Familial and sporadic 15q13.3 microdeletions in idiopathic generalized epilepsy: precedent for disorders with complex inheritance (original) (raw)
Abstract
Microdeletion at chromosomal position 15q13.3 has been described in intellectual disability, autism spectrum disorders, schizophrenia and recently in idiopathic generalized epilepsy (IGE). Using independent IGE cohorts, we first aimed to confirm the association of 15q13.3 deletions and IGE. We then set out to determine the relative occurrence of sporadic and familial cases and to examine the likelihood of having seizures for individuals with the microdeletion in familial cases. The 15q13.3 microdeletion was identified in 7 of 539 (1.3%) unrelated cases of IGE using quantitative PCR or SNP arrays and confirmed by array comparative genomic hybridization analysis using probes specific to the 15q13.3 region. The inheritance of this lesion was tracked using family studies. Of the seven microdeletions identified in probands, three were de novo, two were transmitted from an unaffected parent and in two cases the parents were unavailable. Non-penetrance of the microdeletion was identified in 4/7 pedigrees and three pedigrees included other family members with IGE who lacked the 15q13.3 deletion. The odds ratio is 68 (95% confidence interval 29–181), indicating a pathogenic lesion predisposing to epilepsy with complex inheritance and incomplete penetrance for the IGE component of the phenotype in multiplex families.
INTRODUCTION
Idiopathic generalized epilepsy (IGE) accounts for 20–30% of epilepsy in the general population. The disorder usually follows complex inheritance although rare autosomal dominant families have been described (1). The mutations involved in the dominant forms confer very high risk to individuals who carry them but account for very little of the population risk. Candidate gene association studies and linkage studies across multiplex families to detect susceptibility regions have been reported, but their results have been disappointing (2–4).
Copy number variants (CNVs) are increasingly recognized as an important component of human genetic variation and a potentially significant contributor to disease risk. A recent study found a recurrent microdeletion at 15q13.3 present in 1% of IGE cases (5). This microdeletion was previously identified in intellectual disability (6) and then in schizophrenia (7,8) and autism (9,10). We aimed to confirm the initial finding in independent IGE cohorts and to examine the pattern of inheritance associated with 15q13.3 microdeletions. Specifically, we asked how this lesion was distributed across IGE subsyndromes and to what extent it was vertically transmitted in multiplex families. By an analysis of families associated with each of the index cases, we sought to resolve whether the microdeletion behaved as a highly penetrant mutation or conferred susceptibility for IGE with complex inheritance.
RESULTS
Detection of 15q13.3 microdeletions in IGE patients
We examined 539 patients diagnosed with an IGE syndrome according to the classification of the International League Against Epilepsy (11). All patients had at least two seizures, generalized spike wave on EEG and onset under 30 years of age. They did not have schizophrenia, autism or intellectual disability. The relatives of probands with a microdeletion underwent phenotyping including epilepsy syndrome, developmental history and outcome. The 293 Australian cases and the 246 mixed European cohort (from Turkey, Germany, Italy, Belgium, Netherlands, Denmark, Finland and Austria) were further divided into the IGE subsyndromes childhood absence epilepsy (CAE, 186 patients), juvenile absence epilepsy (JAE, 94 patients), juvenile myoclonic epilepsy (JME, 183 patients) and epilepsy with generalized tonic–clonic seizures alone (GTCSA, 76 patients) (Table 1). The country of origin and the number of subjects with each IGE subtype in the European cohort are shown in Table 2. The 494 Australian controls were drawn from anonymous blood donors and the 3283 German controls were from subjects with various common diseases (atopy n = 802, primary sclerosong cholangitis n = 253, sarcoidosis n = 602, ulcerous colitis n = 972) and a cohort of individuals of high age (n = 654), where epilepsies and other neurodevelopmental disorders are expected to occur at population frequencies.
Table 1.
Phenotypes of the extended EPICURE sample, gender breakdown
| Phenotype | n | Male | Female |
|---|---|---|---|
| CAE | 88 | 29 | 59 |
| JAE | 37 | 11 | 26 |
| JME | 79 | 19 | 60 |
| EGTCS | 42 | 19 | 23 |
| Total | 246 | 78 | 168 |
Table 2.
