Medicinal plants extracts affect virulence factors expression and biofilm formation by the uropathogenic Escherichia coli (original) (raw)
Abstract
Medicinal plants are an important source for the therapeutic remedies of various diseases including urinary tract infections. This prompted us to perform research in this area. We decided to focus on medicinal plants species used in urinary tract infections prevention. The aim of our study was to determine the influence of Betula pendula, Equisetum arvense, Herniaria glabra, Galium odoratum, Urtica dioica, and _Vaccinium vitis_-idaea extracts on bacterial survival and virulence factors involved in tissue colonization and biofilm formation of the uropathogenic Escherichia coli rods. Qualitative and quantitative analysis of plant extracts were performed. Antimicrobial assay relied on the estimation of the colony forming unit number. Hydrophobicity of cells was established by salt aggregation test. Using motility agar, the ability of bacteria to move was examined. The erythrocyte hemagglutination test was used for fimbriae P screening. Curli expression was determined using YESCA agar supplemented with congo red. Quantification of biofilm formation was carried out using a microtiter plate assay and a spectrophotometric method. The results of the study indicate significant differences between investigated extracts in their antimicrobial activities. The extracts of H. glabra and V. vitis_-idaea showed the highest growth-inhibitory effects (p < 0.05). Surface hydrophobicity of autoaggregating _E. coli_ strain changed after exposure to all plant extracts, except _V. vitis_-_idaea_ (_p_ _> 0.05). The B. pendula and U. dioica extracts significantly reduced the motility of the E. coli rods (p < 0.05). All the extracts exhibited the anti-biofilm activity.
Keywords: UPEC, Plant extracts, Biofilm, Virulence factors
Introduction
Urinary tract infections (UTIs) are among the most common bacterial infectious diseases in human population. E. coli are the most predominant pathogens responsible for 80–90 % of community-acquired and 30–50 % of hospital-acquired UTIs [1]. Uropathogenic E. coli strains are equipped with a particular set of virulence determinants allowing them to colonize distinct sites in the urinary system. Hydrophobic cell surface, fimbriae P, curli fiber, ability to move allow them to successfully initiate infections. Bacterial cells after initial attachment to host tissues begin to grow and spread as a monolayer on the surface to form microcolonies that can disaggregate or create biofilm. E. coli biofilms are frequently described for catheter-associated, chronic and recurring UTIs. These structures protect the bacteria against the mechanical flow of urine, host and antibiotics [2, 3].
It is well known that herbal remedies are used by different human cultures since 1,000 of years. Some of those plant natural products are essential in prevention and treatment of UTIs. The most commonly known in this field are cranberry products; however, antibacterial properties of many other plants are also well known [4]. In the present study, we focused on leaf extracts of plants used traditionally in prevention of UTIs. Up to our knowledge, reports describing their antibacterial activities, especially against E. coli and urinary tract infections induced by these bacteria are highly limited and this fact prompted us to perform current study.
The purpose of this study was to evaluate the activity of Betula pendula (silver birch), Equisetum arvense (common horsetail), Herniaria glabra (smooth rupturewort), Galium odoratum (sweet woodruff), Urtica dioica (common nettle), and _Vaccinium vitis_-idaea (lingonberry) leaves extracts against uropathogenic E. coli rods as well as their impact exerted on virulence factors and biofilm formation.
Materials and methods
Plant materials
Six plant species commonly used in folk medicine in Poland were selected. Herbs were purchased from two herbs confectioning factories: “FLOS”, general partnership (Mokrsko, Poland) with marketing authorization numbers as follows: _H. glabra_—13736, _G. odoratum_—13561, and “KAWON-HURT”, general partnership (Gostyń, Poland) with marketing authorization numbers as follows: _B. pendula_—IL-3332/LN, _V. vitis_-_idaea_—IL-3333/LN, _E. arvense_—IL-3347/LN, _U. dioica_—R/2183.
