Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles (original) (raw)
Abstract
Paraspeckles are unique subnuclear structures built around a specific long noncoding RNA, NEAT1, which is comprised of two isoforms produced by alternative 3′-end processing (NEAT1_1 and NEAT1_2). To address the precise molecular processes that lead to paraspeckle formation, we identified 35 paraspeckle proteins (PSPs), mainly by colocalization screening with a fluorescent protein-tagged full-length cDNA library. Most of the newly identified PSPs possessed various putative RNA-binding domains. Subsequent RNAi analyses identified seven essential PSPs for paraspeckle formation. One of the essential PSPs, HNRNPK, appeared to affect the production of the essential NEAT1_2 isoform by negatively regulating the 3′-end polyadenylation of the NEAT1_1 isoform. An in vitro 3′-end processing assay revealed that HNRNPK arrested binding of the CPSF6–NUDT21 (CFIm) complex in the vicinity of the alternative polyadenylation site of NEAT1_1. In vitro binding assays showed that HNRNPK competed with CPSF6 for binding to NUDT21, which was the underlying mechanism to arrest CFIm binding by HNRNPK. This HNRNPK function led to the preferential accumulation of NEAT1_2 and initiated paraspeckle construction with multiple PSPs.
Keywords: alternative RNA processing, noncoding RNA, nuclear body, RNA-binding proteins
Introduction
Recent postgenomic transcriptome analyses reveal that many nonprotein-coding transcripts, so-called noncoding RNAs (ncRNAs), are transcribed from large portions of mammalian genomes (Carninci et al, 2005; Kapranov et al, 2007). The limited numbers of long ncRNAs that have been characterized exhibit diverse functions, as well as cell type-specific expression and localization to subcellular compartments (Prasanth and Spector, 2007; Mercer et al, 2009; Wang and Chang, 2011). Most of the newly discovered ncRNAs are likely transcribed by RNA polymerase II. However, extensive analyses of the subcellular localization of human transcripts reveal that ncRNAs are enriched in the cell nucleus, suggesting that they play diverse roles in nuclear events (Kapranov et al, 2007; Prasanth and Spector, 2007).
The mammalian cell nucleus is highly organized. It is composed of distinct nuclear bodies that contain proteins or RNAs characteristic of particular nuclear processes. To date, ∼10 different nuclear bodies have been characterized (Spector, 2006). Several long ncRNAs, such as Xist, Gomafu (Miat), Malat1 (NEAT2), NEAT1 (MENε/β), TUG1, and GRC-RNAs, localize to specific nuclear bodies (Clemson et al, 1996, 2009; Hutchinson et al, 2007; Sone et al, 2007; Sasaki et al, 2009; Sunwoo et al, 2009; Zheng et al, 2010; Yang et al, 2011). In particular, Malat1 localizes to nuclear speckles, where it regulates alternative splicing by modulating the phosphorylation status of Serine/Arginine (SR)-splicing factors (Tripathi et al, 2010). Malat1 controls growth signal-responsive gene expression through its association with unmethylated polycomb 2 protein (Yang et al, 2011).
Paraspeckles are recently discovered nuclear bodies that are usually detected in cultured cell lines as a variable number of foci found in close proximity to the nuclear speckles. Paraspeckles contain characteristic RNA-binding proteins, including paraspeckle protein 1 (PSPC1), RBM14, NONO, CPSF6, and SFPQ (Fox et al, 2002; Dettwiler et al, 2004; Prasanth et al, 2005). PSPC1, NONO, and SFPQ share common domain structures comprised of two RNA-recognition motifs (RRMs). Collectively, these three proteins comprise the Drosophila melanogaster behaviour and human splicing (DBHS) protein family (Bond and Fox, 2009).
The discovery of the specific paraspeckle localization of NEAT1 ncRNA opened a new window in paraspeckle research (Chen and Carmichael, 2009; Clemson et al, 2009; Sasaki et al, 2009; Sunwoo et al, 2009). NEAT1 ncRNA are transcribed from a genetic locus called familial tumour syndrome multiple endocrine neoplasia (MEN) type I on human chromosome 11 (Guru et al, 1997) and are comprised of two isoform transcripts, 3.7-kb NEAT1_1 (MENε) and 23-kb NEAT1_2 (MENβ). Both RNAs are produced from the same promoter. Alternatively, they can be processed at the 3′-end to produce a canonically polyadenylated NEAT1_1 and a noncanonically processed NEAT1_2. RNase P recognizes the tRNA-like structure and cleaves it to form the nonpolyadenylated 3′-end of NEAT1_2 (Sunwoo et al, 2009). The knockdown of NEAT1 ncRNA leads to the disintegration of paraspeckles, suggesting that these ncRNAs serve as a core structural component (Chen and Carmichael, 2009; Clemson et al, 2009; Sasaki et al, 2009; Sunwoo et al, 2009). However, the biological function of paraspeckles and the role(s) of NEAT1 ncRNA remain to be elucidated.
We recently found that paraspeckles were not essential for viability and development in a mouse model under normal conditions, suggesting that they play roles under certain stress conditions (Nakagawa et al, 2011). It has been noted that CTN-RNA, an isoform of mCat2 mRNA, is retained specifically in the paraspeckle. Intriguingly, the long 3′-untranslated region (UTR) of CTN-RNA is cleaved by an unidentified endoribonuclease upon exposure to certain stresses, which leads to the export of processed mCat2 mRNA for cytoplasmic translation (Prasanth et al, 2005). The CTN-RNA 3′-UTR contains a long inverted-repeat sequence that is capable of forming intramolecular double-stranded RNAs that are A-to-I edited. The hyperedited CTN-RNAs are enriched in the paraspeckles. Thus, paraspeckles are thought to suppress the expression of hyperedited transcripts through nuclear retention (Prasanth et al, 2005). Inverted Alu repeat sequences are commonly found in the 3′-UTRs of multiple mRNAs in human cells (Chen et al, 2008). This finding suggests that the expression of these transcripts is suppressed by a nuclear retention mechanism.
We previously reported that two paraspeckle-localized DBHS family proteins, SFPQ and NONO, are required for paraspeckle integrity and for the accumulation of NEAT1_2 but not NEAT1_1 (Sasaki et al, 2009). These results suggest that NEAT1_1 alone is unable to maintain paraspeckle integrity. By contrast, overexpressed NEAT1_1 is reportedly capable of increasing the number of paraspeckles, which suggests that it is the functional isoform for paraspeckle formation (Clemson et al, 2009). An electron microscopic study revealed the location of the NEAT1_2 and NEAT1_1 isoforms. The common NEAT1 region and NEAT1_2 3′-terminal region were located at the paraspeckle periphery, whereas the NEAT1_2 middle region was located in the paraspeckle interior. These findings suggest the importance of NEAT1_2 for the maintenance of paraspeckle integrity (Souquere et al, 2010).
In this study, the essential components for paraspeckle formation were determined. Plasmid rescue experiments revealed that NEAT1_2 but not NEAT1_1 is a necessary RNA for de novo paraspeckle formation. To analyse the detailed process of paraspeckle formation, we sought to identify unknown paraspeckle components. RNAi analyses identified additional factors, each with distinct roles, which were indispensible for paraspeckle formation. One of the essential PSPs was involved in the alternative 3′-end processing of NEAT1. This protein arrested the canonical NEAT1_1 3′-end processing, which led to preferential selection for the noncanonical processing of the NEAT1_2 3′-end. Our data provide important insights into the process of paraspeckle formation on the specific nuclear-retained long ncRNAs.
Results
The NEAT1_2 ncRNA isoform is essential for paraspeckle formation
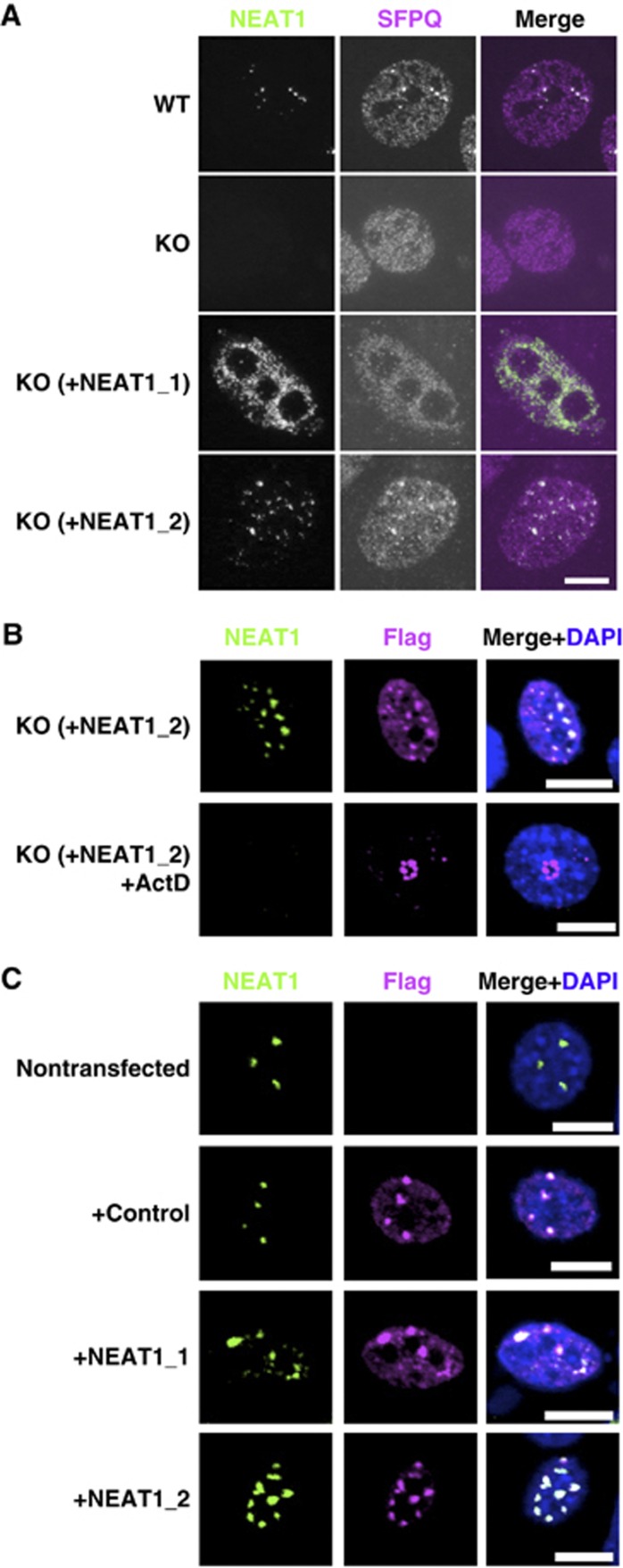
We first attempted to clarify which NEAT1 isoform(s) were required for de novo paraspeckle formation. MEFs were prepared from NEAT1 knockout mice (MEF−/−) (Nakagawa et al, 2011), in which paraspeckles were absent, for rescue experiments with the expression plasmid of either the NEAT1_1 or NEAT1_2 isoform. The expression levels of NEAT1_1 and NEAT1_2 from the plasmids were comparable (Supplementary Figure S1C). Many of the paraspeckle-like foci that were detectable with both anti-SFPQ antibody immunostaining and NEAT1 RNA-FISH appeared when NEAT1_2 but not NEAT1_1 was transiently expressed from the plasmid (Figure 1A; Supplementary Figure S1A and B). This result indicates that NEAT1_2 is an authentic RNA component that is capable of de novo paraspeckle formation.
