Defining the Status of RNA Polymerase at Promoters (original) (raw)
. Author manuscript; available in PMC: 2012 Oct 30.
Summary
Recent genome-wide studies in metazoans have shown that RNA Polymerase II (Pol II) accumulates to high densities on many promoters at a rate-limited step in transcription. However, the status of this Pol II remains an area of debate. Here, we compare quantitative outputs of GRO-seq and ChIP-seq assays and demonstrate the majority of the Pol II on Drosophila promoters is transcriptionally-engaged - very little exists in a preinitiation or arrested complex. These promoter-proximal polymerases are inhibited from further elongation by detergent sensitive factors, and knockdown of negative elongation factor, NELF, reduces their levels. These results not only solidify that pausing occurs at most promoters, but demonstrate that it is the major rate-limiting step in early transcription at these promoters. Finally, the divergent elongation complexes seen at mammalian promoters are far less prevalent in Drosophila, and this specificity in orientation correlates with directional core promoter elements, which are abundant in Drosophila.
Introduction
Transcription regulation is a major and primary mode by which developmental, nutritional, and environmental signals control gene expression. This regulation must ultimately target the activity of RNA Pol II, which encodes all mRNAs and many critical non-coding RNAs. Chromatin Immuno-Precipitation (ChIP) studies in Drosophila and mammals have shown that Pol II accumulates disproportionately at a large fraction of promoters relative to downstream gene regions (Baugh et al. 2009, Guenther et al. 2007, Muse et al. 2007, Zeitlinger et al. 2007); thereby identifying what appears to be a rate-limiting step in transcription. At least a portion of the accumulated Pol II at promoters has initiated transcription (Core et al. 2008, Nechaev et al. 2010), but whether this polymerase is predominantly bound and uninitiated in a pre-initiation complex (PIC) with general transcription factors (Juven-Gershon et al. 2008) or exists as an elongation complex proximal to the promoter requires a quantitative analysis. Additionally, accumulated Pol II at promoters could be either paused, transcribing and undergoing rapid cycles of initiation and termination, or backtracked to an arrested state that is incapable of elongation. A quantitative determination of which of these forms of polymerase predominates at a given gene promoter would provide a basis for understanding how that gene is regulated; however, no single assay determines this in vivo.
Two assays that are commonly used to examine the density of polymerases along DNA are the Chromatin immunoprecipitation (ChIP) assay and the nuclear run-on (NRO) assay. ChIP assays can quantify Pol II levels across the genome, but they cannot distinguish whether Pol II is transcriptionally engaged, backtracked and arrested, or bound in a PIC; nor can it assess the orientation of engaged polymerases. NRO assays measure polymerases that are transcriptionally engaged and competent to elongate and can determine the direction of transcription (Lis 1998), but, on their own cannot determine what fraction of the total polymerase present at a given location is in this form. Also, engaged polymerases could be transiently passing through the promoter or could be stably held in a paused state as seen at the extensively-characterized Drosophila Hsp70 gene (Lis 1998), and the human c-myc gene (Krumm et al. 1995, Strobl and Eick 1992). At these promoters, the paused Pol II is thought to be physically held back since conditions that disrupt protein-protein and protein-DNA interactions, but do not affect transcriptionally engaged polymerases (i.e. high concentrations of salt or addition of the detergent Sarkosyl) are required for efficient run-on transcription of promoter-proximal Pol II (Hawley and Roeder 1985, Rougvie and Lis 1988). These inhibitory interactions led to the hypothesis that this step is likely to be regulated in vivo (Rougvie and Lis 1988), and is now consistent with our current knowledge of the mechanism of promoter-proximal pausing: Pol II is held paused by the cooperative action of Spt5 and negative elongation factor (NELF) protein complexes. Regulated recruitment of positive elongation factor-b (PTEF-b), alleviates this negative block, resulting in escape of Pol II from the pause site and entry into productive elongation (Nechaev and Adelman 2011). However, not all promoters have been characterized to extent of the Hsp70 gene, making it difficult to extrapolate these characteristics of the Hsp70 promoter to other genes.
We developed a sensitive Global Run-On Sequencing assay (GRO-seq) that maps the position, amount and orientation of transcriptionally engaged polymerases genome wide (Core et al. 2008). Application of GRO-seq to a human primary cell line showed transcription occurring within 70% of genes, with 40% of these genes experiencing a significant accumulation of promoter-proximal polymerase that has properties of transcriptionally paused Pol II. We also observed that the majority of active promoters in human cells have a peak of transcriptionally-engaged polymerase that is upstream and divergent relative to the annotated gene. This finding has initiated a debate over whether these upstream divergent transcripts are functional, or if they instead represent aberrant, “sloppy” transcription initiation events that result from open promoter chromatin (Buratowski 2008, Seila et al. 2009).
Here, we used GRO-seq in Drosophila S2 cells to assess the genome-wide transcription pattern and characterize promoters. Our GRO-seq data shows that transcription is tightly associated with annotated genes, with very little evidence of complete genomic transcription or initiation at 3′-ends of genes. We also report, as suggested elsewhere (Nechaev et al. 2010), that Drosophila promoters generally lack divergently-engaged Pol II seen at the majority of human promoters. In this work, we show evidence that a well-known DNA element can specify increased directionality at human promoters, thereby providing a simple explanation for the strong directionality in Drosophila promoters, which are inherently rich in orientation specific elements (FitzGerald et al. 2006). To then quantify the status of polymerase at promoters, we use a normalized comparison of the polymerase densities at promoters as seen by ChIP-seq and GRO-seq, to conclude that the majority of polymerases at promoters are transcriptionally engaged and competent for elongation under steady state conditions. Moreover, we find that paused polymerases are physically tethered or blocked at promoters as they transcribe efficiently only in the presence of the anionic detergent sarkosyl. These observations establish not only that pausing occurs at most promoters, but that the predominant form of Pol II at promoters is paused in a manner that is similar to pausing at the Drosophila Hsp70 gene. Altogether, these observations provide a framework with which to study transcription factor function during basal and activated states.