Preparation of extracts
Purchased dry herbs were ground into powder in an electric blender. 20 g of each leaf powder was dissolved in 180 mL of distilled water in a glass bottles, heated to 85 °C in a water bath and kept at this temperature with shaking for 8 h. After cooling, the liquid was filtered through the Whatman No. 1 filter paper. The filtrates were then condensed and dried in smaller glass bottles at 37 °C for 48 h. Then, the dried extracts were dissolved in distilled water to obtain concentrations ranging from 0.125 to 20.0 mg/mL.
UPLC–Q-TOF–MS conditions
Compounds identification was performed on an Acquity ultra-performance liquid chromatography (UPLC) system coupled with a quadrupole-time of flight (Q-TOF) MS instrument (UPLC/Synapt Q-TOF MS, Waters Corp., Milford, MA, USA) with an electrospray ionization (ESI) source. Separation was achieved on a Acquity™ BEH C18 column (100 mm × 2.1 mm i.d., 1.7 μm; Waters). Detection wavelengths were set at 254, 280, 320, 380 and 520 nm. Mobile phase was a mixture of 4.5 % formic acid (A) and acetonitrile (B). The gradient program was as follows: initial conditions—99 % (A), 12 min—75 % (A), 12.5 min—100 % (B), 13.5 min—99 % (A). The flow rate was 0.45 mL/min and the injection volume was 5 μL. The column was operated at 30 °C. The major operating parameters for the Q-TOF MS were set as follows: capillary voltage 2.0 kV, cone voltage 45 V, cone gas flow of 11 L/h, collision energy 50 eV, source temperature 100 °C, desolvation temperature 250 °C, collision gas, argon; desolvation gas (nitrogen) flow rate, 600 L/h; data acquisition range, m/z 100–1,000 Da; ionization mode, negative. The data were collected by Mass-Lynx™ V 4.1 software.
Bacterial strain
Escherichia coli clinical strain was isolated from the urine of patient with pyelonephritis, hospitalized in the Academic Hospital in Wrocław. The species affiliation of the examined strain was confirmed using the API-20E test kit (BioMérieux, Warsaw, Poland). The strain was maintained on Mueller–Hinton agar slopes (Oxoid) at 4 °C.
Phylogenetic classification and virulence-associated genes carriage
The presence of selected nucleotide sequences was verified by PCR on total DNA isolated from bacterial overnight culture using GeneMATRIX Bacterial & Yeast Genomic DNA Purification Kit (EURx, Poland). All PCR analyses were performed using DreamTaq™ DNA polymerase (Fermentas, Germany). Phylogenetic group was determined using primers specific for two genes (chuA and yjaA) and an anonymous DNA fragment (TspE4.C2) according to the method of Clermont et al. [5]; however, instead multiplex PCR, yjaA sequence was amplified separately. Strain was screened for the presence of adhesins (papC, sfa, afa, csgA), siderophore (aer), toxins (hlyA, cnf1) and biofilm-related genes (luxS, mcbA, mqsR, sdiA, and ant43). Sequence coding for 16SrRNA was used as a positive control. The characteristics of all used primers, as well as amplicons length, are listed in Table 1.
Table 1.