Figure 1.
NEAT1_2 is a potent RNA component for paraspeckle formation. (A) NEAT1_2 but not NEAT1_1 rescues paraspeckle formation. Intact paraspeckles were detected by RNA-FISH with the antisense probe of mouse NEAT1 ncRNA and coimmunostaining of endogenous SFPQ. Paraspeckles, which were observed in WT MEF cells, were undetectable in MEF cells prepared from NEAT1-knockout mice (KO). Paraspeckles were detected in KO-MEF cells transfected with a plasmid expressing NEAT1_1 (KO+NEAT1_1) or NEAT1_2 (KO+NEAT1_2). (B) Effect of actinomycin D treatment on the reformed paraspeckle-like foci. KO-MEF cells were cotransfected with plasmids expressing NEAT1_2 ncRNA and SFPQ–Flag. Transfected cells were treated with 0.3 μg/ml actinomycin D for 4 h. Reformed paraspeckle-like foci were visualized with RNA-FISH of NEAT1 and coimmunostained with anti-Flag M2 antibody. (C) NEAT1_2 ncRNA is more competent than NEAT1_1 ncRNA at elevating the number of paraspeckles. NIH3T3 cells were cotransfected with expression plasmids of control (+control), NEAT1_1 (+NEAT1_1), or NEAT1_2 (+NEAT1_2), together with SFPQ–Flag. The counted paraspeckle numbers are shown in Supplementary Figure S1D. Scale bar, 10 μm.
To prove that the rescued foci exhibited characteristics common to endogenous paraspeckles, transfected MEF(−/−) were treated with actinomycin D. Rescued foci did not appear with actinomycin D treatment. Instead, SFPQ–Flag (as a cotransfected marker) and endogenous PSPs relocated to perinucleolar caps, from which NEAT1_2 ncRNA was absent (Figure 1B). Paraspeckles reportedly display actinomycin D-induced disruption and the concomitant relocation of protein components (Fox et al, 2002; Shav-Tal et al, 2005). In the present study, the overexpression of either NEAT1_2 or NEAT1_1 in NIH3T3 cells led to elevated nuclear paraspeckle numbers; however, NEAT1_2 was more stimulatory than NEAT1_1 (Figure 1C; Supplementary Figure S1D). Taken together, these results indicate that the NEAT1_2 isoform is an essential RNA component for paraspeckle formation, and the NEAT1_1 isoform is not essential but can contribute to paraspeckle formation only when the NEAT1_2 isoform is present.
Identification of new paraspeckle components
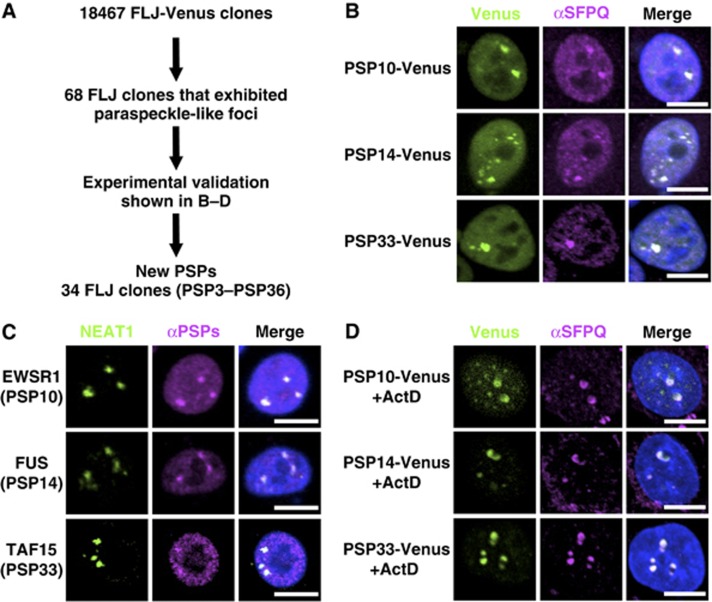
We previously reported that two RNA-binding proteins that are essential for paraspeckle formation, NONO and SFPQ, preferentially bind to and stabilize the NEAT1_2 isoform (Sasaki et al, 2009). To obtain further insights into the paraspeckle structure, additional PSPs were searched for by employing the human full-length cDNA resource (FLJ Clones) available from the authors’ affiliate (Maruyama et al, 2012). In this cDNA collection, the intact protein-coding regions of 18 467 human proteins are fused with Venus fluorescent protein. FLJ Clones provides information concerning the intracellular localization of >18 000 human proteins, through the transfection of each cDNA clone (Figure 2A).
Figure 2.
Identification of novel PSP components. (A) Experimental strategy to identify new PSPs. (B) Selection of FLJ-Venus clones that localize to paraspeckle-like nuclear foci. Paraspeckles were visualized by the immunostaining of SFPQ. Three representatives (PSP10, PSP14, and PSP33) of the new PSPs are shown. (C) Confirmation of the paraspeckle localization of endogenous PSP counterparts. Antibodies against each counterpart of PSP10, PSP14, and PSP33 (EWSR1, FUS and TAF15, respectively) were employed to monitor the localization (see Supplementary Table S4). NEAT1 ncRNA was used as a paraspeckle marker. (D) Effect of actinomycin D treatment on the localization patterns of selected PSPs. Localization of the Venus clones in B was monitored after actinomycin D treatment. SFPQ is an endogenous PSP control. Data regarding other PSPs are shown in Supplementary Figures S2 and S3. Scale bar, 10 μm.
Initially, 68 cDNA clones whose products exhibited the typical localization pattern of paraspeckle-like nuclear foci were selected. The identities of the foci were determined by immunostaining the endogenous SFPQ, to see if the Venus signals overlapped with the SFPQ signals (Figure 2B; Supplementary Figure S2A; Supplementary Table S1) but not with the signals of other nuclear bodies (Supplementary Figure S3B). This screening led to the eventual selection of 34 cDNA clones. Endogenous proteins corresponding to the identified cDNA clones were immunostained with their respective antibodies, when available (Figure 2C; Supplementary Figure S2B; Supplementary Table S1). The correct paraspeckle localization of all 27 examined proteins was confirmed, and no false positives were identified.
As a second screen, we confirmed the relocation of each Venus-fusion protein upon actinomycin D treatment. All 34 fusion proteins relocated to the perinucleolar caps (Supplementary Table S1). These caps corresponded to the destination of the endogenous PSPs (Figure 2D; Supplementary Figure S3A), but were distinct from those of the nucleolar or Cajal body proteins (Supplementary Figure S3B).
Therefore, 34 cDNA clones, designated PSP3 through PSP36, were confirmed as new PSPs. Additionally, TARDBP (TDP43), which was recently reported to interact prominently with NEAT1 ncRNA in the brain from frontotemporal lobar degeneration (FTLD) patients (Tollervey et al, 2011), was confirmed to localize to the paraspeckle in HeLa cells by immunostaining of endogenous protein (Supplementary Figure S2B) and detection of TARDBP-Venus localization (Supplementary Figure S2A).
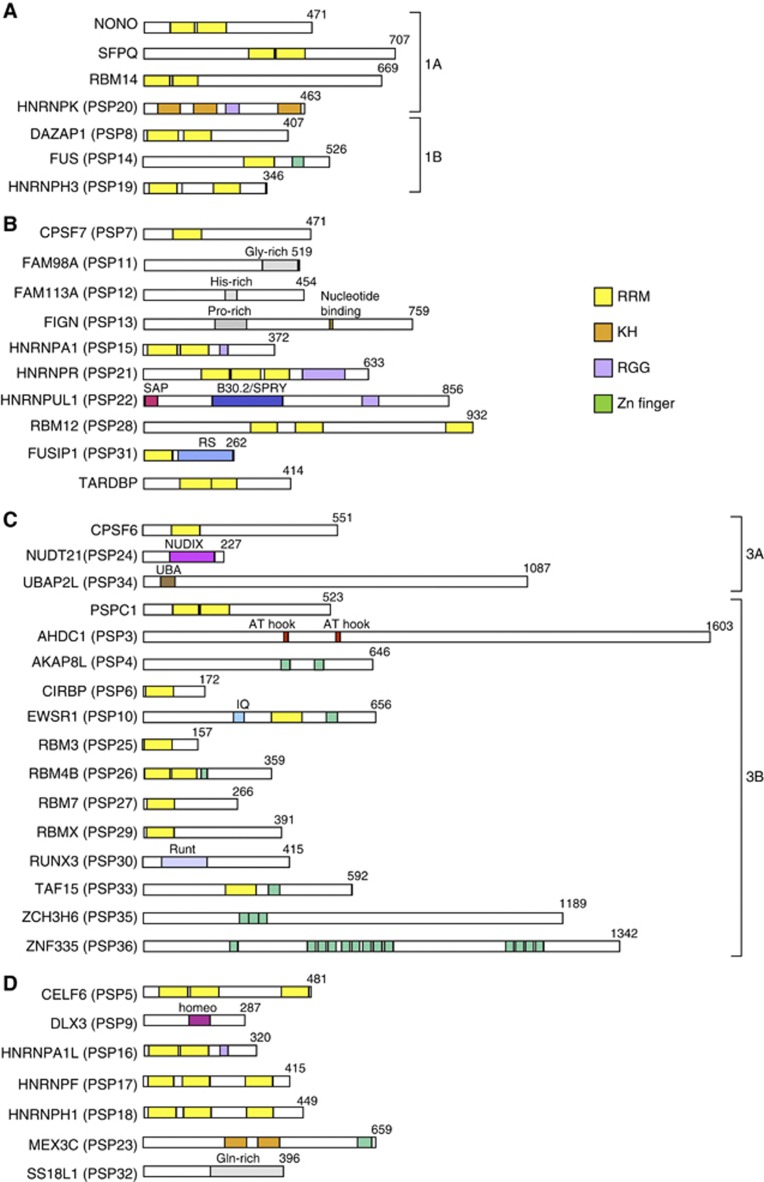
A comparison of all of the PSPs (Table I; Figure 4) indicated that most possessed canonical RNA-binding domains (Burd and Dreyfuss, 1994): 20 proteins with RRMs, two proteins with KH motifs, and five proteins with RGG boxes. Eight proteins possessed one or more zinc-finger motifs, which are involved in RNA binding (Brown, 2005). Three related RNA-binding proteins, FUS, EWSR1, and TAF15, which are known as significant disease-related proteins (Law et al, 2006), were all identified as PSPs. We also identified CPSF6, NUDT21, and CPSF7, which are components of the CFIm complex that regulates the 3′-end processing of mRNA, as PSPs.
