The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna (original) (raw)
Abstract
Molecular tools have revolutionized the exploration of biodiversity, especially in organisms for which traditional taxonomy is difficult, such as for microscopic animals (meiofauna). Environmental (eDNA) metabarcode surveys of DNA extracted from sediment samples are increasingly popular for surveying biodiversity. Most eDNA surveys use the nuclear gene-encoding small-subunit rDNA gene (18S) as a marker; however, different markers and metrics used for delimiting species have not yet been evaluated against each other or against morphologically defined species (morphospecies). We assessed more than 12,000 meiofaunal sequences of 18S and of the main alternatively used marker [Cytochrome c oxidase subunit I (COI) mtDNA] belonging to 55 datasets covering three taxonomic ranks. Our results show that 18S reduced diversity estimates by a factor of 0.4 relative to morphospecies, whereas COI increased diversity estimates by a factor of 7.6. Moreover, estimates of species richness using COI were robust among three of four commonly used delimitation metrics, whereas estimates using 18S varied widely with the different metrics. We show that meiofaunal diversity has been greatly underestimated by 18S eDNA surveys and that the use of COI provides a better estimate of diversity. The suitability of COI is supported by cross-mating experiments in the literature and evolutionary analyses of discreteness in patterns of genetic variation. Furthermore its splitting of morphospecies is expected from documented levels of cryptic taxa in exemplar meiofauna. We recommend against using 18S as a marker for biodiversity surveys and suggest that use of COI for eDNA surveys could provide more accurate estimates of species richness in the future.
Keywords: DNA barcodes, species delimitation, microinvertebrates, environmental DNA
Species are a fundamental unit of biological diversity and their delimitation is central in ecology and evolution. Species delimitation using DNA (DNA taxonomy) has the potential to bypass many of the difficulties associated with traditional morphological taxonomy (1). With developments in high-throughput sequencing technologies and bioinformatics pipelines, it is now possible to sequence a single genetic locus en masse from the environment and delimit species in large scale environmental (eDNA) metabarcoding surveys (2, 3). Such eDNA surveys have been used to describe the diversity of microorganisms (4), macrofauna (5), fungi (6), and plants (7) in previously understudied habitats, such as the soil biota (8), sediments (9), and water (5, 10). Faced with these technological advances, it is now more important than ever to evaluate the reliability of the genetic loci used for DNA taxonomy, and to assess the congruence of their results with morphological taxonomy. Without such an assessment, biodiversity analyses may be misleading.
Given this potential, there have been surprisingly few broad-scale attempts to calibrate the use of different markers and metrics in eDNA surveys. Here, we use a clade-targeted sampling regime to test different loci and metrics used in eDNA surveys, relative to morphologically defined species (morphospecies). Our study is, taxonomically, one of the broadest surveys to date. By identifying diversity from systematic samples of clades, we provide guidelines that can be used with eDNA surveys to better understand the evolution and ecology of animal diversity, using the example of meiofauna, a major reservoir of biodiversity that contains most of the animal phyla (11).
Meiofaunal organisms (defined as animals that can pass through a 500-μm mesh) cannot be reliably treated using traditional taxonomic methods because of their size (<2 mm), morphological homogeneity (12), noninformative species descriptions (13), and lack of expert taxonomists (14). It is therefore expected that morphospecies will underestimate the true diversity of these organisms (12). The use of eDNA surveys can potentially make species delimitation more efficient and cost-effective (3). Current meiofaunal eDNA surveys rely mainly on the nuclear small subunit 18S rDNA gene (10, 15, 16). Cytochrome c oxidase subunit I (COI) mtDNA, although used for global barcoding initiatives (i.e., Consortium for the Barcode of Life, www.barcodeoflife.org, and the International Barcode of Life project, www.ibol.org), is less widely used for eDNA. One of the main reasons is that high variability of COI can necessitate the use of taxon-specific amplification and sequencing primers (e.g., refs. 4 and 17), although universal primer sets for next generation sequencing (NGS) have been designed and successfully implemented recently (18).
