Genetic and environmental factors modify bovine spongiform encephalopathy incubation period in mice (original) (raw)
Abstract
The incubation period (IP) and the neuropathology of transmissible spongiform encephalopathies (TSEs) have been extensively used to distinguish prion isolates (or strains) inoculated into panels of inbred mouse strains. Such studies have shown that the bovine spongiform encephalopathy (BSE) agent is indistinguishable from the agent causing variant Creutzfeldt–Jakob disease (vCJD), but differs from isolates of sporadic CJD, reinforcing the idea that the vCJD epidemic in Britain results from consumption of contaminated beef products. We present a mouse model for genetic and environmental factors that modify the incubation period of BSE cross-species transmission. We have used two mouse strains that carry the same prion protein (PrP) allele, but display a 100-day difference in their mean IP following intracerebral inoculation with primary BSE isolate. We report genetic effects on IP that map to four chromosomal regions, and in addition we find significant factors of host environment, namely the age of the host's mother, the age of the host at infection, and an X-cytoplasm interaction in the host.
Transmissible spongiform encephalopathies (TSEs) or prion diseases are fatal neurodegenerative disorders of the central nervous system (CNS). They comprise a class of diseases that include sheep scrapie, bovine spongiform encephalopathy (BSE), and the human diseases Creutzfeldt–Jakob disease (CJD), Gerstmann–Straussler–Scheinker syndrome, Kuru, and Fatal Familial Insomnia. Prion diseases may present as familial, acquired, or sporadic disorders, all of which involve an aberrant metabolism of the prion protein (PrP) (1). The conversion of cellular prion protein, PrPC, to the disease-specific protease-resistant isoform, PrPSc, involves a conformational change, whereby the α-helical content decreases and the amount of β-sheet increases (2). The hallmark of prion diseases, whether sporadic, familial, or acquired by infection, is the accumulation of PrPSc in the extracellular space in the brains of humans and animals that have contracted the disease.
Experimental transmission of prions in inbred mouse strains has enabled the genetic analysis of prion disease expression. Animals devoid of the gene encoding PrP (_Prnp_-deficient) do not develop disease after transmission (3–5) and in transgenic mice with higher copy numbers of Prnp, progression of the disease is faster (6). All laboratory mouse strains tested contain one of two forms of PrP—PrP A has leucine at residue 108 and threonine at 189, whereas PrP B is 108 phenylalanine and 189 valine. The incubation period of the disease depends on the Prnp genotype of the mouse (7), but the size and direction of this effect differ according to the prion strain used (8, 9). However, the host Prnp genotype does not account for all of the variation seen in the incubation period, indicating that other genes are involved in the control of the disease. An H-2D–linked locus, named Pid1, and two loci on chromosomes 9 and 11, Pid2 and Pid3, have been reported to affect the length of incubation period in mouse passaged scrapie-infected mice (10, 11), but the nature of the underlying genes remains to be determined.
Human PrP is also polymorphic in sequence. Homozygosity for methionine at residue 129 seems to associate with disease phenotype in acquired TSE. All tested victims of vCJD have been homozygous for methionine at codon 129 of the PRNP gene (12). In other acquired prion diseases, such as Kuru, homozygosity for methionine is overrepresented in the younger age groups, whereas heterozygosity with valine 129 is more prevalent in the older age groups (13). Sporadic cases of CJD have been reported to be more common in homozygotes for either valine or methionine at codon 129 than in heterozygotes (14). The codon 129 polymorphism also influences the phenotype of familial disease associated with an aspartic acid to asparagine mutation at codon 178 of the human PRNP gene (15).
One of the main characteristics of prion diseases is the long incubation period, which could be explained by a slow but controlled proliferation and transport of the infectious agent. Accumulating evidence suggests that, following exposure to infection by a peripheral route, replication usually takes place in the host's lymphoid organs preceding replication in the CNS. For transmission of BSE from cattle to mice, and some other interspecies transmissions, a replication phase in peripheral tissues appears to be obligatory before neuroinvasion can occur, even when infection is introduced directly into the brain (16, 17). Whether modifier genes of prion incubation period affect the replication rate of the infectious agent, the susceptibility of the host to the pathogenic effects of prion replication, or both remains unclear.