Table 1. Paraspeckle proteins.
| Proteins | Accession | RNA-binding motifs | Other motifs | Category | ||
|---|---|---|---|---|---|---|
| PSP # | HUGO | Synonyms | ||||
| New paraspeckle proteins | ||||||
| PSP3 | AHDC1 | Q5TGY3 | AT hook | 3B | ||
| PSP4 | AKAP8L | Q9ULX6 | 2 ZnF C2H2s | 3B | ||
| PSP5 | CELF6 | BRUNOL6 | Q96J87 | 3 RRMs | ND | |
| PSP6 | CIRBP | Q14011 | RRM | 3B | ||
| PSP7 | CPSF7 | CFIm59 | Q8N684 | RRM | 2 | |
| PSP8 | DAZAP1 | Q96EP5 | 2 RRMs | 1B | ||
| PSP9 | DLX3 | O60479 | homeodomain | ND | ||
| PSP10 | EWSR1 | Q01844 | RRM | ZnF RanBP2 | 3B | |
| PSP11 | FAM98A | Q8NCA5 | 2 | |||
| PSP12 | FAM113A | Q9H1Q7 | 2 | |||
| PSP13 | FIGN | Q5HY92 | 2 | |||
| PSP14 | FUS | TLS | P35637 | RRM | ZnF RanBP2 | 1B |
| PSP15 | HNRNPA1 | P09651 | 2 RRMs | 2 | ||
| PSP16 | HNRNPA1L2 | Q32P51 | 2 RRMs | ND | ||
| PSP17 | HNRNPF | P52597 | 3 RRMs | ND | ||
| PSP18 | HNRNPH1 | P31943 | 3 RRMs | ND | ||
| PSP19 | HNRNPH3 | P31942 | 2 RRMs | 1B | ||
| PSP20 | HNRNPK | P61978 | 3 KHs | 1A | ||
| PSP21 | HNRNPR | O43390 | 3 RRMs | 2 | ||
| PSP22 | HNRNPUL1 | Q9BUJ2 | SAP, SPRY | 2 | ||
| PSP23 | MEX3C | Q5U5Q3 | 2 KHs | ZnF RING | ND | |
| PSP24 | NUDT21 | CFIm25 | O43809 | NUDIX hydrolase | 3A | |
| PSP25 | RBM3 | P98179 | RRM | 3B | ||
| PSP26 | RBM4B | RBM30 | Q9BQ04 | 2 RRMs | ZnF CCHC | 3B |
| PSP27 | RBM7 | Q9Y580 | RRM | 3B | ||
| PSP28 | RBM12 | Q9NTZ6 | 3 RRMs | 2 | ||
| PSP29 | RBMX | P38159 | RRM | 3B | ||
| PSP30 | RUNX3 | Q13761 | 3B | |||
| PSP31 | SRSF10 | FUSIP1, SRp38 | O75494 | RRM | RS | 2 |
| PSP32 | SS18L1 | O75177 | ND | |||
| PSP33 | TAF15 | Q92804 | RRM | ZnF RanBP2 | 3B | |
| PSP34 | UBAP2L | Q14157 | 3A | |||
| PSP35 | ZC3H6 | P61129 | 3 ZnF C3H1s | 3B | ||
| PSP36 | ZNF335 | Q9H4Z2 | 13ZnF C2H2s | 3B | ||
| TARDBP | TDP43, ALS10 | Q13148 | 2 RRMs | 2 | ||
| Known paraspeckle proteins | ||||||
| CPSF6 | CFIm68 | Q16630 | RRM | 3A | ||
| NONO | p54nrb | Q15233 | 2 RRMs | 1A | ||
| PSPC1 | PSP1 | Q8WXF1 | 2 RRMs | 3B | ||
| RBM14 | PSP2, CoAA | Q96PK6 | 2 RRMs | 1A | ||
| SFPQ | PSF | P23246 | 2 RRMs | 1A |
Although SR-rich splicing factors are commonly enriched in nuclear speckles, SRSF10 was exceptionally enriched in paraspeckles rather than nuclear speckles. The set of PSPs included several abundant heterogenous nuclear ribonucleoproteins (hnRNPs) (A1, A1L, F, H1, H3, K, R, and UL1). Although the numbers were limited, some of the PSPs possessed putative DNA-binding domains, including the AT-hook, Zn finger, homeodomain, and SAP domain. Several of the PSPs possessing RNA-binding domains (e.g., NONO, SFPQ, EWSR1, FUS, TAF15, and HNRNPUL1) are known to be involved in transcription. This observation suggests that the paraspeckle may serve as a platform of transcription and subsequent RNA processing events (see Discussion).
Functional assignments for each PSP identified additional essential factors for paraspeckle formation
To investigate the respective roles of the newly identified and known PSPs in paraspeckle construction, each PSP was knocked down with at least two independent siRNAs (Supplementary Table S2). The resultant changes in paraspeckle appearance (i.e., proportion [%] of cells possessing intact paraspeckles) and in the levels of each NEAT1 isoform (summarized in Supplementary Table S3) were examined.
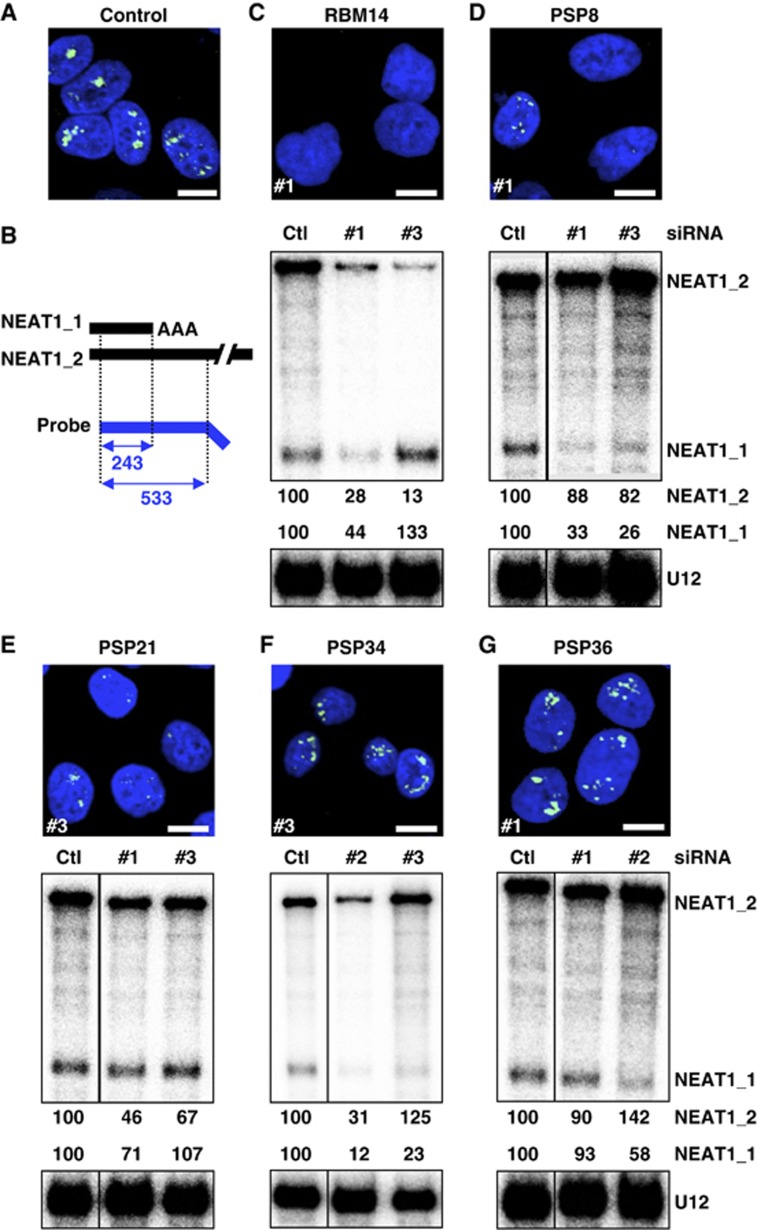
The PSPs were classified into three distinct categories according to the proportion of paraspeckle-possessing cells after RNAi. After treatment with control siRNA, 88% of the cells examined possessed paraspeckles; this group was defined as the control (Ctl) group (100%). Categories 1, 2, and 3 included PSPs whose RNAi led to a marked decrease (⩽30% of Ctl), substantial decrease (30–75% of Ctl), or no obvious change (⩽75% of Ctl) in the proportion of paraspeckle-possessing cells, respectively (Supplementary Table S3; Supplementary Figure S4). Investigation of the NEAT1 levels by RNase protection assays (RPAs) (Figure 3B) revealed that category 1 could be divided into two subcategories, in which the NEAT1_2 levels were markedly diminished to ⩽30% (category 1A) or were unchanged (category 1B). Similarly, category 3 was divided into two subcategories, in which NEAT1_1 levels were either diminished to ⩽30% (category 3A) or were unchanged (category 3B). The PSP categorization is summarized in Figure 4 and Table I. Representative data from each of the five categories are shown in Figure 3C–G.
Figure 3.