Results
Transcription is predominantly associated with annotated genes
We performed GRO-seq assays under several conditions in Drosophila S2 cells (Table S1). Under standard conditions that detect all transcriptionally competent polymerases, 67% of engaged polymerases occupy the sense strand of gene annotations, and 15% occupy the antisense strand of annotated genes (82% of total)(Figure S1). These numbers increase to 78% and 19% (98% of total), respectively, if gene boundaries are expanded by 0.5kb. Thus, as we reported with a human primary lung fibroblast line (Core et al. 2008) and mouse embryonic stem cells (Min et al. 2011), the vast majority of transcription in Drosophila is associated with annotated gene regions.
Debate of whether or not genomes are ‘pervasively’ transcribed depends on different assays of accumulated RNAs (Kapranov et al. 2007, van Bakel et al. 2010) and on semantics. The GRO-seq assay, which has high sensitivity and low background (libraries are estimated to be >99% pure), measures the distribution of transcriptionally competent polymerases. The snapshot of transcriptome activity provided by GRO-seq does not depend on RNA processing rates or transcript stability. The assay reveals that the vast majority, 98%, of transcriptionally-competent RNA polymerases are focused within or near currently annotated genes and these genes cover ∼46% of the genome. Thus, while our GRO-seq data do not deal with the sum of transcripts produced in multiple cell types, they do argue that any ‘pervasive’ transcription of the genome in Drosophila S2 cells must occur at levels that are indistinguishable from the low background of our assay.
Drosophila promoters are more directional than mammalian counterparts
Alignment of all reads relative to observed transcription start sites (TSSs) (Nechaev et al. 2010), or plotting of the distribution of sense vs. antisense reads at promoters, revealed a prominent lack of divergent transcription at Drosophila promoters compared to human promoters (Figures 1B, 1C and S1F). In support of this, 95% of promoter–associated reads map in the direction of the annotated gene at Drosophila promoters compared to 58% for human promoters. ChIP-seq, and ChIP-chip datasets in human and mouse cells show that Pol II and histone marks associated with initiation coincide with divergent initiation upstream of TSSs (Seila,A.Cet al. 2008, Core, Waterfall, and Lis, 2008). Consistent with this, Pol II ChIP-seq and the H3K4me3 initiation mark are strongly associated with the direction of transcription at Drosophila TSSs (Figures S1D-S1E). In addition, in Drosophila, only unidirectional profiles are evident in datasets comprised of small, 5′-OH or 5′-capped RNAs (Nechaev et al. 2010, Taft et al. 2009). The GRO-seq data confirms that the inability to detect divergent transcription in small RNA pools is not due to preferential capping or processing of the nascent RNA in one direction versus the other since GRO-seq will detect nascent RNAs regardless of how the RNA end is modified. Likewise, failure to detect divergent transcription in GRO-seq is not due to an alternative form of Pol II that is undetectable by nuclear run-on. Combined, these results reinforce the notion that marks of initiation, such as H3K4me3, coincide with promoter direction (Seila,A.C. 2008, Core,L.J. 2008).
Figure 1. RNA polymerase distribution on mRNA-encoding genes using GRO-seq.
(A) A representative view of GRO-seq data from S2 cells in the UCSC genome browser (Kent WJ, Sugnet CW, et al, 2002). GRO-seq reads (reads/base) aligning to the plus strand are shown in red; minus strand in blue. ChIP-seq for total Pol II (a-Rpb3) is shown in green (reads/25bp bin), and gene annotations are shown at the bottom in blue. (B) GRO-seq data aligned to Transcription Start Sites (TSSs). For all genes, reads aligning to the sense strand of the gene are in red; antisense strand in blue. For non-bidirectional genes (head-to-head promoters within 1 kb removed), reads aligning to the sense strand of the gene are in green; antisense strand in orange. (C) Comparison of directionality of Drosophila and human promoters. The distribution of the ratios of sense and antisense reads around promoters (log2) is plotted for active promoters (>25 reads) in IMR90 cells (green) and Drosophila S2 cells (blue). How different types of directionality of transcription from promoters are reflected in the ratio are indicated in italicized lettering. (D) GRO-seq profiles from −/+ 1.5 kb relative to TSS are shown for all human promoters (green, sense; orange, antisense) or human promoters that contain a TATA box (red, sense; blue, antisense). (E) GRO-seq data aligned to gene end for all genes (red, sense; blue, antisense), and after convergent genes within 1.5 kb are removed (green, sense; orange, antisense). See Also Figure S1.