Given the likely link between vCJD and cattle BSE, we attempted to detect and map modifier genes for BSE incubation period by means of experimental transmission to mice. To avoid any effect of the coding region of the Prnp gene, we used C57BL/Fa/Dk and RIII/Fa/Dk mouse strains, both of which carry the _Prnp_a allele. Following intracerebral (i.c.) challenge with BSE inoculum, C57BL and RIII mouse strains have a 100-day difference in incubation period (16, 18). It has been shown, however, that C57BL and RIII mice differ in the Prnp promoter region (H. Baybutt and J.M., unpublished observation), leaving open the possibility that different Prnp expression levels may be associated with incubation period. In addition, the two strains differ in their haplotypes on the H-2D region, enabling us to test whether there are detectable H-2D linked effects on BSE incubation period.
Materials and Methods
Crosses.
Reciprocal crosses between C57BL and RIII strains were performed. The F1 animals are named CR or RC depending on whether the maternal strain was C57BL or RIII, respectively. F1 females were backcrossed to either strain giving four backcross populations (CRC, CRR, RCC, and RCR). In total, 1,200 backcross animals were generated, 300 in each backcross.
Inocula Preparation.
Brain tissue was collected from seven terminally affected cows with histopathologically confirmed BSE. The tissue was homogenized in physiological saline at 10% concentration.
Infections.
All 1,200 backcross mice and 30 animals from each parental strain and from reciprocal F1s were challenged intracerebrally with BSE inoculum, in three cohorts. Mice were anaesthetized by halothane inhalation and injected intracerebrally with 0.02 ml of inoculum at 3 to 8 weeks of age. All animals were housed under SPF conditions, monitored daily for signs of intercurrent illness, and, from 250 days after BSE challenge, were subject to a formal clinical monitoring system (19). Animals then were checked daily and scored weekly by an experienced observer for clinical signs of TSE-like disease, using well established criteria that have previously been shown to give precise and reproducible measurements of incubation period in an extensive range of TSE studies. Animals were killed after three consecutive weekly scores of “definitely affected” or when there was a significant deterioration between scoring days. About 85% of the animals in each injection group eventually developed a TSE disease. The remaining 15% were killed or died with intercurrent disease throughout the course of the experiment. Because the maximum incubation periods seen in mice that did develop BSE were very long, few, if any, of the 15% can be regarded as true survivors. Clinical diagnosis was, in all cases, confirmed by histopathological examination of the brains of the mice.
Incubation period measurement was calculated as the interval between injection and a standard clinical end-point when the mice were showing clear signs of neurological disease, as described (19).
DNA Isolation and Genotyping.
Genomic DNA was isolated from tail snips. Approximately 1/2 inch of tail was removed during the electronic tagging session of the animals. Tail tips were incubated overnight at 55°C in 1 ml of extraction buffer (0.05 M Tris⋅HCl/0.1 M EDTA/1% SDS) containing 500 μg/ml proteinase K. The samples were then extracted with phenol-chloroform. High molecular weight DNA was obtained after ethanol precipitation and redissolved in 200 μl TE (10 mM Tris/1 mM EDTA, pH 7.5). DNA for genotyping was resuspended in distilled water at 30 ng/μl. The most polymorphic microsatellite markers among inbred mouse strains were selected, based on the MIT database (http://www.genome.wi.mit.edu). Absolute and relative distance between the markers on the chromosomes was estimated from the Mouse Genome Database (http://www.informatics.jax.org), and a panel of 150 markers was tested on DNA from the parental strains. Final genotypes were obtained for 90 markers spread throughout the genome. PCR products were amplified by using optimized multiplex conditions. Genotypes were assayed and scored in an ABI 310 capillary system (Applied Biosystems).
Linkage analysis was performed by using the GeneLink package (20). Retyping of double recombinants was performed for all intervals. The average distance between markers is 17 cM. A list of the markers used and genetic maps are available from the authors.
Statistical analysis was carried out by using the software package MINITAB, version 13.1 (Minitab, State College, PA). The regression of the variance on the means across groups at this scale of measurement gave x = 8 + 0.101_y_, where x is the standard deviation of the group and y is the mean incubation period. To establish independence of the variance and the means, the transformation ln(y + 8/0.101) was used to calculate the individual data points that were entered for further analysis. The age of mice at injection (B week) and the age of the mother at birth of those animals (M week) were fitted as covariates for the regression model. Goodness of fit was judged based on a lack of fit test calculated by the program.
Interaction between maternal and X-chromosome effects were obtained as I ij = zij − G i − E j, where i represents the columns and j the rows of Table 2. G i is the mean within each column and E j is the deviation of the mean within rows from the F1 population mean (μ_G_) (21). E j represents the maternal effects, and the deviation of G i from μ_G_ represents the X-chromosome effects.