Functional assignment of new PSPs in paraspeckle formation by extensive RNAi treatment. Paraspeckle appearance and NEAT1 levels were monitored by RNA-FISH and RPA to detect NEAT1_1 and NEAT1_2. (A) Paraspeckles in cells treated with control siRNA. (B) Schema for the RPA probe used and the protected fragments (with size, nt) of NEAT1_1 and NEAT1_2. Data for a representative from each category (1A: RBM14, 1B: PSP8/DAZAP1, 2: PSP21/HNRNPR, 3A: PSP34/UBAP2L, 3B: PSP36/ZNF335) are shown in (C–G), respectively. The siRNA numbers (#) used in the RNA-FISH analysis are shown at the lower left of each photo. For RPA, the ratio of band intensities of the two isoforms, normalized by those of U12 snRNA, is shown below (Ctl was defined as 100%). RNAi data regarding all PSPs are compiled in Supplementary Figure S4. Their quantified data are shown in Supplementary Table S3. The siRNAs used are shown in Supplementary Table S6. Scale bar, 10 μm. Figure source data can be found with the Supplementary data.
Figure 4.
Compilation of PSPs. Schematics of the major domains of the PSPs that belong to category 1 (A), 2 (B), and 3 (C) are grouped and shown. Uncategorized PSPs are shown in (D). Subcategories in categories 1 and 3 are shown as 1A and 1B in (A) and 3A and 3B in (C). The amino-acid length of each PSP is shown in the right corner. The colour codes of four putative RNA-binding domains are shown on the right.
Several PSPs were unable to be categorized because their expression was either undetectable in HeLa cells (PSP5, 9, and 23) or they showed inconsistent paraspeckle phenotypes between treatments with the two siRNAs (PSP16, 17, 18, and 32). Several PSPs were tentatively categorized according to the consistent paraspeckle phenotypes between treatments with two siRNAs, although the RPA data were highly variable (PSP10, 15, 25, and 27).
Three PSPs are involved in NEAT1 isoform synthesis by modulating alternative 3′-end processing
Because the NEAT1 isoforms share an identical 5′-terminus, they are likely produced by alternative 3′-end processing. The 3′-ends of NEAT1_1 and NEAT1_2 are formed by two distinct mechanisms: canonical polyadenylation and RNase P cleavage, respectively. The above RNAi experiments identified factors that are involved in this alternative 3′-processing event.
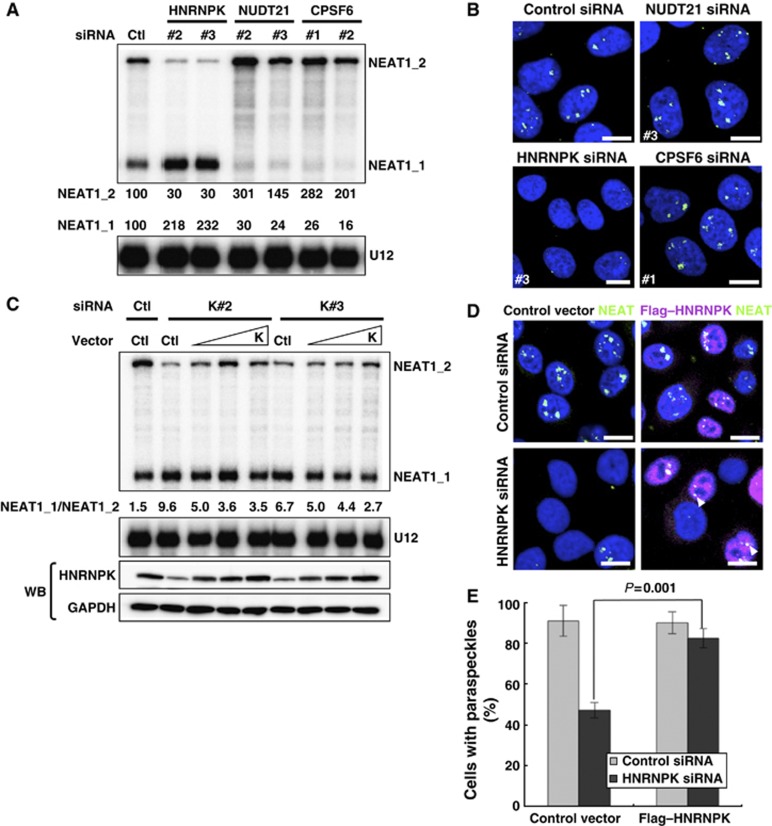
Two PSPs, CPSF6 and PSP24/NUDT21, form a heterodimer (CFIm complex) to facilitate the 3′-end processing of alternatively processed mRNAs (Kim et al, 2010). These PSPs also appear to act in NEAT1_1 3′-end processing. We observed that the RNAi of NUDT21 or CPSF6 markedly diminished NEAT1_1 levels and simultaneously increased the NEAT1_2 level (Figure 5A; Supplementary Figure S5B).
Figure 5.
Alternative 3′-end processing of NEAT1 is the initial essential step underlying paraspeckle formation. (A) PSPs required for the alternative 3′-end processing of NEAT1. RPA was performed as in Figure 3. Data are shown for NUDT21 and CPSF6, which are required for NEAT1_1 3′-end processing, and HNRNPK, which is required for NEAT1_2 synthesis by interfering with NEAT1_1 3′-end processing. (B) Paraspeckle appearance in HeLa cells treated with siRNAs against NUDT21, CPSF6, or HNRNPK. (C) Plasmid rescue of a defect of NEAT1_2 synthesis in HNRNPK-eliminated cells. Two siRNAs against HNRNPK (K#2 and K#3) were used to eliminate endogenous HNRNPK, and HNRNPK rescue plasmid (K) was introduced at three concentrations (1–5 μg). NEAT1 ncRNA levels were measured by RPA as in Figure 3. The ratios of NEAT1_1 to NEAT1_2 (NEAT1_1/NEAT1_2) are shown below the upper panel. GADPH and HNRNPK were detected by western blotting (WB). (D) Paraspeckle formation is rescued by plasmid expression of HNRNPK. The siRNAs and rescue plasmids used are shown on the left and top, respectively. Paraspeckles were detected by RNA-FISH of NEAT1. Transfected cells were visualized by immunostaining with αFlag. Arrowheads indicate paraspeckles that formed in the rescued cells. Scale bar, 10 μm. (E). Quantification of the results in (D). Cells possessing more than one paraspeckle-like focus were counted. Total cell numbers counted are the siRNAs used are shown in Supplementary Table S6. _P_-value was calculated by Student’s _t_-test. The cell numbers counted for control and HNRNPK-eliminated cells were 152 and 136, respectively.
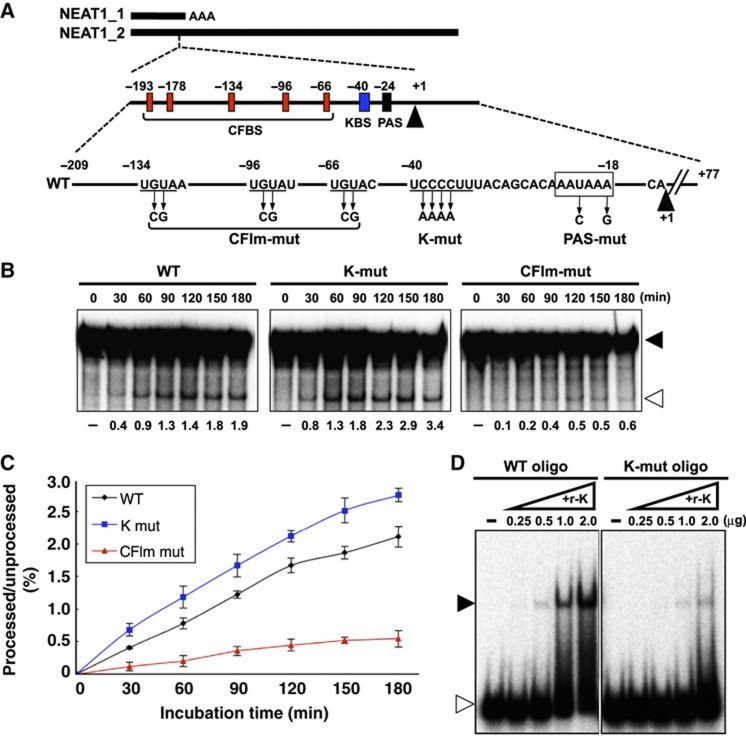
The CFIm complex binds to UGUA sequences located upstream of the canonical polyadenylation signal (PAS) and recruits the general 3′-end processing machinery to polyadenylation sites (Venkataraman et al, 2005). Sequence searches revealed that five UGUA sequences are clustered 42–169 nt upstream of the PAS (AAUAAA) for NEAT1_1 3′-end processing (Figure 6A). This result strongly suggests that CFIm facilitates the 3′-end processing of NEAT1_1 through binding to the UGUA sequences.
Figure 6.
Roles of CFIm and HNRNPK in 3′-end processing of NEAT1_1. (A) Schematic representation of the substrate RNAs for the in vitro processing reaction that contains the region spanning the 3′-end of NEAT1_1. Numbers indicate distance from the polyadenylation site of NEAT1_1. Middle scheme represents putative sequences around the NEAT1_1 3′-end processing site that are recognized by the CFIm complex (CFBS: red boxes) or HNRNPK (KBS: blue box). Mutated positions on the mutant substrates (CFIm-mut, K-mut, and PAS-mut) are indicated. (B) Recapitulation of CFIm-dependent 3′-end processing of NEAT1_1 in vitro. Incubation time is shown above each panel. Substrate RNAs are represented on the top. Unprocessed and processed bands are shown with closed and open triangles, respectively, on the right. Processing efficiencies (%) are shown below each panel. (C) Average values of the processing efficiencies obtained from three independent experiments. (D) Detection of sequence-specific RNA binding of HNRNPK. Gel mobility shift assay to detect binding of recombinant HNRNPK protein (r-K) with RNA fragments (30 nt) derived from WT and K-mut, WT oligo, and K-mut oligo, respectively, are shown. The RNA–protein complex and free RNA are shown with closed and open triangles, respectively. Amounts of supplemented r-K (μg) are shown above each panel.
Intact paraspeckles remained detectable after treatment with RNAi for either NUDT21 or CPSF6 (Figure 5B), even though NEAT1_1 was obliterated (Figure 5A; Supplementary Figure S5B). This result confirms that NEAT1_1 is dispensable for paraspeckle formation. Although RNAi elimination of PSP10/EWSR1 upregulated NEAT1_1, this condition did not affect NEAT1_2 (Supplementary Figure S4E). This observation suggests that EWSR1 may control the stability of NEAT1_1.