Primer sequences used in PCR
| Gene | Primer name | Sequence (5′–3′) | Amplicon size (bp) | Reference or gene bank accession no (genome region) |
|---|---|---|---|---|
| chuA | ChuA.1 | GACGAACCAACGGTCAGGAT | 279 | [5] |
| ChuA.2 | TGCCGCCAGTACCAAAGACA | |||
| yjaA | YjaA.1 | TGAAGTGTCAGGAGACGCTG | 211 | [5] |
| YjaA.2 | ATGGAGAATGCGTTCCTCAAC | |||
| TspE4.C2 | TspE4C2.1 | GAGTAATGTCGGGGCATTCA | 152 | [5] |
| TspE4C2.2 | CGCGCCAACAAAGTATTACG | |||
| papC | pap1 | GACGGCTGTACTGCAGGGTGTGGCG | 328 | [6] |
| pap2 | ATATCCTTTCTGCAGGGATGCAATA | 336 | ||
| pap3 | GCAACAGCAACGCTGGTTGCATCAT | |||
| pap4 | AGAGAGAGCCACTCTTATACGGACA | |||
| sfa | sfa1 | CTCCGGAGAACTGGGTGCATCTTAC | 410 | [6] |
| sfa2 | CGGAGGAGTAATTACAAACCTGGCA | |||
| afa | afa1 | GCTGGGCAGCAAACTGATAACTCTC | 750 | [6] |
| afa2 | CATCAAGCTGTTTGTTCGTCCGCCG | |||
| csgA | csgAF | GTAGCAGCAATTGCAGCAATCG | 383 | AE014075 |
| csgAR | TTAGATGCAGTCTGGTCAACAG | (1247944..1248402) | ||
| aer | aer1 | TACCGGATTGTCATATGCAGACCG | 602 | [6] |
| aer2 | AATATCTTCCTCCAGTCCGGAGAAG | |||
| hlyA | hlyA1.10f | GCTGCAAATAAATTGCACTCAG | 665 | [7] |
| hlyA2.10r | CCCTGCACCGATATTATCAAG | |||
| cnf1 | cnf1 | AAGATGGAGTTTCCTATGCAGGAG | 498 | [6] |
| cnf2 | CATTCAGAGTCCTGCCCTCATTATT | |||
| ant43 | ant43_F | TGGCACCATCAGCCTGCGTG | 127 | AE014075 |
| ant43_R | CGTACCACTGTTGCCGGCGT | (1225454..1228729) | ||
| luxS | luxS_F | CGGCAGCCCATTGGCGAGAT | 178 | AE014075 |
| luxS_R | TGAACACCCCGCATGGCGAC | (3096814..3097329) | ||
| mcbA | mcbA_F | CGCCTTGTTCGCGCGCTTTT | 138 | NC_000913 |
| mcbA_R | TCACGGCTTATGCCGCGCAA | (841019..841279) | ||
| mqsR | mqsR_F | GCCTGTAACAAGCCTGGGTCTGT | 187 | U00096 |
| mqsR_R | TGTCAATGCCGGGCAAGTTCGT | (3166270..3166566) | ||
| sdiA | sdiA_F | ATGGTACCGGGTGGCGGACA | 130 | AE014075 |
| sdiA_R | TGGCGTCGCACGATGCTGTT | (2144786..2145520) | ||
| 16srRNA | rRNA16SF | AGAGTTTGATCATGGCTCAG | 919 | [8] |
| rRNA16SR | CCGTCAATTCATTTGAGTTT |
PCR products visualization and analysis
PCR amplification of the DNA was confirmed by running 20 μL of the PCR products on a 2 % agarose gel. Gel images were visualized and analyzed using the Quantity One system (Bio-Rad).
Antibacterial activity of plant extracts
The antimicrobial activity of plant extracts was determined as described below. Briefly, the strain was grown overnight, and then bacterial cells were transferred to fresh Mueller–Hinton broth (MHB, BIOCORP, Warsaw, Poland) and incubated at 37 °C for 30 min. Following incubation, the bacterial cells were centrifuged (4,000 rpm for 20 min) and suspended in phosphate-buffered saline (PBS) to reach the final density 0.5 in McFarland scale. Bacterial suspension and plant extracts were mixed together to obtain following concentrations: 0.125, 0.25, 0.5, 1.0, 5.0, 10.0, 15.0, and 20.0 mg/mL of extract in sample. All samples were incubated at 37 °C for 24 h, then diluted and cultured on nutrient agar plates (BIOMED, Warsaw, Poland). After 24-h incubation at 37 °C, the number of colony forming units (c.f.u.) was counted. Control sample contained no plant extracts was taken as 100 % survival. The experiment was repeated three times. In each experiment, six repeats for control as well as examined, samples were taken.
Effect of plant extracts on hydrophobicity of bacterial cells
Bacterial cells were incubated with plant extracts for 24 h at 37 °C. After incubation, they were washed three times in PBS. After last centrifugation, samples were diluted to obtain final optical density (measured at 470 nm) of 1.0. Untreated bacterial strain was assessed as a control. The salt aggregation test (SAT) of ammonium sulfate was used [9]. The control and treated suspensions (20 μL) were mixed with a series of dilutions of ammonium sulfate (20 μL) ranging from 0 to 3.2 M. The lowest concentration of ammonium sulfate at which bacterial aggregation was visible was determined. Each test was repeated three times. Based on the SAT values, the bacterial cell surface was classified as: <0.2 M—very strong hydrophobic, 0.4–1.0 M—strong hydrophobic, 1.2–1.6 M—hydrophobic, >1.8 M—hydrophilic.