Phenotypes of the extended EPICURE sample, country of origin
| Country | Total | CAE | JAE | JME | EGTCS |
|---|---|---|---|---|---|
| Turkey | 62 | 41 | 9 | 10 | 2 |
| Germany | 57 | 6 | 11 | 18 | 22 |
| Italy | 40 | 15 | 5 | 9 | 11 |
| Belgium | 35 | 12 | 4 | 15 | 4 |
| Netherlands | 28 | 8 | 3 | 17 | |
| Denmark | 12 | 2 | 8 | 2 | |
| Finland | 10 | 6 | 3 | 1 | |
| Austria | 2 | 1 | 1 | ||
| Total | 246 | 88 | 37 | 79 | 42 |
In total, 405 IGE patients and 494 Australian controls were screened by quantitative PCR (qPCR), and 134 IGE patients and 3283 European controls were screened by Affymetrix Human SNP Array 6.0 to detect deletions and their reciprocal duplications. The micro-chromosomal abnormalities in the 15q13.3 region detected by these screening technologies were confirmed by array comparative genomic hybridization (CGH) using customized arrays covering the critical region (Fig. 1). Where copy number changes were identified, parents and first degree relatives where available were also screened by qPCR and array CGH. Six 15q13.3 microdeletions were found in the Australian IGE cases (Families 2–6) and one microdeletion was found in a European (Turkish) IGE proband (Family 1), thus a total of seven microdeletions were found from a total of 539 IGE cases. The frequency of the 15q13.3 microdeletion in our IGE cohort was 1.3%. A microdeletion was found in three cases with the IGE subsyndrome JME, three cases with CAE (one with low normal intellect, one evolving to JME) and one with JAE. No deletions were identified in the 494 Australian or the 3283 European controls. The distribution of 15q13.3 microdeletions among IGE sub-syndromes appears random based on current ascertainments (Fisher's exact, all P > 0.4). Association between the IGE phenotype and the 15q13.3 microdeletion was investigated using Fisher's exact test. In total, 15q13.3 microdeletions were identified in 7/539 cases with IGE and 0/3777 controls (P = 4.6 × 10−7).
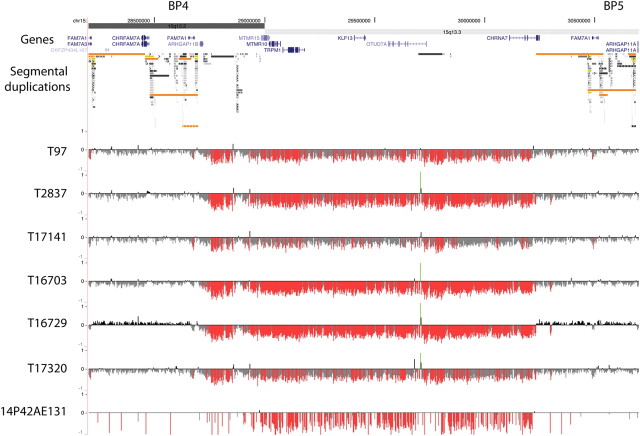
Figure 1.
High-density oligonucleotide array CGH analysis of 15q13.3 microdeletions in patients with idiopathic generalized epilepsy. The 15q13.3 microdeletions were confirmed using high-density oligonucelotide array CGH (NCBI Build 36, chr15: 28 200 000–30 700 000 shown). The minimal deletion region lies between the segmental duplication clusters at breakpoints 4 (BP4) and 5 (BP5) and contains six RefSeq genes, shown in blue. Segmental duplications of increasing similarity (90–98%, 98–99% and >99%) are represented by grey/yellow/orange bars, respectively. For each individual, deviations of probe log2 ratios from zero are depicted by grey/black lines, with those exceeding a threshold of 1.5 standard deviations from the mean probe ratio colored green and red to represent relative gains and losses, respectively.
Segregation analysis of the 15q13.3 microdeletions
The 15q13.3 microdeletions arose de novo in three probands (Fig. 2, Family 1, 2 and 5) as it was not present in either parent. Two probands inherited the microdeletion from an unaffected parent (Families 4 and 7) and the parental status was not able to be determined for two probands (Families 3 and 6). Parentage in Families 2, 4 and 5 was confirmed using eight highly polymorphic microsatellite markers. The microdeletion did not account for all the epilepsy risk in these families as there were both multiple affected individuals without 15q13.3 microdeletions and unaffected individuals with the deletion; this signifies segregation of the microdeletion as a susceptibility variant. Overall, wherever there was at least one relative carrying the microdeletion, apart from the proband, non-penetrance was demonstrated (four of seven cases) and in only one of four multiplex families did all affected members carry the microdeletion (Family 4). This finding presents a more accurate perspective than the impression given by the strong association detected with the 15q13.3 microdeletion in the case–control approach.
