ELAV mediates 3′ UTR extension in the Drosophila nervous system (original) (raw)
In previous work, the authors identified 30 or so genes that showed coordinate lengthening of their 3′ UTRs during embryogenesis. Here Hilgers et al. investigate the mechanistic basis for the regulation of extended 3′ UTRs in neural tissues. They find that the RNA-binding protein ELAV (embryonic-lethal abnormal visual system) is responsible for this 3′ lengthening and regulates the alternative cleavage and polyadenylation of many mRNAs in the developing nervous system.
Keywords: ELAV, alternative polyadenylation, 3′ UTR, nervous system, extension, post-transcriptional regulation
Abstract
Post-transcriptional gene regulation is prevalent in the nervous system, where multiple tiers of regulatory complexity contribute to the development and function of highly specialized cell types. Whole-genome studies in Drosophila have identified several hundred genes containing long 3′ extensions in neural tissues. We show that ELAV (embryonic-lethal abnormal visual system) is a key mediator of these neural-specific extensions. Misexpression of ELAV results in the ectopic synthesis of long messenger RNAs (mRNAs) in transgenic embryos. RNA immunoprecipitation assays suggest that ELAV directly binds the proximal polyadenylation signals of many target mRNAs. Finally, ELAV is sufficient to suppress 3′ end formation at a strong polyadenylation signal when tethered to a synthetic RNA. We propose that this mechanism for coordinating 3′ UTR extension may be generally used in a variety of cellular processes.
Regulatory elements located within 3′ untranslated regions (UTRs) provide an assembly platform for RNA-binding proteins and microRNAs, which control stability, subcellular localization, and translatability of messenger RNAs (mRNAs) (Martin and Ephrussi 2009; Fabian et al. 2010). Alternative 3′ end formation—i.e., usage of different polyadenylation [poly(A)] sites—results in the synthesis of multiple mRNA isoforms, differing only in the lengths of their 3′ UTRs. Alternative cleavage and poly(A) (APA) is a major mechanism for the correct spatial and temporal control of gene expression during development (Di Giammartino et al. 2011; Proudfoot 2011). Approximately half of all expressed genes are thought to be subject to APA in humans, mice (Tian et al. 2005), and Drosophila (Smibert et al. 2012), often in a tissue-specific manner (Zhang et al. 2005; Wang et al. 2008).
APA-mediated 3′ UTR shortening has been linked to cell proliferation, oncogenic transformation, pluripotency, lymphocyte activation, and neuronal activation (Takagaki et al. 1996; Flavell et al. 2008; Sandberg et al. 2008; Ji and Tian 2009; Mayr and Bartel 2009). Proximal poly(A) sites are thought to enhance gene expression by minimizing destabilizing elements (e.g., microRNA recognition sites) that are likely to occur in extended 3′ UTRs. Conversely, longer 3′ UTRs have been observed in later developmental stages and differentiated tissues (e.g., Ji et al. 2009; Hilgers et al. 2011). Recent studies have shown that hundreds of genes undergo APA-mediated 3′ UTR extension during the course of Drosophila development. Many of these genes exhibit staggered 3′ extensions, with intermediate mRNA isoforms appearing during early stages of neurogenesis, and longer isoforms appearing at later stages. Strikingly, expression of the extended transcripts is predominantly restricted to the nervous system (Hilgers et al. 2011; Smibert et al. 2012).
The function of the 3′ UTR extensions is currently unknown. Extended transcripts are enriched in conserved regulatory signals, including binding sites for microRNAs and RNA-binding proteins such as Pumilio. Long mRNA isoforms possess a larger post-transcriptional potential than their short counterparts, which might reflect their role in neural-specific processes. Many of the genes that are subject to neural-specific APA are required for nervous system function. It has been proposed that 3′ extensions mediate a specific function within the neuron, such as mRNA transport along axons (Hilgers et al. 2011).
In this study, we investigate the basis for the coordinate appearance of extended 3′ UTRs in neural tissues. The pan-neuronal RNA-binding protein ELAV (embryonic-lethal abnormal visual system) regulates mRNA processing in a neural-specific manner. ELAV is required for alternative splicing of armadillo (arm), neuroglian (nrg), and erect wings (ewg), resulting in neural-specific isoforms (Lisbin et al. 2001; Soller and White 2003). ELAV also binds to its own mRNA to regulate ELAV levels (Samson 1998; Borgeson and Samson 2005). Here, we show that ELAV regulates APA of multiple mRNAs in the developing nervous system. We found that extended mRNA isoforms are specifically expressed in cells where ELAV protein is present. There is a dramatic reduction of these isoforms in elav mutants. Furthermore, ectopic expression of ELAV in nonneural embryonic tissues induces ectopic 3′ extension. RNA immunoprecipitation (RIP) assays identify ELAV binding in the vicinity of the proximal poly(A) signal of extended mRNAs. Finally, tethering ELAV to an exogenous mRNA triggers transcriptional readthrough several kilobases beyond a strong poly(A) signal. These data provide the first evidence that ELAV coordinates the expression of 3′ UTR extensions during neural development. We propose that similar mechanisms are used in other metazoan processes.
