehyde in 250 mM Hepes, pH 7.6 (10 min and 2 h, respectively), according to previously established protocols (Guillot et al., 2004). Permeabilization was performed in 0.5% saponin/0.5% Triton X-100/20 mM glycin/PBS for 1 h. After extensive PBS washes, cells were stored in 20% glycerol/PBS at −80°C. Indirect immunofluorescence was performed as described in the previous section, and cells were fixed again in 4% formaldehyde/0.2% Triton X-100/PBS followed by 20 nm glycin/PBS incubation for 30 min, and another 30-min incubation in 0.1 M HCl. Cellular RNA was digested in a solution containing 500 µg/ml RNase A (Roche) at 37°C for 2 h. DNA was denatured in 70% deionized formamide in 2× SSC for 8 min at 75°C. Hybridization using a MHC II–specific probe was performed at 37°C for 48 h in 50% deionized formamide/2× SSC/10% dextrane sulfate. The probe to detect the MHC II locus (bacterial artificial chromosome clone RP11-10A19; BACPAC Resources, Children’s Hospital Oakland Research Institute) was characterized previously (Müller et al., 2004). The probe was labeled with digoxigenin-11–deoxy-UTP using a Nick Translation kit according to the manufacturer’s protocol (Roche), denatured for 10 min at 70°C, and coprecipitated with human Cot1 and herring sperm DNA. Posthybridization washes were as follows: 50% formamide in SSC (1 h at 37°C), 0.1× SSC (30 min at 60°C), and 4× SSC/0.1% Tween 20 (20 min at 42°C). The probe was detected using a sheep antidigoxigenin antibody (Roche). Blocking and washing steps were performed in 0.5% blocking solution (Tris-HCl/150 mM NaCl at 37°C; Roche). Nuclei were counterstained with 2 mM TOTO-3 (Invitrogen). Coverslips were mounted and polymerized in MobiGLOW (MoBiTec).
Immunoprecipitation
HEp-2 cells were transfected and treated as indicated. After 2 d, ∼107 cells were harvested, shock frozen in liquid nitrogen, suspended in 200 µl lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, freshly added 40 mM _N_-ethylmaleimide, 0.1% benzonase nuclease, 2 mM Pefabloc, 2 µg/ml aprotinin, 2 µg/ml leupeptin, and 2 µg/ml pepstatin) for ∼15 min on ice, and subjected to sonication (10 cycles of 15 s on/off at high intensity and 4°C; Bioruptor Plus; Diagenode). After centrifugation for 15 min at 16,100 g and 4°C, the supernatant was diluted with 800 µl of ice-cold washing buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, freshly added 2 mM Pefabloc, 20 mM _N_-ethylmaleimide, and protease inhibitor cocktail [Roche]), keeping 25 µl of the supernatant as input for Western blotting. For each precipitation, ∼25 µl GFP- or RFP-Trap bead slurry (ChromoTek) was equilibrated in washing buffer, added to the dilution, and incubated for ∼3 h with gentle end-over-end mixing at 4°C. After centrifugation (2 min at 2,700 g and 4°C) and discarding the supernatant, the beads were washed twice, resuspended in 50 µl SDS sample buffer, and boiled for 10 min at 95°C.
FRAP
FRAP experiments were performed at 37°C in Hepes-buffered medium on a confocal microscope (LSM 710/ConfoCor3) essentially as described before (Weidtkamp-Peters et al., 2008; Brand et al., 2010), using a 63× oil objective (numerical aperture of 1.3) and ZEN software. 5–10 images were taken before the bleach pulse, and 50–200 images were taken after bleaching of regions of interest containing one nuclear body each at 0.05% laser transmission to minimize scan bleaching. Image acquisition frequency was adapted to the recovery rate of the respective GFP fusion protein. The pinhole was adjusted to 1 airy unit. Quantitation of relative fluorescence intensities was performed according to Weidtkamp-Peters et al. (2008) using Excel (Microsoft) and Origin software (OriginLab). Recovery halftimes and relative amounts of the two differently mobile populations of molecules exchanging at PML bodies were determined from the fitted two-component exponential functions (Sprague et al., 2004). Assuming that FRAP of the slower exchanging component is unaffected by free diffusion and therefore reaction dominant, the dissociation rate (_k_off) of the slower component is identical to the decay constant (_k_slow) of its exponential term (Sprague et al., 2004). Therefore, the residence time was calculated as (_k_slow)−1.