Table 2.
Relative magnitudes of maternal and X-linked effects associated with each parental strain
| X chromosome | Maternal effect | ||
|---|---|---|---|
| R | C | ||
| Maternal environment | |||
| R | 506 | 462 | +13.75 |
| C | 425 | 488 | −13.75 |
| X effect | −4.75 | +4.75 | |
| Interactions: maternal effect + X-chromosome effect | |||
| R+R or C+C | +26.75 days | ||
| R+C or C+R | −26.75 days |
Qantitative trait loci (QTL) mapping was performed by using the software package QTL CARTOGRAPHER, version 1.01 (http://statgen.ncsu.edu/qtlcart/). Interval mapping results were compared with the results obtained by MAPMANAGERQTX7B (http://mcbio.med.buffalo.edu/mmQTX.html) and QTL CAFÉ (http://web.bham.ac.uk/g.g.seaton/). Composite interval mapping results were compared between QTL CARTOGRAPHER and QTL CAFÉ, using the option for linked QTL. Estimates of locations and effects from the different packages were in agreement. Genome-wide significance levels were obtained by running 1,000 permutations on MAPMANAGERQTX7B. Epistatic interactions between pairs of loci were assessed by using the program EPISTAT (http://www.larklab.4biz.net/epistat.htm).
Results
The mean incubation period in these experiments was 541 days for the C57BL strain and 441 days for the RIII strain. This is in accord with previous studies showing the two strains to have one hundred days difference in their mean IP following primary BSE transmission (16, 18). However, in our present study the mean IP in these strains is longer than previously reported, indicating a relatively low titer of BSE in the inoculum. Table 1 summarizes the mean IP data for the parental strains, F1 progeny and the backcross populations. The parental strains and three of the four backcross populations do not have a significant difference in mean IP between males and females. However, the F1 animals do have a significant sex difference, with males having a significantly longer mean IP than females. The F1 male IP was not significantly different from the value midway between the two parental strains. However, F1 females, from crosses in either direction, had a mean IP significantly shorter than the equivalent males. One of the four backcross populations also had significant sex-specific differences. In addition, the data indicate that the phenotypic variance across the whole backcross generation is not statistically different from the variance of the parental strains and F1 animals.
Table 1.
Sex specific effects on IP
| Population | Sex | n | Mean | 95% c.i. SD | P |
|---|---|---|---|---|---|
| C57BL | F | 23 | 545 | 42.5–84.9 | |
| M | 12 | 535 | 44.3–121.1 | ns | |
| RIII | F | 19 | 440 | 34.8–75.2 | |
| M | 14 | 443 | 38.4–96.1 | ns | |
| CxR | F | 17 | 425 | 33.9–79.3 | |
| M | 15 | 488 | 43.2–104.2 | 0.002 | |
| RxC | F | 12 | 462 | 29.1–79.5 | |
| M | 14 | 506 | 33.0–82.6 | 0.02 | |
| RCxC | F | 118 | 517 | 58.8–67.4 | |
| M | 140 | 518 | 53.8–70.4 | ns | |
| CRxC | F | 116 | 526 | 53.9–61.9 | |
| M | 141 | 513 | 51.9–68.0 | ns | |
| CRxR | F | 121 | 465 | 56.5–75.5 | |
| M | 132 | 461 | 48.6–64.2 | ns | |
| RCxR | F | 121 | 465 | 54.9–73.5 | |
| M | 138 | 446 | 48.7–63.9 | 0.01 |
F1 Sex Differences: X Chromosome and Cytoplasm Interaction.
