Expansive Evolution of the TREHALOSE-6-PHOSPHATE PHOSPHATASE Gene Family in Arabidopsis (original) (raw)
Abstract
Trehalose is a nonreducing sugar used as a reserve carbohydrate and stress protectant in a variety of organisms. While higher plants typically do not accumulate high levels of trehalose, they encode large families of putative trehalose biosynthesis genes. Trehalose biosynthesis in plants involves a two-step reaction in which trehalose-6-phosphate (T6P) is synthesized from UDP-glucose and glucose-6-phosphate (catalyzed by T6P synthase [TPS]), and subsequently dephosphorylated to produce the disaccharide trehalose (catalyzed by T6P phosphatase [TPP]). In Arabidopsis (Arabidopsis thaliana), 11 genes encode proteins with both TPS- and TPP-like domains but only one of these (AtTPS1) appears to be an active (TPS) enzyme. In addition, plants contain a large family of smaller proteins with a conserved TPP domain. Here, we present an in-depth analysis of the 10 TPP genes and gene products in Arabidopsis (_TPPA_-TPPJ). Collinearity analysis revealed that all of these genes originate from whole-genome duplication events. Heterologous expression in yeast (Saccharomyces cerevisiae) showed that all encode active TPP enzymes with an essential role for some conserved residues in the catalytic domain. These results suggest that the TPP genes function in the regulation of T6P levels, with T6P emerging as a novel key regulator of growth and development in higher plants. Extensive gene expression analyses using a complete set of promoter-β-glucuronidase/green fluorescent protein reporter lines further uncovered cell- and tissue-specific expression patterns, conferring spatiotemporal control of trehalose metabolism. Consistently, phenotypic characterization of knockdown and overexpression lines of a single TPP, AtTPPG, points to unique properties of individual TPPs in Arabidopsis, and underlines the intimate connection between trehalose metabolism and abscisic acid signaling.
The presence of trehalose in a wide variety of organisms and the existence of different biosynthesis pathways suggest a pivotal and ancient role for trehalose metabolism in nature. The most widely distributed metabolic pathway consists of two consecutive enzymatic reactions, with trehalose-6-phosphate (T6P) synthase (TPS) catalyzing the transfer of a glucosyl moiety from UDP-Glc to Glc-6-phosphate to produce T6P and UDP, and T6P phosphatase (TPP) catalyzing dephosphorylation of T6P to trehalose (Cabib and Leloir, 1958; Avonce et al., 2006). Apart from operating as a (reserve) carbon source and structural component in bacteria, fungi, and invertebrates, trehalose also functions as a major stress protectant of proteins and membranes during adverse conditions such as dehydration, high salinity, hypoxia, and nutrient starvation (Elbein et al., 2003). Trehalose accumulation is also observed in a few lower vascular resurrection plants (e.g. Selaginella lepidophylla). Until about a decade ago, higher vascular plants were believed to have lost the ability to produce trehalose, but with the emergence of more sensitive assays, genome sequencing, and the use of yeast (Saccharomyces cerevisiae) mutant complementation, minute amounts of trehalose and T6P, and functional plant enzyme orthologs were found (Goddijn et al., 1997; Vogel et al., 1998; Lunn et al., 2006). In addition, heterologous expression and disruption of trehalose metabolism in plants conferred pleiotropic effects, ranging from altered stress tolerance, leaf morphology, and developmental timing to embryo lethality (Holmström et al., 1996; Goddijn et al., 1997; Romero et al., 1997; Eastmond et al., 2002; Schluepmann et al., 2003; Avonce et al., 2004; Satoh-Nagasawa et al., 2006; Miranda et al., 2007; Chary et al., 2008), pointing to an important regulatory function. The intermediate T6P has been highlighted as a novel signal for carbohydrate status (for review, see Paul, 2008), positively correlating with Suc levels, redox-regulated ADP-Glc pyrophosphorylase activity, and starch biosynthesis (Lunn et al., 2006). Recently, it was reported that T6P inhibits the activity of the SnRK1 protein kinase to activate energy-consuming biosynthetic processes in growing tissue (Zhang et al., 2009) and that it is required for the onset of leaf senescence (Wingler et al., 2012).
In most bacterial and eukaryotic species, the TPS and TPP activities are found on separate proteins. Recent phylogenetic and biochemical analyses showed that some archaea and bacteria, such as Cytophaga hutchinsonii, express proteins that have both active TPS and TPP domains resulting from gene fusion, suggesting that such prokaryotic bifunctional proteins are the evolutionary ancestors of the large eukaryotic trehalose biosynthesis enzymes in which one or both domains have subsequently lost their catalytic activity (Avonce et al., 2010). The yeast TPP enzyme Tps2, for example, harbors an inactive N-terminal TPS domain and an active C-terminal TPP domain. In contrast to the single TPS and TPP genes in most microorganisms, the genomes of higher plants encode a remarkably large family of putative trehalose biosynthesis enzyme homologs. These are commonly classified in three distinct subgroups, according to their similarity to the microbial TPS and TPP proteins and/or presence of specific motifs (e.g. conserved phosphatase boxes; Thaller et al., 1998; Leyman et al., 2001; Eastmond et al., 2003). Even primitive plants such as the alga Ostreococcus tauri and the moss Physcomitrella patens already contain members of each of these gene families, pointing to the early establishment and conservation of these proteins in plant evolution (Lunn, 2007; Avonce et al., 2010). In Arabidopsis (Arabidopsis thaliana), the class I TPS proteins (AtTPS1-4) show most similarity to the yeast TPS Tps1, but also have a C-terminal domain with limited similarity to TPPs. However, only one of these, AtTPS1, appears to have heterologous enzymatic TPS activity in yeast (Blázquez et al., 1998; Vandesteene et al., 2010). Strikingly, AtTPS1 is the only class I enzyme with an N-terminal extension that seems to operate as an autoinhibitory domain (Van Dijck et al., 2002). The class II TPS proteins (AtTPS5-11) are similar bipartite proteins with a TPS-like domain but a more conserved TPP domain. They appear to lack both heterologous TPS and TPP activity (Ramon et al., 2009). The high level of conservation of putative substrate-binding residues in class I and class II proteins, however, suggests that substrates might still bind (Avonce et al., 2006; Lunn, 2007; Ramon et al., 2009; Vandesteene et al., 2010). Together with the specific expression patterns of the class I genes (van Dijken et al., 2004; Geelen et al., 2007; Vandesteene et al., 2010) and the extensive expression regulation of all class II members by plant carbon status (Baena-González et al., 2007; Usadel et al., 2008; Ramon et al., 2009), this suggests tissue-specific regulatory functions for these proteins in metabolic regulation of plant growth and development. Finally, Arabidopsis also harbors a family of 10 smaller proteins (AtTPPA-J; 320–385 amino acids) with limited similarity to the class I and class II proteins (795–942 amino acids). Like class II proteins, they contain the phosphatase box consensus sequences, characteristic of the l-2-haloacid dehalogenase (HAD) super family of enzymes, which includes a wide range of phosphatases and hydrolases (Thaller et al., 1998). It has been suggested that the origin of these plant TPP genes is different from the origin of the class I and II genes (Avonce et al., 2010) and that plants recruited the TPP genes after their divergence from fungi, most probably from proteobacteria or actinobacteria. Consistently, homologous TPP proteins are present in proteobacteria such as Rhodopherax ferrireducens (Avonce et al., 2010). To date, only a few of these single-domain plant TPP proteins have been subject to biochemical characterization, e.g. TPPA and TPPB from Arabidopsis (Vogel et al., 1998), OsTPP1 and OsTPP2 from rice (Oryza sativa; Pramanik and Imai, 2005; Shima et al., 2007), and RAMOSA3 (RA3) from maize (Zea mays; Satoh-Nagasawa et al., 2006).
The phenotypic alterations observed in plants fed with trehalose or genetically modified in trehalose biosynthesis, suggest a pivotal role for trehalose metabolism in integrating the metabolic status with growth and development. Disruption of the only known active TPS enzyme in Arabidopsis (AtTPS1) results in embryo lethality (Eastmond et al., 2002) and, when rescued to bridge embryogenesis, causes a strong disruption of vegetative and generative development and abscisic acid (ABA) hypersensitivity (van Dijken et al., 2004; Gómez et al., 2010). Overexpressing AtTPS1 on the other hand renders seedlings sugar and ABA insensitive (Avonce et al., 2004, 2005). These observations strongly link trehalose metabolism with ABA signaling. Interestingly, a mutation of a TPP gene in maize, RA3, results in a distinct phenotype, with incorrect axillary meristem identity and determinacy in both male and female inflorescences (Satoh-Nagasawa et al., 2006). Arabidopsis plants with overall increased T6P levels, such as OtsA (Escherichia coli TPS) overexpression plants, similarly show increased inflorescence branching (Schluepmann et al., 2003; van Dijken et al., 2004).
To better understand why higher plants harbor such a large number of putative TPP proteins, we have made a comprehensive study of the 10 Arabidopsis TPP genes and gene products, combining phylogenetic approaches and yeast growth complementation assays, together with a detailed analysis of all 10 TPP gene expression profiles in Arabidopsis, and a more detailed single AtTPP mutant phenotypic analysis.
