A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer (original) (raw)
. Author manuscript; available in PMC: 2012 Nov 8.
Published in final edited form as: Nat Genet. 2007 May 27;39(7):870–874. doi: 10.1038/ng2075
Abstract
We conducted a genome-wide association study (GWAS) of breast cancer by genotyping 528,173 single nucleotide polymorphisms (SNPs) in 1,145 cases of invasive breast cancer among postmenopausal white women, and 1,142 controls. We identified a set of four SNPs in intron 2 of FGFR2, a tyrosine kinase receptor previously shown to be amplified and/or over-expressed in some breast cancers, as highly associated with breast cancer and we confirmed this association in 1,776 cases and 2,072 controls from three additional studies. In both association testing and ancestral recombination graph analysis, FGFR2 haplotypes were associated with risk of breast cancer. Across the four studies the association with all four SNPs was highly statistically significant (Ptrend for the most strongly associated SNP, rs1219648 = 1.1 × 10−10; population attributable risk = 16%). Four SNPs at other chromosomal loci most strongly associated with breast cancer in the initial GWAS were not associated with risk in the three replication studies. Our summary results from the GWAS are freely available online in a form that should speed the identification of additional loci conferring risk.
Family history is an established risk factor for breast cancer, yet estimates of the inherited component of the disease are uncertain1. Investigation of multiple-case families segregating breast cancer with Mendelian pattern inheritance led to the identification of the tumor suppressor genes BRCA1 and BRCA2 that account for a substantial proportion of early-onset breast cancer, but a much smaller proportion of late-onset disease2,3. Most late-onset cases occur in the absence of a first-degree family history of breast cancer and are often called “sporadic” cases. Previously, family-based studies have been the primary focus of study in the search for genetic determinants, but with new technologies that enable analysis of hundreds of thousands of SNPs, together with new insights into the structure of genomic variation in the human genome, it is now possible to scan across the genome in an agnostic manner in search of common genetic variants associated with disease risk4. One strategy for conducting a GWAS is to analyze early-onset cases, often enriched with cases with a positive family history of the disease, in order to maximize the opportunity to detect inherited causal variants. Already, two such studies, one of diabetes and one of breast cancer have successfully identified and replicated common genetic variants5,6. Alternatively, GWAS analysis of older subjects may identify common genetic variants associated with sporadic disease, as has been successfully demonstrated for prostate cancer7,8.
We initially genotyped 1183 cases of postmenopausal invasive breast cancer and 1185 individually-matched controls from the Nurses’ Health Study (NHS) cohort, using the Illumina HumanHap500 array, as part of the National Cancer Institute CGEMS (Cancer Genetic Markers of Susceptibility) Project. Cases were identified from a consecutive series of postmenopausal women, unselected for any other characteristics, who were among the 32,826 cohort members who gave a blood sample in 1989–1990 and had not been previously diagnosed with breast cancer, but who were subsequently diagnosed prior to June 1, 2004. Controls were women who were not diagnosed with breast cancer during follow-up, were matched to cases on year of birth and post-menopausal hormone use at blood draw, and were postmenopausal. All cases and controls were self-described Caucasians. Fifty-nine (30 cases and 29 controls) samples were removed from the analysis because of completion rates less than 90% for the 528,173 SNPs that passed quality control. An additional 18 (5 cases and 13 controls) were removed because of unclear identity (or possible contamination) and 4 (3 cases and 1 control) were removed from the analysis due to evidence of intercontinental admixture (Supplemental Figure 1). Thus, the GWAS analysis was performed on 1,145 cases and 1,142 controls.
We analyzed each locus in a logistic regression model using a two-degree of freedom score test with indicator variables for heterozygous and homozygote carriers of the variant allele (for rare variants we collapsed heterozygote and homozygote carrier categories, see Methods). The distribution of the observed P values shows no suggestion of distortion due to population stratification or other sources of bias or distortion of Type I error rates (Supplemental Figures 2a,2b). When we adjusted for matching factors and the top three principal components from an analysis of genetic covariance, the overall distribution of p-values did not significantly change (Komolgorov-Smirnov p=0.34). All loci with unadjusted p<2×10−5 maintained this level of significance after adjustment. This suggests that these associations are not due to population stratification.