Each F1 population is genetically identical, apart from X-chromosome dosage in females and the Y chromosome in males. The sex differences in IP in the F1 populations may be due to an effect of the X chromosome. It should normally be possible to measure this effect simply by comparing F1 males with different X chromosomes (i.e., with different strain mothers, from the reciprocal crosses), but these males also have a different maternally derived cytoplasm and maternal environment. We can demonstrate that there is a statistically significant maternal effect, because genetically identical female F1 animals show a 37-day difference in mean IP depending on the maternal strain (females with RIII mother and C57BL father have an IP of 462 days, those with C57BL mother and RIII father have an IP of 425 days). The effect of the X chromosome, then, can only be determined after subtraction of the maternal effect. In Table 2 we show the mean IP of reciprocal F1s and dissect out the relative maternal and X-linked effects associated with each parental strain (see Materials and Methods). Both effects are clearly present, but their magnitudes are subject to large statistical error due to the relatively small sample size. The maternal effect associated with the RIII strain prolongs the IP by ≈14 days, whereas the RIII strain X-chromosome (XR)-linked effect acts in the opposite direction and shortens the IP by about 5 days. However, when both the X chromosome and the maternal effect are of the same strain of origin, the effect seen is not an addition of the two effects (which would be a longer IP by ≈9 days), rather their interaction prolongs the IP by almost 27 days. Hence, when the X chromosome and cytoplasm are from the same strain, the F1 animals have a longer IP. F1 females have a significantly shorter IP because there is a deleterious interaction between the maternal effect of one strain with an X chromosome of a different strain. F1 males always inherit both the X chromosome and the cytoplasm from the mother (same inbred strain), escape the deleterious effect of this interaction, and have prolonged IP in comparison to their sisters. It is possible that the X-linked effect reflects a parent of origin rather than a strain of origin effect. The F1 data cannot distinguish between the two possibilities, but the data in the backcross generation are compatible with the simpler explanation of a strain-of-origin hypothesis. According to this scenario no sex-specific differences are expected in the backcross generation because, during F1 female meiosis, recombination and Mendelian segregation of the X chromosome will ensure in the progeny that the same proportion of both sexes experience the effects of the deleterious interaction. Three of the four backcrosses showed no sex-specific effect. The RCR backcross did have an effect, the reverse of that seen in the F1, such that females had a prolonged mean IP compared with males. In this cross, transmission ratio distortion resulted in an excess of maternally inherited XR alleles among the females. These alleles, combined with a cytoplasm of RIII origin, led to an excess of prolonged IP among females on average, because fewer female progeny experienced the deleterious X-cytoplasm interaction in comparison to males.
Age Effects.
Segregation of genes during F1 meiosis should normally increase the phenotypic variance in the backcross generation. Unexpectedly, we find that this is not the case. Two explanations could account for the restricted variance in the backcross populations. Either the heritability of the trait is low or there is negative correlation between particular individuals (short/long IP) with respect to macroenvironment (age groups, maternal effects, litter size, etc.). In the latter case, such a nonrandom distribution can create a negative covariance term, which masks the true genetic variance present in the backcross populations (21). In fact, we detect significant effects of age on the IP. Both the age of the mice at the time of injection and, surprisingly, the maternal age at birth of these animals were negatively correlated with the length of the IP (P < 0.001). Our data suggest that the purely genetic component of the trait is only 3% greater than the negative covariance term, made up of the genetic/environmental correlation, and thus the variance resulting from genetic factors is effectively masked.
Genetic Factors.
Taking into account the effects described above, we can then model the genetic factors acting on the backcross population. Table 1 shows that groups having shorter IP tend to also have smaller phenotypic variance. This phenomenon makes the original measurement scale statistically inappropriate and we used a transformation (see Materials and Methods) to treat the data. Table 3 shows the results of the regression analysis on the individual IP data collected from all populations. As expected, the X-cytoplasm interaction, the age of the mice at the time of injection, and the age of the mother at birth of the offspring are all statistically significant and must be included in the final model.
Table 3.
Coefficients for predictor variables by regression analysis on the individual transformed IP data collected from all populations
| Predictor | Coefficient | SE coefficient | T-statistic | P |
|---|---|---|---|---|
| Parametric mean | 6.74641 | 0.04897 | 137.77 | <0.0005 |
| α | 0.08796 | 0.01235 | 7.12 | <0.0005 |
| Mw | −0.0069118 | 0.0009951 | −6.95 | <0.0005 |
| Bw | −0.010302 | 0.001350 | −7.63 | <0.0005 |
| d | −0.32255 | 0.05198 | −6.21 | <0.0005 |
| αxα | −0.33601 | 0.05293 | −6.35 | <0.0005 |
| I | 0.05143 | 0.01337 | 3.85 | <0.0005 |
| αxd | 0.05433 | 0.02844 | 1.91 | 0.056 |
| cyt | −0.007184 | 0.003701 | −1.94 | 0.052 |
We find that single gene effects due to both additive gene action and gene dominance are significant, but that dominance contributes 4-fold more to the phenotype. Epistasis (gene interaction) between additive genes contributes as much as dominance to the phenotype, and the fact that the epistatic term is negative indicates that genes of opposite effects (long and short IP genes) are present in each parental strain (i.e., there is gene dispersion). Finally, effects linked to the cytoplasm and to more complex epistatic interactions, although of marginal significance, are required for model fitting.