RESULTS
Collinearity Analysis Shows That the TPP Gene Family in Eudicots Mainly Expanded through Genome Duplication
It has been established that many, if not most plant lineages are paleopolyploids (Masterson, 1994; Cui et al., 2006; Soltis et al., 2009; Van de Peer et al., 2009; Jiao et al., 2011). Current evidence that a genome duplication occurred in the common ancestor of extant seed plants, and another one at the base of the angiosperm lineage (Jiao et al., 2011). In addition, a hexaploidization event (referred to as γ) occurred around the base of the eudicots (Jaillon et al., 2007; Tang et al., 2008). Many eudicot lineages underwent additional genome duplication events, many of them around the Cretaceous-Tertiary boundary, 65 million years ago (mya; Fawcett et al., 2009; Van de Peer et al., 2009). For instance, two additional genome duplications have occurred within the Brassicales lineage, referred to as β and α in Arabidopsis (Jaillon et al., 2007; Tang et al., 2008). Remnants of ancient genome duplications have also been discovered in monocot genomes, e.g. an event shared by Sorghum spp. and Oryza spp. probably dating back to the base of the Poaceae clade (Paterson et al., 2004), and in the moss P. patens (Rensing et al., 2008).
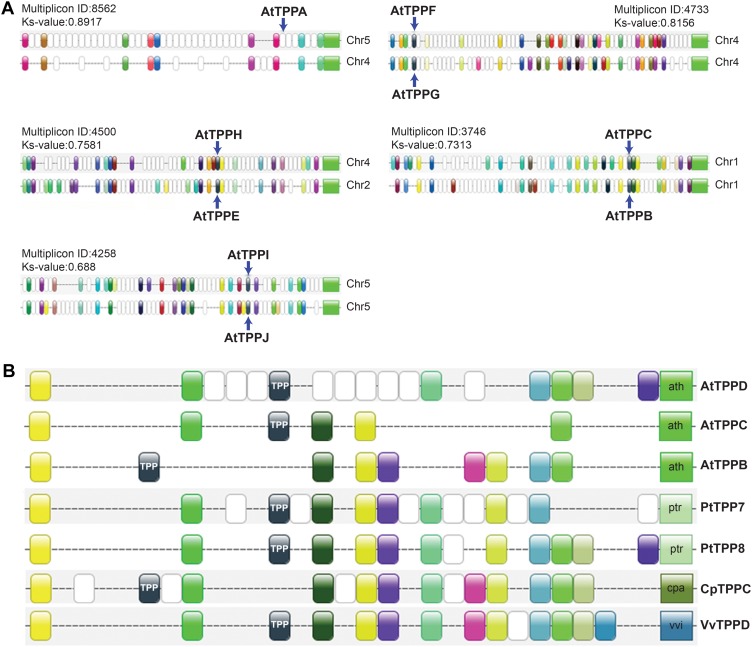
Genome duplications have had a profound impact on the expansion of certain gene families in plants, in particular regulatory gene families (Maere et al., 2005; Freeling, 2009; Maere and Van de Peer, 2010). We investigated their impact on the TPP gene family by searching for collinearity of genomic segments containing TPP genes, using the PLAZA v1.0 platform (Proost et al., 2009). Strikingly, all TPP genes in eudicots appear to be linked through collinearity relationships, indicating that the TPP gene family in eudicots expanded exclusively through genome duplication. In Arabidopsis, nine out of 10 AtTPP genes, all except TPPD, were found in duplicated blocks remaining from the most recent genome duplication (α, _K_S around 0.7–0.8; Fig. 1A). Moreover, all TPPs were retained in duplicate after the α event, except for TPPA and TPPD. Whereas TPPA still lies in a duplicated block remaining from α, for TPPD we could not find back any syntenic relationships dating back to the α event, indicating that the whole duplicated stretch, including the duplicate of TPPD, was lost or has degraded beyond recognition. Deeper collinearity relationships caused by previous genome duplication events were also evident. TPPD, for instance, has a syntenic relationship with TPPB, but this duplicated block is much older (_K_S = 1.6) and originates from an earlier genome duplication (data not shown). Extending our analysis to other species, we found clear collinearity relationships linking all TPPs across the eudicots (see for example Fig. 1B), indicating that all eudicot TPP duplicates created after the monocot-dicot split have been derived from genome duplications. Several collinearities among monocot TPP genes were also apparent but their phylogenetic relationship is less clear.
Figure 1.
Collinearity of the Arabidopsis TPP genes. A, Collinearity information extracted from the PLAZA v1.0 platform. For each multiplicon, the PLAZA v1.0 multiplicon ID and corresponding _K_S value are depicted. The _K_S values for all multiplicons shown here are typical of duplicated blocks remaining from the most recent genome duplication in Arabidopsis (Fawcett et al., 2009). B, Example of collinearity relationships among TPPs across the eudicots in muliplicon 894, identified from the PLAZA v1.0 database using _At_TPPC as the seed gene in the Skyline Plot utility. Genes with the same color represent members of the same gene family that are present in several of the homologous blocks (also known as anchor points). TPP genes are colored black. The ratio of collinear regions expected for Vitis:Poplar:Carica:Arabidopsis on the basis of their genome duplication history is 1:2:1:4 (Fawcett et al., 2009), very close to the observed ratio of 1:2:1:3, indicating that very little loss has occurred in this clade. Only the collinear region of TPPD created by the most recent whole-genome duplication in Arabidopsis appears to have been lost.
Collinearity Constraints Improve the Phylogenetic Reconstruction of the TPP Gene Family
We used the TREEFINDER package (Jobb et al., 2004) to reconstruct a naive phylogeny of the TPP gene family in the green plant lineage (see “Materials and Methods”). The resulting tree is depicted in Supplemental Figure S1A. It is in good agreement with the tree calculated with different methods by Avonce et al. (2010). The tree multifurcates in four branches, the P. patens TPP genes and three clades of angiosperm TPP genes, all having average to high bootstrap support (78.6–98.3) except for the branch containing the Arabidopsis TPPF, TPPG, and TPPA genes that had a much lower bootstrap support value of 52.5.
Next, we sought to impose topological constraints on the tree by taking into account the genome duplication relationships among the TPPs. Based on the collinearity relationships found with PLAZA v1.0 (Proost et al., 2009) and the available knowledge on the timing and phylogenetic position of genome duplications and speciations in angiosperm plants (Fawcett et al., 2009), we were able to severely constrain the topology of the TPP tree (Supplemental Fig. S1B). Because the phylogenetic interpretation of the collinearities among monocot TPP genes was less clear, we did not use them to constrain the tree topology. The constrained tree was again fed into the TREEFINDER package to resolve the remaining unconstrained branches, using a bootstrap Maximum Likelihood analysis with 1,000 iterations. The resulting phylogenetic tree of the TPP gene family in the land plants is presented in Supplemental Figure S1B. The constrained phylogenetic relationships fit relatively well with the unconstrained phylogeny (see Supplemental Fig. S1A). Three major subclades of eudicot TPPs are apparent in both trees, and the within-species phylogenetic relationships among TPP genes are identical. Within the subclades, the between-species phylogenetic relationship of the TPPs is more variable. Papaya (Carica papaya) TPPs are, for instance, often put outside Arabidopsis-Populus spp. clades in the unconstrained topology, possibly an artifact of slower evolution of these genes in papaya. On two occasions, grapevine (Vitis vinifera) TPPs are similarly put erroneously close together in the unconstrained topology. The fact that all nodes in the unconstrained tree that do not match the constrained tree have relatively low bootstrap support is also an indication that constraining the topology on the basis of collinearity relationships significantly improves the tree.