RNAi
HEp-2 cells at low density in 6-well plates were transfected with SMARTpool siRNAs against PML, Sp100, CIITA, or a control siRNA (Thermo Fisher Scientific). Transfection was performed using HiPerFect (QIAGEN) at 50-nM RNAi concentration. 3 d after transfection, siRNA-treated cells were harvested. Each experiment was performed at least three times.
RNA extraction and cDNA synthesis
Total RNA was extracted from whole cells using an isolation system (peqGOLD RNAPure; Peqlab). RNA samples were resuspended in sterile diethylpyrocarbonate pyrogen-treated, RNase/DNase-free water (Invitrogen). RNA concentration was measured using the spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific). Reverse transcription synthesis of cDNA was performed on 1 µg of total RNA using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen) with random hexamer primers following the manufacturer’s instructions.
Quantitative real-time PCR
5 ng cDNA was used for quantitative real-time PCR of a 25-µl total volume. Each independent PCR reaction contains 12.5 µl Platinum SYBR Green qPCR SuperMix uracil DNA glycosylase (Invitrogen) and 0.2 µM of each primer. The run and analysis were performed using a real-time PCR instrument and software (iQ5; Bio-Rad Laboratories). Primer sequences are as follows: LMP7 forward, 5′-CAGCTATTCTGGAGGCGTTG-3′, and reverse, 5′-GCTGAGCCCGTACTCTCTCTT-3′; TAP1 forward, 5′-CAGGAGACGGAGTTTTTCCA-3′, and reverse, 5′-CAGAGCATGATCCCCAAGAG-3′; HLA-F forward, 5′-AGGAACAGACCCAGGACACA-3′, and reverse, 5′-ACCACAGCTCCAAGGACAAC-3′; HLA-DRA-A forward, 5′-TAAGGCACATGGAGGTGATG-3′, and reverse, 5′-GTACGGAGCAATCGAAGAGG-3′; HLA-DMB forward, 5′-CTCTCACAGCACCTCAACCA-3′, and reverse, 5′-TAGAAGCCCCACACATAGCA-3′; and CIITA forward, 5′-AGCCTTTCAAAGCCAAGTCC-3′, and reverse, 5′-TTGTTCTCACTCAGCGCATC-3′. For the normalization of the qPCR values, glyceraldehyde 3-phosphate dehydrogenase was used as a housekeeping gene (forward, 5′-GTCAGTGGTGGACCTGACCT-3′, and reverse, 5′-AAAGGTGGAGGAGTGGGTGT-3′). All primers were validated, and serial dilutions of cDNA were run to generate a standard curve to analyze the efficiency of the primers.
Primer for semiquantitative RT-PCR (Fig. 2 D)
Primers were as follows: CIITA forward, 5′-AACATCACTGACCTGGGTGCCTAC-3′, and reverse, 5′-CCCACGTCGCAGATGCAGTTATTG-3′; TAP1 forward, 5′-AGGGCTATGACACAGAGGTAGAC-3′, and reverse, 5′-CTGGTGGCATCATCCAGGATAAG-3′; LMP7 forward, 5′-GGTCCTACATTAGTGCCTTACG-3′, and reverse, 5′-AGTACAGCCTGCATTCCTTG-3′; HLA-DRA forward, 5′-TCCGCAAGTTCCACTATCTC-3′, and reverse, 5′-ACACCACGTTCTCTGTAGTC-3′; HLA-DMB forward, 5′-TGTCCTCTCACAGCACCTCAAC-3′, and reverse, 5′-ACATAGCAGGCCAGCATCACAG-3′; and glyceraldehyde 3-phosphate dehydrogenase forward, 5′-TGCACCACCAACTGCTTAG-3′, and reverse, 5′-GATCGCAGGGATGATGTTC-3′.