Here, we compare putative species counts obtained using either COI or 18S across 55 meiofaunal datasets comprising three taxonomic ranks (15 species complexes, 26 genera, and 14 higher taxa above the genus level, including orders, classes, and phyla) and totaling more than 12,000 sequences. A number of analytical metrics exist for delimiting species (1, 19–23), which focus on different biological properties, require different data types (DNA, morphology, and so forth), and have different minimal sampling requirements (1, 20–23). The relative performance of these different metrics has not yet been fully evaluated. Estimates are therefore compared between the two genes using four different delimitation metrics: the nucleotide divergence threshold (NDT) (1), the automatic barcode gap discovery (ABGD) (21), the K/θ method (22), and the generalized mixed Yule-coalescent model (GMYC) (23). These metrics were chosen because they can be implemented when DNA sequence data are the sole information available, as is the case in eDNA surveys.
Results
Choice of Marker.
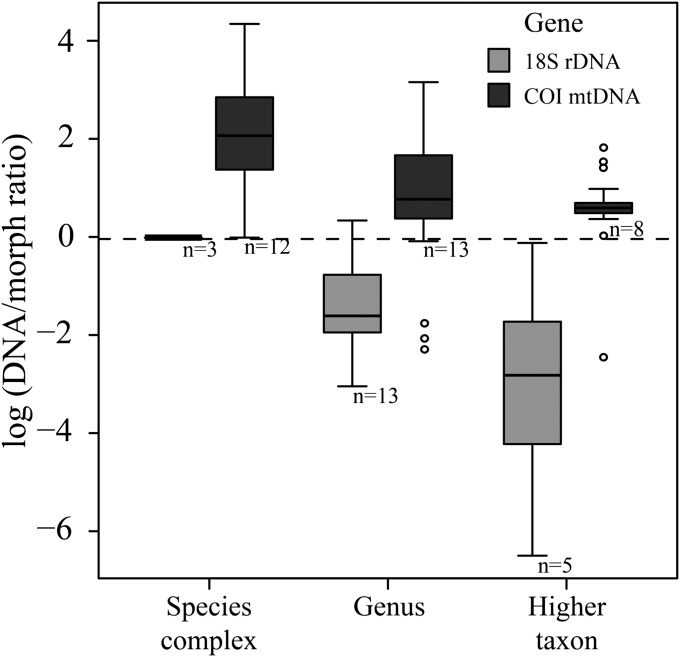
Diversity estimates in meiofauna were significantly different between the two markers (COI and 18S) and between morphospecies and both genes in turn (Fig. 1 and Table 1). COI yielded increased estimates of diversity relative to morphospecies, whereas 18S yielded decreased estimates of diversity (Fig. 1).
Fig. 1.
Ratio of species diversity estimated using DNA taxonomy compared with morphological taxonomy (dotted line) using either COI mtDNA (dark gray) or 18S rDNA (light gray) over three taxonomic ranks (species complex, genus, and higher taxon). DNA/morph ratios were averaged across both datasets and delimitation metric and log transformed. Open circles represent outlier values. The number of datasets (n) is shown under the boxes.
Table 1.
ANOVA used to explain differences in DNA/morph ratios
| Factor | Sum sq. | df | F value | P value | % Var. |
|---|---|---|---|---|---|
| Gene | 428.7 | 1 | 352.79 | <2.2e−16 | 53.10 |
| Metric | 21.0 | 3 | 5.76 | 0.00087 | 2.60 |
| Tax. | 110.0 | 2 | 45.26 | <2.2e−16 | 13.62 |
| Sampling | 0.1 | 1 | 0.07 | 0.79 | 0.01 |
| Metric × tax. | 11.2 | 6 | 1.53 | 0.17 | 1.38 |
| Metric × sampling | 18.8 | 3 | 5.16 | 0.0019 | 2.33 |
| Residuals | 217.5 | 179 |
Sampling effort differed between the two genes, being greater for COI, but this did not account for the differences in the ratio of the species count based on DNA relative to the species count for morphology (referred to as the DNA/morph ratio) between the genes (Table 1). Furthermore, the number of datasets was balanced between COI and 18S, with 31 and 24 datasets, respectively (Table S1), and geographic coverage for each dataset did not differ significantly between the genes (Table S1).