All 10 Putative Arabidopsis TPP Enzymes Have Heterologous TPP Activity in Yeast
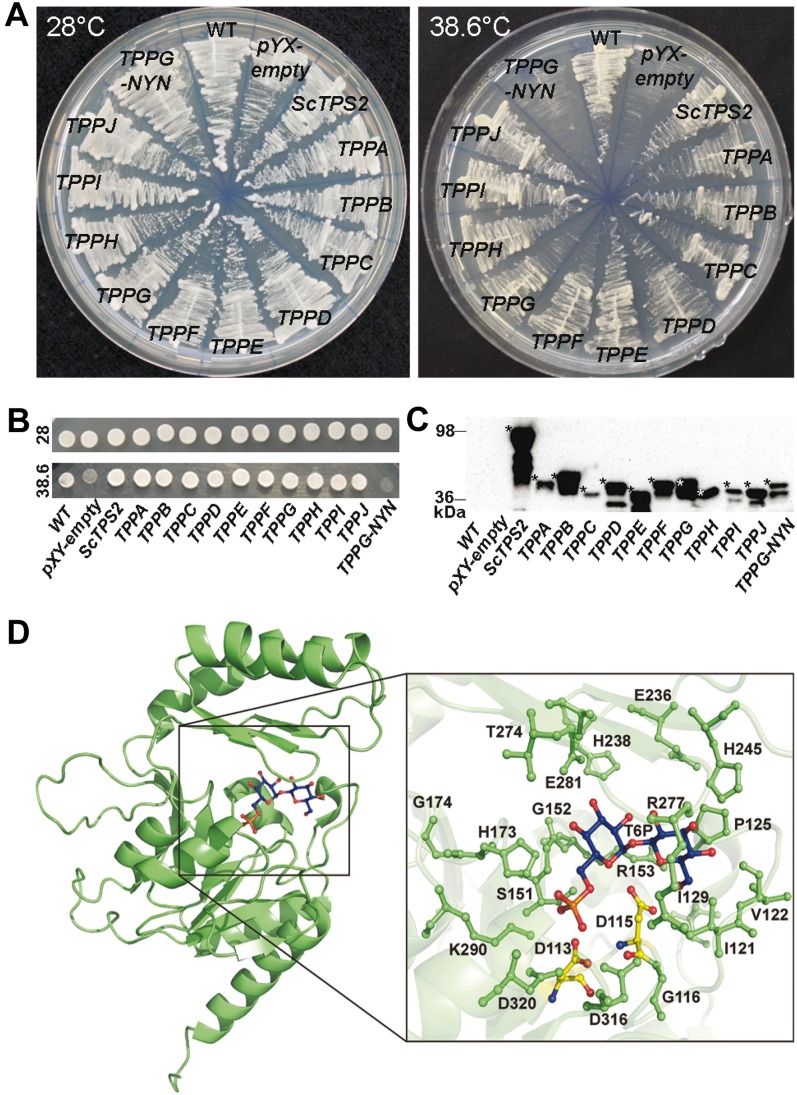
The TPP gene family in higher plants appears to have expanded mainly by whole-genome duplication, but it is not clear whether all have maintained enzymatic activity. To assess TPP activity, we have used a yeast TPP mutant complementation assay. Yeast _tps2_Δ mutants exhibit thermosensitive growth due to accumulation of high levels of T6P at increased temperature (De Virgilio et al., 1993). Previously, complementation of this thermosensitive growth phenotype was used to isolate the TPPA and TPPB genes from an Arabidopsis complementary DNA (cDNA) library (Vogel et al., 1998). We have now analyzed all putative Arabidopsis TPP enzymes in the yeast _tps2_Δ mutant strain with efficient expression of the 10 proteins confirmed by western-blot analysis. All 10 TPP members were able to complement growth deficiency of the yeast _tps2_Δ mutant strain at elevated temperature (Fig. 2, A–C). Based on sequence alignment and the crystal structure of the TPP-related TPP protein from the archaean Thermoplasma acidophylum (TaT6PP; Rao et al., 2006), a strikingly conserved active site is found in all 10 Arabidopsis TPP proteins (Avonce et al., 2006; Rao et al., 2006; Lunn, 2007). We made an overall representation model with T6P modeled in the active site of Arabidopsis TPPG (Fig. 2D). TaT6PP uses the common catalytic reaction mechanism typical for the HAD superfamily, with the active site located within the cavity at the interface of two major domains (Rao et al., 2006). Three sequence motifs make up the active site, with the N-terminal DXDX(T/V) motif (motif I, amino acids 7–11 in TaT6PP) being the most conserved. In TaT6PP, a nucleophilic attack of Asp-7 (equivalent to D113 in TPPG) on the phosphorus atom of T6P is thought to result in a phospho-Asp intermediate, followed by hydrolysis of an activated water molecule, generated by Asp-9 (D115 in TPPG). Asp-9 might also assist as a general acid catalyst, protonating the oxygen atom of the leaving group (Rao et al., 2006). Here, we provide experimental evidence for the importance of these two conserved Asp residues for TPP catalytic activity. Mutation of these Asp residues in TPPG (D113 and D115, TPPG-DYD) to Asn (similar spatial structure as Asp but harboring an uncharged polar R group; TPPG-NYN) abolished yeast _tps2_Δ mutant growth complementation (Fig. 2, A–C; confirmed by bioscreen growth analysis, data not shown).
Figure 2.
The 10 Arabidopsis TPP candidates display heterologous TPP activity. A, Yeast _tps2_Δ growth complementation assay of TPP Arabidopsis proteins at 38.6°C with strains grown at a permissive temperature of 28°C as control treatment. _tps2_Δ yeast strains expressing the _TPPG_-NYN mutant (Asp-113 and Asp-115 of wild-type TPPG were mutated to Asn) showed heat-sensitive growth phenotype, indicating the double mutation causes loss of TPPG catalytic activity. B, For the spot assay, cells of all samples were diluted to OD600 1 and incubated at 28°C and 38.6°C. C, Expression of the HA-tagged proteins was confirmed by western-blot analysis. Asterisks indicate the full-length tagged protein, based on the predicted mass (The Arabidopsis Information Resource) and the prestained protein standard. D, Overall representation model of the TPPG protein (green), and detail of T6P (blue) modeled in the active site of TPPG. Catalytic important residues near the active site are reported in green ball and stick figures. The two Asp of motif I, which are predicted to play a crucial role in the substrate binding and the catalytic mechanism, are indicated in yellow. Asp-113 and Asp-115 of TPPG (TPPG wild type) were mutated to Asn (TPPG-NYN). Figures prepared with PYMOL (DeLano, 2002).
Spatiotemporal Expression Profiles Suggest Functional Diversity among the Arabidopsis TPP Family
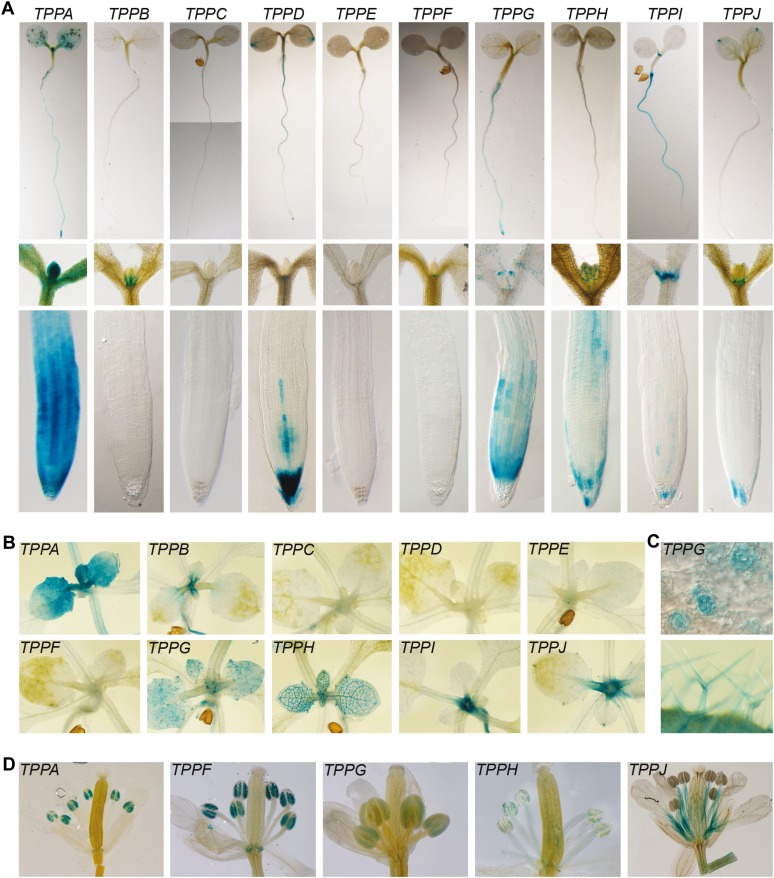
In multicellular organisms, protein function is often tightly linked to tissue- and cell-type-specific expression. As all Arabidopsis TPP proteins seem to have T6P-phosphatase activity, it is important to determine their spatiotemporal expression in planta to get an initial clue about their functional specificity. We therefore fused a 2-kb promoter sequence of each Arabidopsis TPP gene to a GUS/GFP reporter, and introduced these constructs into Arabidopsis ecotype Columbia-0 (Col-0). For each construct, at least three independent homozygous lines (with single insertion) were selected in the T3 generation, and results of homozygous lines with representative expression patterns are shown. A global overview of the expression patterns of the 10 TPP genes in various organs at different developmental stages was obtained with GUS staining and microscopic analyses (Figs. 3 and 4; Supplemental Fig. S2). TPP gene expression occurred in various and distinct organs, from root tips to leaves. At the seedling level, already soon after germination clear differences in TPP gene expression could be observed, with six TPPs expressed in the shoot apical meristem and four TPPs found expressed in the root apical meristem (Figs. 3A and 4). At the tissue level, we can see TPPD expression in the root cap, whereas TPPA and TPPG are expressed in the protoderm (Fig. 3A). In older plants, some TPPs are more highly expressed in young leaves than in mature ones as seen for TPPA, TPPB, and TPPG promoter-GUS lines (Fig. 3B; Supplemental Fig. S2). At the cellular level, some TPPs have particular expression profiles in specific cell types, e.g. TPPG in the stomata and trichomes of actively growing young leaves, TPPH in the vasculature of all leaves and in stomata, and TPPJ in hydathodes (Fig. 3, B and C). During the reproductive stage, five TPPs are found in flowers, especially in the stamens (Fig. 3D; Supplemental Fig. S2). Specific expression in the anthers is found for TPPA, TPPF, TPPG, and TPPH, while TPPJ is expressed in the filament.
Figure 3.
Differential expression profiles of Arabidopsis TPP genes. A, Overview of TPP expression patterns in young seedlings, with images of whole seedlings, and detailed views of the shoot meristemic zones and the root tips. Specific expression profiles of each TPP in seedlings can be noticed. B, Expression patterns of the TPP genes in shoots. Some TPP genes are expressed in the shoot apical meristem as well as in the leaves, e.g. TPPA and TPPG have an epidermal pattern, TPPH is present in vasculature, TPPJ in hydathodes. C, Detailed TPPG expression profile in leaves. TPPG is expressed in trichomes and stomata. D, Overview of TPP gene expression in flowers. Only TPP promoter-GUS lines with detectable GUS expression in flowers are shown.
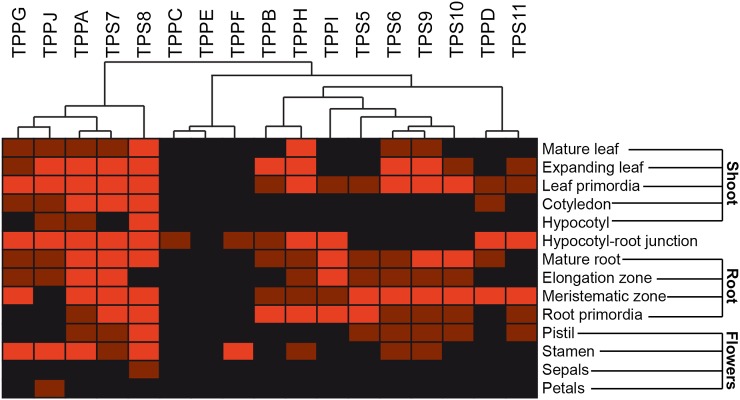
Figure 4.