Theoretical association probability
To calculate the theoretical probability for a colocalization event between sphere-shaped objects (PML bodies and gene loci) in the nucleus, the volume of the nucleus and the number and volume of the objects were taken into account. These data were determined from the 3D image stacks of FISH experiments. In case of a colocalization (or association) event of a genomic locus and a PML body, the distance between the centers of the two colocalizing distinct spheres within the nucleus is equal or less than the sum of the sphere’s radii: d < rg + rp, in which rg is the radius of the genomic locus, and rp is the radius of a PML body. Considering one single nucleus of radius R with m PML bodies of radius rp each, and n genomic loci with radius rg each, we assume that the positions of all genomic loci are equally distributed within the nucleus and that no two genomic loci are closer than 2 rg + 2 rp, which guarantees that a specific PML body does not colocalize with more than one genomic locus at a time. Then, considering the volume of the nucleus and the total volume of the spheres around all genomic loci, which define a colocalization event, the colocalization probability p for one PML body is given by p = n (rg + rp)3/R3. As for m PML bodies, the probability pm that no PML body colocalizes with any genomic locus is pm = (1 − p)m. We get the final result for the probability pA to find at least one colocalization between a PML body and a genomic locus within the whole nucleus with m PML bodies and n genomic loci: pA = 1 − (1 − n (rg + rp)3/R3)m. This model contains the explicit assumption that two genomic loci have a minimum distance of 2(rp + rg), which is a plausible assumption regarding the size of genomic elements and of PML bodies. On the contrary, PML bodies are allowed to intersect in this probability model, which does not coincide with our experimental findings. Nevertheless, for the numbers of PML bodies in our experiments, the effect of this approximation on the true probability is very small.
From the colocalizing probability pA within one nucleus, one can compute the probability pk(j) for the event to find among k investigated cells j cells with at least one colocalization caused by chance: pk(j) = (k/choose j) pAj (1 − pAj)(k − j). If in an experiment with k cells there are q cells with at least one colocalization, the p-value for this event is the sum of the probabilities pk(j) to find q cells or more with at least one colocalization: p-value = SUM(q ≤ I ≤ k) pk(i).
Online supplemental material
Fig. S1 shows by immunostaining that a transient siRNA-mediated knockdown of PML in HEp-2 cells leads to reduced cellular IFN-γ–induced MHC class II molecule expression. Fig. S2 shows by Western blotting that lack of PML protein does not affect the protein level of the MHC class II enhanceosome components RFX5 or NFY-C. Fig. S3 shows by immunostaining that a subfraction of CIITA accumulates at all PML bodies at the endogenous level. Fig. S4 shows by qPCR and immunofluorescence that the fusion protein GFP-CIITA is functional with respect to induction of MHC II mRNA and protein expression, respectively. Table S1 shows the theoretical probability of association between PML bodies and genomic loci based on our self-developed software model. Table S2 provides a complete list of all dynamic parameters obtained from FRAP analyses of GFP-CIITA at PML bodies in nuclei of living cells dependent on coexpression of individual PML isoforms. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201112015/DC1.
Acknowledgments
We are grateful to J. Boss, D. Doenecke, and H. Will for providing expression plasmids and/or antibodies. P. Hemmerich and A. Horch wish to thank Ana Pombo for extensive help with FISH experiments.
This work was supported by grants HE 2484/3-1 and SFB796 from the Deutsche Forschungsgemeinschaft.