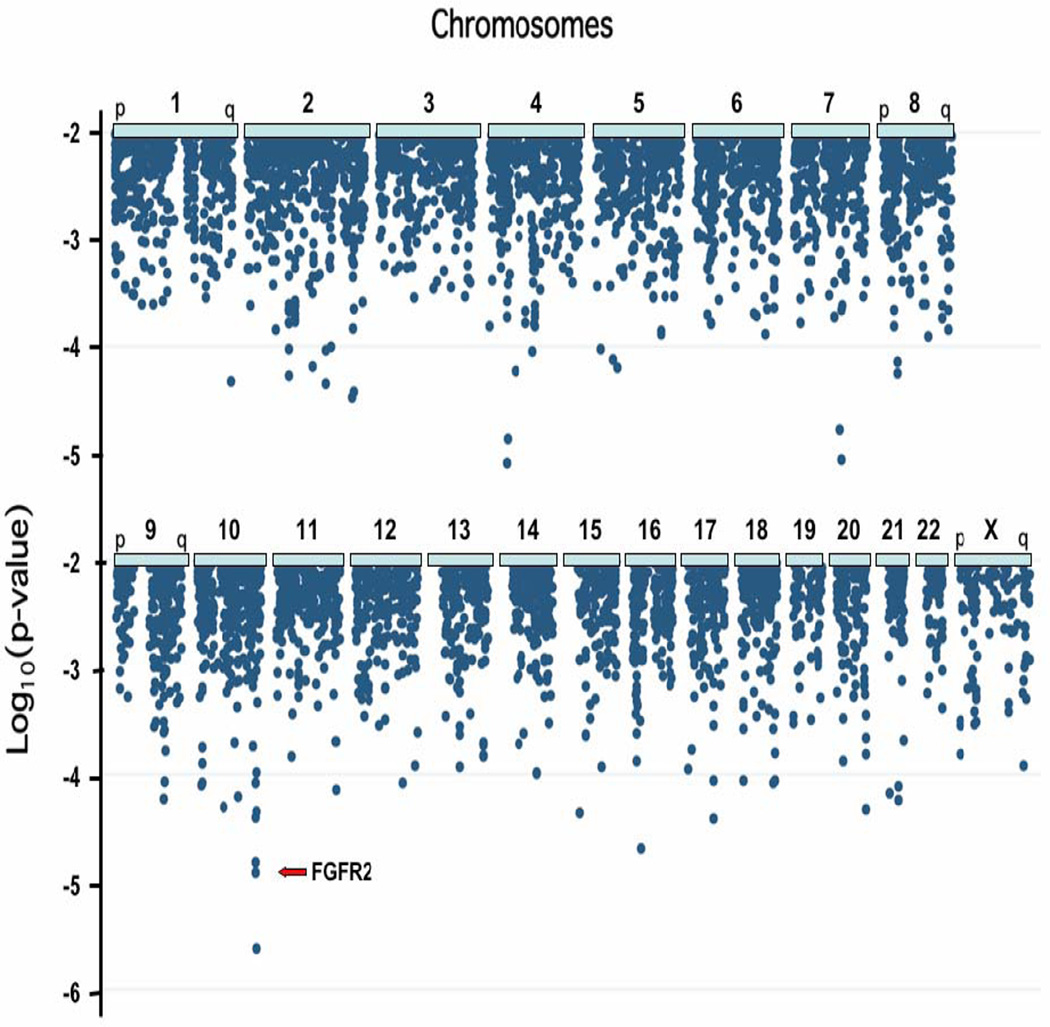
The GWAS identified several genomic locations as potentially associated with breast cancer (Fig 1). Of 528,173 SNP’s tested, two of the most significant P values (rs1219648 and rs2420946, Table 1) were in intron 2 (Fig 2) of the gene FGFR2, a tyrosine kinase receptor previously shown to be important in mammary gland development and neoplasia9; an additional two SNPs in FGFR2 (rs11200014 and rs2981579) were among the 16 most extreme P values from the unadjusted analysis. Modeling all pairwise combinations of the four SNPs and their interactions, as well as haplotypes of the four SNPs, suggested that all four were similar with respect to their association with breast cancer risk, consistent with the very high degree of linkage disequilibrium (each pairwise D’ >0.95 and r2>0.84). None of the other 31 SNPs at the FGFR2 locus were in strong linkage disequilibrium (r2) with the SNPs in intron 2 and none were associated with breast cancer risk (Fig 2). In silica determination of haplotypes (see Methods) indicated 4 common haplotypes, with the AAGT haplotype the most common risk haplotype, present in 43.58% of case chromosomes and 36.87% of control chromosomes in the Nurses’ Health Study (Table 3).
Figure 1. Summary of genome-wide association study results by chromosome.
Association with breast cancer was determined for 528,173 SNPs among 1,145 cases of postmenopausal breast cancer, and 1,142 controls. The x axis represents position on each chromosome from pter (left) to qter (right) ; the y axis shows the P value on a logarithmic scale. Only P values <10−2 are displayed.
Table 1.
The six SNPs with the smallest P values of the 528,173 tested among 1,145 cases of postmenopausal invasive breast cancer and 1,142 controls (full results available at http://cgems.cancer.gov).