QTL Mapping.
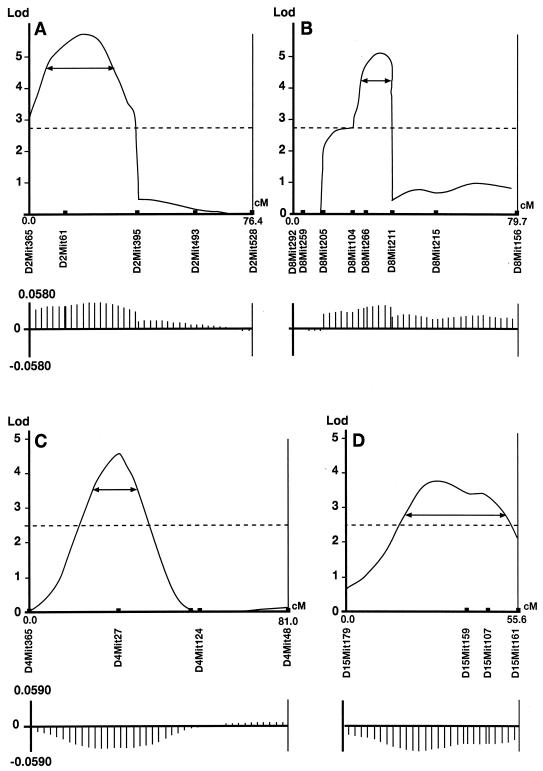
The genetic model has allowed us to go on to map QTLs in the backcross populations. Each backcross animal was genotyped at 90 loci distributed throughout the genome on all chromosomes. QTL analysis was initially performed separately for each backcross. Because similar results were obtained for backcrosses to the same inbred parent (data not shown), we grouped them and present the results of the combined analysis. Fig. 1 shows the QTLs detected in the backcrosses to C57BL (A and B) and RIII (C and D). All detected QTLs are dominant, because they were detected only in one direction of cross, with C57BL alleles contributing to a longer and RIII alleles to a shorter IP. All detected QTLs were either significant (chromosome 15) or highly significant (chromosomes 2, 4, and 8) for a genome-wide probability. The effects of individual QTLs on the phenotype are only moderate, each explaining 4–7% of the total phenotypic variance. After correction for dominance, the cumulative effects from the detected QTLs account for 50% of the initial difference between the parental strains. The relatively weak effects of the individual QTLs produce rather poor genetic localization (22), with 95% confidence intervals ranging from 10 to 25 cM.
Figure 1.
Lod score plots for BSE incubation period in backcross populations. (A and B) Animals backcrossed to C57. (B and C) Animals backcrossed to RIII. The x axes indicate the chromosomal linkage maps in centimorgans, based on recombination frequencies calculated by using Gene-Link. (A, chromosome 2; B, chromosome 8; C, chromosome 4; D, chromosome 15). The first marker analyzed on each chromosome is positioned at 0 cM. The y axes indicate lod scores for intervals containing putative QTLs affecting BSE incubation period based on composite interval mapping by using QTL CARTOGRAPHER. One lod, 95% confidence intervals, are indicated by the double-headed arrows. The dotted lines intersecting the y axes indicate the lod score representing statistical significance for the respective data sets (A and B, 2.71; C and D, 2.50). Additive effects calculated by QTL CARTOGRAPHER are plotted beneath (not corrected for dominance).
The regression model indicated that epistatic interactions between loci are an important component of the length of IP. However we could not detect them in our sample, which suggests that, although a number of interactions might be present, their effects do not reach statistical significance when pairs of loci are considered separately. This lack of detectable epistatic interactions includes markers on chromosome 2, in the proximity of the Prnp gene.
Discussion
The four QTLs we detect only explain 50% of the 100-day difference between the parental strains and none accounts for more than 7% of the total phenotypic variance. With our sample size, a QTL explaining less than 3% of the phenotypic variance would not have been detected as significant (23). Hence, the remaining 50% of the difference between the parental strains must be due to epistatic interactions that do not reach significance, to a number of undetected single QTLs, or to a combination of these. If there are undetected single QTLs that determine the length of IP, which display no dominance and explain about 3% of the phenotypic variance each, then there must be no fewer than four of them. In this case about 700 F2 animals will be required to map them in 25 cM 95% confidence intervals (24). If they display dominance, the number of undetected QTLs is higher, and at least 700 backcross animals will be needed to detect them. However, because the direction of dominance is not known a priori, 700 animals in each backcross direction would be required. If epistatic interactions determine most of the remaining difference, and given that they are weak, then a very large F2 cross (>1,000 animals) is required to detect them, but it is doubtful that we will be able to identify the individual genes implicated.