The position and direction of transcription initiation are specified by a variety of core promoter sequence motifs. Drosophila promoters are enriched for several directional motifs, whereas human promoters appear to be enriched mainly for non-directional motifs and CpG islands (FitzGerald et al. 2006). To test the hypothesis that directional motifs in Drosophila may be responsible for specifying uni-directional transcription, we generated an orientation index (OI) for all human promoters. The OI is defined as the fraction of GRO-seq density at promoters that is orientated in the sense direction. We then compared the OI of human promoters that contain directional and non-directional motifs identified in a comparative analysis between Drosophila and human promoters (FitzGerald et al. 2006). Of these motifs, the TATA box (TATAWAAR) (Juven-Gershon et al. 2008), is the only one to show a clear bias toward unidirectional transcription at human promoters (OI = 0.86 compared to OI = .57 for all promoters)(Figure 1D, Table S2). Interestingly, the composite profile at human TATA-containing promoters more closely resemble Drosophila promoters (Figure S1F). We also found that promoters with a TATA box embedded within a CpG island also produce directional transcription (Figure S1G), suggesting that the TATA box can act dominantly in the context of human CpG islands to enhance initiation in the direction of the gene. However, because only 5-20% of Drosophila and mammalian promoters contain an identifiable TATA box (FitzGerald et al. 2006, Kutach and Kadonaga 2000, Sandelin, Carninci, et al, 2007), it is likely that other DNA elements or protein factors that specify unidirectional transcription in Drosophila are either not present or not functional in the context of mammalian promoters.
RNA Pol II accumulates at the majority of active promoters and to a lesser extent at gene ends
Alignment of reads to the 3′-end of genes showed much smaller peaks in both the sense and antisense directions (Figure 1E,). Neither peak at the 3′-end appears to be associated with genuine initiation at the 3′-end of genes, because there is no corresponding enrichment of small, capped RNAs (Figures S1H - S1K). Thus, this 3′-sense peak likely represents Pol II that slows down after the poly-adenylation signal is exposed. In support of this, the antisense peak is dramatically reduced when convergent genes are removed from the analysis (Figure 1E).
The striking accumulation of GRO-seq density in the promoter-proximal region indicates the existence of a rate-limiting step following transcription initiation. Accordingly, when we define active genes based on GRO-seq signal in gene bodies (pval < .01, fisher's exact test; supplemental methods), we find that 6,044 of 9,544 (63%) of these genes have significantly enriched GRO-seq signal at the 5′-end (p-val < .01, fisher's exact test). This fraction is likely an underestimate since overlapping transcription from neighboring genes can result in a false positive call for gene transcription when the actual promoter is not active. When we use 7,336 promoters defined as active by sequencing small, capped RNAs from nuclei (>10 reads within +/− 50 bases from TSS) (Nechaev et al. 2010), or 3,168 promoters called bound by Pol II from a ChIP-seq experiment (Nechaev et al. 2010), we find that 5,166, (70%) and 2,784 (89%) of promoters, respectively, show significantly enriched Pol II in our GRO-seq analysis. Thus, post-initiation regulation occurs at the majority of promoters that show signs of Pol II binding or transcription activity. These polymerases that accumulate at promoters could be in the form of stably paused polymerases or polymerases that are actively transcribing within the promoter region, for example, undergoing cycles of initiation and rapid early termination. Thus, we sought to distinguish these two forms.
Engaged Pol II is physically tethered to promoters
A genuinely paused Pol II that is held near the promoter by pause stabilizing factors requires the disassociation of these factors by the addition of high salt or the anionic detergent sarkosyl to resume transcription in a run-on assay (Rougvie and Lis 1988). In contrast, Pol II that is undergoing active elongation transcribes efficiently with or without high salt or sarkosyl (Hawley and Roeder 1985, Rougvie and Lis 1988). We therefore produced matched GRO-seq data sets in the presence or absence of sarkosyl to test for pausing genome-wide. Our results show that run-on signal at nearly all promoters is dependent on sarkosyl, with the average promoter showing a ∼4 fold increase in signal in the presence of sarkosyl (Figure 2A-C,F). In contrast, read densities in gene bodies are unaffected by sarkosyl (Figure 2D,F). The stimulation by sarkosyl at gene ends (1.12 fold, Figure 2E,F) was much less pronounced than at promoters, indicating that the slowing down of polymerase near gene ends immediately prior to termination occurs through a different mechanism than pausing at promoters.
Figure 2. Sarkosyl-dependent run-ons identify distinct forms of polymerase at promoters relative to downstream gene regions.
(A) Composite profile of GRO-seq data showing the density reads in 10bp windows from −200bp to +500bp relative to TSSs for Run-ons performed with or without sarkosyl. Y-axis represents read/window/million reads sequenced. The number of genes shown = 11,800. (B) Schematic showing how GRO-seq signal was quantified at promoters, the gene body, or at gene ends. After removing genes based on overlaps and filtering for genes that have active promoters, the number of genes for this analysis = 4,652. (C-E) Scatter plots showing the effects of sarkosyl on run-on signal in promoters (C) or genes (D), or at gene ends (E). (F) Cumulative distribution plots showing the differential effect of sarkosyl at promoters (blue) versus within genes (orange), or at gene ends (green). The average effect in the gene, promoter, and the Hsp70 promoter are denoted by the hashed vertical lines. The effect at the Hsp70 promoter is shown as a hashed vertical line in red. The non-logged value for the fold effect after sarkosyl stimulation is shown in the legend. See Also Figure S2.
Interestingly, the effect of Sarkosyl at the Hsp70 gene in the GRO-seq data set is equivalent to the genome-wide average (Figure 2F). Thus, the majority of promoters in the Drosophila genome behave in a manner similar to the Hsp70 gene, which has served as a classic gene model for regulation through Pol II pausing. These results indicate not only that a high degree of stable pausing likely occurs at most promoters, but also that transcription elongation is inherently different at promoters versus downstream regions (Pal et al. 2001, Saunders et al. 2006). This implies that regulatory mechanisms are in place to control the level of pausing, presumably by modulating interactions that retain stably paused Pol II or release it into productive elongation.