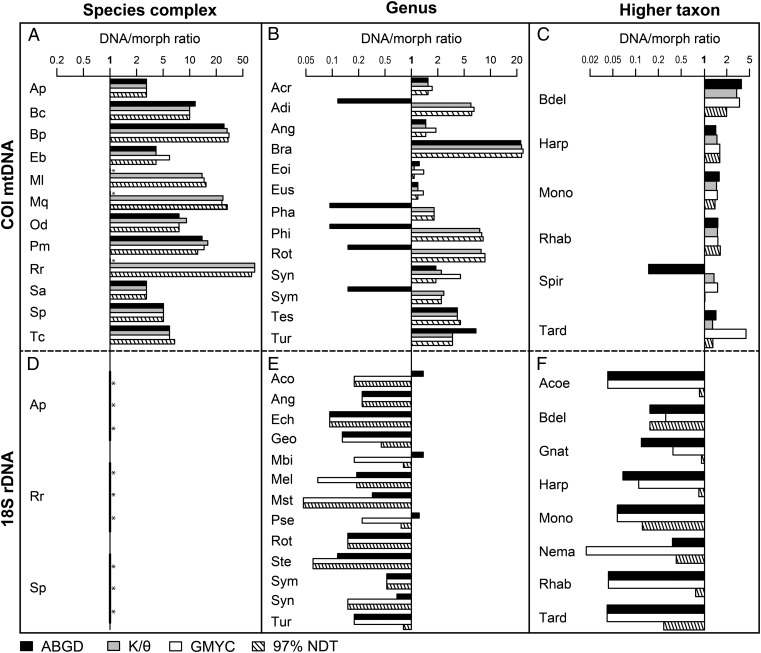
The average number of taxa estimated for COI and 18S using the different metrics was 7.6- (± 2.1 SE) and 0.4- (± 0.1) times the morphological species estimate, respectively. Cryptic taxa were identified in all COI datasets (Fig. 2) across all taxonomic ranks. A higher number of taxa was expected from the COI dataset than the 18S dataset, because most of the sequences come from investigations into cryptic species. Nevertheless, the estimated number of taxa using 18S was even lower than the number indicated by morphological taxonomy (Fig. 2). Thus, 18S was not able to reach the level and detail of identification of diversity that taxonomists can reach with morphology alone.
Fig. 2.
The ratio numbers of entities estimated using DNA taxonomy compared with morphological taxonomy (the y axis intersecting the x axis at 1) using different delimitation metrics and taxonomic ranks. Four different delimitation metrics (ABGD, black; K/θ method, gray; GMYC, white; 97% NDT, hatched) were used to compare DNA estimates to morphospecies, inferred from two different genes: COI (A_–_C) and 18S (D–F) and across three taxonomic ranks: species complexes (A and D), genera (B and E), and higher taxa (C and F). The x axis is log-transformed for ease of comparison. The K/θ method can be used in conjunction with mtDNA only, and is absent from the plots using 18S. Species estimates and full names are shown in Table S1. Acronyms have two letters for species complexes, three for genera, and four for higher taxa. Asterisks (*) specify instances where no additional species are estimated by DNA taxonomy compared with morphological taxonomy.
Controlling for potential confounding effects of differing sampling, by using datasets with COI and 18S sequenced from the same organisms, confirmed the general trend, with an average of 3.2- (± 1.0) and 0.4- (± 0.1) times the morphological species estimate for COI and 18S, respectively (Fig. S1).
Taxonomic Rank.
Taxonomic rank had a strong influence on the DNA/morph ratio for both COI and 18S (Table 2), with larger ratios for species complexes (15.3 ± 2.7 and 1.0 ± 0.0 times more entities for COI and 18S, respectively) than for genera (4.4 ± 0.8 and 0.3 ± 0.1, respectively) and higher taxa (2.2 ± 0.3 and 0.2 ± 0.1, respectively) (Fig. 1).
Table 2.