PSP20/HNRNPK is a new member of category 1A that is required for NEAT1_2 accumulation. Treatment with HNRNPK RNAi disrupted the paraspeckles and decreased the NEAT1_2 level, but simultaneously elevated the NEAT1_1 level (>2-fold) (Figure 5A and B; Supplementary Figure S5B). This finding was not observed with an RNAi of any other category I protein (Supplementary Figure S4A), which suggests that HNRNPK facilitates NEAT1_2 synthesis, rather than stabilization, by modulating NEAT1_1 3′-end processing.
RT–qPCR measurement of NEAT1 ncRNA coimmunoprecipitated with anti-NUDT21 antibody (αNUDT21) revealed that the NEAT1_1/NEAT1_2-overlapped region but not the NEAT1_2-specific region was markedly increased with HNRNPK RNAi (Supplementary Figure S5C and D). This finding indicates that the association of NUDT21 with NEAT1_1 was accelerated by HNRNPK elimination in vivo.
The HNRNPK-eliminated cells were transfected with a plasmid for siRNA-resistant HNRNPK (K#2 and K#3 in Figure 5C), and the ratio of the NEAT1_1 and NEAT1_2 isoforms was measured (NEAT1_1/NEAT1_2, referred to as the NEAT1 ratio). As the amount of exogenous HNRNPK increased (lower panels of Figure 5C), the NEAT1 ratio proportionally decreased (Figure 5C). Moreover, exogenous HNRNPK rescued the defect of paraspeckle formation (Figure 5D and E). These results indicate that HNRNPK is responsible for NEAT1_2 synthesis, which determines the NEAT1 ratio and consequent paraspeckle formation.
Previous SELEX analyses have identified CU-rich stretches as preferred binding sequences for HNRNPK (Thisted et al, 2001). We identified a CU-rich stretch (UCCCCUU) that perfectly matched a SELEX-derived sequence, which was present in a region adjacent to the canonical PAS (blue box in Figure 6A) that is conserved in rodents. Therefore, HNRNPK likely binds to the CU-rich stretch and interferes with NEAT1_1 3′-end processing, resulting in the preferential synthesis of NEAT1_2. These data indicate that HNRNPK modulates the alternative 3′-end processing for NEAT1_2 synthesis that initiates paraspeckle formation.
HNRNPK binding arrests the CFIm-dependent 3′-end processing of NEAT1_1 in vitro
To investigate how HNRNPK controls alternative 3′-end processing, the NEAT1_1 3′-end processing reaction was recapitulated in HeLa cell nuclear extract (HNE). A 32P-radiolabelled RNA substrate that contained 303 nt spanning the processing site of NEAT1_1 (Figure 6A) was incubated in HNE. Because no Mg2+ was added to the in vitro reaction, the endonucleolytic cleavage solely produced the 209-nt processed RNA without subsequent polyadenylation. The cleavage product was detectable after incubation for 30 min (Figure 6B; Supplementary Figure S6A). Processed RNA was not generated from an RNA substrate with a mutated PAS (PAS-mut in Supplementary Figure S6A), which indicated that accurate RNA processing was recapitulated in vitro.
To check the roles of CFIm and HNRNPK in the 3′-end processing of NEAT1_1 in vitro, RNA substrates in which the putative CFIm-binding sequences (CFBS) or HNRNPK-binding sequence (KBS) were mutated (CFIm-mut and K-mut, respectively, in Figure 6A) were applied to the in vitro processing reaction. Time-course experiments revealed that CFIm-mut exhibited marked deceleration of the processing compared with the wild-type substrate (WT) (Figure 6B and C), confirming the reported evidence that CFIm facilitates 3′-end processing through its association with CFBS (Venkataraman et al, 2005). By contrast, K-mut accelerated in vitro processing (Figure 6B and C), which supported our in vivo result that HNRNPK acts to suppress the 3′-end processing of NEAT1_1. The results of a gel mobility shift assay with recombinant HNRNPK protein (r-K) confirmed that r-K binding was mostly abolished by the KBS mutation (Figure 6D), indicating that HNRNPK binds to KBS and arrests the CFIm-dependent 3′-end processing of NEAT1_1.
HNRNPK arrests the RNA binding of CFIm for NEAT1_1 3′-end processing
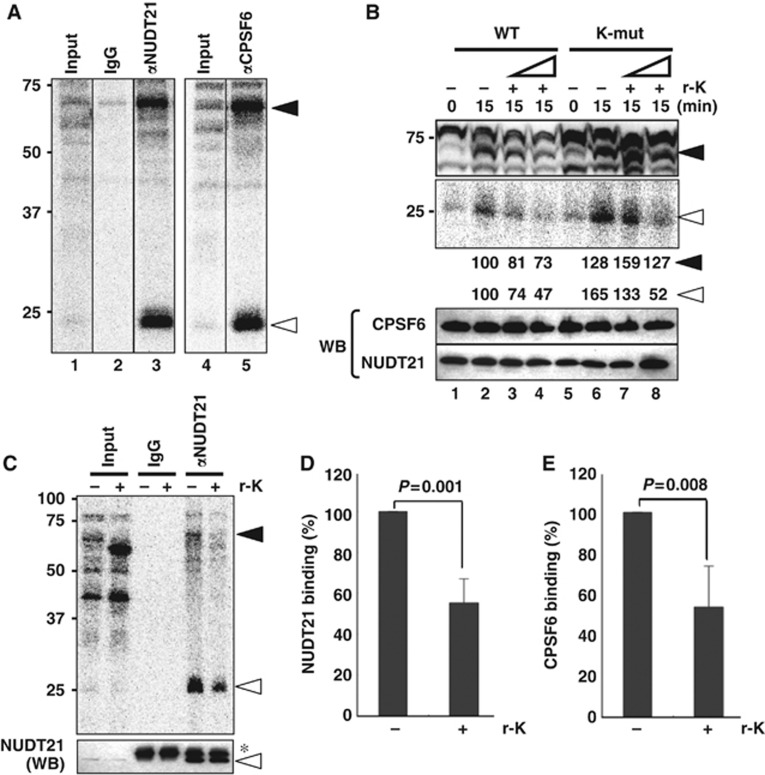
We examined the effect of HNRNPK on the RNA binding of CFIm during in vitro processing. Proteins that bound to WT during in vitro processing were visualized by UV-crosslinking (Figure 7A, lanes 1 and 4). Immunoprecipitation with αNUDT21 and αCPSF6 revealed that the UV-crosslinked ∼68- and ∼25-kDa proteins were efficiently precipitated by each antibody (Figure 7A), indicating that they corresponded to CPSF6 and NUDT21, respectively.
Figure 7.
Molecular mechanism of the alternative 3′-end processing of NEAT1. (A) Detection of RNA binding of CFIm during in vitro processing by UV-crosslinking. UV-crosslinked WT substrate RNA-binding proteins were detected as 32P-labelled proteins on SDS–PAGE. The 68- and 25-kDa UV-crosslinked RNA-binding proteins (closed and open arrows, respectively) were immunoprecipitated with antibodies against CPSF6 and NUDT21. (B) RNA bindings of CPSF6 and NUDT21 are affected by the addition of r-K. The two RNA substrates (WT and K-mut) employed are shown on the top. Addition of r-K (+) at two concentrations (10 × and 30 × excess of endogenous HNRNPK in HNE) and the incubation time (min) are shown above. Closed and open triangles are as in (A). Intensities of the 68- and 25-kDa bands were quantified and normalized by the levels of CPSF6 and NUDT21, respectively, which were detected by the western blot (WB) shown below. Molecular weight marker is shown on the left. (C) Confirmation of r-K-dependent inhibition of RNA binding of CPSF6 and NUDT21. Presence (+) or absence (−) of r-K (30 × excess) is indicated above the panel. UV-crosslinking of r-K (∼55 kDa) was detected in the Input lane (+r-K) and in Supplementary Figure S6B. Closed and open triangles are as in (A). Amounts of NUDT21 in the input samples and immunoprecipitated samples were detected by the WB shown below the panel. Asterisk represents IgG light chain. (D) Quantification of immunoprecipitated, UV-crosslinked NUDT21 in the presence (+) or absence (−) of r-K (open triangle in the upper panel in C). Data were normalized with the total amounts of NUDT21 in each immunoprecipitation sample (lower panel in C). Graph shows the average (with s.d.) of three independent experiments. _P_-value was calculated by Student’s _t_-test. (E) Quantification of immunoprecipitated CPSF6, as in (D). Antibodies are shown in Supplementary Table S4. Figure source data can be found with the Supplementary data.
To assess whether the extra HNRNPK interfered with the RNA binding of CFIm, r-K was added to the in vitro processing. On the WT substrate, r-K markedly interfered with the UV-crosslinking of CPSF6 and NUDT21 in a concentration-dependent manner (Figure 7B, lanes 1–4). Immunoprecipitation of UV-crosslinked CPSF6 and NUDT21 clearly revealed that the RNA bindings of CPSF6 and NUDT21 were diminished (∼50%) in the presence of r-K (Figure 7C–E). The mutation of KBS (K-mut) substantially elevated the UV-crosslinking of CPSF6 and NUDT21 (Figure 7B, lanes 2 and 6). The interfering effect of r-K on CPSF6 binding was milder on K-mut compared with WT (Figure 7B, lanes 6–8). These data indicate that HNRNPK binding to KBS results in the arrest of CFIm binding.