Effect of plant extracts on swimming motility
Bacterial cells were incubated with plant extracts and washed as described in previously. The final density of bacterial suspension was adjusted to 0.5 in McFarland scale. 10 μL of suspension was inoculated onto motility plates (1 % tryptone, 0.25 % NaCl, and 0.3 % agar). The plates were incubated at 37 °C for 24 h and the diameters of the swimming zone were measured [10]. Presented results are the mean of three experiments. In each experiment, four repeats for control as well as examined samples were taken.
Effect of plant extracts on hemagglutination and expression of P fimbriae
Assays were performed on each strain grown overnight with plant extracts. After washing thrice in PBS, the final density of bacterial suspension was adjusted to 0.5 in McFarland scale. P fimbriae expression was confirmed by the hemagglutination of 3 % erythrocytes from human with blood group 0 in the presence or absence d-mannose [11]. The experiment was repeated three times.
Effect of plant extracts on curli expression
Effect of plant extracts on curli expression was assessed according to Hammar et al. [12]. Bacterial suspension was prepared as described in paragraph devoted to hydrophobicity determination. 10 μL of suspension was inoculated onto plate containing YESCA agar supplemented with congo red (CRI) and the same subMICs. Curli-producing E. coli bound congo red dye and formed red colonies, whereas curli-negative bacteria formed white colonies. Control culture contained no plant extracts. The experiment was repeated three times.
Biofilm formation assay and quantification
The capacity to form biofilms was assayed in microtiter plates essentially as described by O’Toole and Kolter [13] with slight modification. Briefly, cells were initially grown for 24 h in MHB at 37 °C. Subsequently, 150 μL overnight culture was added to 96-well polystyrene microtiter plates and incubated for 24 h at 37 °C. Unattached bacterial cells were then removed from the culture medium, and the biofilm was stained with 0.1 % (w/v) crystal violet for 15 min (this dye stains the cells but not the polystyrene). The excess crystal violet dye was washed out, and this was followed by washing the samples three times with distilled water. To release the dye, 200 μL 96 % ethanol was added to the wells. Subsequently, 125 μL sample was transferred to another well, and the optical density (OD) was measured at 495 nm using a plate reader (ANALCO-GBG STAT-FAX 2100). In each plate, four wells were used as blanks containing MHB medium. On the basis of ODs of bacterial biofilms, E. coli strains were classified into four categories [14]. The cut-off OD (ODc) was defined as three standard deviations (SD) above the mean OD of the negative control. Strains were classified as follows: OD ≤ ODc no biofilm producer, ODc < OD ≤ 2 × ODc weak biofilm producer, 2 × ODc < OD ≤ 4 × ODc moderate biofilm producer; 4 × ODc < OD strong biofilm producer. In our study the ODc value was 0.003.
Effect of plant extracts on biofilm formation
Effect of plant extracts on biofilm formation was performed according to Di Bonaventura et al. [15]. Due to the smallest differences in survival of treated bacteria, the extracts concentrations 0.125 mg/mL were used in this experiment. Samples were prepared in microtiter plate wells by adding the appropriate volume of plant extract to 200 μL of MHB containing 20 μL of culture of bacteria (0.5 in McFarland scale). After 1–10 days of incubation, biofilm formation was measured as described in the previous section. Plant extract-free medium was used as a control. The results are given as mean values from three separate experiments. In each experiment, seven repeats for control as well as examined samples were taken.
Statistical analysis
The differences in growth, motility and biofilm formation between rods exposed to plant extracts and unexposed were analyzed by a parametric t test for independent samples. Non-parametric Chi square test was used to correlate the concentrations of tested plant extracts and cell surface hydrophobicity. All tests were analyzed at the significance level p < 0.05 using Statistica 7.1.