Our study of primary BSE transmission to mice gives contrasting results with a recent study on the incubation period in mice challenged intracerebrally with mouse-passaged scrapie. In both studies parental strains were used that both carry the _Prnp_a allele. The results for mouse-passaged scrapie indicate a few genes of additive gene action contributing to the phenotype, whereas ours indicate that IP is a polygenic trait subject to dominance. None of the mapped QTLs in the two studies is found on the same chromosome. However, because our study crosses a species barrier between cattle and mouse (whereas the other study does not), differences in pathogenesis are likely to underlie the different results obtained. In intraspecies transmissions (mouse-passaged scrapie to mice), an intracerebral injection establishes infection directly in the brain, resulting in a relatively short IP (approximately two thirds of the IP following i.p. challenge) (8); genetic effects are therefore likely to operate within the CNS. In contrast, in some interspecies transmissions, including the transmission of cattle BSE to mice, the IPs following intracerebral and peripheral challenge are approximately equal (16). Furthermore, immunodeficient mice are relatively resistant to BSE challenge, even by the intracerebral route (17). These studies suggest that, in the presence of a species barrier, the BSE agent must be processed in peripheral tissues before it can infect the nervous tissue. Therefore, genetic effects in our BSE study could operate in both peripheral tissues and the CNS.
Previous studies have shown that RIII and C57BL mice are equally susceptible to BSE infection and that the same strain of agent is isolated in each mouse strain (16, 18). Infectivity can accumulate in all components of the lymphoreticular system (LRS) and experimental evidence points toward the follicular dendritic cells as the likely cell type that could support prion replication in the LRS (25). The spread of infectivity from the LRS to CNS is probably accomplished via peripheral nerves, to the spinal cord from where it finally ascends to the brain. Other studies (C.F., unpublished work) indicate that the difference in IP seen between C57BL and RIII strains after i.c. inoculation results from a difference in the timing of BSE replication in these tissues.
Our study indicates that there are a number of chromosomes with minor effects on the phenotype rather than a few major ones. Given the relatively weak effects of the detected QTLs and the rather large confidence intervals, it is not straightforward to pinpoint candidate genes in these regions. Based on the observations mentioned above involving the two parental strains, we can speculate about the mode of action of these genes. It is interesting to note that the mouse homologue of the human intestinal DPP4 gene that encodes a T lymphocyte activating molecule, which is a cell-surface antigen with peptidase activity, maps within the 95% C.I. on chromosome 2. The IFN α and β families are located near to the most likely position for the QTL on chromosome 4. Scya17, which encodes for a chemokine that specifically attracts CD4+ T helper type 2 lymphocytes in lymphoid and nonlymphoid organs, is located at the most likely position for the QTL on chromosome 8. Finally, at the most likely position of the QTL on chromosome 15 is found the Ly6 complex (_Ly6A_-Ly6D). Antigens encoded by this complex are present on peripheral T and B cells of spleen and lymph nodes and bone marrow. However, there is no independent support from other studies that any of these genes are indeed involved in prion replication and/or the length of incubation period of the disease. Given the rather large confidence intervals, the possibility cannot be excluded that other genes located in these intervals are underlying the mapped QTLs. For instance, on chromosome 8, within the 95% confidence interval, maps a family of metallothioneins (_Mt1_-Mt3), which are thought to play an important role in heavy-metal detoxification and metabolism, such as Cu++. Early studies showed that cuprizone, a copper chelating agent, induces neuropathological changes in mice similar to those found in prion diseases (26), suggesting a role for copper in the pathology of prion diseases. More recently, an octapeptide repeat in the N-terminal region of the prion protein has been shown to bind copper (27), and upon binding the protein appears to have a more defined structure in this region, which promotes the conformational shift from an α-helical to a β-sheet structure (28). There is also evidence from _Prnp_-deficient mice that their cerebellar cells have an increased sensitivity to the toxicity of copper-containing salts (29), a marked decrease in membrane copper content, and decreased activity of superoxide dismutase 1 (30, 31), suggesting that the prion protein may play a role in copper homeostasis in the CNS. The role of copper in other neurodegenerative diseases raises the possibility of a common neurodegenerative process linked to copper homeostasis and transport (32).