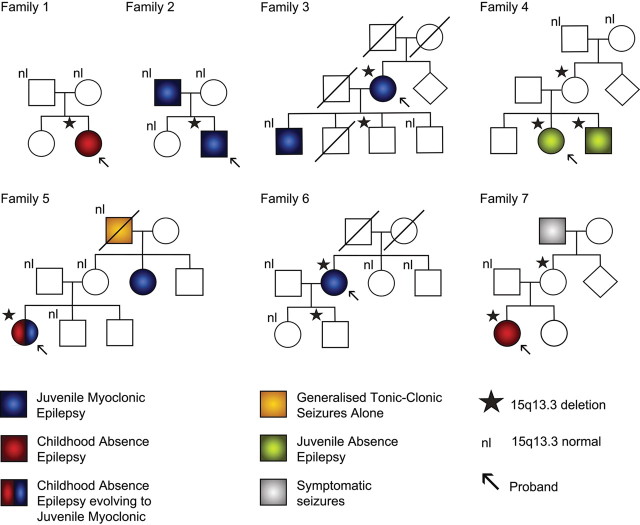
Figure 2.
Family pedigrees of probands with 15q13.3 microdeletions showing only partial segregation with affected status. Family 1 is Turkish and Families 2–7 are Australian. Three microdeletions arose de novo in the proband in three families (1, 2 and 5), two were inherited (4 and 7) and in two families the parents of the proband were not available for study (3 and 6). Transmission of the microdeletion occurred in four families (3, 4, 6 and 7) and in all cases at least one unaffected individual carries the microdeletion. Four families have multiple individuals affected with IGE (2, 3, 4 and 5) and in one family (4) the microdeletion is present in all affected members of the family. Overall 8/12 subjects with the deletion are affected while 8/12 of those affected with IGE have the microdeletion. nl: signifies subjects who have been tested and who do not have the 15q13.3 microdeletion.
DISCUSSION
This study confirms in the independent cohorts examined that the 15q13.3 microdeletion is overrepresented among cases diagnosed with IGE and is present at frequencies of 1–2%. The frequencies in other non-epilepsy phenotypes associated with the 15q13.3 microdeletion are lower than in IGE, the highest being 0.3% in autism (9). IGE is thus the most frequent phenotype associated with the 15q13.3 microdeletion. A reliable estimate of the frequency of the microdeletion in the general population of 0.02% can be deduced from control cohorts in various previous publications investigating the 15q13.3 microdeletion, in which more than 50 000 controls have been genotyped in total (Table 3).
Table 3.
15q13.3 deletions in different cohorts
| IGE | Origin | Total | 15q13.3 del | Frequency |
|---|---|---|---|---|
| Australiaa | 293 | 6 | 0.02 | |
| Turkeya | 62 | 1 | 0.02 | |
| Germanya | 57 | 0 | 0 | |
| Italya | 40 | 0 | 0 | |
| Belgiuma | 35 | 0 | 0 | |
| Netherlandsa | 28 | 0 | 0 | |
| Denmarka | 12 | 0 | 0 | |
| Finlanda | 10 | 0 | 0 | |
| Austriaa | 2 | 0 | 0 | |
| Western Europeb | 647 | 7 | 0.01 | |
| Switzerlandb | 205 | 2 | 0.01 | |
| USAb | 133 | 1 | 0.01 | |
| Northern Europeb | 238 | 2 | 0.01 | |
| This papera | 539 | 7 | 0.01 | |
| Total | 1762 | 19 | 0.01 | |
| Controls | ||||
| Australiaa | 494 | 0 | 0 | |
| Germanya | 3283 | 0 | 0 | |
| Germanyb | 1202 | 0 | 0 | |
| USAb | 2497 | 0 | 0 | |
| Icelandc | 32 442 | 7 | 0.0002 | |
| Scotlandc | 670 | 0 | 0 | |
| Netherlandsc | 4039 | 1 | 0.0002 | |
| Germanyc | 1034 | 0 | 0 | |
| Denmarkc | 501 | 0 | 0 | |
| Norwayc | 272 | 0 | 0 | |
| Finlandc | 200 | 0 | 0 | |
| UKc | 96 | 0 | 0 | |
| Italyc | 91 | 0 | 0 | |
| Scotlandd | 1097 | 0 | 0 | |
| Irelandd | 914 | 0 | 0 | |
| Bulgariad | 646 | 0 | 0 | |
| Swedend | 437 | 0 | 0 | |
| Portugald | 200 | 0 | 0 | |
| This papera | 3777 | 0 | 0 | |
| Total | 50 115 | 8 | 0.0002 |
If our data are combined with a previous report (5) (19 of 1762 IGE cases), this gives an odds ratio of 68.2 for IGE susceptibility in subjects with the 15q13.3 microdeletion (95% confidence interval 28.5–181.2) (Table 4). In interpreting the odds ratio, the population risk of disease needs to be considered. The prevalence of IGE is relatively low with about 1 in 200 people (0.5%) developing it during their lifetime. The odds ratio is a measure of the increase in risk conveyed by a risk factor, in this case the 15q13.3 microdeletion. Although a 68-fold increase in risk appears very high, this would increase the risk for an individual with a 15q13.3 deletion from 1 in 200 to around 1 in 3. Although the upper end of the confidence interval for the odds ratio might be consistent with dominant inheritance, the likely value is suggestive of the complex inheritance demonstrated by the segregation in families.