NELF increases promoter occupancy of paused Pol II
If promoter-proximal pausing is a rate-limiting step in transcription governed by the interactions of pausing factors with the transcribing complex, then we expect that disruption of a factor involved in stabilizing the paused complex would reduce the accumulation of Pol II in the promoter-proximal region (Muse et al. 2007, Wu et al. 2003, Yamaguchi et al. 1999). Indeed, RNAi knock-down of NELF leads to a general decrease in the GRO-seq signal on promoters (Figure 3A and B) relative to gene bodies and 3 ′ends (Figures 3B-D, and S3). This moderate decrease in Pol II at promoters following NELF knock-down is not surprising, because residual NELF, or its partner DSIF, could still be sufficient to induce pausing (Figure S3A).
Figure 3. Use of GRO-seq to examine function of NELF at promoters and identification of genes affected by NELF knockdown.
(A) Composite profile of GRO-seq data showing the density reads in 10bp windows from +/−100bp relative to TSSs for untreated (green), mock- (blue), and NELF-depleted (red) cells. The number of genes shown = 11,800. (B -D) Cumulative distribution plots showing the overall effect of NELF RNAi on polymerase density in promoters (B) or genes (C), or at gene ends (D). Figure panels and legends are displayed as in Figure 2. (E) Scatter plot showing the comparison of GRO-seq signal in the gene body region in mock – or RNAi – treated cells. Genes were identified as significantly affected (red) using edgeR (Robinson MD, McCarthy DJ, and Smyth GK, 2010), with an FDR of 0.01. The green line represents a 1:1 fit. (F) MNase-seq patterns relative to TSSs in mock – or RNAi – treated cells for genes that are either up or down regulated after NELF-RNAi (identified in (E)). Genes that are up-regulated after NELF RNAi (orange and blue) have overall lower nucleosome density around their promoters than genes that are down-regulated (red and green). As seen previously (Gilchrist et al, 2010) genes that are down regulated by NELF RNAi have increased encroachment of nucleosomes over the TSS after NELF RNAi. See Also Figure S3.
The reduction of Pol II at promoters after NELF RNAi could be accounted for by either increased escape of polymerase into the gene without immediate entry of a new polymerase into the pause site, or by decreased initiation due to increased nucleosome occupancy at promoters (Gilchrist et al. 2010). Previous studies relying on ChIP-chip have been unable to determine conclusively at which genes the reduced amount of Pol II at promoters is due to increased escape of Pol II into the gene, or decreased initiation (Gilchrist et al. 2010). The highly sensitive of GRO-seq assay can detect both significant increases and decreases in the polymerase density in the downstream portion of genes (Figure 3E, Table S3). Since GRO-seq measures nascent RNA transcription, the significantly changed genes are more likely to be directly affected by NELF RNAi, than those identified by microarray, providing a high confidence gene list with which to investigate the molecular phenotypes of NELF knockdown, and the effects on promoter chromatin. Therefore, we examined the effect of NELF RNAi on MNase-seq pattern around promoters of genes that were identified as up- or down- regulated by GRO-seq. As seen previously, down-regulated genes have increased nucleosome density at the promoter (Figure 3F), consistent with the model that a paused polymerase competes with nucleosomes for occupancy of some promoters (Gilchrist et al. 2010). In contrast, the MNase pattern at up-regulated genes does not change after NELF knockdown, and these promoters have an overall lower level of nucleosome occupancy before or after NELF RNAi (Figure 3F). These data indicate that each promoter has an inherent propensity to displace or position nucleosomes around the promoter and this influences the net effect on transcription caused by removing a pausing factor.
Pol II pauses at enhancers
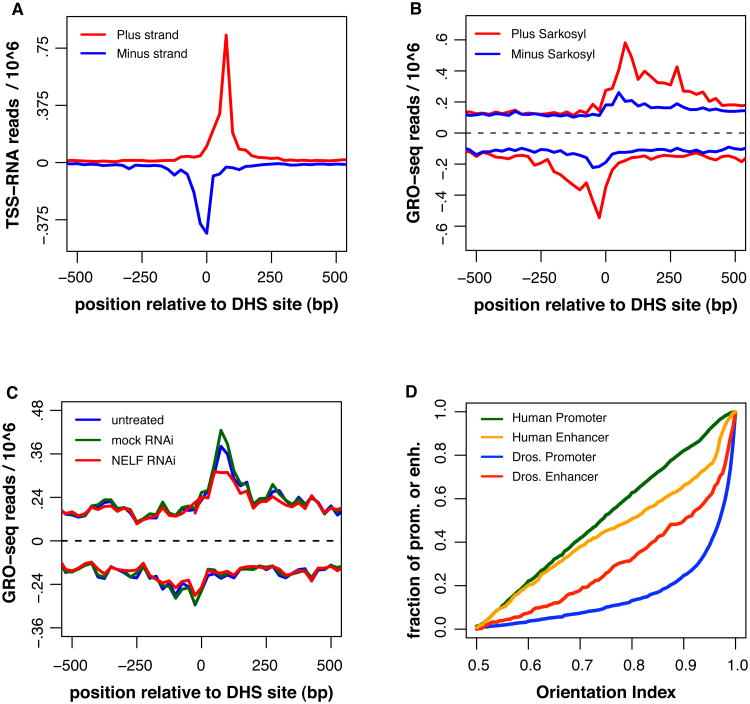
Transcripts originating from enhancers, or eRNAs, are a newly identified class of RNAs with unknown regulatory functions (Kim, Hemberg, et al, 2010). Transcription at enhancers is associated with active enhancers, and the resulting eRNAs can emanate bidirectionally from enhancers; can be spliced and polyadenylated, but have little coding potential (Kim, Hemberg, et al, 2010, Wang, Garcia-Bassets, et al, 2011). Enhancers themselves can be found within or outside of genes, and are enriched in mono-methylation of histone 3 at lysine 4 (H3K4me1) but have lower levels of tri-methylation at the same site (H3K4me3)(Heintzman, Stuart, et al, 2007). In contrast, active promoters are highly enriched with H3K4me3) (Kim, Barrera et al. 2005). To characterize the status and directionality of polymerase at Drosophila enhancers, we examined putative intergenic enhancers as identified by the ModENCODE group (Kharchenko, Alekseyenko, et al, 2011). 5′-RNA sequencing (Nechaev et al, 2010) provides evidence of initiation and pausing at these sites (Figure 4A). In addition, the polymerase at enhancers appears similar to that at promoters in that it is stimulated by sarkosyl during the run-on (Figure 4B), colocalizes with NELF (Figure S4), and has reduced occupancy after NELF RNAi (Figure 4C).