Results and Discussion
We showed previously that long 3′ UTR extensions are first synthesized 4–6 h after egg laying (AEL) and specifically localize to the nervous system. Computational analyses revealed that these extensions are enriched in conserved U-rich sequences; i.e., putative ELAV-binding sites (EBSs) (Hilgers et al. 2011; Smibert et al. 2012). The panneuronal protein ELAV is a known regulator of nuclear pre-mRNA processing mediated by degenerate, low-complexity sequences (Soller and White 2004). We therefore sought to determine whether ELAV might be involved in the regulation of APA-mediated 3′ extensions.
Coexpression of ELAV and 3′ extensions
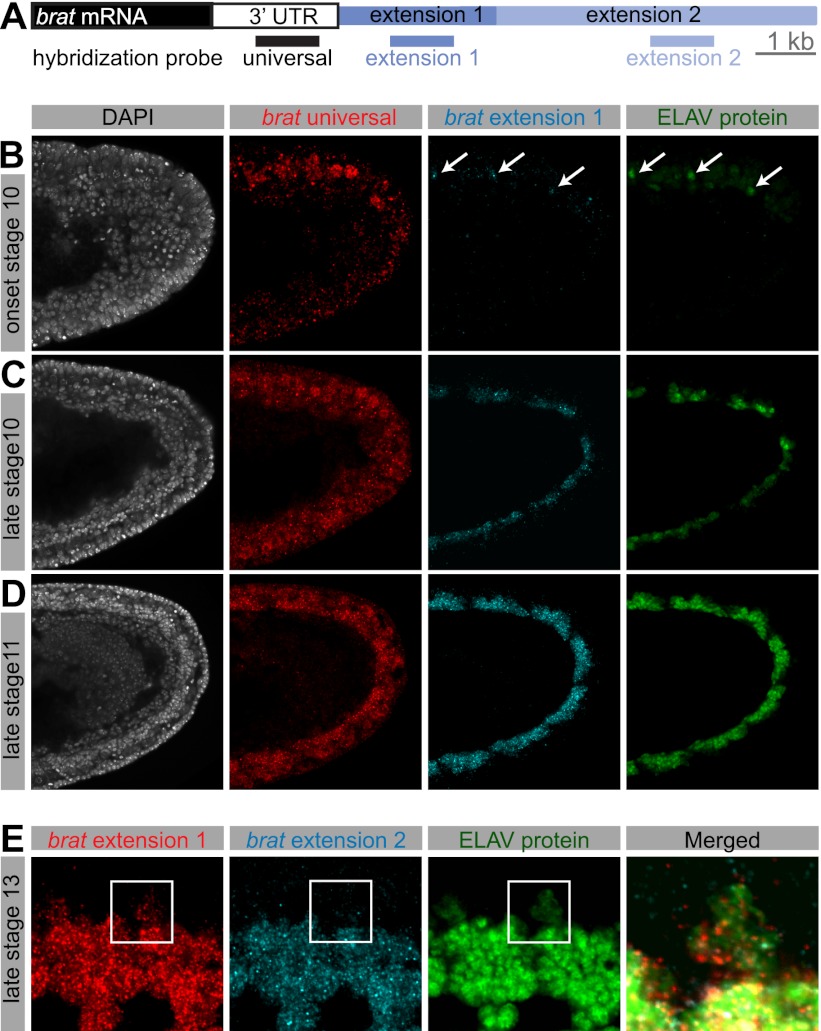
We performed RNA in situ hybridization coupled to antibody staining to visualize the distribution of ELAV protein and extended 3′ UTRs. Particular efforts focused on brain tumor (brat), which is subject to two distinct APA processes, producing three brat isoforms: a short 4.5-kb transcript bearing a 1.3-kb 3′ UTR (universal), an intermediate 6.4-kb transcript that is produced after 4 h AEL (extension 1), and finally, a fully extended 11.7-kb transcript that appears several hours later in development (extension 2) (Fig. 1A; Hilgers et al. 2011).
Figure 1.