Table 4.
Association results in current cohort and previous data
| Cases | Controls | OR (95% CI) | _P_-value | |
|---|---|---|---|---|
| Current study | 7/539 | 0/3777 | Infinite (10.2–inf.) | 4.6 × 10−7 |
| Combined previous data | 12/1223 | 8/46 338 | 57.3 (21.5–163.3) | 8.3 × 10−15 |
| All data combined | 19/1762 | 8/50 115 | 68.2 (28.5–181.2) | <2.2 × 10−16 |
The 15q13.3 microdeletions encompass seven genes including the alpha7 nicotinic receptor subunit gene (CHRNA7). This gene may play a role in epilepsy genetics as mutations in other members of the nicotinic receptor subunit gene family (CHRNA2, CHRNA4 and CHRNB2) cause the familial idiopathic epilepsy autosomal dominant nocturnal frontal lobe epilepsy. Whether it is the loss of the CHRNA7 gene alone, or the combined loss of several genes constituting a contiguous gene syndrome, or the alteration of regulatory mechanisms that contributes to the interplay of mechanisms causing IGE or other morbidities is yet to be determined (12). The 15q13.3 microdeletion can be non-penetrant where it segregates in families, as observed in this and a previous report (5). Moreover, the microdeletion is not necessarily present in all affected family members either, which is consistent with how the genetic architecture for the epilepsies with complex inheritance can be conceptualized (13).
Overall, although the 15q13.3 microdeletion shows a very strong association with IGE in case–control studies, it does not segregate as a dominant Mendelian allele in multiplex families. It behaves as a susceptibility component in a polygenic model where a combination of susceptibility alleles contributes to the phenotype in any one patient. While it is possible to formally predict using population data, the proportion of subjects with the microdeletion likely to develop IGE, such estimates of penetrance based on small numbers of cases are imprecise. Simple pedigree inspection of the few families so far known is equally instructive at this point (Fig. 2). Clearly, despite the high odds ratio associated with this susceptibility allele, once detected in a family, not all individuals with the microdeletion are affected clinically.
The 15q13.3 microdeletion in IGE is acting as a risk factor consistent with the common-disease, multiple rare-variant hypothesis of complex disease causation. Since the breakpoints map within segmental duplications that flank the critical region, non-allelic homologous recombination continues to create individuals with nearly identical genetic variation despite that variation being subject to selective pressure. The confirmed association of the 15q13.3 microdeletion with an IGE phenotype and other common brain disorders suggests that structural genomic variation represents an emerging entity contributing to human disorders with complex inheritance.
MATERIALS AND METHODS
Patient diagnoses
The 539 patients included in this study were diagnosed with IGE and its subsyndromes CAE, JAE, JME and epilepsy with GTCSA according to the Commission on Classification and Terminology of the International League Against Epilepsy (1989). The 494 Australian controls were drawn from anonymous blood donors and the 3283 German controls were from subjects with various common diseases and a cohort of individuals of high age.