Clustering of the qualitative expression levels of the trehalose metabolism genes in several tissues of Arabidopsis. TPS7-8 cluster more closely with TPPA-J-G, TPS5-6-9-10 with TPPB-H-I, and TPS11 with TPPD, reflecting similarities in tissue specificity. Clustering was performed with the Genesis program (Sturn et al., 2002). Light red = strong expression; dark red = weak expression; black = no expression.
Publicly available expression data (Genevestigator; Zimmermann et al., 2004; Hruz et al., 2008), and eFP browser (Winter et al., 2007) largely fit with the GUS expression profiles. For example, in accordance with Genevestigator, we detected TPPA, TPPF, TPPG, and TPPH expression in pollen grains (Supplemental Fig. S2). Also consistent with Genevestigator expression levels below background, we did not detect TPPC and TPPE GUS expression. No GUS staining was observed in developing seeds from any of the reporter lines (data not shown). This broadly agrees with the absence of detectable TPP transcripts in developing seeds, except for very low expression of TPPB, TPPG, and TPPH, which was restricted to the chalazal endosperm.
Previously, we have performed similar promoter-GUS/GFP analyses for the seven class II TPS proteins (Ramon et al., 2009). To determine whether there is any overlap in the spatiotemporal expression profiles of the different TPS and TPP genes, we have generated a combined expression map where we clustered expression patterns (Fig. 4). Whereas the class II TPS genes have largely overlapping expression profiles, the TPP genes have more specific expression patterns. We can observe some overlap between the expression profiles of specific TPS and TPP genes. TPS7 and TPS8 cluster more closely with TPPA, TPPJ, and TPPG. They are all broadly expressed with high expression in the leaf primordia, the cotyledons, at the hypocotyl-root junction, and in the stamens. From these genes, TPPA and TPS7 are more or less expressed throughout the whole plant. There is also some overlap between the expression profiles of TPS5, TPS6, TPS9, TPS10, and TPPB, TPPH, and TPPI. Also TPPD and TPS11 seem to be coexpressed in the same tissue types, more specifically in the columella cells in the root meristematic zone.
Functional Analysis of TPPs in Arabidopsis
To assess the functionality of the TPP proteins in Arabidopsis, we analyzed the phenotypes of Arabidopsis plants disrupted in a single TPP by means of available transferred DNA (T-DNA) insertion lines. Screening of 23 lines from different T-DNA insertion collections led to the identification of knockout or knockdown lines of eight TPPs, tppa, tppb, tppd, tppf, tppg, tpph, tppi, and tppj (Supplemental Fig. S3). Seeds of homozygous insertion lines germinated like wild-type Col-0 when sown on soil, and the resulting plants show comparable vegetative and reproductive growth to the wild type, with the exception of tppb mutants producing a larger rosette (Supplemental Fig. S4). Overexpression of the TPPA, TPPF, TPPG, or TPPJ genes, driven by the cauliflower mosaic virus 35S promoter, did not alter the seed germination or vegetative growth characteristics of the plants when grown on soil (Supplemental Fig. S4A). However, overexpression of several of the TPP genes did affect floral morphology, leading to a shorter perianth, the floral structure including petals and sepals (as exemplified by 35S::TPPB plants shown in Supplemental Fig. S4B). The result is that the reproductive organs (pistil and stamen) are not protected or covered by the perianth. Apart from TPPB, we also observed this for TPPC and TPPI overexpression lines (data not shown). As growth phenotypes in the mutant TPP lines were not obvious we investigated the function of the TPP enzymes in the biosynthesis of trehalose by measuring trehalose and T6P contents, along with other sugars and metabolites, in whole seedlings of some of these lines (Table I).
Table I. Levels of T6P, sugar phosphates, sugars, trehalose, and starch in TPP mutants.
Measurements were done in 12-d after germination plants grown on vertical plates supplemented with 1% Suc under continuous light. n = 4 independent experiments, ±sd. *, Data of shoot only; FW, fresh weight.
| Genotype/Metabolite | Col-0 | tppa | tppf | tppg | tppj | Col-0* | tppb* | tppi* | tpph* |
|---|---|---|---|---|---|---|---|---|---|
| T6P (nmol/g FW) | 0.104 ± 0.02 | 0.084 ± 0.01 | 0.079 ± 0.02 | 0.097 ± 0.01 | 0.104 ± 0.02 | 0.262 ± 0.02 | 0.180 ± 0.03 | 0.201 ± 0.03 | 0.272 ± 0.04 |
| Suc6P (nmol/g FW) | 0.365 ± 0.07 | 0.300 ± 0.04 | 0.323 ± 0.06 | 0.327 ± 0.03 | 0.329 ± 0.03 | 0.575 ± 0.09 | 0.516 ± 0.03 | 0.521 ± 0.06 | 0.443 ± 0.05 |
| Glc6P (nmol/g FW) | 199.4 ± 33.5 | 184.8 ± 9.2 | 177.4 ± 10.5 | 178.3 ± 26.5 | 187.1 ± 14.5 | 160.9 ± 18.7 | 192.1 ± 7.6 | 136.7 ± 11.7 | 167.7 ± 3.8 |
| Fru6P (nmol/g FW) | 57.6 ± 12.1 | 49.4 ± 3.8 | 46.8 ± 5.1 | 50.0 ± 6.6 | 52.7 ± 3.4 | 68.4 ± 1.3 | 73.7 ± 2.5 | 58.6 ± 3.3 | 57.7 ± 4.4 |
| UDPG (nmol/g FW) | 110.7 ± 16.7 | 109.8 ± 3.3 | 101.7 ± 1.6 | 102.8 ± 5.8 | 111.7 ± 10.2 | 54.1 ± 7.4 | 50.7 ± 2.4 | 43.3 ± 3.7 | 51.7 ± 7.1 |
| Suc (µmol/g FW) | 2.9 ± 0.4 | 2.7 ± 0.3 | 3.5 ± 0.7 | 3.3 ± 0.5 | 2.5 ± 0.4 | 1.9 ± 0.2 | 1.9 ± 0.2 | 4.5 ± 0.8 | 3.4 ± 0.7 |
| Glc (µmol/g FW) | 4.1 ± 0.3 | 4.5 ± 0.3 | 4.3 ± 0.2 | 4.7 ± 0.3 | 4.0 ± 0.2 | 5.2 ± 1.1 | 3.6 ± 0.1 | 6.6 ± 0.1 | 6.0 ± 0.2 |
| Fru (µmol/g FW) | 1.6 ± 0.1 | 1.9 ± 0.2 | 1.8 ± 0.2 | 1.8 ± 0.3 | 1.8 ± 0.3 | 2.7 ± 0.4 | 1.7 ± 0.5 | 4.2 ± 0.3 | 2.1 ± 0.8 |
| Trehalose (nmol/g FW) | 9.3 ± 1.9 | 7.9 ± 2.7 | 9.5 ± 2.4 | 7.7 ± 1.1 | 6.7 ± 1.3 | 22.9 ± 2.7 | 14.3 ± 3.4 | 21.2 ± 6.1 | 39.1 ± 8.2 |
| Starch (μmol gluc equivalents/g FW) | 14.6 ± 2.1 | 15.4 ± 1.9 | 11.4 ± 9.5 | 16.9 ± 7.7 | 15.5 ± 2.2 | 36.3 ± 3.9 | 29.7 ± 4.9 | 67.0 ± 11.6 | 42.7 ± 14.0 |
Remarkably few differences in the levels of T6P, trehalose, and other metabolites were observed in the tpp mutants compared with the wild type. The most notable changes were a lower level of trehalose in the tppb plants and the higher Suc and starch content in tppi plants. As the TPPs have cell- and tissue-specific expression in planta, it is plausible that the effect of single TPP down-regulation on metabolite contents is diluted in whole-seedling extracts. It is also conceivable that metabolic changes in the cells where TPP expression has been down-regulated might have systemic effects that led to compensatory metabolic adjustments in cells that do not normally express the affected TPP gene.
Mutation of a Single TPP in Arabidopsis, AtTPPG, Causes ABA-Related Phenotypes
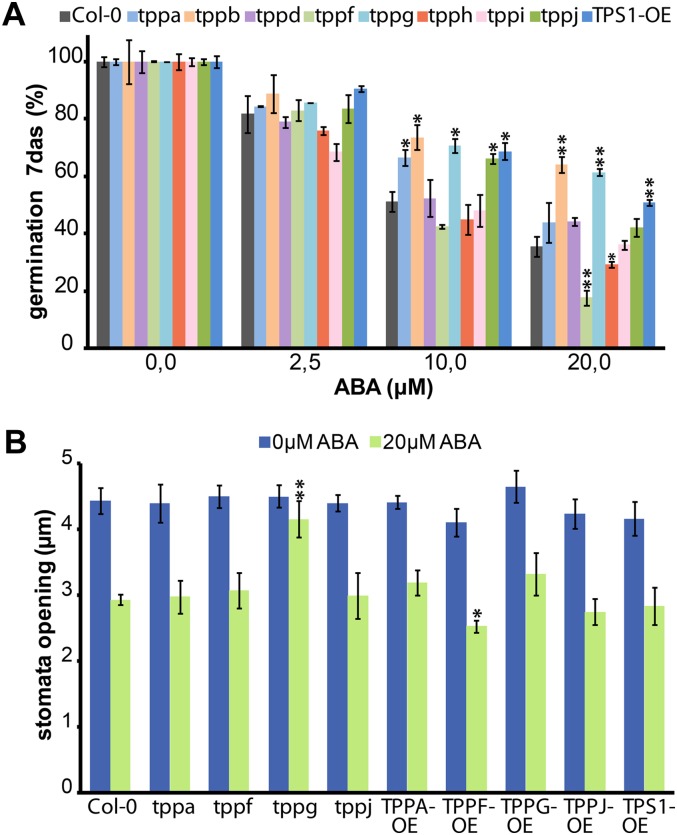
Early reports have linked ABA signaling to trehalose metabolism, specifically with the function of TPS1. To investigate whether TPPs are also involved in this response, we investigated the effects of ABA treatments on germination of tpp mutant seeds (Fig. 5A). At 7 d after sowing, some tpp mutants responded differentially to increasing concentrations of ABA when compared with the wild-type Col-0 control and a 35S::AtTPS1 line, whose germination is not inhibited by ABA (Avonce et al., 2004). The tppb and tppg mutants are insensitive to increasing concentrations of ABA, showing even higher germination rates than the 35S::AtTPS1 positive control line with 20 μm ABA and comparable germination at lower concentrations. The tppf mutant, on the other hand, is hypersensitive to ABA.