Coexpression of 3′ extensions and ELAV. (A) Schematic representation of the 3′ end of the gene brat. In situ hybridization probes were designed to detect all brat RNA isoforms (universal), both extended isoforms (extension 1), or exclusively the longest brat isoform (extension 2). Bar, 1 kb. (B–E) Double fluorescent in situ hybridization for brat mRNA using probes indicated in A, combined with antibody staining against ELAV protein. (B–D) Single confocal sections of the posterior end of stage 10 (B,C) and stage 11 (D) embryos. Nuclei were stained with DAPI. Lateral view; anterior is to the left, and dorsal is to the top. While the universal probe detects brat mRNA throughout the presumptive nerve cord, brat extensions first appear in cells at the onset of ELAV protein expression (B, arrows), and expression of extensions continues to coincide with ELAV expression as it expands in the ventral nerve cord (C,D). (E) Single confocal section of the developing CNS in a stage 13 embryo. Ventral view; anterior is to the left. In cells in which ELAV protein is weakly expressed (magnified cutout), extension 1 expression is reduced in comparison with neighboring cells showing high ELAV expression, and extension 2 is altogether absent. Note that Elav protein is mostly restricted to the nucleus, whereas the detected RNAs are also present in the cytoplasm.
ELAV protein is present in newborn neurons at the onset of embryonic stage 10 (Fig. 1B; Robinow and White 1991). A hybridization probe detecting all brat isoforms (universal) (Fig. 1A) showed expression throughout the ventral nerve cord primordium in stages 10 and 11 (Fig. 1B–D). A probe specific to the intermediate brat 3′ isoform (extension 1) detected nascent transcripts only in those few neurons containing ELAV at the onset of stage 10 (Fig. 1B, arrows). At later stages, ELAV protein localized to clusters of neurons as they were born. There was a coincident appearance of brat extensions in the same pattern (Fig. 1C,D). At embryonic stage 13, short brat mRNA isoforms are found throughout the embryonic brain and ventral nerve cord. As seen at earlier stages, brat extensions were detected only in ELAV-expressing neurons (Supplemental Fig. S1A).
ELAV exhibits weak and transient expression in embryonic neuroblasts and glial cells (Berger et al. 2007). brat extension 1 was detected (at low levels) in these cells, but not the full-length mRNA containing extension 2. Rather, the longest form of brat was restricted to neurons that express high levels of ELAV (Fig. 1E). These observations suggest a dosage relationship between the levels of ELAV and the extent of APA-mediated 3′ UTR extension. Similar results were observed for additional APA-regulated genes (hrb27C, musashi [_msi_], scratch, and elav) (Supplemental Fig. S1B–E), suggesting a broad correlation between ELAV and 3′ extensions.
ELAV mutants lack 3′ extensions
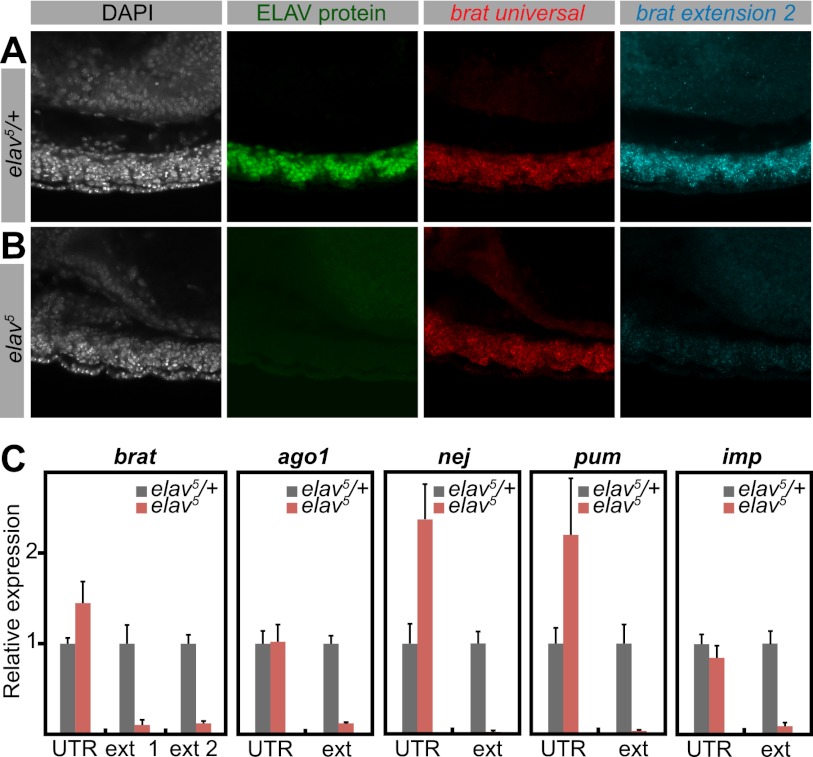
To investigate the causal link between ELAV and 3′ extensions, we examined brat expression in elav mutants. We used the null allele elav5 (Yao et al. 1993) as well as a deficiency encompassing elav [Df(elav)]. ELAV was absent in both mutants. Short (universal) brat isoforms were present and not detectably affected. However, extended brat isoforms were barely detectable in elav mutants (Fig. 2B) in comparison with their heterozygous siblings (Fig. 2A; Supplemental Fig. S2A). We obtained similar results for hrb27C and scratch (Supplemental Fig. S2B,C).