| SNP ID | χ2* | P* | ORhet* | ORhomo* | Chromosome | Gene |
|---|---|---|---|---|---|---|
| 1. rs10510126 | 25.92 | 0.0000024 | 0.59 | 0.59 | 10 | |
| 2. rs12505080 | 23.45 | 0.0000081 | 1.22 | 0.51 | 4 | |
| 3. rs17157903 | 23.28 | 0.0000088 | 1.60 | 0.77 | 7 | RELN |
| 4. rs1219648 | 22.63 | 0.000012 | 1.23 | 1.80 | 10 | FGFR2 |
| 5. rs7696175 | 22.40 | 0.000013 | 1.39 | 0.86 | 4 | TLR1,TLR6 |
| 6. rs2420946 | 22.17 | 0.000015 | 1.24 | 1.79 | 10 | FGFR2 |
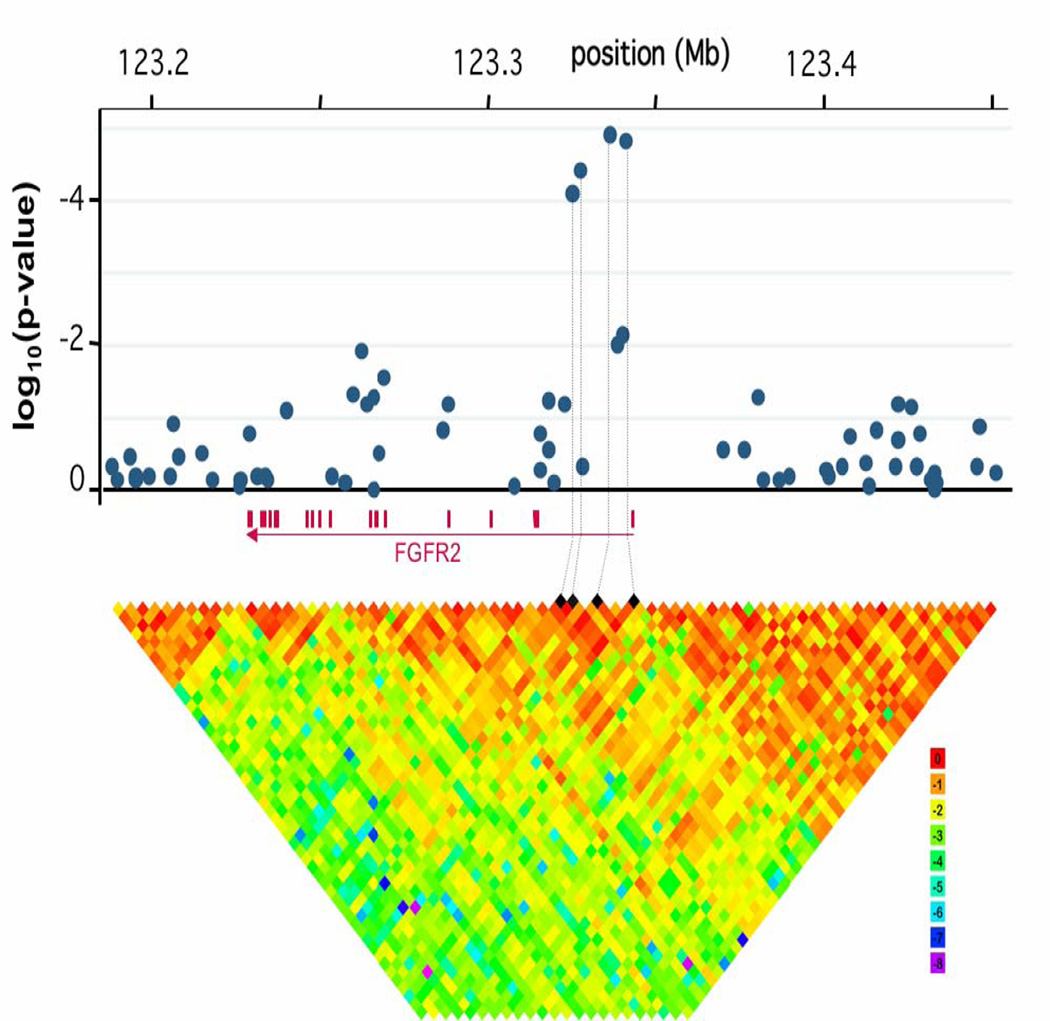
Figure 2. Association Analysis of SNPs Across the FGFR2 gene.
Upper Panel. P-values for association testing drawn from the genome wide association scan covering the FGFR2 gene and 100 Kb 5’ upstream. The presented analysis is based on the two degrees of freedom test corrected for age and the three first principal components of population stratification (see Supplemental materials and Methods).
Lower Panel. Estimates of the squared correlation coefficient, r2, were calculated for each pairwise comparison of SNPs. Log10 r2 was color coded according to the scale shown on the left. The 4 black diamonds indicate the 4 SNPs most strongly associated with breast cancer risk.
Table 3.
Haplotypes for 4 SNPs in intron 2 of FGFR2 (rs11200014, rs2981579, rs1219648 rs2420946) and breast cancer risk, in the individual studies and pooled across studies.
| Nurses Health Study | ||||||
|---|---|---|---|---|---|---|
| Haplotype | cases | controls | OR | 95% | CI | P |
| G-G-A-C | 1195 | 1348 | 1.0 | |||
| A-A-G-T | 998 | 842 | 1.33 | 1.18 | 1.50 | 3×10−6 |
| A-A-A-C | 40 | 47 | 0.96 | 0.62 | 1.48 | 0.85 |
| A-A-G-C | 24 | 19 | 1.40 | 0.76 | 2.57 | 0.28 |
| Rare <1% | 33 | 28 | 1.36 | 0.81 | 2.29 | 0.24 |
| Nurses Health Study 2 | ||||||
|---|---|---|---|---|---|---|
| Haplotype | cases | controls | OR | 95% | CI | P |
| G-G-A-C | 295 | 667 | 1.0 | |||
| A-A-G-T | 276 | 474 | 1.34 | 1.09 | 1.65 | 0.0061 |
| A-A-A-C | 7 | 21 | 0.76 | 0.30 | 1.90 | 0.55 |
| A-A-G-C | 13 | 20 | 1.50 | 0.70 | 3.21 | 0.30 |
| Rare <1% | 12 | 16 | 1.96 | 0.90 | 4.28 | 0.09 |
| PLCO Study | ||||||
|---|---|---|---|---|---|---|
| Haplotype | cases | controls | OR | 95% | CI | P |
| G-G-A-C | 994 | 1140 | 1.0 | |||
| A-A-G-T | 795 | 728 | 1.13 | 0.99 | 1.29 | 0.064 |
| A-A-A-C | 32 | 24 | 1.44 | 0.83 | 2.47 | 0.19 |
| A-A-G-C | 11 | 24 | 0.47 | 0.23 | 0.97 | 0.041 |
| Rare <1% | 21 | 34 | 0.63 | 0.36 | 1.10 | 0.11 |
| ACS CPS-II | ||||||
|---|---|---|---|---|---|---|
| Haplotype | cases | controls | OR | 95% | CI | P |
| G-G-A-C | 583 | 664 | 1.0 | |||
| A-A-G-T | 482 | 406 | 1.38 | 1.16 | 1.65 | 0.00040 |
| A-A-A-C | 21 | 22 | 1.07 | 0.59 | 1.94 | 0.82 |
| A-A-G-C | 6 | 10 | 0.67 | 0.23 | 1.89 | 0.45 |
| Rare <1% | 18 | 10 | 1.98 | 0.91 | 4.31 | 0.084 |
| Pooled across studies | ||||||
|---|---|---|---|---|---|---|
| haplotype | cases | controls | OR | 95% | CI | P |
| G-G-A-C | 3068 | 3718 | 1.0 | |||
| A-A-G-T | 2551 | 2450 | 1.26 | 1.17 | 1.35 | 6×10−10 |
| A-A-A-C | 102 | 114 | 1.09 | 0.83 | 1.43 | 0.55 |
| A-A-G-C | 54 | 74 | 0.88 | 0.61 | 1.27 | 0.50 |
| Rare <1% | 107 | 151 | 0.87 | 0.68 | 1.12 | 0.28 |
To further explore the association signal observed on FGFR2 in the NHS, we performed analyses using inferred ancestral recombination graphs (ARGs). This involves estimating a simple approximation to the distribution of possible genealogies relating the haplotypes of the cases and controls. Using the Margarita program10, we inferred ARGs for 81-SNP haplotypes spanning the FGFR2 gene and its flanking regions (from position 123225862 to position 123471190 on NCBI build35, as shown in Supplemental Figure 3). For every ARG, a putative risk mutation was placed on the marginal genealogy at each SNP position by