Figure 5.
Response of TPP Arabidopsis mutants to ABA. A, Germination inhibition assay with ABA. Seeds of available TPP mutants, _TPS1_-OE, and Col-0 wild-type plants were sown in different ABA concentrations; the percentage of seed germination was calculated 7 d after sowing. tppb and tppg seedlings were able to germinate at high levels of ABA, showing resistance to the inhibition of germination by ABA as seen with the positive control line _TPS1_-OE. The opposite is observed for the tppf line. B, Stomatal aperture size in epidermal strips isolated from some tpp mutants and overexpressing plants in the absence or presence of 20 μm of ABA. Among all the TPP mutants tested only tppg stomata show resistance to ABA promotion of closure.
To investigate further the effect of ABA on the tpp mutants, we looked at the effect of this hormone in stomatal aperture and closure. In the aboveground organs, TPPG is rather specifically expressed in guard cells, and the tppg mutant showed resistance to ABA inhibition of germination. Therefore, we focused our attention on tppg and mutants in its duplicate gene (tppf) and a close homolog (tppa), with one representative from a distant clade (tppj) for comparison (see Supplemental Fig. S1). We also generated and included the overexpressing lines of these TPPs (_TPP_-OE) using a constitutive promoter. Epidermal peels of fully expanded leaves of the above-mentioned tpp mutants and overexpressing lines were incubated under conditions that promote stomatal opening and then treated with 20 μm ABA to induce stomatal closure. As seen in the germination assay, tppg was resistant to ABA, showing no stomatal closure in response to ABA, while the other mutants behaved as the wild type. The OE line responses were similar to the wild type with the interesting exception of _TPPF_-OE, which is slightly more sensitive to the ABA treatment (Fig. 5B). The decreased sensitivity of seed germination and stomatal opening to ABA in the tppg mutant suggests that TPPG might have a particular role in linking trehalose metabolism to ABA signaling in a cell-type-specific way.
DISCUSSION
In Arabidopsis, there appears to be only a single enzyme, TPS1, capable of synthesizing T6P, but a great diversity of TPP enzymes able to hydrolyze T6P to trehalose. As many of the TPP proteins show specific spatiotemporal expression patterns, they clearly have great potential to exert control over T6P levels according to the needs of specific cell and tissue types. Given the substantial and growing evidence that T6P is an essential signal metabolite in plants, with far-reaching influence on plant metabolism, growth, and development, there is an urgent need to understand the origins and physiological significance of the large number of plant TPP proteins.
Collinearity analysis of the TPP gene family strongly indicates that all eudicot and many monocot TPP genes originate from whole-genome duplications. Maere et al. (2005) and Freeling (2009) found that only particular classes of genes preferentially expand through genome duplications in plants, mainly regulatory genes (transcription factors, signal transducers) and genes encoding complex-forming proteins. Similar observations have been made in mammals (Blomme et al., 2006) and other organisms (Maere and Van de Peer, 2010 and refs. therein). A likely explanation for this phenomenon is offered by the dosage balance hypothesis, which postulates that the stoichiometric quantities of proteins functioning within protein complexes and regulatory/signaling cascades must be preserved to ensure proper function (Papp et al., 2003; Birchler et al., 2005; Veitia et al., 2008; Maere and Van de Peer, 2010). This requirement is violated by small-scale gene duplications but fulfilled by whole-genome duplications, where all components of such complexes and/or cascades are simultaneously duplicated. Moreover, after genome duplication, balance-sensitive gene duplicates cannot easily be lost, as this would again give rise to dosage balance effects. The particularly strong genome duplication bias in the history of the TPP gene family therefore hints at a potential regulatory role for the TPP proteins.
The next step to further elucidate the functionality of these proteins was to analyze if these 10 TPP genes in Arabidopsis still encode active T6P-phosphatase enzymes. Heterologous expression in yeast showed that all 10 candidate TPP genes encode heterologously active TPP enzymes; apparently all 10 proteins have conserved their enzymatic function during evolution. We also confirmed the essential role of some conserved residues in the catalytic domain as predicted by an archaean crystal structures, by specific site-directed mutagenesis.
Analyzing promoter GUS/GFP lines, we found very diverse spatiotemporal expression patterns for the 10 Arabidopsis TPP genes, indicating possible functional diversification. In general, our data correlate well with public available gene expression datasets, although there were some differences from the spatiotemporal root expression maps published by Brady et al. (2007), for example, they did not find detectable levels of expression TPPD in the root cap, or TPPA and TPPG in the epidermis. The apparent discrepancies between our promoter-reporter lines and the transcript profiling data could be explained by differences in GUS/GFP and TPP transcript stability, with some TPP transcripts being rapidly turned over in certain cell or tissue types, raising the possibility that the pattern of TPP gene expression in roots is subject to posttranscriptional regulation.
Since trehalose metabolism and especially the intermediate T6P seem to have profound effects on global plant performance, and interfere with plant growth and development, differential spatiotemporal regulation of the trehalose metabolic genes is plausible. Its connection to sugar sensing/signaling suggests that cellular monitoring of, and responses to, subtle fluctuations of T6P levels are essential for carbon homeostasis and the integration of the nutritional status with plant growth and development. The phylogenetic analysis and the differential expression patterns indicate distinct functionality among the different TPPs; a hypothesis supported by available microarray datasets showing TPP expression regulation by various environmental signals (Kreps et al., 2002; Wang et al., 2003; Contento et al., 2004; Scheible et al., 2004; Thimm et al., 2004; Bläsing et al., 2005; Brenner et al., 2005; Liu et al., 2005; Roth et al., 2006), and by published coexpression analyses that cluster every individual TPP in separate networks associated with genes that identify different functional categories (Ma et al., 2007; Li et al., 2008). Since partial overlap is present in the promoter-GUS expression patterns, some redundancy between the TPP isoforms cannot be excluded. Further research is needed to clarify if the TPPs operate by spatiotemporal regulation of T6P levels and/or have a regulatory function uncoupled from their enzymatic TPP activity. To investigate the later possibility, it will be interesting to investigate whether catalytically inactive forms of the enzyme can complement the phenotypes of the tpp loss-of-function mutants, however, this is outside the scope of this study. The class II TPS proteins appear to have been present in the green plant lineage from its earliest evolutionary origins and are structurally related to the yeast TPP (Avonce et al., 2006; Lunn, 2007). While the Arabidopsis class II proteins still contain TPP-like domains, suggesting that they might once have had TPP activity, none of these proteins confers heterologous TPP activity (Ramon et al., 2009). We might speculate that the recruitment of the 10 TPP genes encoding active TPP enzymes may have released the class II proteins from their putative function as TPP enzymes, allowing them to evolve new, apparently beneficial, functions, which may or may not be linked to trehalose biosynthesis or T6P signaling. This might explain why there is little similarity in the expression patterns between TPS and TPP genes.
As is often the case for multigene families, single-mutant phenotypes are hard to find, plausibly because of redundancy among family members. Growth phenotypes in normal conditions were not evident for most of the tpp mutants, but smaller rosette leaves and later flowering were observed in different _TPP_-OE lines (Supplemental Fig. S4). Metabolic phenotypes were also elusive, at least at a whole-plant or whole-rosette level. Even for abundant metabolites, measuring the levels in specific cell or tissue types, for example by laser capture microdissection, is still technically very challenging, and for low-abundance metabolites such as T6P and trehalose is not yet feasible. Emerging technologies such as scanning matrix-assisted laser-desorption ionization time-of-flight mass spectrometry or Fluorescence Resonance Energy Transfer-based reporters promise to increase the resolution of metabolite analysis, even down to the single-cell level, and might eventually reveal metabolic differences in the specific cell or tissue types where TPP expression has been disrupted.
Interestingly, however, a single TPP knockdown did have a relevant effect when subjected to ABA treatment. The phenotype of Arabidopsis plants deficient in TPPG, links its expression in guard cells with a regulatory function in stomatal closure. Although TPPH is also expressed in leaf stomata in some growth conditions, the ABA-related stomatal closure phenotype of the tppg mutant indicates a unique role for TPPG in this response. Whether the stomatal kinetics of the tppg plants are caused by the tissue-specific accumulation of T6P levels in the guard cells (through the loss of the enzyme activity), or by the loss of the TPPG protein itself (which might have a regulatory function, uncoupled from its catalytic activity) will be the subject for further research. Interestingly, former reports with transgenic plants overexpressing yeast TPS1 also link stomatal regulation during drought stress with trehalose metabolism (Gaff, 1996; Stiller et al., 2008). A coexpression analysis linked AtTPPG to DREB2A and DREB2B expression (involved in drought response; Li et al., 2008). The AtTPPG promoter also contains an ABA response element binding site and DRE core motifs (ATHENA; O’Connor et al., 2005; AGRIS, Palaniswamy et al., 2006).