Footnotes
Abbreviations used in this paper:
CIITA
class II transactivator
HFF
human foreskin fibroblast
HLA
human leukocyte antigen
IFN
interferon
JAK
Janus kinase
MHC
major histocompatibility
MLL
mixed lineage leukemia
PML
promyelocytic leukemia
PTM
posttranslational modification
qPCR
quantitative PCR
RFX
regulatory factor X
shRNA
small hairpin RNA
STAT
signal transducer and activator of transcription
References
- Aoto T., Saitoh N., Ichimura T., Niwa H., Nakao M. 2006. Nuclear and chromatin reorganization in the MHC-Oct3/4 locus at developmental phases of embryonic stem cell differentiation. Dev. Biol. 298:354–367 10.1016/j.ydbio.2006.04.450 [DOI] [PubMed] [Google Scholar]
- Ascoli C.A., Maul G.G. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785–795 10.1083/jcb.112.5.785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R., Pandolfi P.P. 2007. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8:1006–1016 10.1038/nrm2277 [DOI] [PubMed] [Google Scholar]
- Bernardi R., Scaglioni P.P., Bergmann S., Horn H.F., Vousden K.H., Pandolfi P.P. 2004. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat. Cell Biol. 6:665–672 10.1038/ncb1147 [DOI] [PubMed] [Google Scholar]
- Bernassola F., Salomoni P., Oberst A., Di Como C.J., Pagano M., Melino G., Pandolfi P.P. 2004. Ubiquitin-dependent degradation of p73 is inhibited by PML. J. Exp. Med. 199:1545–1557 10.1084/jem.20031943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., Hendzel M.J., Bazett-Jones D.P. 2000. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 148:283–292 10.1083/jcb.148.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F.M., Kruhlak M.J., Box A.K., Hendzel M.J., Bazett-Jones D.P. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099–1106 10.1083/jcb.152.5.1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontron S., Ucla C., Mach B., Steimle V. 1997. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol. Cell. Biol. 17:4249–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branco M.R., Pombo A. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138 10.1371/journal.pbio.0040138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand P., Lenser T., Hemmerich P. 2010. Assembly dynamics of PML nuclear bodies in living cells. PMC Biophys. 3:3 10.1186/1757-5036-3-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Carvajal M.M., Klingler S., Schnappauf F., Hake S.B., Steimle V. 2004. Importance of class II transactivator leucine-rich repeats for dominant-negative function and nucleo-cytoplasmic transport. Int. Immunol. 16:65–75 10.1093/intimm/dxh010 [DOI] [PubMed] [Google Scholar]
- Chen M., Singer L., Scharf A., von Mikecz A. 2008. Nuclear polyglutamine-containing protein aggregates as active proteolytic centers. J. Cell Biol. 180:697–704 10.1083/jcb.200708131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching R.W., Dellaire G., Eskiw C.H., Bazett-Jones D.P. 2005. PML bodies: a meeting place for genomic loci? J. Cell Sci. 118:847–854 10.1242/jcs.01700 [DOI] [PubMed] [Google Scholar]
- Cho Y., Lee I., Maul G.G., Yu E. 1998. A novel nuclear substructure, ND10: distribution in normal and neoplastic human tissues. Int. J. Mol. Med. 1:717–724 [DOI] [PubMed] [Google Scholar]
- Choi N.M., Majumder P., Boss J.M. 2011. Regulation of major histocompatibility complex class II genes. Curr. Opin. Immunol. 23:81–87 10.1016/j.coi.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christova R., Jones T., Wu P.J., Bolzer A., Costa-Pereira A.P., Watling D., Kerr I.M., Sheer D. 2007. P-STAT1 mediates higher-order chromatin remodelling of the human MHC in response to IFNgamma. J. Cell Sci. 120:3262–3270 10.1242/jcs.012328 [DOI] [PubMed] [Google Scholar]
- Collins T., Korman A.J., Wake C.T., Boss J.M., Kappes D.J., Fiers W., Ault K.A., Gimbrone M.A.J., Jr, Strominger J.L., Pober J.S. 1984. Immune interferon activates multiple class II major histocompatibility complex genes and the associated invariant chain gene in human endothelial cells and dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 81:4917–4921 10.1073/pnas.81.15.4917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine W., Takahashi Y., Zhu J., Puvion-Dutilleul F., Guegan S., Janin A., de Thé H. 2006. Characterization of endogenous human promyelocytic leukemia isoforms. Cancer Res. 66:6192–6198 10.1158/0008-5472.CAN-05-3792 [DOI] [PubMed] [Google Scholar]