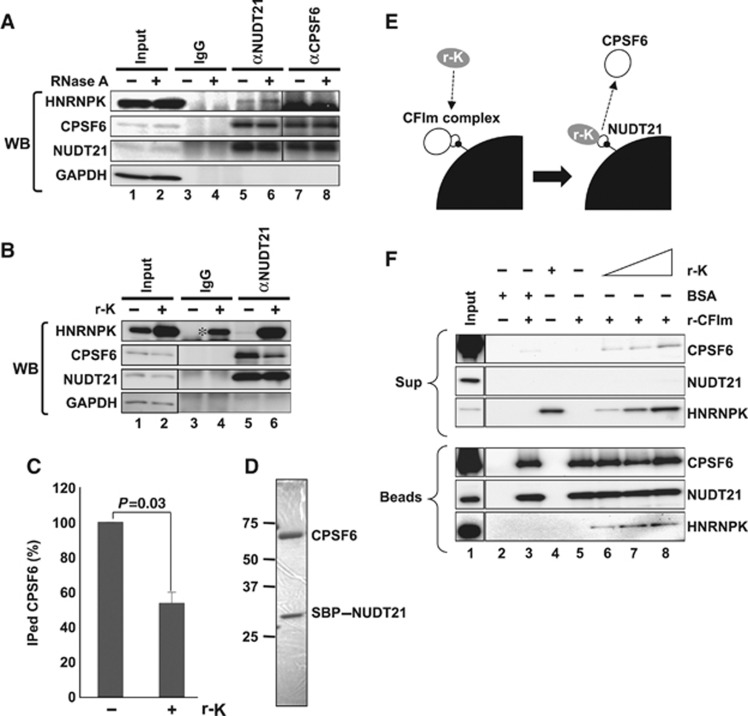
To obtain further mechanistic insights into the HNRNPK-dependent arrest of the 3′-end processing of NEAT1_1, the interaction between HNRNPK and CFIm was investigated. Endogenous HNRNPK was coimmunoprecipitated with NUDT21 in the presence of RNase A (Figure 8A, lanes 5 and 6) but not with CPSF6 (Figure 8A, lanes 7 and 8). This interaction was confirmed during in vitro processing, in which the supplemented r-K prominently coprecipitated with αNUDT21 (Figure 8B). This result was supported by protein-interaction data in the public database, in which NUDT21 but not CPSF6 was listed as an HNRNPK interactor (http://www.genecards.org). Importantly, the NUDT21–CPSF6 interaction was markedly weakened (∼50%) in the presence of excess r-K (Figure 8B and C), suggesting that HNRNPK competes for the binding of NUDT21 with CPSF6.
Figure 8.
HNRNPK captures NUDT21 from the functional CFIm complex. (A) Interaction between HNRNPK and NUDT21 in vivo. Immunoprecipitations with αNUDT21 and αCPSF6 were performed in the presence (+) or absence (−) of RNase A. CPSF6, NUDT21, and HNRNPK were detected by WB. (B) The CPSF6–NUDT21 interaction is affected by HNRNPK. HNE was incubated with r-K in the presence of WT RNA. Immunoprecipitation with αNUDT21 was performed in the presence (+) or absence (−) of r-K. CPSF6, NUDT21, HNRNPK, and GAPDH were detected by WB. Immunoprecipitation of CPSF6 was diminished in the presence of r-K. Asterisk represents nonspecific binding of excess r-K to IgG. (C) Quantification of immunoprecipitated CPSF6 in the presence or absence of r-K. Relative amount (%) of CPSF6 was normalized by the immunoprecipitated NUDT21 level (B). Graph shows the average (with SD) of three independent experiments. _P_-value was calculated by Student’s _t_-test. (D) Purified CFIm complex. The SDS–PAGE gel was stained with Coomassie Brilliant Blue. Both CPSF6 and SBP–NUDT21 possess Flag- and HA-tags. Size maker is shown on the left. (E) Schematics of the competitive binding assay carried out in F. The CFIm complex (30 pmol NUDT21–CPSF6 complex, white circles) was immobilized on streptavidin-conjugated beads (black quarter circle) through SBP-tag (small black circle). Recombinant HNRNPK (0–90 pmol r-K, grey circle) was mixed with the beads. Binding of r-K with NUDT21 is expected to lead to dissociation of CPSF6 from the beads. BSA (90 pmol) was used as a control. (F) Detection of CPSF6, NUDT21, and HNRNPK in bead (Beads) and supernatant (Sup) fractions. Proteins in the binding reaction are shown by+on the top of the panels. Each protein was detected by western blot, as shown on the right of the panels. Input lanes were loaded with 1/30 of the proteins used. Antibodies used are shown in Supplementary Table S4. Figure source data can be found with the Supplementary data.
To verify the competition between HNRNPK and CPSF6 for binding with NUDT21, the recombinant CFIm complex (NUDT21–CPSF6) was purified from HEK293T cells that were contransfected with two expression plasmids for the streptavidin-binding peptide (SBP)-tagged NUDT21 and CPSF6 (Figure 8D). Purified CFIm complex was immobilized on streptavidin-conjugated beads through the SBP-tag on NUDT21 (Figure 8E). The beads were mixed with r-K, and the binding of r-K to the beads was detected simultaneously with CPSF6 dissociation from the beads (Figure 8E).
As shown in Figure 8F, r-K specifically associated with the beads only when the CFIm complex was conjugated to the beads (compare lanes 4 and 8 in the bottom panel of Figure 8F). The addition of r-K caused the marked dissociation of CPSF6 from the beads, whereas the addition of BSA caused much less release of CPSF6 (compare lanes 8 and 3 in the top panel in Figure 8F). These results strongly support our argument that HNRNPK competes for the binding of NUDT21 with CPSF6, which is a possible underlying mechanism for the arrest of CFIm binding by HNRNPK (Figure 9A).
Figure 9.
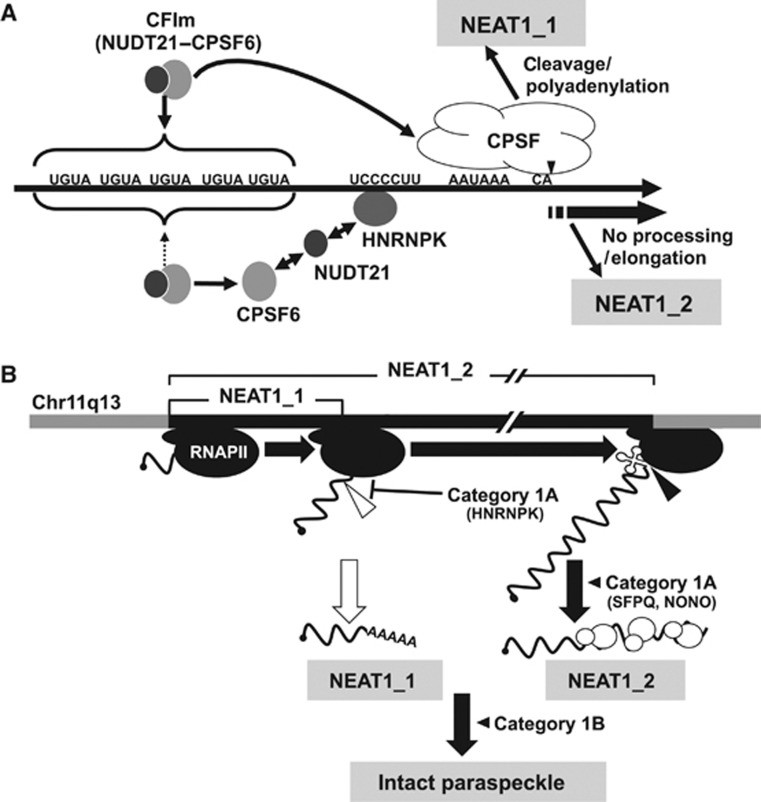
(A) Models of NEAT1 isoform synthesis. NEAT1 border region is shown with the critical sequence elements. Pathways for NEAT1_1 and NEAT1_2 synthesis are shown above and below the NEAT1 scheme. For NEAT1_2 synthesis, HNRNPK binds with the UCCCCUU sequence, captures NUDT21 from the functional CFIm (CPSF6–NUDT21), and arrests CFIm binding to upstream UGUA sequences. (B) Current model of intact paraspeckle formation. The essential steps, including 1) ongoing transcription of NEAT1 by RNA polymerase II (RNAPII), 2) NEAT1_2 synthesis by alternative 3′-end processing, 3) NEAT1_2 stabilization by category 1A proteins, such as SFPQ and NONO, and 4) subsequent assembly step(s), are schematized and represented with bold black arrows. Category 1B proteins act in an essential step other than NEAT1_2 accumulation. NEAT1_1 synthesis is dispensable; therefore, it is shown with a white arrow. The 3′-ends of NEAT1_1 and NEAT1_2 are formed by distinct mechanisms: canonical polyadenylation (open triangle) and RNase P cleavage (closed triangle). The significance of the noncanonical 3′-end processing of NEAT1_2 remains uncertain.
Discussion
Expansion of paraspeckle components
In the current study, the FLJ cDNA-based localization screening revealed 34 new PSPs. Many of the new PSPs are likely present throughout the nucleoplasm, and subsets may be concentrated in paraspeckles. An analysis of the compilation of all of the PSPs (Figure 4; Table I) indicated that most possess canonical RNA-binding domains. NUDT21, which was found to possess no canonical RNA-binding motif, has a NUDIX hydrolase domain that acts like an authentic RNA-binding protein (Yang et al, 2010). Some of the paraspeckle-localized RNA-binding proteins (e.g., NONO, SFPQ, RBM14, EWSR1, FUS, TAF15, and TARDBP) mediate both transcription and RNA processing (Auboeuf et al, 2005). Several of the new PSPs (e.g., AHDC1, DLX3, and ZNF335) are likely to be DNA-binding proteins that are involved in transcriptional control. This finding raises the possibility that paraspeckles may integrate tightly coupled transcription and post-transcriptional events.
Yang et al (2011) reported that nuclear body-localized ncRNAs mediate the attachment of the specific chromosomal locus to the nuclear bodies that control the epigenetic status of the gene loci. NEAT1 may play a similar role in the attachment of the specific chromosomal locus to the paraspeckles, where multiple regulatory PSPs are enriched for modulating gene expression from the specific locus. Alternatively, paraspeckles may be involved in RNA-dependent epigenetic regulation. Indeed, two of the identified PSPs, FUS and HNRNPK, participate in epigenetic regulation through their interactions with long ncRNAs (Wang et al, 2008; Huarte et al, 2010).
Several PSPs are disease-related. The genes for nine PSPs (NONO, SFPQ, CPSF6, EWSR1, FUS, TAF15, DAZAP1, RBM3, and SS18L1) are the breakpoints of chromosomal translocation that result in the production of abnormal fusion proteins responsible for various cancers (Kim et al, 2006). Four of them (SFPQ, NONO, DAZAP1, and FUS) belong to category 1, suggesting that the paraspeckle structure is altered in tumour cells in which the genes have undergone translocations. FUS and TARDBP are commonly associated with a neurodegenerative disease, familial amyotrophic lateral sclerosis (ALS) (Lagier-Tourenne and Cleveland, 2009). TARDBP was found to associate prominently with NEAT1 in the brain of patients with FTLD-TDP, which is an ALS-related neurodegenerative disease with TARDBP inclusions (Tollervey et al, 2011). These evidences suggest that TARDBP associated with NEAT1 is sequestered in paraspeckles, where it is functionally modulated.