The opposing effects of ABA on germination of mutant and transgenic plants, with tppg mutants being resistant and _TPPG_-OE plants hypersensitive to ABA (Supplemental Fig. S5), further corroborate the strong cross talk present between ABA and trehalose metabolism, previously reported by Avonce et al. (2004) and Gómez et al. (2010). Surprisingly, the duplicated homolog of TPPG, TPPF, appears to display an opposite activity during the germination process, suggesting that control of T6P levels might not be its unique function, but that the enzyme functionally diverged. Another possibility is that TPPF and TPPG may differ in their _K_m for T6P or other kinetic and regulatory properties, and thus have different effects on T6P and/or trehalose levels when up- or down-regulated independently. ABA is known to inhibit the induction of cell wall hydrolases that weaken and break the micropylar endosperm, allowing the radicle to emerge from the seed during the germination process (Müller et al., 2006; Gimeno-Gilles et al., 2009). Consistent with the observed phenotype, TPPG expression is localized in the micropylar endosperm of the developing seed (http://www.seedgenenetwork.net). The differences in ABA sensitivity of the tppg and _TPPG_-OE mutants also fit with former observations of TPS1 Arabidopsis mutant lines, with contrasting ABA sensitivity correlating with the differences in T6P content. Three TILLING alleles of tps1 with reduced T6P levels confer hypersensitivity to ABA in germination assays and defects in ABA regulation of stomatal pore aperture regulation (Gómez et al., 2010), whereas _AtTPS1_-overexpressing Arabidopsis seedlings exhibit ABA insensitivity in germination assays and abolished Glc-induced ABA accumulation on 6% Glc (Avonce et al., 2004, 2005). These observations all indicate that there is a strong link between T6P levels and sensitivity to ABA.
In conclusion, our analyses point to the functional divergence of the large multigene family of TPPs in the model plant Arabidopsis. All 10 genes originate from whole-genome duplications, rather than from small-scale gene duplications, suggesting that this family may exhibit important regulatory functions. Detailed gene expression profiles indicate that tissue-specific regulation of T6P levels might be the key function of this class of 10 catalytically active proteins. The ABA-related stomatal phenotype of a single TPP mutant in Arabidopsis, finally, illustrates the functional divergence and specificity of a single TPP protein.
MATERIALS AND METHODS
Phylogenetic and Collinearity Analyses
The genes used for the collinearity analysis and for the phylogenetic reconstruction of the TPP gene family in the green plant lineage were collected using the PLAZA v1.0 platform (Proost et al., 2009). Sequences were available for the following land plants: Arabidopsis (Arabidopsis thaliana), papaya (Carica papaya), Populus trichocarpa, grapevine (Vitis vinifera), rice (Oryza sativa), Sorghum bicolor, and the moss Physcomitrella patens. The sequence of an Ostreococcus lucimarinus TPP gene belonging to the Chlorophyta was added as an outgroup. Amino acid level multiple sequence alignment was performed with MUSCLE (Edgar, 2004). For phylogenetic reconstruction, we used the TREEFINDER program (Jobb et al., 2004), a program that can infer maximum-likelihood phylogenies based under a variety of models of sequence evolution with the possibility to specify topological constraints and perform tree calibration. We used the JTT protein substitution model (Jones et al., 1992) in combination with the discrete γ heterogeneity model (Yang, 1994) with five rate categories, as suggested by the TREEFINDER model Proposer. Bootstrap analyses were performed with 1,000 maximum-likelihood iterations.
Modeling of T6P in the Active Site of AtTPPG
We modeled binding of the ligand T6P to the active site of Arabidopsis TPPG. A model of TPPG was constructed with MODELER 9v8 (http://salilab.org/modeller/) using the crystal structure of the TPP-related protein from the archaean Thermoplasma acidophylum (TaT6PP, Protein Database ID: 1U02), an Mg2+-dependent phosphatase belonging to the HAD superfamily (Rao et al., 2006) as a template. T6P (taken from Protein Database ID: 1BYK; Hars et al., 1998) was manually placed into the active site of TPPG with COOT (Emsley et al., 2010) according to Rao et al. (2006) and afterward was energy minimized using Crystallography Nuclear magnetic resonance System (Brünger et al., 1998).
Yeast Growth Complementation Assay
For the yeast (Saccharomyces cerevisiae) growth complementation assay, the yeast W303-1A wild-type strain (_Mat_α _leu2_-3, _112 ura3_-_1 trp1_-_1 his3_-11,_15 ade2_-_1 can1_-100 GAL SUC2), the _tps1_Δ deletion strain YSH290 (W303-1A, _tps1_Δ::TRP1), and the _tps2_Δ deletion strain YSH448 (W303-1A, _tps2_Δ::HIS3; Hohmann et al., 1993) were used (see also Ramon et al., 2009; Vandesteene et al., 2010). The different TPP coding sequence (CDS; confirmed by, and if necessary corrected to, the sequence alignment to The Arabidopsis Information Resource Arabidopsis Genome Initiative Coding Sequence dataset), were amplified from Arabidopsis cDNA and cloned in a modified yeast multicopy pYX212 plasmid with an HXT7 promoter and URA3 marker, without stop codon and in frame with C-terminal double hemagglutinin (HA) tag (primers, see Supplemental Table S1). Primers used for AtTPPG-NYN mutagenesis were 5′ATAGCTGTGTTTCTAAATTATAATGGAACACTTTCTC3′ and 5′GAGAAAGTGTTCCATTATAATTTAGAAACACAGCTAT3′. As a control for the complementation assay, the _tps2_Δ strain was also transformed with the empty vector and the yeast TPP TPS2 (Vogel et al., 1998; Zentella et al., 1999; Ramon et al., 2009). Yeast transformation was performed using the one-step method as described by Chen et al. (1992). Cultures of the transformed _tps2_Δ and wild-type control strains were grown overnight at 28°C on synthetic defined without uracyl (SD-ura) containing 2% Glc and drop assays were performed on SD-ura containing 2% Glc and Gal. Transformants were spotted at an optical density at 600 nm (OD600) of 1 and incubated at 28°C (control) or 38.6°C and analyzed after 2 d. For growth curves, precultures were grown in SD-ura with 2% Glc at 28°C. Samples were then diluted to OD600 0.100 in 200-μL SD-ura in 100-well Honeycomb2 plates (Thermo Electron Corporation) in three replicates. Blanks and appropriate growth controls were also included. The transformed _tps2_Δ strains and wild-type strain were grown in SD-ura with 2% Glc at 28°C (control) and 38.6°C. Growth was followed by optical density measurements every 15 min with continuous shaking in the automated microbiology growth analysis system (Bioscreen C; Oy Growth Curves AB Ltd).
Western-Blot Analysis of Heterologous Protein Expression
After overnight growth, 50-mL yeast cell cultures were harvested and resuspended in ice-cold lysis buffer (500 μL 1× phosphate-buffered saline, 0.1% Triton X-100, 10% glycerol, 2.5 mm MgCl2, 1 mm EDTA, pH 8) containing protease inhibitors (complete EDTA-free, Roche). Cells were lysed by vortexing (0.5-mm glass beads). After centrifugation at 4°C for 10 min at maximum speed, protein concentrations in the supernatant were determined using the dye-binding assay (Bradford, 1976) and the soluble extract was subjected to immunoprecipitation using anti-HA rat antibodies 3F10 (Roche) and protein G-agarose beads (Roche). Bead-bound proteins were collected by centrifugation at 4°C (30 s, 2,000 rpm), and washed several times with lysis buffer. Amounts of immunoprecipitated protein did not vary more than 3-fold between samples. SDS-PAGE sample buffer was added, followed by boiling for 5 min. After centrifugation, equal amounts of proteins were loaded on a Mini-Protean precast gel (Bio-rad) together with the protein standard (Invitrogen, SeeBlue prestained protein standard) in a Tris/Gly/SDS running buffer and analyzed using immunoblotting. For detection of HA-tagged proteins, we used horseradish peroxidase-coupled anti-HA high-affinity rat antibodies (1/1,000; Roche). Protein bands close to the predicted size of the respective TPP-HA fusion protein were considered to be full length.
Promoter GUS/GFP Constructs
To study the expression patterns of all TPPs in vivo, we amplified and fused the promoter/5′ untranslated region sequences of around 2 kb of all TPPs to GUS/GFP reporter genes in the pHGWFS7 vector (Karimi et al., 2002) using the Gateway technology (Invitrogen) according to manufacturer’s instructions. Vectors were obtained from the plant systems biology department (VIB, University of Ghent; http://www.psb.ugent.be/gateway). Primers used are listed in Supplemental Table S1. Wild-type Arabidopsis accession Col-0 plants were transformed by floral dip with Agrobacterium tumefaciens (C58C1) containing the promoter constructs (Clough and Bent, 1998). Homozygous plants for at least three (independent and single-insertion) transformed Arabidopsis lines were selected for each TPP promoter construct. Seeds were surface sterilized using chlorine and sown in petri dishes on full-strength Murashige and Skoog salt mixture including vitamins medium (4.4 g/L) with MES (0.5 g/L) and solidified with Phytagar (8 g/L; Duchefa) with HygromycinB following a modified protocol from Harrison et al. (2006).