Molecular genetic analyses
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Maxi Kit (Qiagen). Four hundred and five IGE patients and 494 controls were screened by quantitative PCR to detect deletions and their reciprocal duplications. The forward (5′-CCATCATACATTGCCTCCTTTCCAA-3′) and reverse (5′-AGCATGATTCTGTCTGTAGGAGGAA-3′) primers were designed to hybridize to intron 2 of CHRNA7 and the TaqMan (Applied Biosystems) assay used the probe CACTCGCCTCAAATGT-FAM. CNV analysis of 134 European IGE patients and 3283 German controls were performed using the Affymetrix Genome-Wide Human SNP array 6.0. The microchromosomal abnormalities detected using these approaches were confirmed by array CGH using customized array CGH covering the critical region as previously reported (5).
FUNDING
This work was supported by the National Health and Medical Research Council of Australia; SA Pathology; Thyne Reid Charitable Trusts; The European Community [FP6 Integrated Project EPICURE, LSHM-CT-2006-037315]; the German Ministry of Education and Research (NGFNplus, EMINet); The Netherlands National Epilepsy Fund, and The Netherlands Organization for Scientific Research.
ACKNOWLEDGEMENTS
The authors are grateful to the patients and families for their participation in this study. We thank Bev Johns for performing DNA extractions.
Participating Centers of the EPICURE Integrated Project EPILEPSY COHORT: Department of Molecular Genetics, University of Antwerp, Antwerpen, Belgium (P. De Jonghe, A. Suls); Department of Neurology, Danish Epilepsy Centre, Dianalund, Denmark (H. Hjalgrim, J.M. Madsen, R.S. Møller); Neuroscience Center, University of Helsinki, Finland (A.E. Lehesjoki, A. Siren); Department of Neurology, Charité University Medicine, Campus Virchow Clinic, Humboldt University of Berlin, Berlin, Germany (V. Gaus, D. Janz, B. Schmitz, T. Sander); Department of Epileptology, University of Bonn, Bonn, Germany (C.E. Elger, K. Hallmann, A.A. Kleefuß-Lie, W.S. Kunz, A. Raabe); Department of Neuropediatrics University Medical Center Schleswig-Holstein, Kiel Campus, Kiel, Germany (I. Helbig, H. Muhle, P. Ostertag, T. Obermeier, S. von Spiczak, U. Stephani); Neurological Clinic, University of Ulm, Ulm, Germany (H. Lerche, Y.G. Weber); Department of Neuroscience, Institute G. Gaslini, Genova, Italy (P. Striano, F. Zara); Child Neurology Unit, Children's Hospital A. Meyer, University of Florence, Florence, Italy (C. Marini); Section Complex Genetics, Department of Medical Genetics, University Medical Center, Utrecht, The Netherlands (E.H. Brilstra, D. Kastelijn-Nolst Trenité, B.P.C. Koeleman, C.G.F. de Kovel, D. Lindhout, M.E.M. Swinkels); SEIN Epilepsy Institute in the Netherlands, Heemstede, The Netherlands (D. Lindhout); Department of Molecular Biology and Genetics, Bogazici University, Istanbul, Turkey (H. Caglayan, O. Yalcin); Departments of Child Neurology and Neurology, Istanbul Medical School, Istanbul University, Istanbul, Turkey (Z. Yapici, B. Baykan); Department of Neurology, Sisli Etfal Education Hospital, Sisli, Istanbul, Turkey (D. Yalcin); Institute of Neurological Sciences, Marmara University, Maltepe, Istanbul, Turkey (D. Turkdogan); Ministry of Health Tepecik Education and Research Hospital, Tepecik, Izmir, Turkey (G. Dizdarer); Department of Neurology, Cerrahpasa Medical School, Istanbul University, Istanbul, Turkey (C. Ozkara).