ANOVA used to explain DNA/morph ratio (COI and 18S analyzed separately)
| Factor | Sum sq. | df | F value | P value | % Var. |
|---|---|---|---|---|---|
| COI mtDNA | |||||
| Metric | 27.8 | 3 | 12.03 | <0.0001 | 13.5 |
| Tax. | 54.4 | 2 | 35.27 | <0.0001 | 26.4 |
| Country | 14.8 | 1 | 19.14 | <0.0001 | 7.2 |
| Undersampling | 0.2 | 1 | 0.26 | 0.61 | 0.1 |
| Metric × tax. | 2.0 | 6 | 0.44 | 0.85 | 1.0 |
| Tax. × country | 2.2 | 2 | 1.40 | 0.25 | 1.1 |
| Metric × undersampling | 9.5 | 3 | 4.11 | 0.0088 | 4.6 |
| Tax. × undersampling | 4.1 | 2 | 2.68 | 0.074 | 2.0 |
| Country × undersampling | 0.1 | 1 | 0.12 | 0.73 | 0.04 |
| Metric × tax. × undersampling | 15.4 | 6 | 3.33 | 0.0052 | 7.5 |
| Tax. × country × undersampling | 5.8 | 2 | 3.76 | 0.027 | 2.8 |
| Residuals | 69.4 | 90 | |||
| 18S rDNA | |||||
| Metric | 16.6 | 2 | 11.63 | <0.0001 | 9.8 |
| Tax. | 64.8 | 2 | 45.42 | <0.0001 | 38.2 |
| Country | 15.3 | 1 | 21.38 | <0.0001 | 9.0 |
| Metric × Tax. | 24.2 | 4 | 8.47 | <0.0001 | 14.3 |
| Metric × Country | 5.9 | 2 | 4.10 | 0.021 | 3.5 |
| Residuals | 42.8 | 60 |
Delimitation Metrics.
Congruence among the different delimitation metrics was greater using COI than 18S (Fig. 2 and Table S2). In COI, this pattern was not correlated to number of countries or taxonomic rank. Only species estimates from ABGD (4.4 ± 1.2-times the morphological estimate) were significantly different from the others (9.5 ± 1.5) (Fig. S2 and Table S3). The ABGD metric failed to delimit additional taxa in 26% of the datasets (Table S1). The DNA/morph ratio was influenced by the interaction between delimitation metric and degree of undersampling (i.e., the observed diversity compared with the expected diversity as determined using Chao estimators) (Table 2), and so the small differences in diversity estimates among the metrics may be a function of how they behave with undersampling.
For 18S, there was very little congruence among results using different delimitation metrics (Fig. 2 and Table S2). The GMYC estimated significantly fewer taxa (0.2 ± 0.1-times the morphological estimate) than did both the nucleotide divergence 97% threshold (0.5 ± 0.1) and ABGD metrics (0.4 ± 0.1) (Fig. S2). The estimates from the nucleotide divergence threshold became increasingly disparate from the other metrics with increasing threshold values. A 99% or a 99.5% threshold produced species estimates that were closer to the morphological estimate (average estimates were 0.8 ± 0.1- and 1.1 ± 0.2-times the morphological estimate, respectively).
Misidentification of Morphospecies.
To test whether misidentification of sequenced organisms (i.e., errors in morphospecies counts) could explain these patterns, we identified the subset of sequences generated by studies including a coauthor who has published a taxonomic paper describing a recently identified species (86.3% of COI and 55.1% of 18S sequences) (Table S1). Our reasoning is that morphological identifications in those studies should be correct. Reanalysis using only those data confirmed the same scenario that 18S yields lower diversity estimates and COI yields higher diversity estimates than morphospecies (Table S4). Furthermore, choice of metric, taxonomic rank, and sampling effect still affect the magnitude of estimates as identified for the entire dataset (Table S5).