Figure 2.
Depletion of 3′ extensions in elav mutants. (A,B) Double fluorescent in situ hybridization for short (universal) and long (extension 2) brat mRNA isoforms, combined with antibody staining against ELAV protein. Nuclei were stained with DAPI. Shown are projections of consecutive confocal sections of the ventral nerve cord of stage 13 embryos. Lateral view; anterior is to the left and dorsal is to the top. (A) In heterozygous control embryos, ELAV protein is expressed in neurons throughout the ventral nerve cord. (B) In a hemizygous elav mutant, ELAV protein is absent. Although the bulk of brat mRNA (universal) is not detectably affected in the mutant, the longest brat isoform (extension 2) is depleted in elav5 compared with the control. (C) Quantification of indicated transcripts by qPCR using primer combinations detecting all isoforms (UTR) or specific to the long isoforms (ext). RNA was extracted from heterozygous control (elav5/+) or elav mutant (elav5) embryos. Levels were normalized to rp49 RNA. For each primer pair, expression in control embryos was set to the value 1. In elav mutants, the overall expression of the genes brat, ago1, nej, pum, and imp is higher or not affected compared with control embryos. However, longer isoforms are strongly depleted in the mutants. Error bars represent mean ± SD of six embryos for each genotype.
We next performed RT-qPCR on single 10- to 12-h elav5 mutant embryos. elav mRNA levels were measured in each embryo to identify null mutants and heterozygous control embryos. Total mRNA levels of brat, argonaute1 (ago1), nejire (nej), pumilio (pum), and IGF-II mRNA-binding protein (imp) were unchanged or elevated in mutant embryos. In contrast, extended isoforms were dramatically reduced for each of these genes in elav mutants (Fig. 2C; Supplemental Fig. S2D). We therefore conclude that ELAV is necessary for the synthesis of many of the previously identified 3′ UTR extensions.
Misexpression of ELAV results in ectopic 3′ extensions
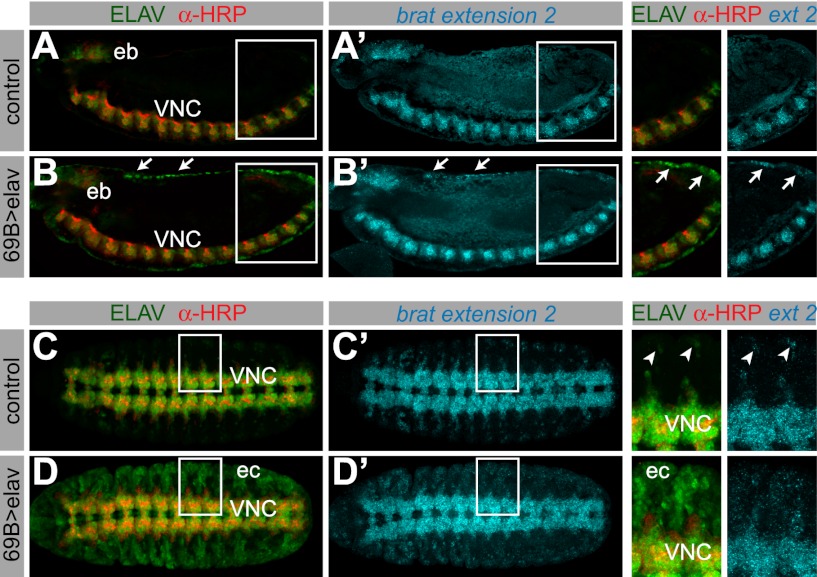
We examined the consequences of misexpressing ELAV on the APA of target transcripts. We used the pan-ectodermal 69B-Gal4 driver in combination with either UAS-elav or UAS-GFP. Neurons were distinguished from nonneuronal cells that ectopically express ELAV using the anti-horseradish peroxidase (anti-HRP) antibody, which specifically stains neural membranes (Fig. 3A–D; Snow et al. 1987).
Figure 3.
Misexpression of ELAV results in ectopic 3′ extensions. In situ hybridization against the longest brat isoform, combined with antibody staining in embryos expressing UAS-GFP (A,_A_′,C,_C_′) or UAS-elav (B,_B_′,D,_D_′) under control of the 69B-Gal4 driver. The top and bottom panels show a lateral and a ventral view of stage 13 embryos, respectively. (B) Nonneural cells ectopically expressing ELAV can be distinguished by the absence of anti-HRP signal (e.g., arrows). (A,_A_′) brat extension 2 is only detected in the ventral nerve cord (VNC) and the embryonic brain (eb) in control embryos. (B,_B_′) In embryos ectopically expressing ELAV, ectopic brat extensions can be detected in the ectoderm (arrows, and arrows in the magnified cutout). (C,_C_′) brat extension 2 is detected in the ventral nerve cord and peripheral neurons in control embryos (arrowheads in the magnified cutout). In embryos where ELAV is expressed throughout the ectoderm (D; ec), brat extension is ectopically expressed in cells that contain high levels of ELAV protein (_D_′ and magnified cutout).