maximizing the association between the mutation and disease status. We evaluated the significance of this observed association using a maximum of 106 permutations on the phenotypes. Supplemental Figure 3 shows that the permutation p-value was consistently higher than 0.05 over the entire region with the exception of the 20 Kb segment located between 123.32 Mb and 123.34 Mb in intron 2 of FGFR2. In this segment, all marginal trees demonstrated association with a P value lower than 3 ×10−3. The permutation p-values for four notable SNPs were all smaller than 2×10−5, and the frequency of the inferred mutation was similar for all four SNPs. (Supplemental Table 1). These results suggest that there is a single risk locus in the FGFR2 region.
For the six most significant SNPs in the GWAS (two in FGFR2, four at other loci), and for the two additional SNPs that appeared to define the FGFR2 risk haplotype, we attempted to replicate the initial associations in the GWAS in an additional 1,776 cases and 2,072 controls from breast cancer case-control studies nested in three prospective cohorts: the Nurses’ Health Study 2 (NHS2), the Prostate, Lung, Colorectal, and Ovary Cancer Screening Trial (PLCO) Cohort, and the American Cancer Society Cancer Prevention Study-II (CPS-II) (Table 2). For the SNP in FGFR2 most strongly associated with breast cancer in the GWAS (rs1219648), the pooled P value across all four studies = 4.2 × 10−10 for the 2 d.f. model, and 1.1 × 10−10 for the Cochran-Armitage test for trend. Both P values are lower than a threshold for genome-wide significance based on the conservative Bonferroni correction for 528,173 tests with a nominal α=0.05. There was no statistical evidence of heterogeneity of the genotype-specific odds ratios across the studies. The pooled estimate of the odds ratios across studies, compared to wild-type homozygotes, was 1.20 (95% CI 1.07 – 1.34) for heterozygotes, and 1.64 (95%CI 1.42–1.90) for homozygote variants. Across the four studies the SNP with the strongest association (rs1219648) was associated with a Population Attributable Risk (PAR) of 16%11. In each of the three replication studies, and in all studies combined, the AAGT haplotype was the only common haplotype significantly associated with risk of breast cancer (Table 3). The associations were not significantly different across categories of age at diagnosis of breast cancer. In the NHS2, a study involving mainly premenopausal women, the associations were equivalent to those in the other three studies comprised of postmenopausal cases. None of the 4 SNPs at other chromosomal loci was associated with increased risk in the pooled replication studies.
Table 2.
Association of rs1219648 in the Nurses Health Study, Nurses’ Health Study 2, the PLCO study, the ACS CPS-II study, and pooled across studies.
| Study Population(N cases/N controls) | Allele Frequency | ORhet(95% CI) | ORhomo(95% CI) | Ptrend | |
|---|---|---|---|---|---|
| Cases(%) | Controls(%) | ||||
| NHS (1,145/1,141) | 45.54 | 38.47 | 1.24(1.04–1.50) | 1.81(1.43–2.31) | 2.0 × 10−6 |
| NHS2 (302/594) | 48.18 | 40.57 | 1.29(0.95–1.75) | 1.93(1.31–2.86) | 0.002 |
| PLCO (919/922) | 44.50 | 41.49 | 1.06(0.86–1.30) | 1.22(0.94–1.58) | 0.13 |
| ACS CPS-II (555/556) | 44.95 | 37.41 | 1.32(1.02–1.72) | 2.06(1.42–2.97) | 0.0002 |
| Pooled estimates (2,921/3,213) | 1.20(1.07–1.34) | 1.64(1.42–1.90) | 1.1 × 10−10 |
FGFR2 is a tumor suppressor gene that is amplified and over-expressed in breast cancer9,12. Furthermore, alternatively spliced variants of this gene result in differential signal transduction and transformation of mammary epithelial cell lines. Further work is needed to identify the causal variant at this locus.