Discussion
A key limitation inherent to most biodiversity studies is the arduous task of identifying organisms in the wild. Reliance upon good taxonomy and species identification goes beyond biodiversity surveys because it has wide implications for a number of ecological and evolutionary disciplines (for a perspective, see ref. 24). Here we demonstrate that biodiversity surveys of meiofaunal taxa will be greatly impacted by the choice of molecular marker. Using data covering a broad diversity of meiofaunal taxa, sampled across multiple taxonomic ranks, and applying four widely used delimitation metrics, we have shown that both 18S and morphology underestimate species diversity relative to COI (Fig. 1), independently of degree of undersampling. Furthermore, the disparity in congruence among metrics for both genes was not driven by differences in the discreteness of geographical sampling (Table 2) or by errors in morphospecies identification.
Although COI leads to higher species counts than morphology, COI provides a better indicator of the true diversity than morphology for several reasons. First, cryptic taxa have previously been documented in many of the morphospecies assessed here (e.g., refs. 12 and 25), and species boundaries have been confirmed within an integrative taxonomic framework (i.e., DNA taxonomy with cross-mating or additional morphological assessment) (e.g., refs. 26 and 27). Second, increased diversity based on COI is not simply because of higher variability, but associated with a sharp distinction in genetic distance within versus between evolutionary species (Table S1), which is indeed the signature that the evolutionary methods of K/θ and GMYC use to delimit species entities. There exist genetic clusters separated by larger “gaps” in genetic variation than expected if all individuals belonged to a single interacting population. In sexual lineages, these clusters reflect reproductively isolated species. In asexual lineages, they are caused by geographical isolation or specialization on distinct ecological niches (28), as demonstrated by studies of exemplar clades (e.g., refs. 12, 29, and 30). Patterns of genetic variation and differences between COI and 18S apply equally to asexual (i.e., bdelloid rotifers) and related sexual lineages (i.e., monogonont rotifers) in our sample (Tables S6 and S7).
We conclude that taxonomic entities estimated from analyses using 18S are not reliable proxies for diversity and very likely underestimate true species richness. Only a fixed threshold of 99.5% based on 18S (approaching the rate of potential PCR errors) can produce eDNA estimates comparable to morphological estimates, but these are known to be underestimates because of the prevalence of cryptic species. The use of 18S might be appropriate to compare levels of relative diversity at higher taxonomic scales, but species-level patterns, such as ecological distributions and geographical turnover, should not be inferred from these data. Furthermore, the degree of lumping per morphospecies estimated from 18S varies among different taxonomic groups (Fig. S1) and thus diversity might be very different depending on the composition of taxonomic groups present in an eDNA sample.
The four metrics used require a different minimum number of sequences per entity to detect putative species, and thus in principle intraspecific undersampling might affect the metrics in different ways (20). Intraspecific undersampling did not affect our results; however, we observed a high degree of congruence for COI in terms of both species number and identity, particularly among the nucleotide divergence 97% threshold, K/θ, and GMYC methods (Fig. 2). Interestingly, the simple rule of pair-wise similarity (1) gave results congruent to those provided by the two methods detecting signatures of independent evolution predicted by population genetic theory (22, 23). The consistency among different delimitation metrics was much lower for 18S than for COI (Fig. 2 and Table S3).
Although these results provide strong reasons against using 18S for species delimitation, there are also difficulties associated with COI, which include high substitution rates, excessive saturation, biased substitution patterns, high AT content, and poorly conserved priming sites (15, 17). These issues are known to reduce the efficacy of COI in certain meiofauna [e.g., nematodes (31) and proseriates (17)] and this may be one of the main reasons why COI has rarely been used for meiofaunal eDNA surveys (but see ref. 4). Nonetheless, COI has been successfully used in an integrative taxonomic framework to delimit nematode cryptic species (e.g., refs. 26 and 32). Here we included 11 nematode datasets and in each case diversity was split using COI and lumped using 18S, relative to morphological species, indicating that COI can be used to survey nematode diversity more reliably than with 18S.
An increasing number of next-generation DNA metabarcoding surveys aim to quantify biodiversity (2), but this is a field in its infancy, which is expanding largely in tandem with evolving sequencing technologies and the wider integration into bioinformatic pipelines (3). Use of COI with NGS is not yet popular. However, it has already been shown that a short minibarcode fragment of COI can be amplified en masse with 454 pyrosequencing (18) and used to identify individual species with high accuracy across many eukaryotic groups. Combining the variability of COI with NGS technologies and a reference database of genetic sequences identified to a species level could massively enhance the taxonomic resolution and rate of diversity exploration in our changing world.