Results
Qualitative and quantitative analysis of plant extracts
Identification of compounds was performed on the basis accurate mass searching, fragmentation analysis (MS/MS), comparison of accurate mass and matching of MS/MS pattern with standards and with data published in literature. The identified compounds and their LC/MS data are shown in Table 2. In the extract of B. pendula, 3,4′-dihydroxypropiophenone-3-β-d-glucoside (DHPPG), belonging to the group of propiophenone derivatives, was the main constituent. The other components present in significant amounts in this extract were quercetin derivatives belonging to flavonols (quercetin-3-galactoside, quercetin-3-glucuronide). Caffeic acid derivatives and _p_-coumaric acid derivatives were also identified. In the extract of E. arvense, three flavonols (quercetin dihexoside, kaempherol dihexoside, kaempherol-dirhamnosyl-hexoside) and four phenolic acids (protocatechuic, caftaric, ferulic, caffeic acids) were detected. The predominant compounds are caftaric acid and its derivatives (dimer and hexoside). In the extract of H. glabra, we found caffeoylquinic and feruloylquinic isomers, flavonols (quercetin, kaempherol and isorhamnetin derivatives) and iridoids. Phenolic acids (protocatechuic acid, caffeoylquinic isomers), flavonols (quercetin and kaempherol derivatives) and iridoids were the main compounds of G. odoratum extract. The dominant components of the U. dioica extract were phenolic acids (protocatechuic, ferulic, _p_-coumaric, and dicaffeoylquinic acids). Flavonols (quercetin derivatives), phenolic acids (derivatives of caffeoylquinic, caffeoyl-hexose-hydroxyphenol and coumaroyl-hexose-hydroxyphenol acids), procyanidins (A and B dimmers) and iridoids were the three most dominant compounds extracted from _V. vitis_-idaea.
Table 2.
Compounds identified in plant extracts by using negative ions in LC–MS and MS/MS
| Parent ion [M−H]− (m/z) | Daughter ion MS/MS (m/z) | Compound | B. pendula | E. arvense | H. glabra | G. odoratum | U. dioica | _V. vitis_-idaea |
|---|---|---|---|---|---|---|---|---|
| Flavonols and derivatives | ||||||||
| 269.1342 | Apigenin derivative | x | ||||||
| 433.065 | 300.0171/271.0211/255.0303/179.0020 | Quercetin-3-arabinopyranoside | x | x | ||||
| 433.1033 | 300.049 | Quercetin-xyloside | x | x | ||||
| 447.0743 | 301.0319/300.0207 | Quercetino-3-rhamnoside | x | x | ||||
| 463.0887 | 30.0319/300.0313 | Quercetin-glucoside | x | x | ||||
| 463.0931 | 300.0242/301.0354 | Quercetin-galactoside | x | |||||
| 477.1022 | 301.0354 | Quercetin-glucuronide | x | |||||
| 477.1263 | 175.0372/301.0273/300.0348 | Quercetin derivative | x | |||||
| 591.1436 | 529.1354/489.1132/447.1003/301.0461/300.0242 | Quercetin-3-_O_-(4 bis-3-hydroxy-3-methylglutaryl)-α-rhamnoside | x | |||||
| 593.1606 | 285.0429 | Kaempherol ramnohexoside | x | |||||
| 609.1331 | 447.0916/285.0083 | Kaempherol diglycoside | x | |||||
| 609.1533 | 463.0931/301.0319 | Rutin | x | x | x | |||
| 623.1342 | 497.1168/315.0440/107.4882 | Isorhamnetin rhamnose-hexose | x | |||||