- de Thé H., Riviere M., Bernhard W., Bernhard W. 1960. [Examination by electron microscope of the VX2 tumor of the domestic rabbit derived from the Shope papilloma]. Bull. Assoc. Fr. Etud. Cancer. 47:570–584 [PubMed] [Google Scholar]
- Dong Y., Rohn W.M., Benveniste E.N. 1999. IFN-gamma regulation of the type IV class II transactivator promoter in astrocytes. J. Immunol. 162:4731–4739 [PubMed] [Google Scholar]
- El Bougrini J., Dianoux L., Chelbi-Alix M.K. 2011. PML positively regulates interferon gamma signaling. Biochimie. 93:389–398 10.1016/j.biochi.2010.11.005 [DOI] [PubMed] [Google Scholar]
- Everett R.D., Earnshaw W.C., Pluta A.F., Sternsdorf T., Ainsztein A.M., Carmena M., Ruchaud S., Hsu W.L., Orr A. 1999. A dynamic connection between centromeres and ND10 proteins. J. Cell Sci. 112:3443–3454 [DOI] [PubMed] [Google Scholar]
- Everett R.D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80:7995–8005 10.1128/JVI.00734-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabunmi R.P., Wigley W.C., Thomas P.J., DeMartino G.N. 2001. Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J. Cell Sci. 114:29–36 [DOI] [PubMed] [Google Scholar]
- Fagioli M., Alcalay M., Pandolfi P.P., Venturini L., Mencarelli A., Simeone A., Acampora D., Grignani F., Pelicci P.G. 1992. Alternative splicing of PML transcripts predicts coexpression of several carboxy-terminally different protein isoforms. Oncogene. 7:1083–1091 [PubMed] [Google Scholar]
- Fuchsová B., Novák P., Kafková J., Hozák P. 2002. Nuclear DNA helicase II is recruited to IFN-α–activated transcription sites at PML nuclear bodies. J. Cell Biol. 158:463–473 10.1083/jcb.200202035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambacorta M., Flenghi L., Fagioli M., Pileri S., Leoncini L., Bigerna B., Pacini R., Tanci L.N., Pasqualucci L., Ascani S., et al. 1996. Heterogeneous nuclear expression of the promyelocytic leukemia (PML) protein in normal and neoplastic human tissues. Am. J. Pathol. 149:2023–2035 [PMC free article] [PubMed] [Google Scholar]
- Gialitakis M., Arampatzi P., Makatounakis T., Papamatheakis J. 2010. Gamma interferon-dependent transcriptional memory via relocalization of a gene locus to PML nuclear bodies. Mol. Cell. Biol. 30:2046–2056 10.1128/MCB.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande M.A., van der Kraan I., van Steensel B., Schul W., de Thé H., van der Voort H.T., de Jong L., van Driel R. 1996. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J. Cell. Biochem. 63:280–291 [DOI] [PubMed] [Google Scholar]
- Guillot P.V., Xie S.Q., Hollinshead M., Pombo A. 2004. Fixation-induced redistribution of hyperphosphorylated RNA polymerase II in the nucleus of human cells. Exp. Cell Res. 295:460–468 10.1016/j.yexcr.2004.01.020 [DOI] [PubMed] [Google Scholar]
- Hager G.L., McNally J.G., Misteli T. 2009. Transcription dynamics. Mol. Cell. 35:741–753 10.1016/j.molcel.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake S.B., Masternak K., Kammerbauer C., Janzen C., Reith W., Steimle V. 2000. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol. Cell. Biol. 20:7716–7725 10.1128/MCB.20.20.7716-7725.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P., Weidtkamp-Peters S., Hoischen C., Schmiedeberg L., Erliandri I., Diekmann S. 2008. Dynamics of inner kinetochore assembly and maintenance in living cells. J. Cell Biol. 180:1101–1114 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmerich P., Schmiedeberg L., Diekmann S. 2011. Dynamic as well as stable protein interactions contribute to genome function and maintenance. Chromosome Res. 19:131–151 10.1007/s10577-010-9161-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K., Shiels C., Freemont P.S. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene. 20:7223–7233 10.1038/sj.onc.1204765 [DOI] [PubMed] [Google Scholar]
- Kießlich A., von Mikecz A., Hemmerich P. 2002. Cell cycle-dependent association of PML bodies with sites of active transcription in nuclei of mammalian cells. J. Struct. Biol. 140:167–179 10.1016/S1047-8477(02)00571-3 [DOI] [PubMed] [Google Scholar]