Histochemical and Histological Analysis
For detailed analysis of seedlings and roots, seedlings were grown for 2 d after germination on vertical plates containing 0.5× Murashige and Skoog and 1% (w/v) Suc in continuous light. For shoot analysis, plants were grown for 15 d after germination, on horizontal plates (as above) with a 16/8-h d/night cycle. For flower analysis, plants were grown on soil for 5 to 6 weeks with a 16/8-h d/night cycle. The GUS-staining assays were performed according to Beeckman and Engler (1994). Seedlings, shoots, and flowers were afterward cleared by mounting them in 90% lactic acid (Acros Organics) on glass microscope slides. Samples were analyzed by differential interference contrast microscopy (Olympus BX51). Qualitative GUS expression levels of the 10 TPP genes in different tissues, were clustered by the Genesis program (Sturn et al., 2002).
Characterization of AtTPP Mutants and AtTPP-OE Lines
T-DNA insertion lines were obtained from European Arabidopsis Stock Centre (Alonso et al., 2003) and genotyped for homozygosity with specific primers according to T-DNA express: Arabidopsis Gene Mapping Tool (http://signal.salk.edu/cgi-bin/tdnaexpress; Supplemental Table S2). _AtTPP_-OE lines were generated by cloning the corresponding TPP CDS with a 2× HA-tagged sequence from the pYX212 vectors generated for the yeast growth complementation assay (primers; Supplemental Table S2), into a pDONR221 donor vector, followed by subcloning in a pK7WG2 expression vector with 35S promoter (Karimi et al., 2007) using the GatewayTM technology (Invitrogen) according to manufacturer’s instructions. Arabidopsis accession Col-0 plants were transformed with the expression vector and selected on kanamycin. Homozygous, independent single-insertion lines were selected by screening of T3 progeny.
TPP expression in the mutant and OE lines was tested with real-time quantitative reverse transcription-PCR. RNA was extracted with an RNeasy kit (Qiagen), and cDNA prepared from 1 μg of total RNA using an iScript cDNA synthesis kit (Bio-Rad) before analysis on a LightCycler 480 real-time PCR instrument (Roche Applied Science) using SYBR Green I master reagent (Roche Applied Science) according to manufacturer’s instructions. All reactions were done in triplicate. Relative expression levels of target genes were quantified with specific primer sets (Supplemental Table S2), and normalized to cDKA 1;1 (At3g48750) and EEF1_α_4 (At5g60390) expression levels.
Metabolite Measurements
Between 10 to 15 plants grown for 11 d after germination in vertical plates on standard one-half-strength Murashige and Skoog supplemented with 1% Suc under continuous light (110 μE m−2 s−1 photosynthetically active radiation) at 22°C, were pooled and frozen immediately in liquid nitrogen. Tre6P, phosphorylated intermediates, and organic acids were measured by liquid chromatography coupled to tandem mass spectrometry as described in Lunn et al. (2006). Suc, Glc, and Fru contents were measured in ethanol extracts as described by Jelitto et al. (1992) and starch was measured in the ethanol-insoluble residue according to Hendriks et al. (2003) Trehalose was measured in the same plant extracts used for Tre6P measurements (Lunn et al., 2006) by fluorimetric assay with coupling via trehalase and Glc oxidase and Amplex Red (Invitrogen).
ABA Germination Assay
AtTPP, p35S::TPS1, and Col-0 wild-type seeds were sown in 0.5× Murashige and Skoog supplemented with 1% Suc and different concentrations (0.0, 2.5, 10.0, and 20.0 μm) of ABA (Sigma). The percentage of germinated seeds was scored daily for 7 d.
Measurements of Stomatal Aperture
The first pair of fully expanded leaves of 3-week-old plants, grown in soil 16/8-h d/night cycle, was detached and incubated in a stomata opening buffer (10 mm MES-KOH pH 6.15, 10 mm KCl) for 2 h under continuous light. Twenty micromoles of ABA (0 μm in controls) was added to the buffer and leaves were incubated 2 h more. Peels of the abaxial side of the leaf were taken by pasting the abaxial side of the leaf to a microscope slide with double-sided sticky tape, the mesophyll cells were removed by gently scraping the leaf with a blade, and the abaxial epidermis could be visualized in a microscope (Olympus BX51). Pictures of stomata were taken and pore aperture was measured using ImageJ software.
Supplemental Data
The following materials are available in the online version of this article.
- Supplemental Figure S1. Phylogenetic tree of plant TPPs.
- Supplemental Figure S2. Expression of TPPs in pollen grains.
- Supplemental Figure S3. Quantitative PCR on TPP mutant and overexpressing lines.
- Supplemental Figure S4. Shoot phenotype of TPP mutant lines.
- Supplemental Figure S5. Germination inhibition assay with ABA in different concentrations shows hypersensitivity to ABA in TPPG-OE.
- Supplemental Table S1. Primers used for cloning of promoter and CDS.
- Supplemental Table S2. T-DNA lines; primers to detect T-DNA insertions; primers to check TPP expression levels in mutant lines of the 10 AtTPP genes.
Acknowledgments
The authors thank Gustavo Gudesblat for his expertise and help with the ABA stomata experiments.
Glossary
T6P
trehalose-6-phosphate
TPS
T6P synthase
TPP
T6P phosphatase
ABA
abscisic acid
HAD
l-2-haloacid dehalogenase
Col-0
Columbia-0
HA
hemagglutinin
OD600
optical density at 600 nm
cDNA
complementary DNA
T-DNA
transferred DNA
CDS
coding sequence
SD-ura
synthetic defined without uracyl
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Mascorro-Gallardo JO, Van Dijck P, Thevelein JM, Iturriaga G. (2004) The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol 136: 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Leyman B, Thevelein JM, Iturriaga G. (2005) Trehalose metabolism and glucose sensing in plants. Biochem Soc Trans 33: 276–279 [DOI] [PubMed] [Google Scholar]
- Avonce N, Mendoza-Vargas A, Morett E, Iturriaga G. (2006) Insights on the evolution of trehalose biosynthesis. BMC Evol Biol 6: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N, Wuyts J, Verschooten K, Vandesteene L, Van Dijck P. (2010) The Cytophaga hutchinsonii ChTPSP: first characterized bifunctional TPS-TPP protein as putative ancestor of all eukaryotic trehalose biosynthesis proteins. Mol Biol Evol 27: 359–369 [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Engler G. (1994) An easy technique for the clearing of histochemically stained plant tissue. Plant Mol Biol Rep 12: 37–42 [Google Scholar]
- Birchler JA, Riddle NC, Auger DL, Veitia RA. (2005) Dosage balance in gene regulation: biological implications. Trends Genet 21: 219–226 [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Gibon Y, Günther M, Höhne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible W-R, Stitt M. (2005) Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell 17: 3257–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Santos E, Flores CL, Martínez-Zapater JM, Salinas J, Gancedo C. (1998) Isolation and molecular characterization of the Arabidopsis TPS1 gene, encoding trehalose-6-phosphate synthase. Plant J 13: 685–689 [DOI] [PubMed] [Google Scholar]
- Blomme T, Vandepoele K, De Bodt S, Simillion C, Maere S, Van de Peer Y. (2006) The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol 7: R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN. (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Brenner WG, Romanov GA, Köllmer I, Bürkle L, Schmülling T. (2005) Immediate-early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J 44: 314–333 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Cabib E, Leloir LF. (1958) The biosynthesis of trehalose phosphate. J Biol Chem 231: 259–275 [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV. (2008) Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT. (1992) One-step transformation of yeast in stationary phase. Curr Genet 21: 83–84 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim SJ, Bassham DC. (2004) Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiol 135: 2330–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui LY, Wall PK, Leebens-Mack JH, Lindsay BG, Soltis DE, Doyle JJ, Soltis PS, Carlson JE, Arumuganathan K, Barakat A, et al. (2006) Widespread genome duplications throughout the history of flowering plants. Genome Res 16: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Bürckert N, Bell W, Jenö P, Boller T, Wiemken A. (1993) Disruption of TPS2, the gene encoding the 100-kDa subunit of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiae, causes accumulation of trehalose-6-phosphate and loss of trehalose-6-phosphate phosphatase activity. Eur J Biochem 212: 315–323 [DOI] [PubMed] [Google Scholar]
- DeLano WL. (2002) The pyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA
- Eastmond PJ, Li Y, Graham IA. (2003) Is trehalose-6-phosphate a regulator of sugar metabolism in plants? J Exp Bot 54: 533–537 [DOI] [PubMed] [Google Scholar]
- Eastmond PJ, van Dijken AJH, Spielman M, Kerr A, Tissier AF, Dickinson HG, Jones JDG, Smeekens SC, Graham IA. (2002) Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J 29: 225–235 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein AD, Pan YT, Pastuszak I, Carroll D. (2003) New insights on trehalose: a multifunctional molecule. Glycobiology 13: 17R–27R [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y. (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci USA 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Gaff D. (1996) Tobacco-plant desiccation tolerance. Nature 382: 502 [Google Scholar]
- Geelen D, Royackers K, Vanstraelen M, Inzé D, Van Dijck P, Thevelein JM, Leyman B. (2007) Trehalose-6-P synthase: AtTPS1 high molecular weight complexes in yeast and Arabidopsis. Plant Sci 173: 426–437 [Google Scholar]
- Gimeno-Gilles C, Lelièvre E, Viau L, Malik-Ghulam M, Ricoult C, Niebel A, Leduc N, Limami AM. (2009) ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in Medicago truncatula embryo axis. Mol Plant 2: 108–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddijn OJM, Verwoerd TC, Voogd E, Krutwagen RWHH, de Graaf PTHM, van Dun K, Poels J, Ponstein AS, Damm B, Pen J. (1997) Inhibition of trehalase activity enhances trehalose accumulation in transgenic plants. Plant Physiol 113: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez LD, Gilday A, Feil R, Lunn JE, Graham IA. (2010) AtTPS1-mediated trehalose 6-phosphate synthesis is essential for embryogenic and vegetative growth and responsiveness to ABA in germinating seeds and stomatal guard cells. Plant J 64: 1–13 [DOI] [PubMed] [Google Scholar]
- Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A. (2006) A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hars U, Horlacher R, Boos W, Welte W, Diederichs K. (1998) Crystal structure of the effector-binding domain of the trehalose-repressor of Escherichia coli, a member of the LacI family, in its complexes with inducer trehalose-6-phosphate and noninducer trehalose. Protein Sci 7: 2511–2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks JHM, Kolbe A, Gibon Y, Stitt M, Geigenberger P. (2003) ADP-glucose pyrophosphorylase is activated by posttranslational redox-modification in response to light and to sugars in leaves of Arabidopsis and other plant species. Plant Physiol 133: 838–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S, Neves MJ, de Koning W, Alijo R, Ramos J, Thevelein JM. (1993) The growth and signalling defects of the ggs1 (fdp1/byp1) deletion mutant on glucose are suppressed by a deletion of the gene encoding hexokinase PII. Curr Genet 23: 281–289 [DOI] [PubMed] [Google Scholar]
- Holmström KO, Mäntylä E, Welin B, Mandal A, Palva ET, Tunnela OE, Londesborough J. (1996) Drought tolerance in tobacco. Nature 379: 683–684 [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- Jelitto T, Sonnewald U, Willmitzer L, Hajitezaei MR, Stitt M. (1992) Inorganic pyrophosphate content and metabolites in leaves and tubers of potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. (2011) Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Jobb G, von Haeseler A, Strimmer K. (2004) TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones DT, Taylor WR, Thornton JM. (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P. (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. (2002) Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 130: 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Van Dijck P, Thevelein JM. (2001) An unexpected plethora of trehalose biosynthesis genes in Arabidopsis thaliana. Trends Plant Sci 6: 510–513 [DOI] [PubMed] [Google Scholar]
- Li P, Ma S, Bohnert HJ. (2008) Coexpression characteristics of trehalose-6-phosphate phosphatase subfamily genes reveal different functions in a network context. Physiol Plant 133: 544–556 [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J. (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE. (2007) Gene families and evolution of trehalose metabolism in plants. Funct Plant Biol 34: 550–563 [DOI] [PubMed] [Google Scholar]
- Lunn JE, Feil R, Hendriks JHM, Gibon Y, Morcuende R, Osuna D, Scheible W-R, Carillo P, Hajirezaei MR, Stitt M. (2006) Sugar-induced increases in trehalose 6-phosphate are correlated with redox activation of ADPglucose pyrophosphorylase and higher rates of starch synthesis in Arabidopsis thaliana. Biochem J 397: 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Gong Q, Bohnert HJ. (2007) An Arabidopsis gene network based on the graphical Gaussian model. Genome Res 17: 1614–1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Van de Peer Y. (2010) Duplicate retention after small- and large-scale duplications. In DA Liberles, K Dittmar, eds, Evolution after Gene Duplication. Wiley-Blackwell, New York, pp 31-56
- Masterson J. (1994) Stomatal size in fossil plants: evidence for polyploidy in majority of angiosperms. Science 264: 421–424 [DOI] [PubMed] [Google Scholar]
- Miranda JA, Avonce N, Suárez R, Thevelein JM, Van Dijck P, Iturriaga G. (2007) A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 226: 1411–1421 [DOI] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ. (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Palaniswamy SK, James S, Sun H, Lamb RS, Davuluri RV, Grotewold E. (2006) AGRIS and AtRegNet: a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol 140: 818–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424: 194–197 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul MJ. (2008) Trehalose 6-phosphate: a signal of sucrose status. Biochem J 412: e1–e2 [DOI] [PubMed] [Google Scholar]
- Pramanik MH, Imai R. (2005) Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol 58: 751–762 [DOI] [PubMed] [Google Scholar]
- Proost S, Van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. (2009) PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell 21: 3718–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramon M, De Smet I, Vandesteene L, Naudts M, Leyman B, Van Dijck P, Rolland F, Beeckman T, Thevelein JM. (2009) Extensive expression regulation and lack of heterologous enzymatic activity of the class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ 32: 1015–1032 [DOI] [PubMed] [Google Scholar]
- Rao KN, Kumaran D, Seetharaman J, Bonanno JB, Burley SK, Swaminathan S. (2006) Crystal structure of trehalose-6-phosphate phosphatase-related protein: biochemical and biological implications. Protein Sci 15: 1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud P-F, Lindquist EA, Kamisugi Y, et al. (2008) The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319: 64–69 [DOI] [PubMed] [Google Scholar]
- Romero C, Bellés JM, Vayá JL, Serrano R, Culiáñez-Macià FA. (1997) Expression of the yeast trehalose-6-phosphate synthase gene in transgenic tobacco plants: pleiotropic phenotypes include drought tolerance. Planta 201: 293–297 [DOI] [PubMed] [Google Scholar]
- Roth U, von Roepenack-Lahaye E, Clemens S. (2006) Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J Exp Bot 57: 4003–4013 [DOI] [PubMed] [Google Scholar]
- Satoh-Nagasawa N, Nagasawa N, Malcomber S, Sakai H, Jackson D. (2006) A trehalose metabolic enzyme controls inflorescence architecture in maize. Nature 441: 227–230 [DOI] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, Fritz C, Osuna D, Palacios-Rojas N, Schindelasch D, Thimm O, Udvardi MK, Stitt M. (2004) Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol 136: 2483–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluepmann H, Pellny T, van Dijken A, Smeekens S, Paul M. (2003) Trehalose 6-phosphate is indispensable for carbohydrate utilization and growth in Arabidopsis thaliana. Proc Natl Acad Sci USA 100: 6849–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S, Matsui H, Tahara S, Imai R. (2007) Biochemical characterization of rice trehalose-6-phosphate phosphatases supports distinctive functions of these plant enzymes. FEBS J 274: 1192–1201 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack JH, Bell CD, Paterson AH, Zheng CF, Sankoff D, Depamphilis CW, Wall PK, Soltis PS. (2009) Polyploidy and angiosperm diversification. Am J Bot 96: 336–348 [DOI] [PubMed] [Google Scholar]
- Stiller I, Dulai S, Kondrák M, Tarnai R, Szabó L, Toldi O, Bánfalvi Z. (2008) Effects of drought on water content and photosynthetic parameters in potato plants expressing the trehalose-6-phosphate synthase gene of Saccharomyces cerevisiae. Planta 227: 299–308 [DOI] [PubMed] [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z. (2002) Genesis: cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Tang HB, Bowers JE, Wang XY, Ming R, Alam M, Paterson AH. (2008) Synteny and collinearity in plant genomes. Science 320: 486–488 [DOI] [PubMed] [Google Scholar]
- Thaller MC, Schippa S, Rossolini GM. (1998) Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci 7: 1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Usadel B, Bläsing OE, Gibon Y, Retzlaff K, Höhne M, Günther M, Stitt M. (2008) Global transcript levels respond to small changes of the carbon status during progressive exhaustion of carbohydrates in Arabidopsis rosettes. Plant Physiol 146: 1834–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10: 725–732 [DOI] [PubMed] [Google Scholar]
- Van Dijck P, Mascorro-Gallardo JO, De Bus M, Royackers K, Iturriaga G, Thevelein JM. (2002) Truncation of Arabidopsis thaliana and Selaginella lepidophylla trehalose-6-phosphate synthase unlocks high catalytic activity and supports high trehalose levels on expression in yeast. Biochem J 366: 63–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijken AJ, Schluepmann H, Smeekens SC. (2004) Arabidopsis trehalose-6-phosphate synthase 1 is essential for normal vegetative growth and transition to flowering. Plant Physiol 135: 969–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesteene L, Ramon M, Le Roy K, Van Dijck P, Rolland F. (2010) A single active trehalose-6-P synthase (TPS) and a family of putative regulatory TPS-like proteins in Arabidopsis. Mol Plant 3: 406–419 [DOI] [PubMed] [Google Scholar]
- Veitia RA, Bottani S, Birchler JA. (2008) Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet 24: 390–397 [DOI] [PubMed] [Google Scholar]
- Vogel G, Aeschbacher RA, Müller J, Boller T, Wiemken A. (1998) Trehalose-6-phosphate phosphatases from Arabidopsis thaliana: identification by functional complementation of the yeast tps2 mutant. Plant J 13: 673–683 [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Delatte TL, O’Hara LE, Primavesi LF, Jhurreea D, Paul MJ, Schluepmann H. (2012) Trehalose 6-phosphate is required for the onset of leaf senescence associated with high carbon availability. Plant Physiol 158: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. (1994) Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39: 306–314 [DOI] [PubMed] [Google Scholar]
- Zentella R, Mascorro-Gallardo JO, Van Dijck P, Folch-Mallol J, Bonini B, Van Vaeck C, Gaxiola R, Covarrubias AA, Nieto-Sotelo J, Thevelein JM, et al. (1999) A Selaginella lepidophylla trehalose-6-phosphate synthase complements growth and stress-tolerance defects in a yeast tps1 mutant. Plant Physiol 119: 1473–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Primavesi LF, Jhurreea D, Andralojc PJ, Mitchell RA, Powers SJ, Schluepmann H, Delatte T, Wingler A, Paul MJ. (2009) Inhibition of SNF1-related protein kinase1 activity and regulation of metabolic pathways by trehalose-6-phosphate. Plant Physiol 149: 1860–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]