The H-2D–linked effect, initially detected in congenic mouse strains infected with mouse-adapted CJD, has failed to be replicated by us or other investigators (33, 34). Finally, although we do know that there are differences in the promoter region of the Prnp gene between the C57BL and RIII mouse strains, we do not detect any effect linked to the Prnp gene. The QTL detected on Chr 2 is situated about 40 cM proximal to Prnp, which is not included in the 95% confidence interval. If expression level differences were associated with the known polymorphism at the promoter region, and this in turn was related to the length of IP, then we would expect to detect a QTL in the vicinity of Prnp.
Experimental prion transmissions in mice have been invaluable for our understanding of prion biology and disease development. However, different studies reveal different genetic contributions to disease susceptibility. Each combination of inbred strains contains only a limited genetic variability compared with natural populations, and it is unlikely that all genes affecting the IP in natural populations will be revealed by studies like these. Additional complexity is added, because development of the disease is already known to depend on the agent used for the infections (prion strain), the host's environment, and the species involved as source and recipient of the infectious particle (species barrier). Extrapolations to acquired human prion disease have to take into account all of these limitations. These caveats notwithstanding, our findings imply that susceptibility to the disease is polygenic and that the population frequency for at least eight genes other than PrP is likely to affect incubation period. This conclusion is important from an epidemiological point of view because predictions about the possible number of future vCJD victims have been made by assuming a sole genetic risk associated with certain PRNP genotypes (MM homozygotes). However, if the incubation period for vCJD is polygenic and assuming Hardy–Weinberg equilibrium for the different alleles, the frequency of the most susceptible genotype will be lower than currently believed.
Acknowledgments
We thank Peter Teague for statistical support and Peter Keightley and Chris Haley for advice. The project was supported by the United Kingdom Medical Research Council and Department of Health initiative on TSEs, Grant G9631616.
Abbreviations
IP
incubation period
TSE
transmissible spongiform encephalopathy
BSE
bovine spongiform encephalopathy
CJD
Creutzfeldt–Jakob disease
vCJD
variant CJD
CNS
central nervous system
PrP
prion protein
QTL
quantitative trait loci
References
- 1.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 2.Pan K-M, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bueler H A, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner S B, Groth D, Serban A, Koehler R, Foster D, Torchia M, Burton D, Yang S-L, DeArmond S. Proc Natl Acad Sci USA. 1993;90:10608–10612. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manson J C, Clarke A R, McBride P A, McConnell I, Hope J. Neurodegeneration. 1994;3:331–340. [PubMed] [Google Scholar]
- 6.Prusiner S B, Scott M R, Foster D, Pan K-M, Groth D, Mirenda C, Torchia M, Yang S-L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 7.Moore R C, Hope J, McBride P A, McConnell I, Selfridge J, Melton D W, Manson J C. Nat Genet. 1998;18:118–125. doi: 10.1038/ng0298-118. [DOI] [PubMed] [Google Scholar]
- 8.Bruce M E, McConnell I, Fraser H, Dickinson A G. J Gen Virol. 1991;72:595–603. doi: 10.1099/0022-1317-72-3-595. [DOI] [PubMed] [Google Scholar]
- 9.Carlson G A, Ebeling C, Yang S-L, Telling G, Torchia M, Groth D, Westaway D, DeArmond S, Prusiner S B. Proc Natl Acad Sci USA. 1994;91:5690–5694. doi: 10.1073/pnas.91.12.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kingsbury D T, Kasper K C, Stites D P, Watson J D, Hogan R N, Prusiner S B. J Immunol. 1983;131:491–496. [PubMed] [Google Scholar]