Materials and Methods
Data Collection.
We concentrated on meiofaunal species complexes, genera, and higher taxa for which a rich collection of sequences was available. Datasets were compiled if a minimum of 10 different sequences per taxon were available. In some cases (e.g., for some groups of nematodes, nemerteans, and flatworms), even organisms that are not strictly meiofauna have been included (e.g., larger or parasitic) if the majority of the taxa of the group belonged to meiofauna.
We obtained sequences of a fragment of COI (on average 623 bp) (Dataset S1) from 8,576 individuals (6,834 downloaded from GenBank; 1,742 generated for this study according to the protocol presented in SI Materials and Methods), and sequences of 18S rDNA (on average 1,647 bp) (Dataset S1) from 3,668 individuals (3,321 obtained from GenBank and 347 sequenced de novo). The 18S sequences were amplified and sequenced directly only for each unique COI haplotype, as it became apparent from initial data collection that individuals with the same COI haplotype invariably had identical 18S sequences. A list of unique type specimens used in the analyses can be found in Dataset S1.
Overall, 12,244 sequences (of which 4,877 were unique) forming 55 datasets were analyzed. In total, 1,484 morphospecies were included; these were identified before sequencing using the most recent morphological revisions for the groups (listed in ref. 14), or by using their GenBank accession identifier. Ambiguous names were conservatively excluded from the morphospecies count. Phylogenetic analyses were obtained using standard procedures (SI Materials and Methods).
Nucleotide Divergence Threshold.
We applied a nucleotide divergence threshold (1) with a “friends of friends” approach (i.e., if divergence between taxa A-B and B-C is more than 3%, but divergence between A-C is less than 3%, then we consider taxa A, B, and C to be a single cluster). The divergence threshold is based on empirically observed gaps and 97% is the most commonly used threshold for COI (e.g., ref. 33). We used a script written in R that clusters sequences from an alignment based on a user-defined divergence threshold, using an uncorrected distance matrix as input (available in SI Materials and Methods).
ABGD.
ABGD uses a range of prior intraspecific divergences to infer from the data a model-based one-sided confidence limit for interspecific divergence (21). The “barcode gap” is identified as the first significant gap beyond this limit. Genetic clusters were inferred using the ABGD online tool and the default settings (available at http://wwwabi.snv.jussieu.fr/public/abgd/abgdweb.html). The correct species estimate was selected, as suggested by ref. 21, using gene specific priors for maximum divergence of intraspecific diversity (0.01 for COI and 0.001 for 18S).
K/θ Method.
The K/θ method (formerly known as the 4X rule) uses population genetic theory to identify sister clades that are too divergent to arise solely by neutral genetic drift within a single population (22). A neighbor-joining tree was constructed for each mtDNA dataset from uncorrected distance matrices using Geneious Pro-5.4.2. Starting from the tips of the tree, the maximum pair-wise distance was recorded for each clade (θ). For each pair of sister clades, the maximum value of θ was then compared with K (minimum pair-wise distance between clades). Clades that have a ratio of K/θ ≥ 4 are considered to be reciprocally monophyletic entities with ≥95% probability (22).
GMYC Method.
The GMYC method (23) tests for a significant shift in the branching rate in an ultrametric tree. Such a shift is indicative of the switch from between-species to within-species processes, expected if a sample comprises multiple individuals from a set of independently evolving species. The outgroups were removed from the ultrametric trees and the GMYC method was implemented using the splits package in R (available at http://r-forge.r-project.org/projects/splits/). Species were identified by a single threshold defined by a significant shift in branching rate.
Assessment of Reliability.
A linear model was used with a log-transformed DNA/morph ratio as the response variable and gene identity, species delimitation metric, and taxonomic rank as fixed effects. The initial linear models included all of the possible explanatory variables and their interactions. These models were subsequently simplified using the step function in R to obtain the minimum adequate model. Model outputs were given as ANOVA tables and post hoc Tukey honestly significant difference tests were performed to determine which different factors were significantly different from each other.