Ectopic expression of ELAV in nonneural ectodermal cells resulted in the appearance of brat mRNAs containing extended 3′ UTRs. Cells containing high levels of ELAV exhibited the longest brat isoform (extension 2) (Fig. 3B′,D′, arrows and magnified cutouts), whereas only the intermediate isoform (extension 1) was observed in cells with low levels of ELAV (Supplemental Fig. S3A). Control embryos that express GFP did not display such changes in brat expression (Fig. 3A′,C′; Supplemental Fig. S3B). Our results suggest that ELAV functions in a dose-dependent fashion to regulate brat APA. This is consistent with the differential expression of the brat extensions 1 and 2 in normal neural tissues containing either low or high levels of ELAV (see Fig. 1).
ELAV binds in the vicinity of proximal poly(A) signals
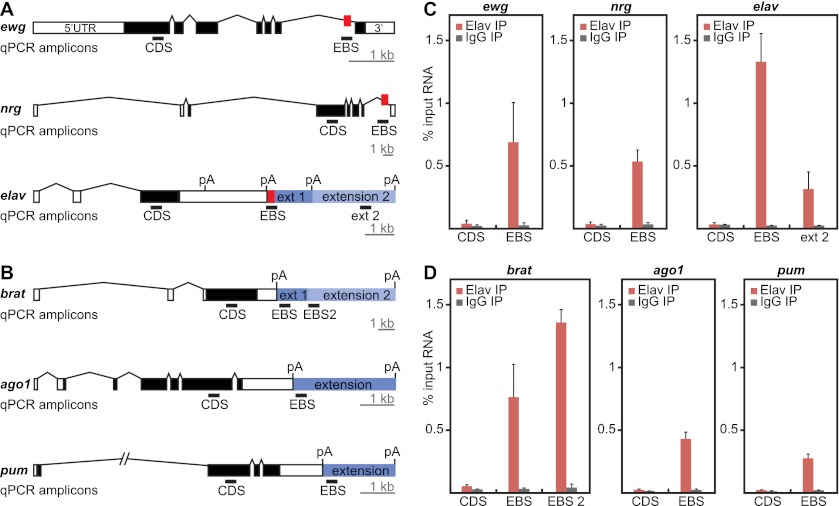
We next ascertained whether ELAV physically associates with transcripts containing 3′ extensions using RIP assays. Nuclear extracts of 8- to 12-h AEL embryos were sonicated in order to obtain RNA fragments measuring 100–300 nucleotides (Supplemental Fig. S4B). ELAV-containing riboprotein complexes were purified using monoclonal anti-ELAV antibodies. As a positive control, we determined whether ELAV binds to known target genes: ewg, nrg, and elav (Lisbin et al. 2001; Soller and White 2003; Borgeson and Samson 2005). qPCR was used to amplify the regions flanking the EBS and coding sequences (CDS) in these genes (Fig. 4A). We observed that EBS-containing regions, but not control CDS regions, were highly enriched for ELAV (Fig. 4C). These results indicate that the RIP assay selectively identifies EBSs in vivo.
Figure 4.
ELAV RIP assays identify 3′ extended mRNAs. (A) Gene models of known ELAV targets. The locations of EBSs are indicated as red boxes. qPCR primers were designed to detect a region in the CDS as a negative control, a region encompassing the EBS, or a region located more distally within the extension (ext 2). (B) Gene models of three candidates for ELAV binding at the 3′ extension. qPCR primers were designed to detect a region in the CDS, a region immediately downstream from the short UTR (EBS), or the first portion of a more distal extension (EBS2). Bar, 1 kb. qPCR amplicons (black bars) are 60–80 base pairs (bp) in length and are not represented to scale. (C,D) ELAV binds specifically to 3′ UTR extension sequences in the vicinity of poly(A) signals. RIP analysis was performed on 8- to 12-h embryos using either anti-ELAV antibodies or control IgGs. RNA from ELAV immunoprecipitates was assayed by qPCR for the regions indicated in A and B and compared with RNA from total sonicated nuclei (input). Error bars represent mean ± SD of four separate immunoprecipitation reactions on independently prepared nuclei.
To determine whether ELAV is directly involved in the synthesis of 3′ extensions, we designed qPCR primers that amplify a region of the mRNA located immediately downstream from proximal poly(A) signals (Fig. 4B, EBS). We found that ELAV associates with the extended mRNA isoforms of brat, ago1, pum, msi, hephaestus (heph), and fasciclin 1 (fas1) near the proximal poly(A) signal. We found little or no association within control regions, including coding, intronic, or short 3′ UTR sequences, or at the distal-most poly(A) signals (Fig. 4D; Supplemental Fig. S4C–E). ELAV was also detected within the 3′ UTR extensions (Fig. 4C; Supplemental Fig. S4C), including significant binding immediately 3′ of the intermediate brat isoform (EBS2) (Fig. 4B,D). This suggests that ELAV controls phased elongation of the brat 3′ UTR in a dose-dependent manner, consistent with the in vivo correlation between the distribution of ELAV and the appearance of the intermediate and long brat isoforms (see Figs. 1, 3).