- Kumar P.P., Bischof O., Purbey P.K., Notani D., Urlaub H., Dejean A., Galande S. 2007. Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9:45–56 10.1038/ncb1516 [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P.P., Puvion E., Freemont P., de Thé H. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor α degradation. J. Exp. Med. 193:1361–1371 10.1084/jem.193.12.1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londhe P., Zhu B., Abraham J., Keller C., Davie J. 2012. CIITA is silenced by epigenetic mechanisms that prevent the recruitment of transactivating factors in rhabdomyosarcoma cells. Int. J. Cancer. 131:E437–E448 10.1002/ijc.26478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louria-Hayon I., Grossman T., Sionov R.V., Alsheich O., Pandolfi P.P., Haupt Y. 2003. The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J. Biol. Chem. 278:33134–33141 10.1074/jbc.M301264200 [DOI] [PubMed] [Google Scholar]
- MacPherson M.J., Beatty L.G., Zhou W., Du M., Sadowski P.D. 2009. The CTCF insulator protein is posttranslationally modified by SUMO. Mol. Cell. Biol. 29:714–725 10.1128/MCB.00825-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P., Boss J.M. 2010. CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Mol. Cell. Biol. 30:4211–4223 10.1128/MCB.00327-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P., Gomez J.A., Chadwick B.P., Boss J.M. 2008. The insulator factor CTCF controls MHC class II gene expression and is required for the formation of long-distance chromatin interactions. J. Exp. Med. 205:785–798 10.1084/jem.20071843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovic-Holm K., Krieghoff E., Jensen K., Will H., Hofmann T.G. 2007. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 26:391–401 10.1038/sj.emboj.7601504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W.G., Rieder D., Kreth G., Cremer C., Trajanoski Z., McNally J.G. 2004. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell. Biol. 24:9359–9370 10.1128/MCB.24.21.9359-9370.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negorev D., Maul G.G. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene. 20:7234–7242 10.1038/sj.onc.1204764 [DOI] [PubMed] [Google Scholar]
- Otten L.A., Steimle V., Bontron S., Mach B. 1998. Quantitative control of MHC class II expression by the transactivator CIITA. Eur. J. Immunol. 28:473–478 [DOI] [PubMed] [Google Scholar]
- Reith W., LeibundGut-Landmann S., Waldburger J.M. 2005. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 5:793–806 10.1038/nri1708 [DOI] [PubMed] [Google Scholar]
- Rockel T.D., Stuhlmann D., von Mikecz A. 2005. Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J. Cell Sci. 118:5231–5242 10.1242/jcs.02642 [DOI] [PubMed] [Google Scholar]
- Saitoh N., Uchimura Y., Tachibana T., Sugahara S., Saitoh H., Nakao M. 2006. In situ SUMOylation analysis reveals a modulatory role of RanBP2 in the nuclear rim and PML bodies. Exp. Cell Res. 312:1418–1430 10.1016/j.yexcr.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Scharf A., Rockel T.D., von Mikecz A. 2007. Localization of proteasomes and proteasomal proteolysis in the mammalian interphase cell nucleus by systematic application of immunocytochemistry. Histochem. Cell Biol. 127:591–601 10.1007/s00418-006-0266-2 [DOI] [PubMed] [Google Scholar]
- Schnappauf F., Hake S.B., Camacho Carvajal M.M., Bontron S., Lisowska-Grospierre B., Steimle V. 2003. N-terminal destruction signals lead to rapid degradation of the major histocompatibility complex class II transactivator CIITA. Eur. J. Immunol. 33:2337–2347 10.1002/eji.200323490 [DOI] [PubMed] [Google Scholar]
- Sharma P., Murillas R., Zhang H., Kuehn M.R. 2010. N4BP1 is a newly identified nucleolar protein that undergoes SUMO-regulated polyubiquitylation and proteasomal turnover at promyelocytic leukemia nuclear bodies. J. Cell Sci. 123:1227–1234 10.1242/jcs.060160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels C., Islam S.A., Vatcheva R., Sasieni P., Sternberg M.J., Freemont P.S., Sheer D. 2001. PML bodies associate specifically with the MHC gene cluster in interphase nuclei. J. Cell Sci. 114:3705–3716 [DOI] [PubMed] [Google Scholar]