To quantify the effect of sampling effort (per taxon sampling) we used the number of sequences per dataset. As an index for degree of undersampling (unsampled diversity), we used the ratio of observed entities to expected number of entities [determined using a Chao estimator (34)]. Chao 1 estimator was used when both singlets and doublets were present in the dataset; Chao 2 estimator was used when either singlets or doublets were absent. These proxies for sampling effort and undersampling were included into a linear model as fixed effects along with gene, metric, and taxonomic rank.
Differing degrees of geographic coverage could inflate the congruency of delimitation metrics if genetic distance increases with geographical distance. We determined whether there was a significant difference in geographical coverage of each of the genes and used a linear model to test whether congruency of delimitation metric species estimates could be explained by geographical coverage, gene, and taxonomic rank. The coefficient of variation among species estimates of different metrics was used as a proxy for their congruence, and number of countries within each dataset as a proxy for geographical coverage.
Misidentification of organisms by the authors submitting sequences to GenBank could underestimate the biodiversity already present in the samples. To address this concern, we identified all authors involved in submitting the sequences and determined whether they were taxonomists based on whether they had published a species description. This method is a crude but true measure of taxonomist integrity, but is likely to underestimate the capabilities of many authors. This subset of data was reanalyzed as described above.
Supplementary Material
Supporting Information
Acknowledgments
We thank the Barralab, Andy Purvis, Beth Okamura, Aelys Humphreys, two anonymous reviewers, and the managing editor for their comments and suggestions on the manuscript; Will Pearse for his help with the programming; and all the meiofauna people who deposited their identified sequences on GenBank. This project was funded by the Natural Environment Research Council, and T.G.B.'s time was funded by Biotechnology and Biological Sciences Research Council Grant B/G004250/1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX494729 and JX494746).
References
- 1.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taberlet P, Coissac E, Hajibabaei M, Rieseberg LH. Environmental DNA. Mol Ecol. 2012;21:1789–1793. doi: 10.1111/j.1365-294X.2012.05542.x. [DOI] [PubMed] [Google Scholar]
- 3.Bik HM, et al. Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol Evol. 2012;27:233–243. doi: 10.1016/j.tree.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robeson MS, 2nd, et al. Environmental DNA sequencing primers for eutardigrades and bdelloid rotifers. BMC Ecol. 2009;9:25. doi: 10.1186/1472-6785-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ficetola GF, Miaud C, Pompanon F, Taberlet P. Species detection using environmental DNA from water samples. Biol Lett. 2008;4:423–425. doi: 10.1098/rsbl.2008.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaalid R, et al. Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol Ecol. 2012;21:1897–1908. doi: 10.1111/j.1365-294x.2011.05214.x. [DOI] [PubMed] [Google Scholar]
- 7.Yoccoz NG, et al. DNA from soil mirrors plant taxonomic and growth form diversity. Mol Ecol. 2012;21:3647–3655. doi: 10.1111/j.1365-294X.2012.05545.x. [DOI] [PubMed] [Google Scholar]
- 8.Wu T, Ayres E, Bardgett RD, Wall DH, Garey JR. Molecular study of worldwide distribution and diversity of soil animals. Proc Natl Acad Sci USA. 2011;108:17720–17725. doi: 10.1073/pnas.1103824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chariton AA, Court LN, Hartley DM, Colloff MJ, Hardy CM. Ecological assessment of estuarine sediments by pyrosequencing eukaryotic ribosomal DNA. Front Ecol Environ. 2010;8:233–238. [Google Scholar]
- 10.Bik HM, et al. Metagenetic community analysis of microbial eukaryotes illuminates biogeographic patterns in deep-sea and shallow water sediments. Mol Ecol. 2012;21:1048–1059. doi: 10.1111/j.1365-294X.2011.05297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giere O. Meiobenthology the Microscopic Motile Fauna of Aquatic Sediments. Heidelberg: Springer; 2009. [Google Scholar]