In a large, three-stage GWAS of breast cancer using the Perlegen platform as the initial genome scan, Easton et al. identified SNPs in FGFR2 as the strongest of their reported associations6. These SNPs were also in intron 2; the association was originally detected with rs2981582, which has an r2 of 1.0 with rs1219648 and rs2420946, 0.97 with rs2981579, and 0.96 with rs11200014 in the HapMap CEU samples, which indicates that we have detected essentially the same association. Substantial resequencing in intron 2 did not discover any SNPs with a more obvious probability of being functionally related to breast cancer risk6. Easton et al. used genotypes from 390 breast cancer cases under age 60, selected to have a strong family history or bilaterality of breast cancer. Our study focused on later-onset, “sporadic” cases. Such consecutive series of cases may be more easily obtained for many diseases, and more generalizable to the most common forms of the disease, so it is reassuring that the use of unselected cases resulted in the identification of the same principal locus.
We focused on the most highly statistically significant associations from our GWAS, identifying variants in FGFR2 as reproducibly associated with breast cancer. Since a subset of true associations would be weakly associated with outcome in any given GWAS, large-scale replication is necessary for confirmation, and some true associations may be missed if they are not carried forward into replication studies13. Multi-stage designs in which potentially associated SNPs from the first stage are carried into additional studies are an economical and scientifically sound approach to cope with the present cost of high throughput genotyping14. In this regard, the precomputed rankings and P values for all the SNPs included in the GWAS conducted in the NHS are freely available from our website (http://cgems.cancer.gov) for others to use in subsequent studies of women with breast cancer.
METHODS
Nurses’ Health Study nested case-control study
The Nurses’ Health Study (NHS) is a longitudinal study of 121,700 women enrolled in 1976. The CGEMS nested case-control study is derived from 32,826 participants who provided a blood sample between 1989 and 1990 and were free of diagnosed breast cancer at blood collection and followed for incident disease until June 1, 2004. Cancer follow-up in the NHS was conducted by personal mailings and searches of the National Death Index. It is estimated that the percentage of true cancers in the cohort captured by this system is greater than 98%15. Permission was requested from all participants diagnosed with cancer to review medical records to confirm the diagnoses and obtain additional information on tumor histology, staging, and other characteristics. All study participants who were menopausal at blood draw with a confirmed diagnosis of invasive breast cancer and had sufficient blood sample available for DNA extraction at the time of case and control selection were included as cases in the CGEMS project. Controls were not diagnosed with breast cancer during follow-up, and were matched to cases based on age at diagnosis, blood collection variables (time of day, season, and year of blood collection, as well as recent (<3 months) use of postmenopausal hormones), ethnicity (all cases and controls are self-reported Caucasians), and menopausal status (all cases were postmenopausal at diagnosis). Menopausal status was defined according to self-reported information on whether a woman’s regular periods had ceased16.
Informed consent was obtained from all participants. The study was approved by the Institutional Review Board of the Brigham and Women’s Hospital, Boston, MA, USA.
Nurses’ Health Study II
The NHS2 began in 1989 when 116,671 female registered nurses (aged 25 to 42) from 14 U.S. states were enrolled17. Blood samples were collected from 29,611 NHS2 participants in 1996–1999. Between blood collection and June 1, 2003 incident breast cancer cases (n=317) were identified via self-report, and confirmed by medical records. To increase power two controls for each case (n=634) were then selected for each case matched case based on year of follow-up, month and year of blood draw, and fasting status at blood draw, as well as age, menopausal status, and ethnicity. Pre-menopausal cases were additionally matched to controls based on luteal day (days before start of next menstrual cycle) to permit analyses of plasma hormone levels.
PLCO Breast Cancer Study
The Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO), a randomized two-arm trial to determine if screening reduced the mortality from these cancers, enrolled during 1993–2001 155,000 men and women, aged 55–74, in 10 U.S. centers18. At study entry, demographic, medical, and lifestyle information and a blood sample were requested from the subjects assigned to the screening arm, including 39,000 women. Incident cancers are ascertained by annual mailed questionnaires (94% completion rate for living Trial participants), as well as searches of the National Death Index. For all reported cancers, medical records were requested from the appropriate hospitals to confirm diagnoses. Of the 37,800 women completing the baseline questionnaire and at least one follow up questionnaire, 83% also provided a blood sample and informed consent. Included in this nested case-control study of breast cancer are all 927 non-Hispanic white women with incident breast cancer reported by June 30, 2005, no previous history of breast cancer, a baseline questionnaire and blood sample, and informed consent. Also selected for this study were 927 non-Hispanic white controls, frequency matched to cases on age at randomization (5-year categories) and year of randomization (two categories), with no report of breast cancer as of June 30, 2005, a baseline questionnaire and blood sample, and informed consent. Because the youngest women in the cohort were 55 y at entry, practically all of the breast cancers diagnosed were postmenopausal.