Mechanism of alternative RNA processing of NEAT1 ncRNA
NEAT1_2 was found to be essential for paraspeckle formation. Therefore, the alternative 3′-end processing event that leads to NEAT1_2 accumulation is a fundamental molecular event for paraspeckle formation. Alternative 3′-end processing, which produces various mRNA isoforms with different 3′-UTR lengths, is utilized mainly for situations in which the produced mRNA isoforms are subjected to differential regulation by 3′-UTR-interacting factors (Lutz, 2008). In the case of NEAT1 ncRNA, this mechanism diversifies the ncRNA functions.
The alternative 3′-end processing of NEAT1 comprises two distinct 3′-end processing mechanisms: canonical polyadenylation for NEAT1_1 and RNase P-mediated cleavage for NEAT1_2. CFIm binds UGUA sequences and facilitates processing and polyadenylation at adjacent sites (Venkataraman et al, 2005). Our RNAi and in vitro analyses indicated a corresponding mechanism for the 3′-end processing of NEAT1_1.
PSP7/CPSF7 was reported to form a heterodimer with NUDT21 to facilitate 3′-end processing (Kim et al, 2010). However, we observed that CPSF7 RNAi markedly decreased the NEAT1_2 level, which was the opposite effect of NUDT21 RNAi (see Supplementary Table S3). This finding suggests that CPSF7 has an additional role in NEAT1_2 processing or stabilization. Alternatively, CPSF7 may play a counteracting role to that of CPSF6–NUDT21 in 3′-end processing under a specific (e.g., paraspeckle-localized) condition.
The multifunctional HNRNPK protein is involved in transcriptional regulation, pre-mRNA splicing, mRNA stability, and translation (Bomsztyk et al, 2004). Our data add a new function to this list: the regulation of 3′-end processing. The results of our RNAi and immunoprecipitation experiments suggest that HNRNPK is required for NEAT1_2 accumulation through its arrest of the 3′-end processing of NEAT1_1. The results of our in vitro processing and UV-crosslinking experiments reveal that HNRNPK interferes with the RNA binding of the CFIm complex through its binding to KBS. The possibility that HNRNPK additionally participates in the stabilization or noncanonical 3′-end processing of the NEAT1_2 isoform cannot be ruled out. Our preliminary results show that HNRNPK is required to maintain the NEAT1_2 level, even in the absence of CPSF6, suggesting additional role(s) of HNRNPK in the accumulation of the NEAT1_2 isoform. Mapping of the HNRNPK-binding sites may uncover additional function(s) of HNRNPK in NEAT1 expression.
Our coimmunoprecipitation results indicated that NUDT21 but not CPSF6 interacted with HNRNPK in vivo and in vitro. The supplemented r-K interacted with NUDT21, which resulted in a diminished interaction with CPSF6 and suggested the underlying mechanism. The binding of HNRNPK to KBS proximal to CFBS would provide an environment in which HNRNPK and CPSF6 effectively compete for association with NUDT21, which eventually determines the NEAT1 isoform ratio. The dissection of HNRNPK and NUDT21 to identify the interacting domain(s) and further interaction studies would solidify this model.
Several RNA-binding proteins (e.g., PTBP1, NONO, and ELAVL1) reportedly bind to upstream sequences implicated in the regulation of mRNA 3′-end processing; however, their detailed mechanisms of action remain to be investigated (Millevoi and Vagner, 2010). It would be intriguing to pursue the generality of the HNRNPK-dependent regulatory mechanism in the 3′-end processing of mRNAs and other ncRNAs. The NEAT1 ratio is controllable because it is variable in different mouse tissues (Nakagawa et al, 2011). The HNRNPK protein is expressed ubiquitously, but its activity is controlled by post-translational modifications, such as phosphorylation, methylation, and ubiquitination, under various conditions, including DNA damage (Chen et al, 2008). This fact raises the possibility that the NEAT1 ratio is controlled through the modification status of HNRNPK. Our experimental system for the functional rescue of HNRNPK (used in Figure 5C and D) would be useful to identify the important modification site(s) of HNRNPK required for the regulation of NEAT1 alternative processing.
CFIm complexes may be involved in the 3′-end processing of other RNAs in paraspeckles. Electron microscopic observation has revealed the localization of CPSF6 in the interior area of the paraspeckle (Cardinale et al, 2007), where the NEAT1_1 isoform is not present (Souquere et al, 2010). Alternatively, the paraspeckle interior area may serve as the storage site of CFIm complexes.
Steps required for paraspeckle formation
Our results provide several important insights into paraspeckle formation. The plasmid rescue experiment clarified that NEAT1_2 but not NEAT1_1 is a necessary RNA component for de novo paraspeckle formation. This evidence supports our previous observations that: (1) SFPQ or NONO RNAi leads to paraspeckle disintegration as a consequence of NEAT1_2 destabilization (Sasaki et al, 2009) and (2) paraspeckles are observable solely in the NEAT1_2-expressing cells of mouse tissues (Nakagawa et al, 2011). By contrast, Shevtsov and Dundr (2011) reported that tethering NEAT1_1 at the specific chromosomal site triggers on-site paraspeckle formation. Clemson et al (2009) reported that NEAT1_1 overexpression in a stable cell line increased the number of nuclear paraspeckles. We observed a similar effect with NEAT1_1 overexpression, although NEAT1_2 overexpression increased the paraspeckle numbers more effectively. Because these experiments were performed in cells possessing intact paraspeckles with endogenous NEAT1_2, it is likely that locally concentrated NEAT1_1 captured the preexisting paraspeckles or their subparticles containing NEAT1_2, which resulted in the formation of paraspeckles containing exogenous NEAT1_1.
The two isoforms of NEAT1 are differentially localized within the paraspeckle: NEAT1_2 is present in the interior core of the paraspeckle, whereas NEAT1_1 is located at the peripheral area (Souquere et al, 2010). This observation is consistent with our hypothesis that the overexpressed NEAT1_1 is efficiently incorporated into the peripheral area of paraspeckles, whose core is constructed around endogenous NEAT1_2.
The existence of category 1B proteins argues that NEAT1_2 accumulation alone is insufficient for paraspeckle formation. An additional step involving category 1B proteins is required for intact paraspeckle formation subsequent to assembly of the primary NEAT1_2 subcomplex with category 1A proteins. Category 1B proteins may be involved in the assembly of a higher-order paraspeckle structure that is built with multiple copies of the NEAT1_2 subcomplex, as well as with the NEAT1_1 subcomplex (Figure 9B). Indeed, DAZAP1 in category 1B has been shown to interact with SFPQ in category 1A (Yang et al, 2009). We cannot rule out the possibility that category 1B proteins bind to unidentified essential RNA component(s) of the paraspeckle, because all category 1B proteins possess RRMs.
The role of NEAT1_1 remains obscure, despite its higher abundance compared with NEAT1_2. The RNAi results indicate that category 3A and 1B proteins contribute to NEAT1_1 accumulation, suggesting that NEAT1_1 forms subcomplexes with these proteins. Paraspeckles presumably are involved in the nuclear retention of specific mRNAs, which raises an interesting possibility: the more conserved NEAT1_1 RNA may serve as a functional unit for paraspeckle-conducting events (such as nuclear mRNA retention), rather than for its structural maintenance. In this way, NEAT1_1 synthesis could account for the amplification of the functional units at the paraspeckle periphery. Our trials to identify PSPs that mediate the nuclear retention of mRNAs by RNAi were unsuccessful, which suggests functional redundancy within the PSPs or the presence of additional unidentified factors.
On the basis of the data presented in this manuscript, we constructed a model of paraspeckle formation (Figure 9B). To understand the details of each process, it is important to map the RNA–protein and protein–protein interactions in this structure. Further studies will identify additional PSPs and RNAs, some of which may be critical for the paraspeckle structure. Indeed, additional paraspeckle-localized proteins that are not included in our list were recently reported (Bond and Fox, 2009). It will be important to investigate the connections among chromatin structure, transcription machinery, and paraspeckle formation. The ongoing transcription of NEAT1 was recently found to be a prerequisite for paraspeckle formation (Mao et al, 2011). Therefore, the initial step of paraspeckle formation may occur cotranscriptionally (Figure 9B). Further mechanistic investigations of NEAT1 ncRNA and PSPs should elicit a novel view of the formation of these tremendously large ribonucleoprotein particles and their linkage to function.
Materials and methods
Cell cultures and transfection
HeLa, HEK293T, MEF, and NIH3T3 cells were grown in DMEM (10% FBS). Some cells were treated with actinomycin D (0.3 μg/ml, 4 h). Transfection of MEF cells was performed with the Nucleofector MEF starter kit and the Nucleofector device (Ronza) or FuGene HD (Promega). Expression from the NEAT1_1 or NEAT1_2 construct in MEF was confirmed by RT–qPCR. Transfection of other cell lines was performed with Lipofectamine 2000 or Lipofectamine LTX (Invitrogen).
In vitro 3′-end processing assay
The 32P-labelled RNA substrate was synthesized with SP6 RNA polymerase (TaKaRa). HNEs were prepared according to Dignam et al (1983). The in vitro RNA processing reaction was performed as described, with minor modifications (Takagaki et al, 1988). Briefly, 32P-labelled RNA (2 × 104 c.p.m., ∼5 fmol) was incubated in a 12.5-μl reaction mixture containing 8 mM HEPES (pH 7.9), 8% glycerol, 40 mM KCl, 0.2 mM PMSF, 0.4 mM DTT, 2.08 mM EDTA, 40 mM creatine phosphate, 40 μg/ml Escherichia coli tRNA, 0.25 U of RNasin (Promega), 2.5% polyvinyl alcohol, and 4 μl of HNE (32%). After the solution was reacted at 30°C for the indicated time, RNA was extracted and separated by 6% PAGE containing 7 M urea.
UV-crosslinking
UV-crosslinking was performed as described (Hirose et al, 2006) after incubating the samples under conditions of in vitro 3′-end processing. UV light (1.8 J/cm2) was applied to an open-top reaction tube on ice with a UV-crosslinking device (CL-1000, UVP). RNase A and RNase T1 were added and incubated for 15 min at 37°C, and precipitation with 50% acetone was performed. Precipitated proteins were fractionated by SDS–PAGE. Substrate RNA was preincubated with r-K for 15 min at 30°C, after which HNE was added for an additional 15 min incubation. The r-K was expressed in E. coli cells [BL21(DE3)-CodonPlus RILP (Stratagene)] and purified with Ni affinity chromatography.