| 625.1591 | 463.0623/301.0176 | Quercetin dihexoside | x | |||||
| 737.1927 | 596.1407/284.0348 | Hexoside-rhamnoside kaempferol and hydroxyferulic acid derivative | x | |||||
| 755.3019 | 593.2354/447.0786/285.0498 | Kaempherol-di-rhamnosyl-hexoside | x | x | ||||
| 771.2246 | 609.1735/285.0049 | Kaempherol-trihexoside | x | |||||
| 771.3156 | 609.2391/463.1460/301.0603 | Quercetin-hexoside-rhamnoside-hexoside | x | |||||
| Flavan-3-ols and procyanidins | ||||||||
| 289.0688 | 245.0659/165.0118/137.0206/125.0218 | Epicatechin | ||||||
| 289.0827 | 245.0787/203.0635/165.0065 | Catechin | ||||||
| 575.1207 | 539.0969/449.0889/407.0727/289.0688/285.0325 | Dimer procyanidin A | x | |||||
| 577.1201 | 407.0645/289.0758 | Dimer procyanidin B | x | |||||
| 577.1594 | 289.0584/245.0370/125.0241 | Dimer procyanidin B | x | |||||
| 577.1933 | 289.0897 | Dimer procyanidin B | x | |||||
| 863.163 | 711.1367/693.1317/573.0953/451.1297/411.0632/289.0723 | Trimer procyanidin A/B2 | x | |||||
| Iridoids | ||||||||
| 389.0818 | 271.0616/124.9967/107.9938 | Iridoide | x | |||||
| 389.0858 | 217.0116/198.9950/155.0041 | Iridoide | x | |||||
| 389.0939 | 191.0100/147.0193 | Aucubioside | x | |||||
| 389.0939 | 209.0213/183.0417/165.0302 | Iridoide | x | |||||
| 499.1633 | 337.1049/235.0395 | Iridoide | x | |||||
| 535.1362 | 371.1109/329.1297/311.0642/191.0326/163.0406 | Coumaroyl iridoide | x | |||||
| 553.1731 | 389.11 | Iridoide | x | |||||
| Phenolic acids | ||||||||
| 153.0027 | 109.0022/116.8969 | Protocatechuic acid | x | x | ||||
| 163.038 | 119.0533 | _p_-Coumaric acid | x | x | ||||
| 173.0419 | 134.0375 | 3-FQA feruloylquinic acid | x | |||||
| 191.0072 | 167.9749/155.9433/110.9921 | Quinic acid derivative | x | |||||
| 191.0524 | 179.0513/113.0108/119.0220/105.0031 | Quinic acid | x | x | ||||
| 311.057 | 227.9305/179.0267/148.9988/135.0298 | Caffeoyl tartrate | x | |||||
| 315.0549 | 152.9976/109.0022 | Protocatechuic acid glucoside | x | |||||
| 325.0358 | 193.0236/135.0131 | Ferulic acid pentose derivative | x | |||||
| 325.1244 | 163.0275/119.0309 | Coumaroylglucose | x | |||||
| 337.0937 | 191.0468/163.0406/119.0533 | 3-_p_-Coumaroylquinic acid | x | x | ||||
| 337.1012 | 173.0419 | 5-_p_-Coumaroylquinic acid | x | |||||
| 337.1049 | 191.0496 | _p_-Coumaric acid derivative | x | |||||
| 337.1162 | 191.0524/163.0197 | Coumaroylquinic acid | x | |||||
| 337.1162 | 173.0365/163.0249 | Coumaroylquinic acid | x | |||||
| 341.1113 | 195.0507/163.0249/119.0354 | _p_-Coumaric acid derivative | x | |||||
| 345.1148 | 193.0521/146.9424 | Ferulic acid derivative | x | x | ||||
| 353.0648 | 191.0298/147.0367 | 4′-Caffeoylquinic acid | x | x | x | x | ||
| 353.0918 | 191.0666/179.0321/173.0419 | Caffeoylquinic acid | x | x | x | |||
| 353.1033 | 191.0439/179.0458 | 5′-Caffeoylquinic acid | x | x | x | x | x | |
| 367.0899 | 173.0419 | 4 FQA tri-feruloylquinic acid trans | x | |||||
| 367.0923 | 193.0464/191.0666 | Feruloylquinic acid isomer | x | |||||