The American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II)
The American Cancer Society Cancer Prevention Study II Nutrition Cohort (CPS-II) was established in 1992; the cohort includes over 86,000 men and 97,000 women from 21 U.S. states who completed a mailed questionnaire in 199219. At baseline, the cohort was 97% white and the median age of participants was 63 (range: 40–92). Starting in 1997, follow-up questionnaires have been sent to surviving cohort members every other year to update exposure information and to ascertain occurrence of new cases of cancer; a >90% response rate has been achieved for each follow-up questionnaire. Incident cancers are verified through medical records, state cancer registries, or death certificates. From 1998 – 2001, blood samples were collected from a subgroup of 39,376 cohort members.
Cases of postmenopausal breast cancer were frequency matched to controls on single year of age, ethnicity, date of sample collection and menopausal status (all postmenopausal). All controls were selected from individuals who were cancer-free (except for non-melanoma skin cancer) at the beginning of the interval preceding the diagnosis of each case. Similarly, cases were not eligible if they had a history of another cancer other than non-melanoma skin cancer prior to their diagnosis of breast cancer. A total of 555 non-Hispanic white cases of invasive breast cancer and 556 non-Hispanic white controls were included in this analysis.
Genotyping and quality control for NHS
DNA samples were received from the NHS biorepository and visually inspected for adequate fluid in individual tubes. Three measurements of quantification were performed according to the standard procedures at the Core Genotyping Facility of the National Cancer Institute7. These include pico-green analysis, optical density spectrophotometry and real time PCR (http://cgf.nci.nih.gov/dnaquant.cfm). Samples were also analyzed for 15 short tandem repeats and the Amelogenin marker in the Identifiler™ Assay (ABI, Foster City, CA). All samples advanced to genotype analysis completed no less than 13 of the 15 micro-satellite markers.
After final review and sample handling, a total of 1188 cases DNAs, and 1183 controls DNAs were selected for genotyping in CGEMS. Ninety three DNAs were aliquoted twice and five DNAs were aliquoted three times, resulting in the addition of 103 redundant DNAs from the NHS used for quality control. Finally, 23 external QC control DNAs were added. Thus, genotyping was attempted on a total of 2,494 DNA samples.
Genotyping of the CGEMS Breast Cancer Study was performed at the NCI Core Genotyping Facility using the Sentrix® HumanHap550 genotyping assay according to a protocol designated by the manufacturer.
Initial Assessment of sample completion rates
A total of 555,352 SNP genotype assays were attempted on the 2,494 DNA samples using the Illumina HumanHap550 chip. Whenever the completion rate for a sample was below 90%, the sample was assayed a second time. Samples that did not meet the 90% completion threshold after a second attempt were excluded from further analysis. Fifty-nine samples from NHS (30 cases and 29 controls) were excluded from further analysis based on these criteria, which left 2,435 DNAs for the subsequent analyses
Assessment of SNP call rates
A total of 8,706 SNPs (~1.57% overall) failed to provide accurate genotype results due to either no calls or low call rates (<90%). Further quality control analysis was performed on the remaining 546,646 SNPs. An additional 18,473 SNPs with an observed low MAF (<1%) were dropped from the association analysis; thus 528,173 SNPs (95.1%), were maintained in the subsequent analyses.
Summary of completion rate for NHS samples
| Sample Completion rate for | 528,173 SNPs(retained) | 546,646 SNPs(attempted) |
|---|---|---|
| Scan 1 study | 99.754 % | 95.754 % |
| Scan 1 case | 99.756 % | 95.704 % |
| Scan1 control | 99.773 % | 95.799 % |
The genotyping of the 516,383 SNPs with high call rate on the 2,412 NHS DNAs with high completion rate generated 1.26 billion genotype calls. For this set of SNPs and samples, the percentage of missing data was less than 1%.
Concordance rate
The genotype concordance rate for SNP assays was evaluated using the 93 pairs of known NHS duplicated DNAs. These pairs of DNAs were separate aliquots from the same DNA preparation; all met quality control criteria required for the other DNAs, thereby, providing reliable data for comparison. Analysis of the discrepancies within these pairs of DNA revealed similar results to the CEPH DNA duplicates reported in the prostate cancer CGEMS genome-wide association scan. An average concordance rate of 99.985% was observed (50,820,003 concordant genotype calls out of 50,827,468 comparisons). No SNP or DNA was removed from the search for association as a result of this analysis.
Deviation from Hardy–Weinberg proportions in control DNAs
Genotype data for all autosomal SNPs were tested for deviation from Hardy-Weinberg proportions. The analysis was conducted in the NHS control group. Significant deviations were observed for 28,710 SNPs (5.57% of 515,302 SNPs) at the level of P = 0.005 and for 2843 SNPs (0.55%) at P = 0.001. However, none of these SNPs were excluded from analysis since the tests for association applied to such data are valid in the presence of departure from Hardy-Weinberg proportions, although with potentially reduced power when these deviations are due to systematic genotyping errors with equal effects among cases and controls.
Final sample selection for association analysis
For all DNAs the frequency of heterozygote loci on the X chromosome was compatible with a female origin. Eighteen DNA samples (5 cases, 13 controls) had unclear identity as they could not be mapped back unambiguously to previous genotype results from these samples and were excluded. Subsequent inspection of the genotype concordance rate between pairs of DNAs did not disclose unexpected duplicates. Finally, based on two analyses with two independent sets of 7,050 and 7,061 randomly selected SNPs with very low linkage disequilibrium (r2<0.01) using the STRUCTURE program20. Four subjects (3 cases, 1 control) were estimated to be of admixed origin with greater than 15% of either Asian or West African ancestry (described in detail below). These 4 subjects were also removed from subsequent analyses. Thus, the search for associations was performed on a final set of 2,287 unique subjects including 1,145 cases and 1,142 controls.