- 11.Stephenson D A, Chiotti K, Ebeling C, Groth D, DeArmond S, Prusiner S B, Carlson G A. Genomics. 2000;69:47–53. doi: 10.1006/geno.2000.6320. [DOI] [PubMed] [Google Scholar]
- 12.Will R G, Zeidler M, Stewart G E, Macleod M A, Isornside J W, Cousens S, Mackenzie J, Estibeiro K, Green A J, Knight R S. Ann Neurol. 2000;47:575–582. [PubMed] [Google Scholar]
- 13.Cervenakova L, Goldfarb L G, Garruto R, Carleton-Gajdusek D, Brown P. Proc Natl Acad Sci USA. 1998;95:13239–13241. doi: 10.1073/pnas.95.22.13239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer M S, Dryden A J, Hughes J T, Collinge J. Nature (London) 1991;352:340–342. doi: 10.1038/352340a0. [DOI] [PubMed] [Google Scholar]
- 15.Goldfarb L G, Petersen R, Tabaton M, Brown P, LeBlanc A C, Montagna P, Cortelli P, Jilien J, Vital C, Pendelbury W, et al. Science. 1992;258:806–808. doi: 10.1126/science.1439789. [DOI] [PubMed] [Google Scholar]
- 16.Bruce M E, Chree A, McConnell I, Foster J, Pearson G, Fraser H. Philos Trans R Soc London B. 1994;343:405–411. doi: 10.1098/rstb.1994.0036. [DOI] [PubMed] [Google Scholar]
- 17.Brown K L, Stewart K, Bruce M E, Fraser H. J Gen Virol. 1997;78:2707–2710. doi: 10.1099/0022-1317-78-10-2707. [DOI] [PubMed] [Google Scholar]
- 18.Fraser H, Bruce M E, Chree A, McConnell I, Wells G A. J Gen Virol. 1992;73:1891–1897. doi: 10.1099/0022-1317-73-8-1891. [DOI] [PubMed] [Google Scholar]
- 19.Dickinson A G, Meikle V M H, Fraser H. J Comp Pathol. 1968;78:293–299. doi: 10.1016/0021-9975(68)90005-4. [DOI] [PubMed] [Google Scholar]
- 20.Montagutelli X. J Hered. 1990;81:490–491. doi: 10.1093/oxfordjournals.jhered.a111033. [DOI] [PubMed] [Google Scholar]
- 21.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 22.Darvasi A, Weinreb A, Minke V, Weller J I, Soller M. Genetics. 1993;134:943–951. doi: 10.1093/genetics/134.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darvasi A. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 24.Darvasi A, Soller M. Behav Genet. 1997;27:125–132. doi: 10.1023/a:1025685324830. [DOI] [PubMed] [Google Scholar]
- 25.Bruce M E, Brown K L, Mabbott N A, Farquhar C F, Jeffrey M. Immunol Today. 2000;21:442–446. doi: 10.1016/s0167-5699(00)01696-0. [DOI] [PubMed] [Google Scholar]
- 26.Kimberlin R H. J Comp Pathol. 1974;84:263–270. doi: 10.1016/0021-9975(74)90067-x. [DOI] [PubMed] [Google Scholar]
- 27.Hornshaw M P, McDermott J R, Candy J M. Biochem Biophys Res Commun. 1995;207:621–629. doi: 10.1006/bbrc.1995.1233. [DOI] [PubMed] [Google Scholar]
- 28.Muira T, Hori I-A, Takeuchi H. FEBS Lett. 1996;396:248–252. doi: 10.1016/0014-5793(96)01104-0. [DOI] [PubMed] [Google Scholar]
- 29.Bueler H A, Ficher M, Lang Y, Bluethmann H, Lipp H P, DeArmond S, Prusiner S B, Aguet M, Weissmann C. Nature (London) 1992;356:577–582. doi: 10.1038/356577a0. [DOI] [PubMed] [Google Scholar]
- 30.Brown D R, Qin K, Herms J W, Madlung A, Manson J C, von Bohlen A, Schulz-Schaeffer W, Giese A, Westaway D, Kretzschmar H. Nature (London) 1997;390:684–687. doi: 10.1038/37783. [DOI] [PubMed] [Google Scholar]
- 31.Brown D R, Besinger A. Biochem J. 1998;334:423–429. doi: 10.1042/bj3340423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waggoner D J, Bartnikas T B, Gitlin J D. Neurobiol Dis. 1999;6:221–230. doi: 10.1006/nbdi.1999.0250. [DOI] [PubMed] [Google Scholar]
- 33.Carlson G A, Kinsbury D T, Goodman P A, Coleman S, Marshall S T, DeArmond S, Westaway D, Prusiner S B. Cell. 1986;46:503–511. doi: 10.1016/0092-8674(86)90875-5. [DOI] [PubMed] [Google Scholar]
- 34.Mohri S, Tateishi J. J Gen Virol. 1989;70:1391–1400. doi: 10.1099/0022-1317-70-6-1391. [DOI] [PubMed] [Google Scholar]