- Spilianakis C., Papamatheakis J., Kretsovali A. 2000. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol. Cell. Biol. 20:8489–8498 10.1128/MCB.20.22.8489-8498.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague B.L., Pego R.L., Stavreva D.A., McNally J.G. 2004. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys. J. 86:3473–3495 10.1529/biophysj.103.026765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle V., Otten L.A., Zufferey M., Mach B. 1993. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome). Cell. 75:135–146 [PubMed] [Google Scholar]
- Steimle V., Siegrist C.A., Mottet A., Lisowska-Grospierre B., Mach B. 1994. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 265:106–109 10.1126/science.8016643 [DOI] [PubMed] [Google Scholar]
- Sternsdorf T., Grötzinger T., Jensen K., Will H. 1997. Nuclear dots: actors on many stages. Immunobiology. 198:307–331 10.1016/S0171-2985(97)80051-4 [DOI] [PubMed] [Google Scholar]
- Sun Y., Durrin L.K., Krontiris T.G. 2003. Specific interaction of PML bodies with the TP53 locus in Jurkat interphase nuclei. Genomics. 82:250–252 10.1016/S0888-7543(03)00075-2 [DOI] [PubMed] [Google Scholar]
- Suter D.M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F. 2011. Mammalian genes are transcribed with widely different bursting kinetics. Science. 332:472–474 10.1126/science.1198817 [DOI] [PubMed] [Google Scholar]
- Tavalai N., Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta. 1783:2207–2221 10.1016/j.bbamcr.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Tavalai N., Papior P., Rechter S., Leis M., Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018 10.1128/JVI.00743-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terris B., Baldin V., Dubois S., Degott C., Flejou J.F., Hénin D., Dejean A. 1995. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 55:1590–1597 [PubMed] [Google Scholar]
- Van Damme E., Laukens K., Dang T.H., Van Ostade X. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int. J. Biol. Sci. 6:51–67 10.7150/ijbs.6.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpi E.V., Chevret E., Jones T., Vatcheva R., Williamson J., Beck S., Campbell R.D., Goldsworthy M., Powis S.H., Ragoussis J., et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565–1576 [DOI] [PubMed] [Google Scholar]
- von Mikecz A., Zhang S., Montminy M., Tan E.M., Hemmerich P. 2000. CREB-binding protein (CBP)/p300 and RNA polymerase II colocalize in transcriptionally active domains in the nucleus. J. Cell Biol. 150:265–273 10.1083/jcb.150.1.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Shiels C., Sasieni P., Wu P.J., Islam S.A., Freemont P.S., Sheer D. 2004. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 164:515–526 10.1083/jcb.200305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidtkamp-Peters S., Lenser T., Negorev D., Gerstner N., Hofmann T.G., Schwanitz G., Hoischen C., Maul G., Dittrich P., Hemmerich P. 2008. Dynamics of component exchange at PML nuclear bodies. J. Cell Sci. 121:2731–2743 10.1242/jcs.031922 [DOI] [PubMed] [Google Scholar]
- Wiesmeijer K., Molenaar C., Bekeer I.M., Tanke H.J., Dirks R.W. 2002. Mobile foci of Sp100 do not contain PML: PML bodies are immobile but PML and Sp100 proteins are not. J. Struct. Biol. 140:180–188 10.1016/S1047-8477(02)00529-4 [DOI] [PubMed] [Google Scholar]
- Wright K.L., Chin K.C., Linhoff M., Skinner C., Brown J.A., Boss J.M., Stark G.R., Ting J.P. 1998. CIITA stimulation of transcription factor binding to major histocompatibility complex class II and associated promoters in vivo. Proc. Natl. Acad. Sci. USA. 95:6267–6272 10.1073/pnas.95.11.6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Kong X., Luchsinger L., Smith B.D., Xu Y. 2009. Regulating the activity of class II transactivator by posttranslational modifications: exploring the possibilities. Mol. Cell. Biol. 29:5639–5644 10.1128/MCB.00661-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S.Q., Pombo A. 2006. Distribution of different phosphorylated forms of RNA polymerase II in relation to Cajal and PML bodies in human cells: an ultrastructural study. Histochem. Cell Biol. 125:21–31 10.1007/s00418-005-0064-2 [DOI] [PubMed] [Google Scholar]
- Xu Z.X., Timanova-Atanasova A., Zhao R.X., Chang K.S. 2003. PML colocalizes with and stabilizes the DNA damage response protein TopBP1. Mol. Cell. Biol. 23:4247–4256 10.1128/MCB.23.12.4247-4256.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S., Salomoni P., Pandolfi P.P. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85–E90 10.1038/35010583 [DOI] [PubMed] [Google Scholar]