Figure 4. Pausing and directionality of RNA polymerase at enhancers.
A) A composite profile of TSS-RNA reads (Nech surrounding putative drosophila enhancers as identified by (Kharchenko, Alekseyenko, et al, 2011), n= 533. Data is plotted relative to the DNase hypersensitive site (DHS) site in 25bp bins, and the y- axis is in reads / bin / million reads sequenced. (B) GRO-seq data around the same sites in the presence and absence of sarkosyl, the positive signal (above dotted line) is from the plus stand, the negative signal; the minus strand. (C) GRO-seq data set after RNAi of the pausing factor NELF. (D) Plots of the distribution of orientation indexes for human promoters (green) and enhancers (orange); and drosophila promoters (blue) and enhancers (red). For ease of comparison between promoters and enhancers, the ‘direction’ of the promoter or enhancer is defined by the strand (plus or minus) with the greatest intensity. Thus, the orientation index here will be equal to or greater than 0.5. See Also Figure S4.
Given that human promoters and enhancers both produce divergent transcripts, we compared the orientations of polymerase for Drosophila and human enhancers (Figure 4D). Since enhancers do not have inherent directionality, we specified the ‘direction’ of the enhancer or gene to be the strand with the highest signal, making all OIs > 0.5 for this analysis. Interestingly, the distribution of OIs at Drosophila enhancers resembles a mixed distribution, with many showing strong directionality and a similar number appearing to be bidirectional (Figure 4D). Since the putative directional enhancers could be a result of non-annotated promoters, it is difficult to say whether this represents the true distribution of enhancer orientations. Nonetheless, it appears that Drosophila enhancers could more closely resemble human enhancers in their directionality (or lack thereof), and emphasizes that there is some difference between Drosophila enhancers and promoters.
Majority of Pol II at promoters is engaged and competent for transcription
Limitations of currently available assays have prevented a quantitative characterization of the form of Pol II at promoters. The ChIP assay cannot distinguish between Pol II that is in a PIC, paused, or backtracked and arrested. Sequencing of small (<100nt) RNAs from nuclei can identify RNAs generated by Pol II, but can't discern between Pol II that have paused, arrested, or terminated. GRO-seq can only detect Pol II that is engaged in transcription with the 3′-end of the nascent RNA in register with the active site and competent to transcribe during a nuclear run-on assay. Notably, promoter signals from each of the three assays correlate very well (Figure S5A-C), but these correlations alone do not explicitly identify the major form of Pol II at promoters. For instance, in a population of cells, a promoter could contain a PIC in some cells and a paused Pol II in others. Thus, in an ensemble-type assay like ChIP- and GRO-seq, it is possible that one could see a peak of Pol II at promoters in GRO-seq, even though in most cells the polymerase was still in a PIC. Determining which is the predominant form is a critical distinction for understanding how gene regulation works.
We reasoned that a more quantitative comparison of ChIP-seq and GRO-seq signals at promoters would reveal what fraction of the ChIP signal at promoters is represented by engaged and elongation-competent Pol II. As an internal standard, we used the ChIP-seq and GRO-seq signal in the body of the gene to normalize the gene-specific signal for each assay (Figures 5A and 5B). Because of the presumably high background in the ChIP-seq data (Figure S5), we focused on genes with highest levels of ser2-P ChIP signal (z score > 3); assuming these will contain the highest densities of transcribing Pol II over background. Good quantitative agreement between GRO-seq and total Pol II ChIP-seq levels in these 1,874 genes suggests that the ChIP-seq signal here represents engaged polymerases complexes that are competent for transcription. (Figure S5D). With this gene set we generated a conversion factor that was then used to calculate the fraction of the total Pol II at promoters that can be accounted for by the GRO-seq signal. We call this fraction the engaged/competent fraction (ECF). Approximately 80% of the polymerase found by ChIP-seq can be accounted for by the signal from the GRO-seq dataset (average ECF = 0.82, Figure 5B-D). We identified candidate promoters that were likely to contain PICs in the leftward tail in the ECF distribution (Figure 5C). However, these promoters are likely false positives, because outliers on both ends of the distribution (top and bottom 2.5%, ECF <.06, ECF>2.5), have low levels of Pol II binding as seen in ChIP-seq (Figure 5D). In cases where the relative ChIP-seq signal is greater than GRO-seq at promoters, the ‘non-competent’ polymerase could be in the process of forming a functional PIC or could be backtracked and arrested. However, since the data fits a normal distribution around the mean and there are theoretically impossible instances where relative GRO-seq signal at promoters is greater than the ChIP-seq, we believe that the major discrepancies between the two assays are due to inherent experimental noise or counting biases associated with next-generation sequencing. We therefore conclude that the major form of Pol II found at promoters by ChIP is engaged and competent for elongation.