Based on RIP assays, seven of nine putative target genes exhibited significant ELAV binding at proximal poly(A) signals (Fig. 4; Supplemental Fig. S4). The absence of binding at the remaining genes might be due to technical issues or reflect a regulatory cascade whereby ELAV controls one or more intermediate genes. We favor the former explanation, since previous studies suggest that neural-specific genes exhibit highly synchronous 3′ extension during development (Hilgers et al. 2011).
Tethering ELAV to a synthetic RNA causes transcriptional readthrough
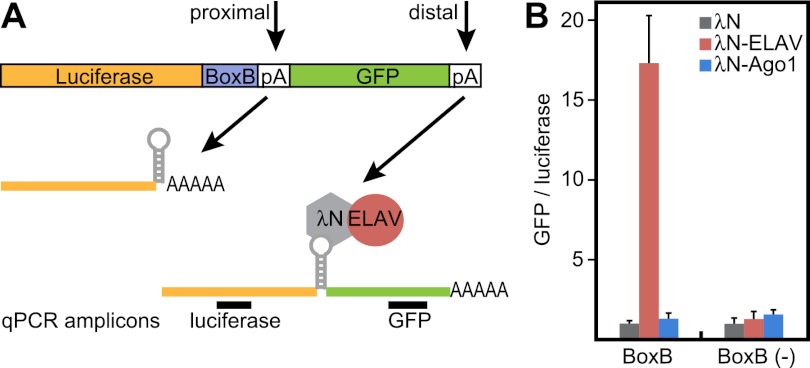
To test whether physical association of ELAV in the vicinity of the proximal poly(A) site is sufficient to trigger 3′ extension, we used a tethering assay in Drosophila S2 cells (Pillai et al. 2004). A reporter mRNA was created that contains the firefly luciferase CDS, followed by the SV40 late poly(A) signal and a GFP CDS. This construct carries five hairpins (BoxB sites) that are bound with high affinity by the N-peptide of bacteriophage λ (λN). ELAV was coexpressed as a fusion protein with the λN peptide to tether ELAV to the luciferase transcript (Fig. 5A).
Figure 5.
ELAV is sufficient to promote 3′ extension in a heterologous system. (A) Schematic of the tethering experiment. A luciferase ORF was placed upstream of BoxB sequences (BoxB) and the SV40 late poly(A) signal. Downstream, a GFP ORF represents an artificial “3′ extension.” The λN peptide, expressed as a λN-ELAV fusion protein, binds to BoxB sequences in the luciferase transcript, thereby tethering ELAV to the transcript in proximity to the poly(A) signal. GFP sequences will only be expressed in the event of polymerase II readthrough beyond the proximal poly(A) site. Primers were designed to amplify target sequences in the luciferase and GFP ORFs. (B) Extent of poly(A) readthrough as measured by levels of GFP RNA. Luciferase and GFP RNA levels were quantified by qPCR using primer combinations shown in A. Tethering λN-ELAV, but not λN-Ago1, causes transcription of downstream GFP sequences. Levels are shown as GFP/luciferase ratios to normalize for transcript stability. Ratios in cells expressing λN alone were set to the value 1. Error bars represent mean ± SD of four independent transfection experiments.
Tethering λN-ELAV, but not λN alone, to the luciferase reporter caused transcription of GFP sequences 3′ of the poly(A) signal. A control mRNA devoid of the BoxB-tethering sites [BoxB (−)] was not significantly extended (Fig. 5B). Furthermore, tethering the RNA-binding protein Ago1 to the reporter had no effect on GFP expression, while Sex lethal (Sxl) promoted only modest extension beyond the poly(A) site (Fig. 5B; Supplemental Fig. S5). These experiments show that ELAV binding is sufficient to bypass a strong poly(A) signal, resulting in the synthesis of a 3′ extension.
Here we presented evidence that the RNA-binding protein ELAV regulates 3′ UTR extension during neuronal differentiation. Past studies have documented a role for ELAV in the regulation of splicing; the present study shows that it is also crucial for the synthesis of a battery of extended 3′ UTRs. A variety of evidence suggests that ELAV functions by suppressing proximal poly(A), thereby permitting readthrough of elongating polymerase II complexes. RIP assays show that ELAV binds at proximal poly(A) signals, and tethering assays indicate that this binding is sufficient to trigger 3′ extension of an exogenous gene in a heterologous system. We propose that ELAV functions in a dose-dependent and promiscuous manner to foster successive extensions in a number of genes. In principle, ELAV can induce 3′ extension in a variety of cell types, but its activity is tightly restricted in vivo (1) spatially, as it is present exclusively in neural tissues; (2) temporally, as expression starts at the onset of stage 10; and (3) in its levels—high in neurons and low in other neural cell types.