Gel mobility shift assay
WT and K-mut RNA oligonucleotides were chemically synthesized by Hokkaido System Science Co., Ltd. The oligonucleotide sequences used were as follows: WT: 5′-AAUCACUUUUCUUCCCCUUUACAGCACAAA-3′ and K-mut: 5′-AAUCACUUUUCUUAAAAUUUACAGCACAAA-3′. 32P-labelled RNA probes were prepared with [γ-32P]ATP and T4 polynucleotide kinase (TaKaRa). 32P-labelled RNA (0.5 × 104 c.p.m., ∼150 fmol) was incubated in a 12.5-μl reaction mixture containing 8 mM HEPES (pH 7.9), 8% glycerol, 40 mM KCl, 2.08 mM EDTA, 0.2 mM PMSF, 0.4 mM DTT, 40 mM creatine phosphate, 40 μg/ml E. coli tRNA, 0.25 U of RNasin (Promega), 2.5% polyvinyl alcohol, and 0.25–2.0 μg of r-K. The mixture was incubated at 30°C for 15 min, and the RNA–protein complexes were separated on 5% native PAGE.
Immunoprecipitation
For protein immunoprecipitation, HeLa cells (1 × 107 cells) were lysed with lysis buffer (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 50 mM NaF, 1 mM Na3VO4, and 0.5% NP40) for 30 min on ice, and the supernatant was recovered by centrifugation at 10 000 g for 10 min. For each immunoprecipitation experiment, cell extract (1 mg protein) was incubated overnight at 4°C with antibody-Dynabead conjugates (25 μl) in the presence of 10 μg/ml RNase A. The beads were washed five times with lysis buffer, and bound proteins were eluted by directly adding SDS loading buffer to the beads. For RNA immunoprecipitation, the cell extract prepared as described above was incubated with antibody-Dynabead conjugates without RNase A for 3 h at 4°C. Bound RNAs were extracted by directly adding Trizol reagent (Invitrogen) to the beads.
Immunoprecipitations from the in vitro processing samples were performed as described (Ideue et al, 2007). Briefly, in vitro processing was performed at a 10-fold scale (125 μl). The reaction mixture was diluted eight-fold with NET2 buffer (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, and 0.5% NP40) immediately after RNase treatment and incubated with antibody-Dynabead conjugates for 16 h at 4°C. The beads were washed five times with NET2 buffer, and bound proteins were eluted by directly adding SDS loading buffer to the beads. Information about the antibodies used is shown in Supplementary Table S4.
Protein-binding assay with purified recombinant proteins
The recombinant CFIm complex was purified basically as described (Arias-Palomo et al, 2011). Briefly, 4 × 107 HEK293T cells were cotransfected with pEF_Flag–HA–SBP–NUDT21 and pEF_Flag–HA–CPSF6 plasmids at a 1:3 ratio using Lipofectamine LTX (Invitrogen). After 48 h, the cells were lysed by sonication in F-lysis buffer (20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 10% [w/v] sucrose, 1% Triton X-100, 0.5% NP40, 1 mM DTT, protease inhibitor cocktail [Roche], and phosphatase inhibitor cocktail [Roche]). They were incubated for 30 min on ice, and the soluble fraction was recovered by centrifugation at 15 000 g for 30 min. The soluble fraction was precleared with Sepharose 4B (Sigma), incubated with streptavidin Sepharose (GE Healthcare) for 2 h at 4°C with gentle rotation, and washed with F-lysis buffer. The affinity-purified SBP-tagged CFIm complex was eluted by incubation with F-lysis buffer containing 2 mM biotin (Sigma) for 30 min at 4°C.
The SBP-tagged CFIm complexes were immobilized to Dynabeads MyOne Streptavidin beads (Invitrogen) for 2 h at 4°C with gentle rotation and washed with NET2 buffer. Beads suspended in 30 μl of NET2 buffer were mixed with 10, 30, or 90 pmol of recombinant His-tagged HNRNPK for 15 min at RT. The bead suspension was further incubated for 2 h at 4°C, and the supernatant was collected. Beads were washed with NET2 buffer five times. Bound proteins were eluted by directly adding SDS loading buffer to the beads.
Supplementary Material
Supplementary Data
Supplementary Data for Figure 3
Supplementary Data for Figure 7
Supplementary Data for Figure 8
Review Process File
Acknowledgments
We thank Y Kisu and H Mochizuki for their help with data mining of the intracellular localization of Venus-fusion proteins, A Yamashita for kindly providing the plasmids and host cell lines for the preparation of recombinant proteins, and Y Hirose for his feedback on the in vitro RNA processing reaction. JA Steitz and K Tycowski are acknowledged for their critical reading of the manuscript. We thank M Nagai and the members of the Hirose laboratory for valuable discussions. This research was supported by the Funding Program for Next Generation World-Leading Researchers (NEXT Program) of the Japan Society for the Promotion of Science (JSPS), and by grants from the New Energy and Industrial Technology Development Organization (NEDO), Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), the Astellas Foundation for Research on Metabolic Disorders, and Takeda Science Foundation.
Author contributions: The study was conceived and supervised by TN and TH; TN performed most of the experiments. SN performed the experiment in Figure 1A. AT and YFS contributed with biochemical and microscope analyses. TN and TH wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arias-Palomo E, Yamashita A, Fernández IS, Núñez-Ramírez R, Bamba Y, Izumi N, Ohno S, Llorca O (2011) The nonsense-mediated mRNA decay SMG-1 kinase is regulated by large-scale conformational changes controlled by SMG-8. Genes Dev 25: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O’Malley BW (2005) A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol Cell Biol 25: 5307–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsztyk K, Denisenko O, Ostrowski J (2004) hnRNP K: one protein multiple processes. Bioessays 26: 629–638 [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH (2009) Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol 186: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RS (2005) Zinc finger proteins: getting a grip on RNA. Curr Opin Struct Biol 15: 94–98 [DOI] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA-binding proteins. Science 265: 615–621 [DOI] [PubMed] [Google Scholar]
- Cardinale S, Cisterna B, Bonetti P, Aringhieri C, Biggiogera M, Barabino SM (2007) Subnuclear localization and dynamics of the Pre-mRNA 3′ end processing factor mammalian cleavage factor I 68-kDa subunit. Mol Biol Cell 18: 1282–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest AR, Zavolan M, Davis MJ, Wilming LG, Aidinis V et al. (2005) The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG (2008) Alu element-mediated gene silencing. EMBO J 27: 1694–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W, Hu G (2008) Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS Lett 582: 1761–1765 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM (2004) Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem 279: 35788–35797 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI (2002) Paraspeckles: a novel nuclear domain. Curr Biol 12: 13–25 [DOI] [PubMed] [Google Scholar]
- Guru SC, Agarwal SK, Manickam P, Olufemi SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y, Emmert-Buck MR, Liotta LA, Spiegel AM, Boguski MS, Roe BA, Collins FS, Marx SJ, Burns L, Chandrasekharappa SC (1997) A transcript map for the 2.8-Mb region containing the multiple endocrine neoplasia type 1 locus. Genome Res 7: 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Ideue T, Nagai M, Hagiwara M, Shu MD, Steitz JA (2006) A spliceosomal intron binding protein, IBP160, links position-dependent assembly of intron-encoded box C/D snoRNP to pre-mRNA splicing. Mol Cell 23: 673–684 [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, Attardi LD, Regev A, Lander ES, Jacks T, Rinn JL (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Sasaki YT, Hagiwara M, Hirose T (2007) Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev 21: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V et al. (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kim N, Kim P, Nam S, Shin S, Lee S (2006) ChimerDB—a knowledgebase for fusion sequences. Nucleic Acids Res 34: D21–D24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Yamamoto J, Chen Y, Aida M, Wada T, Handa H, Yamaguchi Y (2010) Evidence that cleavage factor Im is a heterotetrameric protein complex controlling alternative polyadenylation. Genes Cells 15: 1003–1013 [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Cleveland DW (2009) Rethinking ALS: the FUS about TDP-43. Cell 136: 1001–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG (2006) TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic 5: 8–14 [DOI] [PubMed] [Google Scholar]
- Lutz CS (2008) Alternative polyadenylation: a twist on mRNA 3′-end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 13: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y, Kawamura Y, Nishikawa T, Isogai T, Nomura N, Goshima N (2012) HGPD: Human Gene and Protein Database, 2012 update. Nucleic Acids Res 40: D924–D929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S (2010) Molecular mechanisms of eukaryotic pre-mRNA 3′-end processing regulation. Nucleic Acids Res 38: 2757–2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL (2005) Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the 'genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci USA 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D (2005) Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 16: 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M (2011) Nucleation of nuclear bodies by RNA. Nat Cell Biol 13: 167–173 [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S (2007) The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120: 2498–2506 [DOI] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, Fox A, Pierron G (2010) Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 21: 4020–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL (2006) SnapShot: cellular bodies. Cell 127: 1071. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL (2009) MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Ryner LC, Manley JL (1988) Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell 52: 731–742 [DOI] [PubMed] [Google Scholar]
- Thisted T, Lyakhov DL, Liebhaber SA (2001) Optimized RNA targets of two closely related triple KH domain proteins, heterogeneous nuclear ribonucleoprotein K and alphaCP-2KL, suggest Distinct modes of RNA recognition. J Biol Chem 276: 17484–17496 [DOI] [PubMed] [Google Scholar]
- Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, König J, Hortobágyi T, Nishimura AL, Zupunski V, Patani R, Chandran S, Rot G, Zupan B, Shaw CE, Ule J (2011) Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 14: 452–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, Blencowe BJ, Prasanth SG, Prasanth KV (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HT, Peggie M, Cohen P, Rousseau S (2009) DAZAP1 interacts via its RNA-recognition motifs with the C-termini of other RNA-binding proteins. Biochem Biophys Res Commun 380: 705–709 [DOI] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG (2011) ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147: 773–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Gilmartin GM, Doublié S (2010) Structural basis of UGUA recognition by the Nudix protein CFI(m)25 and implications for a regulatory role in mRNA 3′ processing. Proc Natl Acad Sci USA 107: 10062–10067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Shen Z, Tripathi V, Xuan Z, Freier SM, Bennett CF, Prasanth SG, Prasanth KV (2010) Polypurine-repeat-containing RNAs: a novel class of long non-coding RNA in mammalian cells. J Cell Sci 123: 3734–3744 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data
Supplementary Data for Figure 3
Supplementary Data for Figure 7
Supplementary Data for Figure 8
Review Process File