Figure 5. Pol II at promoters is predominantly engaged and competent for elongation.
(A) Representative browser shot showing Pol II Chip-seq (green) and GRO-seq (red) with Y-axis in reads/bp/10exp6. The regions used for calculating the engaged and competent fraction (ECF) at promoters are indicated below. (B) Schematic explaining the workflow used to calculate the ECF for Pol II at promoters. (C) Histogram showing the distribution of ECF values for significantly bound promoters (n = 3168). The vertical lines represent a 50% (black), the average (red), and the Hsp70 (green) ECFs. Promoters with the lowest ECFs are highlighted in purple. (D) Boxplots showing Pol II ChIP-seq levels at promoters with different ranges of ECF. Promoters with the lowest (purple) and the highest (dark red) ECF values have less Pol II bound at promoters in ChIP-seq experiments than promoters with less extreme ECF values (middle 20% shown), suggesting that the ChIP and GRO discrepancies here could be due to experimental noise. See Also Figure S5.
We also compared the promoter ECF with several other data sets, including level of association of TFIIA, NELF, or SPT5 with promoters as measured by ChIP or levels of TSS RNAs, NELF RNAi sensitivity, sarkosyl sensitivity, or the presence of promoter elements and were unable to identify candidate PICs (Figure S5E-S5P, data not shown). In all datasets, the genes that are the most likely candidates for PICs (i.e. those with the lowest ECF), displayed signals approaching background, further suggesting that these genes are false positives and result from noise inherent to the low signal range. However, if these candidate promoters truly maintain a PIC, they do so at a very low occupancy compared to the occupancy of a paused polymerase. Taken together, these data argue against the notion of a stable pre-initiation complex and indicate that once Pol II is recruited to a promoter, it rapidly initiates RNA synthesis and undergoes pausing.
Discussion
Unlike mammals, Drosophila promoters lack an upstream divergent peak of Pol II
Here we have mapped the nascent transcriptome of Drosophila S2 cells using GRO-seq. A striking difference between the Drosophila and human transcriptomes is the lack of divergent transcription at Drosophila promoters. Drosophila has a collection of directional core promoter elements that serve to direct the transcription complex to the promoter (Juven-Gershon et al. 2008). We searched for several of these directional elements in human promoters and found that the most were either not prevalent or were non-functional because the corresponding protein that binds the element does not exist. Interestingly, the one core element that is present in a subset of human promoters, the TATAWAAR box, does correlate with a subclass of human promoters that show unidirectional transcription. This supports a model where core promoter elements are powerful directors of Pol II direction at a promoter. Human promoters are predominantly characterized by unmethylated CpG islands that by themselves do not specify orientation.
Our analysis of Drosophila enhancers reveals that the polymerase initiates and pauses at these locations. In Drosophila, an interesting difference from promoters is that a higher proportion of enhancers can produce bidirectional transcription. Thus, transcription from human and Drosophila enhancers appears to be more similar than their promoter counterparts. Although the enhancer transcripts themselves may be functional, it seems equally plausible that the act of transcription itself could provide an important function for maintaining enhancer activity. Alternatively, transcription at enhancers could result from non-specific initiation of transcription in a region of chromatin that is both generally accessible and attracting a high localized concentration of polymerases.
Promoter-proximal Pol II is predominantly in an elongationally paused state
Previous ChIP assays have shown that Pol II accumulates at high concentrations on promoters of a large fraction of Drosophila genes in what is apparently a rate-limiting step in transcription (Muse et al. 2007, Zeitlinger et al. 2007). We show here by a quantitative comparison of Pol II in ChIP and GRO-seq assays that the majority of this promoter-associated Pol II seen across the genome is in a paused configuration and thus competent for transcription elongation. The properties of paused Pol II originally uncovered for Drosophila Hsp70 and other heat shock genes: transcription of a short transcript (Rasmussen and Lis, 1993), its CTD phosphorylation state (Boehm et al. 2003, O'Brien et al. 1994), the association of pausing factors (Saunders et al. 2006), and the stimulation of their transcription in nuclear run-on assays by treatments that strip chromatin of repressive factors (Rougvie and Lis 1988, Rougvie and Lis 1990), are shared by a majority of Drosophila genes (Nechaev et al. 2010, and this work). Consistent with this last point and extrapolating from previous data (Gilchrist et al. 2010), we show that knock down of a pausing factor reduces the occupancy of Pol II at promoters and that the overall effect on gene transcription after of disrupting pausing is dependent on whether the promoter itself allows for a competing nucleosome or perhaps another protein complex to occlude the initiation site in the absence of pausing.