Previous studies suggest that ELAV binds its own RNA, and this regulation uncouples ELAV protein levels from elav gene dosage (Samson 1998). It is interesting to note that ELAV protein expression is tightly regulated in that the extent of 3′ extension depends on ELAV levels (Figs. 1, 3). We propose that the temporal appearance of ELAV protein in neural tissues is responsible for the coordinate regulation of hundreds of genes containing 3′ UTR extensions. It might be an important feature of a healthy neuron to maintain a balance between short and long mRNA isoforms. elav mutants are embryonic-lethal, and although neurons are generated, they do not differentiate normally and display numerous axonal defects (Campos et al. 1985). Such mutants contain the short forms of target mRNAs but not the long forms. ELAV is also required later, for neuronal maintenance: Temperature-sensitive mutants display neuronal degeneration when shifted to nonpermissive temperatures (Homyk et al. 1985). Low levels of ELAV may produce insufficient amounts of 3′ extended transcripts or transcripts that have not been fully elongated. This likely contributes to elav mutant phenotypes.
APA is a widespread mechanism for the control of gene expression, and unbalanced APA has been associated with human disease (Jenal et al. 2012). 3′ Extensions have been shown to be associated with neural tissues in Drosophila (Hilgers et al. 2011), humans (Zhang et al. 2005), and zebrafish (Ulitsky et al. 2012). Several factors are involved in neuron-specific APA in mammals. In mice, the splicing factor Nova2 controls APA by both repressing and enhancing poly(A) site use (Licatalosi et al. 2008). Another regulator of APA is the polypyrimidine tract-binding protein (PTB) (Castelo-Branco et al. 2004). Indeed, the Drosophila homologs of Nova2 (pasilla) and PTB (heph) also exhibit APA regulation, just like elav. In humans, three ELAV homologs, HuB, HuC, and HuD, are selectively expressed in neurons, while HuR is ubiquitous. Interestingly, in neurons, all four Hu proteins bind to HuR mRNA to promote alternative poly(A) and generate an extended HuR 3′ UTR (Mansfield and Keene 2012). It is conceivable that the mammalian ELAV homologs also promote coordinate 3′ extension of neural-specific mRNAs, as seen in Drosophila.
Materials and methods
Plasmids and fly strains
The elav deficiency Df(1)ED6396, UAS-GFP, and 69B-Gal4 strains were obtained from the Bloomington Stock Center. The elav5 RNA-null mutant and the transgenic line UAS-elav2e2; UAS-elav3e1 were provided by Matthias Soller. Firefly luciferase reporter plasmids were constructed by inserting the GFP CDS followed by the late SV40 poly(A) signal into the XhoI–BamHI sites of pAc5.1C-FLuc-Stop-5BoxB (Rehwinkel et al. 2005). The control plasmid lacking BoxB sites was made in the same way, using the sites EcoRI–BamHI. To express λN-ELAV, λN-Ago1, and λN-Sxl fusion proteins, CDSs were cloned in-frame into the EcoRI–XhoI sites of the pAc5.1B-λN-HA plasmid. Expression of renilla luciferase was obtained from the pAc5.1C-RLuc-V5His6 construct (Rehwinkel et al. 2005).
Cell transfection
S2 cells were transfected in six-well plates using the Effectene reagent (Qiagen) with 300 ng of λN expression plasmid, 350 ng of firefly luciferase reporter plasmid, and 350 ng of renilla luciferase DNA as a transfection control. Transfections were performed with duplicate technical replicates in four independent experiments. At 60 h post-transfection, cells were pelleted and dissolved in TRIzol reagent (Invitrogen) for RNA extraction.
In situ hybridization and immunocytochemistry
Embryos were collected and fixed following standard procedures. All templates for synthesis of RNA probes were obtained from PCR-amplified genomic fragments cloned into pGEM-T Easy (Promega) and confirmed by sequencing. For each template, antisense RNA probes were transcribed with T7 or SP6 RNA polymerase and digoxigenin-UTP or biotin-UTP (Roche). Embryos were hybridized with riboprobes according to standard protocols. Detection of RNA probes was carried out with anti-digoxigenin and anti-biotin primary antibodies and fluorescent secondary antibodies (Invitrogen). Rat anti-ELAV was obtained from the Developmental Studies Hybridoma Bank (DSHB), and goat anti-HRP was a gift from Nipam Patel. Confocal imaging was performed on a Zeiss LSM 700 microscope.