CONTROL COHORT: Pediatric Pneumology and Immunology, Charité Universitätsmedizin Berlin, Berlin, Germany and Max-Delbrück-Centrum for Molecular Medicine (MDC), Berlin-Buch, Germany (Y. Lee); Department of Pneumology, University of Freiburg, Freiburg, Germany (J. Müller-Quernheim); Clinic for Dermatology, Venerology and Allergology, University Hospital Schleswig-Holstein, Kiel, Germany (R. Fölster-Holst); Institute for Clinical Molecular Biology, Christian-Albrechts-University, Kiel, Germany (A. Franke, S. Hofmann, A. Nebel, S. Schreiber, M. Wittig); Institute of Human Genetics, University of Lübeck, Lübeck, Germany (M. Schürmann); Department of Dermatology and Allergy, Technische Universität München, Munich, Germany and Division of Environmental Dermatology and Allergy, Helmholtz Zentrum Munich and ZAUM-Center for Allergy and Environment, Universität München, Munich, Germany (E. Rodriguez, S. Weidinger); Department of Dermatology and Allergy, Technische Universität München, Munich, Germany and Institute for Medical Statistics and Epidemiology IMSE, Technische Universität München, Munich, Germany (H. Baurecht); Institute of Immunology, The National Hospital, University of Oslo, Oslo, Norway (B.A. Lie); Medical Department, Rikshospitalet University Hospital, Oslo 0027, Norway (K.M. Boberg, T.H. Karlsen), NGFN GWAS Consortium.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Helbig I., Scheffer I.E., Mulley J.C., Berkovic S.F. Navigating the channels and beyond: unraveling the genetics of the epilepsies. Lancet Neurol. 2008;7:231–245. doi: 10.1016/S1474-4422(08)70039-5. [DOI] [PubMed] [Google Scholar]
- 2.Tan N.C.K., Mulley J.C., Berkovic S.F. Genetic association studies in epilepsy: ‘the truth is out there. Epilepsia. 2004;45:1429–1442. doi: 10.1111/j.0013-9580.2004.22904.x. [DOI] [PubMed] [Google Scholar]
- 3.Hempelmann A., Taylor K.P., Heils A., Lorenz S., Prud'homme J.F., Nabbout R., Dulac O., Rudolf G., Zara F., Bianchi A., et al. Exploration of the genetic architecture of idiopathic generalised epilepsies. Epilepsia. 2006;47:1682–1690. doi: 10.1111/j.1528-1167.2006.00677.x. [DOI] [PubMed] [Google Scholar]
- 4.Cavalleri G.L., Weale M.E., Shianna K.V., Singh R., Lynch J.M., Grinton B., Szoeke C., Murphy K., Kinirons P., O'Rourke D., et al. A large-scale multi-centre effort to map genetic variants for sporadic epilepsy syndrome and seizure types. Lancet Neurol. 2007;6:970–980. doi: 10.1016/S1474-4422(07)70247-8. [DOI] [PubMed] [Google Scholar]
- 5.Helbig I., Mefford H.C., Sharp A.J., Guipponi M., Fichera M., Franke A., Muhle H., de Kovel C., Baker C., von Spiczak S., et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 2009;41:160–162. doi: 10.1038/ng.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller D.T., Shen Y., Weiss L.A., Korn J., Anselm I., Bridgemohan C., Cox G.F., Dickinson H., Gentile J., Harris D.J., et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatirc disorders. J. Med. Genet. 2009;46:242–248. doi: 10.1136/jmg.2008.059907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagnamenta A.T., Wing K., Akha E.S., Knight S.J., Bölte S., Schmötzer G., Duketis E., Poustka F., Klauck S.M., Poustka A., et al. A 15q13.3 microdeletion segregating with autism. Eur. J. Hum. Genet. 2009;17:687–692. doi: 10.1038/ejhg.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Commission on Classification and Terminology of the International League Against Epilepsy. Proposal for revised classification of epilepsies and epileptic syndromes. Epilepsia. 1989;30:389–399. doi: 10.1111/j.1528-1157.1989.tb05316.x. [DOI] [PubMed] [Google Scholar]
- 12.Mulley J.C., Dibbens L.M. Chipping away at the common epilepsies with complex genetics: The 15q13.3 microdeletion shows the way. Genome Med. 2009;1:33. doi: 10.1186/gm33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dibbens L.M., Heron S.E., Mulley J.C. A polygenic heterogeneity model for complex epilepsy? Genes Brain Behav. 2007;6:593–597. doi: 10.1111/j.1601-183X.2007.00333.x. [DOI] [PubMed] [Google Scholar]