The fate of promoter proximal paused Pol II
Our quantitative analyses argue that the bulk of promoter-associated Pol II exists largely in a relatively stable paused configuration, and that this polymerase is a target of regulation. We expect that a paused polymerase turns-over both by termination (Brannon, Kim, et al, 2012), and by escape into productive elongation. The rates of either of these processes must be relatively slow to account for the high levels of accumulation of Pol II at pause sites 30-60 bases downstream of the TSS. Although our data does not definitively establish that the paused Pol II is the same Pol II that transcribes through the gene to produce a full mRNA transcript, evidence from our labs support this view. First, the majority of polymerases are engaged and competent for transcription in a nuclear run-on assay; thus, the paused polymerase has the proper alignment to the 3′ end of the RNA and the Pol II active site to transcribe the gene following activation. Second, many genes are firing productive Pol II's into the body of the gene, some quite rapidly, e.g. the induced Hsp70 fires every 4 seconds, yet most active genes still have a peak of promoter paused Pol II. Thus, Occam's razor directs us to propose that the Pol II molecules that undergo pausing subsequently elongate through the gene.
Promoter-proximal pausing as a step in transcription regulation
The biological significance of pausing has both experimental support and compelling speculation. First, some classes of activators directly stimulate pause escape rather than initiation and vice versa (Blau et al. 1996, Rahl et al. 2010, Yankulov et al. 1994), suggesting that different transcription factors could integrate different cellular signals to specify initiation and escape from pausing. Second, pausing of Pol II is accompanied by the capping of its associated short mRNA (Rasmussen and Lis 1993) and by phosphorylation of the CTD of Pol II to a form that provides a scaffold for RNA processing factors that are coupled to transcription elongation (Phatnani and Greenleaf 2006). This suggests that pausing may be a critical checkpoint in metazoans ensuring that RNA capping and the proper maturation of Pol II has an opportunity to occur for efficient transcription elongation and coupled splicing (Mandal et al. 2004, Rasmussen and Lis 1993). Third, the residence time of a paused Pol II allows it to directly compete with nucleosomes for high affinity nucleosome positioning sequences at promoters, thus maintaining promoters in an active state (Gilchrist et al. 2008, Gilchrist et al. 2010), and allowing for regulatory factor binding (Shopland et al. 1995). Fourth, maintenance of promoters in an open configuration provides a means for promoters to be primed for rapid, synchronous regulation in response to a variety of signals (Adelman et al. 2009, Boettiger and Levine 2009). Fifth, the knockdown of factors important for establishing pausing causes defects in both transcription activation and repression, which can be mediated through pausing mechanisms (Adelman et al. 2005, Aida et al. 2006; Missra and Gilmour 2010). Finally, pause site escape is modulated by the recruitment of P-TEFb kinase (Peterlin and Price 2006) that acts to phosphorylate and thereby inactivate paused stabilizing complexes, DSIF and NELF, and phosphorylate Pol II at Ser2 of its CTD to generate the elongationally modified form of Pol II. Evidence that this is a rate-limiting step is supported by the observation that the direct recruitment of P-TEFb to promoters is sufficient to produce high level of activation of Drosophila Hsp70 (Lis et al. 2000) and other genes (Bieniasz et al. 1999, Majello et al. 1999). Together, these observations suggest that pausing serves to potentiate transcription, and at the same time allow a repertoire of transcription factors to fine tune transcript levels both up and down by changing the rate of escape of Pol II from pausing.
Experimental Procedures
RNAi treatment and generation of ChIP-seq data
RNAi in Drosophila S2 cells were performed as described (Gilchrist et al. 2010). Further details regarding the published ChIP-seq data can be found in the supplemental experimental procedures.
Isolation of nuclei for GRO-seq
Nuclei were isolated as described previously (Core, Waterfall, and Lis, 2008), with several modifications. Details regarding the specific protocls used for isolating nuclei from RNAi-treated cells and nuclei for the plus– and minus– sarkosyl datasets can be found in the supplemental experimental procedures.
Preparation of GRO-seq libraries
Untreated, mock and NELF-depleted GRO-seq libraries were prepared as in Core et al. (Core, Waterfall, and Lis, 2008), with the following modifications. Trizol (invitrogen) was used to stop the reaction instead of DNase I and proteinase K treatment. The RNA was further extracted once with acid phenol:chloroform, and once with chloroform before precipitating with 2.5 volumes of −20°C ethanol. Bead binding buffers all contained 4units/ml of SUPERaseIN (ambion) and the following buffers were slightly modified. Bead blocking buffer: 0.25× SSPE, 1mM EDTA, 0.05% tween, 0.1% PVP, and 1mg/ml ultrapure BSA (Ambion); Binding buffer: 0.25×SSPE, 37.5mM NaCl, 1mM EDTA, 0.05% tween; Low salt wash buffer: 0.2× SSPE, 1mM EDTA, 0.05% Tween. High Salt wash buffer: 0.25% SSPE, 137.5mM NaCl, 1mM EDTA, 0.05% Tween. The end repair steps were modified as follows. Pelleted RNA from the first bead binding was resuspended in 20ul, and heated to 70oC for 5min, followed by incubation on ice for 2min. 1.5ul tobacco acid pyrophosphatase (TAP) buffer, 4.5ul water, 1 ul SUPERaseIn, and 1.5ul TAP (Epicentre) were then added and the reaction incubated at 37°C for 1.5 hours. 1ul 300mM MgCl2 and 1ul T4 polynucleotide Kinase (PNK) were added to the reaction for an additional protocols l 30 min. for phosphorylating the 5′-ends, 20ul T4 PNK buffer, 2ul 100mM ATP, 145ul water, 1ul SUPERaseIn, and an additional 2ul of PNK were added for 30 min at 37°C. The reaction was then stopped by addition of 20mM EDTA followed by acid phenol extraction and precipitation.