RIP
Immunoprecipitation on nuclear extracts from 8- to 12-h embryos was performed essentially following the chromatin immunoprecipitation protocol described in Oktaba et al. (2008), with the following modifications. Cross-linking was performed in 1.4% formaldehyde, and nuclei were sonicated for 10 cycles of 30-sec on/30-sec off, kept on ice for 30 min, and sonicated for five additional cycles using a Covaris S220 Ultrasonicator. Nuclear extracts were incubated overnight with 2 μg of either a mixture of rat and mouse anti-ELAV antibodies (DSHB) or a mixture of rat and mouse control IgG. Samples were DNase-treated (Promega) instead of RNase-treated and were not subjected to SDS/proteinase K treatment. RNase inhibitors (RNaseOUT, Invitrogen) were used in all steps of the protocol at a concentration of 40 U/mL. Reversal of cross-links was performed for 1 h at 68°C.
RNA extraction, RT, and qPCR
Total RNA was extracted from individual staged embryos, pelleted cells, or immunoprecipitates using TRIzol (Invitrogen). RNA was treated with RNase-free DNase (Ambion) to eliminate DNA contamination. First strand synthesis used random hexamer primers and SuperScript III reverse transcriptase (Invitrogen). Samples were RNaseH-treated after the RT reaction. For qPCR, samples were used in a 1:100 dilution. qPCR was performed and monitored in a Viia7 real-time PCR system using SYBR Green reagents (Applied Biosystems). Primer sequences are available on request.
Acknowledgments
We thank David Jun for technical assistance. We are grateful to Don Rio and Emma Farley for useful suggestions. V.H. is supported by a fellowship from the International Human Frontier Science Program Organization. S.B.L. is supported by the German National Merit Foundation. This study was funded by a grant from the NIH (GM34431).
Footnotes
Supplemental material is available for this article.
References
- Berger C, Renner S, Lüer K, Technau GM 2007. The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev Dyn 236: 3562–3568 [DOI] [PubMed] [Google Scholar]
- Borgeson CD, Samson M-L 2005. Shared RNA-binding sites for interacting members of the Drosophila ELAV family of neuronal proteins. Nucleic Acids Res 33: 6372–6383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos AR, Grossman D, White K 1985. Mutant alleles at the locus elav in Drosophila melanogaster lead to nervous system defects. A developmental-genetic analysis. J Neurogenet 2: 197–218 [DOI] [PubMed] [Google Scholar]
- Castelo-Branco P, Furger A, Wollerton M, Smith C, Moreira A, Proudfoot N 2004. Polypyrimidine tract binding protein modulates efficiency of polyadenylation. Mol Cell Biol 24: 4174–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W 2010. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379 [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim T-K, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Perry MW, Hendrix D, Stark A, Levine M, Haley B 2011. Neural-specific elongation of 3′ UTRs during Drosophila development. Proc Natl Acad Sci 108: 15864–15869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homyk T, Isono K, Pak WL 1985. Developmental and physiological analysis of a conditional mutation affecting photoreceptor and optic lobe development in Drosophila melanogaster. J Neurogenet 2: 309–324 [DOI] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, van Haaften G, Kühn U, Menzies FM, Oude Vrielink JA, Bos AJ, Drost J, Rooijers K, et al. 2012. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149: 1–16 [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419 doi: 10.1371/journal.pone.0008419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B 2009. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci 106: 7028–7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisbin MJ, Qiu J, White K 2001. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev 15: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield KD, Keene JD 2012. Neuron-specific ELAV/Hu proteins suppress HuR mRNA during neuronal differentiation by alternative polyadenylation. Nucleic Acids Res 40: 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oktaba K, Gutiérrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EEM, Müller J 2008. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell 15: 877–889 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ 2011. Ending the message: Poly(A) signals then and now. Genes Dev 25: 1770–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11: 1640–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K 1991. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J Neurobiol 22: 443–461 [DOI] [PubMed] [Google Scholar]
- Samson M 1998. Evidence for 3′ untranslated region-dependent autoregulation of the Drosophila gene encoding the neuronal nuclear RNA-binding protein ELAV. Genetics 150: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow PM, Patel NH, Harrelson AL, Goodman CS 1987. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. J Neurosci 7: 4137–4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K 2003. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev 17: 2526–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller M, White K 2004. ELAV. Curr Biol 14: R53 doi: 10.1016/j.cub.2003.12.041 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan C, Subtelny AO, Koppstein D, Bell G, Sive H, Bartel D 2012. Extensive alternative polyadenylation during zebrafish development. Genome Res doi: 10.1101/gr.139733.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao KM, Samson ML, Reeves R, White K 1993. Gene elav of Drosophila melanogaster: A prototype for neuronal-specific RNA binding protein gene family that is conserved in flies and humans. J Neurobiol 24: 723–739 [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100 doi: 10.1186/gb-2005-6-12-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]