Statistical analysis
For the initial scan in the Nurses’ Health Study, we analyzed each locus using logistic regression. When the rare homozygote genotype was observed more than fifteen times, we regressed disease status on indicator variables for heterozygous and homozygote carriers of the variant allele. Otherwise, the rare homozygote genotype was collapsed with the heterozygote genotype, and we regressed disease status on variant-allele carrier status. This aggregation of genotypes was performed for 64,589 SNPs.
We calculated unadjusted score tests for genetic association as well as two adjusted score tests. The first adjusted test controlled for age categories (ages <55, 55–59, 60–64, 65–69, 70–74, and >74) and hormone replacement therapy use. The second controlled for age, hormone replacement therapy use, and three eigenvectors from the principal component analysis of genetic covariance (see below). The latter were included in the logistic regression as continuous covariates.
The importance of a second SNP conditional on a given SNP was assessed using nested likelihood ratio tests, comparing the logistic regression model with genotypic indicator variables for the first SNP only to the model with indicator variables for each multi-locus genotype. None of the four FGFR2 SNPs showed any evidence of association with risk of breast cancer after adjusting for any of the other three FGFR2 SNPs.
For the four FGFR2 SNPs, haplotype frequencies and expected haplotype counts for each individual were estimated using a simple expectation-maximization algorithm (implemented in SAS PROC HAPLOTYPE). Haplotype association analyses were performed using the expectation-substitution technique21.
Assessment of population stratification
Two independent sets of 7,050 and 7,061 SNPs with very low linkage disequilibrium (r2<0.01) were analyzed using the STRUCTURE20 program to determine if subjects have an admixed origin greater than 15% of either Asian or West African ancestry (based on HapMap II data)22
The pooled case and control DNAs were analyzed using a set of 14,111 SNPs with very low pair-wise linkage disequilibrium (r2 < 0.01) using the procedure described by Price et al.23. Testing for significance using the Tracy-Widom statistics24 revealed 4 significant principal components at the level of p < 0.05. Inspection of the distribution of the DNAs in the space defined by these components revealed little difference between cases and controls. Nevertheless, statistically borderline significant differences in this distribution for local groups observed in the space defined by the first three components led us to retain these components in the statistical analysis. No difference was observed with the 4th and higher components, which were not retained in the analysis.
Genotyping in replication studies
The same TaqMan assays developed for rs10510126, rs1219648, rs17157903, rs2420946, rs7696175, rs12505080, rs11200014 and rs2981579 were performed at the NCI Core Genotyping Facility and at the DF/HCC Polymorphism Detection Core (Primers and probe sequences available on request).
Statistical analysis of replication studies
For the individual replication studies, we used unconditional logistic regression to fit codominant and additive genetic risk models. For pooled analyses of multiple studies, we used unconditional logistic regression with separate baseline odds parameters for each study. We also adjusted for age in five year intervals. Effect modification by age at diagnosis (comparing 65+ years versus <65, or 55+ versus <55) and by menopausal status at diagnosis was assessed using nested logistic regression models. For these analyses, controls' ages were set to the ages at diagnosis of the matched cases (NHS, NHS2, ACS) or age at censoring (PLCO). We calculated Population Attributable Risk (PAR) using the method of Bruzzi et al11.
ARG analysis with 81 SNPs after 106 Permutations
In order to identify the at-risk haplotypes, the most likely pair of haplotypes present in each case and control was inferred using PHASE and this information was used to generate one hundred ARG. On each ARG, the position of the putative functional mutation that best explains the distribution of cases and controls identifies the haplotypes that harbor the new functional allele. This allele is declared at-risk if its frequency is higher in the cases than in the controls. The new allele was observed deleterious in 46% of the ARG and therefore this analysis was not conclusive. The ARG analysis was however more successful in reproducibly differentiating between the at-risk and protective haplotypes. (Supplemental Figure 3).
Identification of protective and at-risk haplotypes for the FGFR2 susceptibility locus
The most likely pair of haplotypes present in each case and control of the NHS was inferred using PHASE and an ARG analysis was performed to infer the presence of either the protective or of the at-risk allele for each haplotype. For marginal trees at all 4 positions with the lowest permutation p values, the same four of 8 haplotypes were inferred with a probability higher than 97% to carry the at-risk allele (Supplementary Table 1). Similarly, three haplotypes were predicted to carry the protective allele with the same confidence. These two groups of haplotypes systematically differ at 4 of the 9 SNPs positions. One rare haplotype, haplotype-8, with a population frequency of 2%, possibly results from a recombination between one haplotype of each group. Its centromeric region contains 2 alleles specific to the protective group of haplotypes as its telomeric region contains 2 alleles specific to the at-risk group of haplotypes. Unfortunately, the ARG analysis is unable to ambiguously determine whether this rare haplotype carries the at-risk or the protective allele. We note however that haplotype-8 may correspond to haplotype 2111111 of supplementary table 3 of Easton et al6.The latter is shown to be associated with a lower risk of breast cancer. Confirmation of the status of this haplotype would suggest that the functional polymorphisms should lie centromeric (i.e. on the 3’side) to rs12119648. Results of the ARG analysis are shown for all haplotypes with frequencies higher than 1% defined by the following 9 contiguous SNPs : rs3750817, rs11200014, rs2981579 ,rs17542768, rs1219648, rs1219643, rs17102287, rs2420946, rs1047111. The alleles are color coded : red is the ancestral allele, blue is the new allele and black is when this information is unclear. The probability of a haplotype to carry the at-risk allele is evaluated as the ratio of the number of times it is predicted to carry the at-risk allele over the total number of prediction made for this haplotypes for all marginal trees at the indicated position.