Plus- and minus-Sarkosyl matched GRO-seq libraries (cells grown in Lis lab) and the Circ-Ligase libraries (grown in Adelman lab for ECF analysis) were made with three sequential bead enrichment steps as above, but a RNA cloning strategy developed by Ingolia et al. (Ingolia, Ghaemmaghami, et al, 2009), was used to prepare the samples for sequencing with the following modifications. PNK treatment to remove 3′-phosphates was performed after the first bead enrichment. 24.5 ul of NRO-RNA was mixed with 3ul 10X PNK buffer (NEB), 1.5 ul T4-PNK, and 1ul SUPERase Inhibitor (Ambion) for 30 minutes at 37°C. Poly-A tailing of RNAs was performed prior to the third bead enrichment, and performed as described in (Ingolia, 2010). Triple-enriched and poly-A tailed nascent RNAs were then reverse transcribed and circularized as in Ingolia et. al. cDNAs were not linearized or PAGE purified after circularization because the range of sizes (∼150-350bp) of the cDNA prevented efficient separation of the circularized and linearized cDNAs. Samples were amplified and PAGE purified as described (Core, Waterfall, and Lis, 2008), and quantified before submission for sequencing.
Data acquisition and analysis
GRO-seq libraries were sequenced on the Illumina Genome Analyzer II, using standard protocol at the Cornell bioresources center (http//www.BRC.cornell.edu). Bowtie (Langmead, Trapnell, et al, 2009) was used to map 26mers, with up to two mismatches to the DM3 version on the Drosophila genome. Reads were also mapped to a representative of repetitive genes transcribed specifically by Pol I (rRNA gene; GenBank accession #: M21017.1), and Pol III (tRNAs; parsed from flybase gene set described below). The rRNA included the extragenic spacers, and tRNAs, were extended +/− 100 bases to account for nascent transcripts that are processed and not part of the annotated tRNA A summary of sequencing yields and the number of reads mapping uniquely to the genome or other annotations is contained in table S1.
Details on gene and enhancer lists, and the analyses contained throughout the manuscript can be found in the Supplemental information.
Supplementary Material
01
02
03
Highlights.
- Unlike mammals, Drosophila promoters lack an upstream divergent peak of Pol II
- Promoter Pol II are functionally distinct from those undergoing active elongation.
- Transcription at Drosophila enhancers can be bidirectional and involves pausing.
- Majority of Pol II detected at promoters is paused, not un-initiated or arrested
Acknowledgments
We would like to thank Charles Danko and Andre Martins for help with R-programming, Peter Kharchenko for providing a Drosophila enhancer list, and Bing Ren for providing a human enhancer list from IMR90 cells. This research was supported in part by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01 ES101987) to KA and by NIH grants GM25232 and HG004845 to JTL.
Footnotes
Author Contributions: LC and KA and JTL conceived the study and designed the experiments. LC produced the GRO-seq datasets. KA and DAG performed RNAi treatments and produced the ChIP data sets. LC, DAG, JJW, KA, DF and HK performed the data analysis. LC, JTL, KA wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Kennedy MA, Nechaev S, Gilchrist DA, Muse GW, Chinenov Y, Rogatsky I. Immediate mediators of the inflammatory response are poised for gene activation through RNA polymerase II stalling. Proc Natl Acad Sci U S A. 2009;106:18207–18212. doi: 10.1073/pnas.0910177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. doi: 10.1126/science.1169628. [DOI] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc Natl Acad Sci U S A. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Xiao H, McCracken S, O'Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domain. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, Bentley DL. mRNA Decapping Factors and the Exonuclease Xrn2 Function in Widespread Premature Termination of RNA Polymerase II Transcription. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S. Transcription. Gene expression--where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, Li L, Gilmour DS, Adelman K. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley DK, Roeder RG. Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J Biol Chem. 1985;260:8163–8172. [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;3:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT. The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. 2002 doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;7339:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;7052:876–80. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;7295:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- Kutach AK, Kadonaga JT. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol Cell Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;3:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol. 1998;63:347–56. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev. 2000;14:792–803. [PMC free article] [PubMed] [Google Scholar]
- Majello B, Napolitano G, Giordano A, Lania L. Transcriptional regulation by targeted recruitment of cyclin-dependent CDK9 kinase in vivo. Oncogene. 1999;18:4598–4605. doi: 10.1038/sj.onc.1202822. [DOI] [PubMed] [Google Scholar]
- Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A. 2004;101:7572–7. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missra A, Gilmour DS. Interactions between DSIF (DRB sensitivity inducing factor), NELF (negative elongation factor), and the Drosophila RNA polymerase II transcription elongation complex. Proc Natl Acad Sci U S A. 2010;107:11301–11306. doi: 10.1073/pnas.1000681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim Biophys Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Pal M, McKean D, Luse DS. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol Cell Biol. 2001;21:5815–25. doi: 10.1128/MCB.21.17.5815-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci U S A. 1993;90:7923–7. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;6:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–567. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- Shopland LS, Hirayoshi K, Fernandes M, Lis JT. HSF access to heat shock elements in vivo depends critically on promoter architecture defined by GAGA factor, TFIID, and RNA polymerase II binding sites. Genes Dev. 1995;9:2756–2769. doi: 10.1101/gad.9.22.2756. [DOI] [PubMed] [Google Scholar]
- Strobl LJ, Eick D. Hold back of RNA polymerase II at the transcription start site mediates down-regulation of c-myc in vivo. Embo J. 1992;11:3307–14. doi: 10.1002/j.1460-2075.1992.tb05409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;7351:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–14. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–59. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03