- 12.Fontaneto D, Kaya M, Herniou EA, Barraclough TG. Extreme levels of hidden diversity in microscopic animals (Rotifera) revealed by DNA taxonomy. Mol Phylogenet Evol. 2009;53:182–189. doi: 10.1016/j.ympev.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 13.Godfray HCJ. Challenges for taxonomy. Nature. 2002;417:17–19. doi: 10.1038/417017a. [DOI] [PubMed] [Google Scholar]
- 14.Curini-Galletti M, et al. Patterns of diversity in soft-bodied meiofauna: Dispersal ability and body size matter. PLoS ONE. 2012;7:e33801. doi: 10.1371/journal.pone.0033801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Creer S, et al. Ultrasequencing of the meiofaunal biosphere: Practice, pitfalls and promises. Mol Ecol. 2010;19(Suppl 1):4–20. doi: 10.1111/j.1365-294X.2009.04473.x. [DOI] [PubMed] [Google Scholar]
- 16.Fonseca VG, et al. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat Commun. 2010;1:98. doi: 10.1038/ncomms1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanna D, Lai T, Francalacci P, Curini-Galletti M, Casu M. Population structure of the Monocelis lineata (Proseriata, Monocelididae) species complex assessed by phylogenetic analysis of the mitochondrial Cytochrome c Oxidase subunit I (COI) gene. Genet Mol Biol. 2009;32:864–867. doi: 10.1590/S1415-47572009005000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meusnier I, et al. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics. 2008;9:214. doi: 10.1186/1471-2164-9-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiens JJ. Species delimitation: New approaches for discovering diversity. Syst Biol. 2007;56:875–878. doi: 10.1080/10635150701748506. [DOI] [PubMed] [Google Scholar]
- 20.Sites JW, Marshall JC. Delimiting species: A Renaissance issue in systematic biology. Trends Ecol Evol. 2003;18:462–470. [Google Scholar]
- 21.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol Ecol. 2012;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- 22.Birky CW, Jr, Adams J, Gemmel M, Perry J. Using population genetic theory and DNA sequences for species detection and identification in asexual organisms. PLoS ONE. 2010;5:e10609. doi: 10.1371/journal.pone.0010609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pons J, et al. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- 24.Yoccoz NG. The future of environmental DNA in ecology. Mol Ecol. 2012;21:2031–2038. doi: 10.1111/j.1365-294X.2012.05505.x. [DOI] [PubMed] [Google Scholar]
- 25.Suatoni E, Vicario S, Rice S, Snell T, Caccone A. An analysis of species boundaries and biogeographic patterns in a cryptic species complex: The rotifer—Brachionus plicatilis. Mol Phylogenet Evol. 2006;41:86–98. doi: 10.1016/j.ympev.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Fonseca G, Derycke S, Moens T. Integrative taxonomy in two free-living nematode species complexes. Biol J Linn Soc Lond. 2008;94:737–753. [Google Scholar]
- 27.Schröder T, Walsh EJ. Cryptic speciation in the cosmopolitan Epiphanes senta complex (Monogononta, Rotifera) with the description of new species. Hydrobiologia. 2007;593(1):129–140. [Google Scholar]
- 28.Birky CW, Barraclough TG. In: Lost Sex: The Evolutionary Biology of Parthenogenesis. Schön I, Martens K, van Dijk P, editors. London, UK: Springer; 2009. pp. 201–216. [Google Scholar]
- 29.Fontaneto D, et al. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 2007;5:e87. doi: 10.1371/journal.pbio.0050087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schön I, et al. Cryptic species in putative ancient asexual darwinulids (crustacea, ostracoda) PLoS ONE. 2012;7:e39844. doi: 10.1371/journal.pone.0039844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhadury P, et al. Development and evaluation of a DNA-barcoding approach for the rapid identification of nematodes. Mar Ecol Prog Ser. 2006;320:1–9. [Google Scholar]
- 32.Derycke S, et al. Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Mar Ecol Prog Ser. 2005;300:91–103. [Google Scholar]
- 33.Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao A. Nonparametric estimation of the number of classes in a population. Scand J Stat. 1984;11:265–270. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information