ACKNOWLEDGEMENTS
The Nurses’ Health Studies are supported by NIH grants CA 65725, CA87969, CA49449, CA67262, CA50385 and 5UO1CA098233. The authors thank Barbara Egan, Lori Egan, Helena Judge Ellis, Hardeep Ranu, and Pati Soule for assistance, and the participants in the Nurses’ Health Studies.
The PLCO study is supported by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics and contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS. The authors thank Dr Philip Prorok, Division of Cancer Prevention, National Cancer Institute; the Screening Center investigators and staff of the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO); Mr. Tom Riley, Mr. Craig Williams, and staff, Information Management Services, Inc.; Ms. Barbara O’Brien and staff, Westat, Inc.; and Dr. Bill Kopp, Mr. Tim Sheehy, and staff, SAIC-Frederick. Most importantly, we acknowledge the study participants for their contributions to making this study possible.
The ACS study is supported by UO1 CA098710. We thank Cari Lichtman for data management and the participants on the CPS-II.
We thank Mark Minichiello for providing the Margarita program and fruitful discussions.
References
- 1.Pharoah PD, et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat Genet. 2002;31:33–36. doi: 10.1038/ng853. [DOI] [PubMed] [Google Scholar]
- 2.Antoniou A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risch HA, et al. Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst. 2006;98:1694–1706. doi: 10.1093/jnci/djj465. [DOI] [PubMed] [Google Scholar]
- 4.Hirschhorn JN, Daly MJ. Genome-wide association studies for common diseases and complex traits. Nat Rev Genet. 2005;6:95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 5.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 6.Easton D, et al. A genome-wide association study identifies multiple breast cancer susceptibility loci. Nature. (in press). [Google Scholar]
- 7.Daugherty SE, et al. RNASEL Arg462Gln polymorphism and prostate cancer in PLCO. Prostate. 2007 doi: 10.1002/pros.20537. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007 doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 9.Grose R, Dickson C. Fibroblast growth factor signaling in tumorigenesis. Cytokine Growth Factor Rev. 2005;16:179–186. doi: 10.1016/j.cytogfr.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Minichiello MJ, Durbin R. Mapping trait loci by use of inferred ancestral recombination graphs. Am J Hum Genet. 2006;79:910–922. doi: 10.1086/508901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruzzi P, Green SB, Byar DP, Brinton LA, Schairer C. Estimating the population attributable risk for multiple risk factors using case-control data. Am J Epidemiol. 1985;122:904–914. doi: 10.1093/oxfordjournals.aje.a114174. [DOI] [PubMed] [Google Scholar]
- 12.Moffa AB, Ethier SP. Differential signal transduction of alternatively spliced FGFR2 variants expressed in human mammary epithelial cells. J Cell Physiol. 2007;210:720–731. doi: 10.1002/jcp.20880. [DOI] [PubMed] [Google Scholar]
- 13.Chanock S, et al. What constitutes Replication of a Genotype-Phenotpe Association? Summary of an NCI-NHGRI Working Group. Nature. (in press). [Google Scholar]
- 14.Wang H, Thomas DC, Pe'er I, Stram DO. Optimal two-stage genotyping designs for genome-wide association scans. Genet Epidemiol. 2006;30:356–368. doi: 10.1002/gepi.20150. [DOI] [PubMed] [Google Scholar]
- 15.Tworoger SS, Eliassen AH, Sluss P, Hankinson SE. A prospective study of plasma prolactin concentrations and risk of premenopausal and postmenopausal breast cancer. J Clin Oncol. 2007;25:1482–1498. doi: 10.1200/JCO.2006.07.6356. [DOI] [PubMed] [Google Scholar]
- 16.Colditz GA, Rosner B. Cumulative risk of breast cancer to age 70 years according to risk factor status: data from the Nurses' Health Study. Am J Epidemiol. 2000;152:950–964. doi: 10.1093/aje/152.10.950. [DOI] [PubMed] [Google Scholar]
- 17.Eliassen AH, Tworoger SS, Mantzoros CS, Pollak MN, Hankinson SE. Circulating insulin and c-peptide levels and risk of breast cancer among predominately premenopausal women. Cancer Epidemiol Biomarkers Prev. 2007;16:161–164. doi: 10.1158/1055-9965.EPI-06-0693. [DOI] [PubMed] [Google Scholar]
- 18.Hayes RB, et al. Etiologic and early marker studies in the prostate, lung, colorectal and ovarian (PLCO) cancer screening trial. Controlled Clinical Trials. 2000;21:341S–355S. doi: 10.1016/s0197-2456(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 19.Stevens VL, Rodriguez C, Pavluck AL, Thun MJ, Calle EE. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II Nutrition Cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:1226–1228. doi: 10.1158/1055-9965.EPI-05-0930. [DOI] [PubMed] [Google Scholar]
- 20.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol. 2005;28:261–272. doi: 10.1002/gepi.20061. [DOI] [PubMed] [Google Scholar]
- 22.A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Patterson N, et al. Population Structure and Eigenanalysis. PLoS Genet. 2006;2:2076–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]