| 367.1002 | 173.0419 | 4 FQA tri-feruloylquinic acid cis | x | |||||
| 367.108 | 191.0524 | 5 FQA tri-feruloylquinic acid | x | |||||
| 417.1147 | 307.0882/187.0564/163.0302/145.0259/119.0533 | Coumaroyl-hexose hydroxyphenol | x | |||||
| 417.1231 | 307.0739/163.0354/145.0284/119.0465 | Coumaroyl-hexose-hydroxyphenol | x | |||||
| 433.1075 | 323.0721/203.0314/179.0294/161.0241/135.0393 | Caffeoyl-hexose-hydroxyphenol | x | |||||
| 473.065 | 311.0389/179.0431/149.0138/135.0440 | Caftaric acid and hexose derivative | x | |||||
| 475.1194 | 179.0404/161.0293/135.0488 | Caffeic acid derivative | x | |||||
| 475.1328 | 301.0390/179.0349/161.0215 | Caffeic acid derivative | x | |||||
| 515.101 | 353.0802/191.0553/179.0404/173.0419 | Dicaffeoylquinic acid | x | |||||
| 515.1289 | 353.0802/191.0581/179.0349/173.0446 | Dicaffeoylquinic acid | x | |||||
| 515.2682 | 191.0637/179.0515 | Dicaffeoylquinic acid | x | |||||
| 591.1038 | 439.9709/295.0253/179.0075 | Caffeic acid derivative | x | |||||
| 623.078 | 311.0281/179.0075 | Dicaftaric acid | x | |||||
| Propiophenone | ||||||||
| 327.1203 | 147.0367 | DHPPG (3,4′-dihydroxypropiophenone-3-β-d-glucoside) | x |
The results of quantitative analysis of main phenolics (flavonols, phenolic acids, flavanols, procyanidins), iridoids and DHPPG are shown in Table 3. The lingonberry extract had the greatest amount and variety of phenolics.
Table 3.
Quantitative analysis of major compounds identified in extracts from plants (mg/100 g dw)
| Flavonols (mg QG/g) | Phenolic acids (mg CQA/g) | DHPPG (mg CQA/g) | Iridoids (mg LA/g) | Flavanols/procyanidins (mg C/g) | Sum of main phenolics (mg/g) | |
|---|---|---|---|---|---|---|
| B. pendula | 92.9 | 15.4 | 34.4 | Nd | Nd | 142.6 |
| E. arvense | 8.2 | 19.6 | Nd | 11.0 | Nd | 38.8 |
| G. odoratum | 69.7 | 53.2 | Nd | 58.1 | Nd | 181.0 |
| H. glabra | 6.9 | 10.2 | Nd | 11.8 | Nd | 28.8 |
| U. dioica | Nd | 2.1 | Nd | Nd | Nd | 2.1 |
| _V. vitis_-idaea | 134.6 | 10.7 | Nd | 12.6 | 41.1 | 199.1 |
Molecular characterization of bacterial strain
Phylogenetic studies have shown that E. coli strains can be divided into four main phylogenetic groups, designated A, B1, B2, and D [16], depending on the presence/absence of two genes chuA and yjaA and an anonymous DNA fragment TspE4.C2 [5]. On this basis, our _chuA_-positive and _yjaA_-positive strain belongs to the phylogenetic group B2 (Fig. 1a). The most frequently observed E. coli phylogenetic groups among UPEC strains are B2, D and occasionally A, B1 [17]. Thus, the E. coli strain used in our research is classified as UPEC.
Fig. 1.
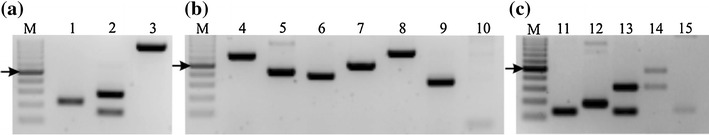
Agarose gel electrophoresis of amplified PCR products. a phylogenetic analysis, b virulence factors genes, c biofilm-related genes. Lanes: M molecular size markers (100 bp, Fermentas), 1_—_yjaA, 2_—_chuA (upper band), TspE4.C2 (lower band), _3_—16SrRNA (control), 4_—_aer, 5_—_sfa, 6_—_csgA, 7_—_cnf1, 8_—_hlyA, 9_—_papC, 10_—_afa, 11_—_ant43, 12_—_luxS, 13_—_sdiA (lower band), 14_—_mqsR, 15_—_mcbA. Arrows indicate 500 bp. Upper band visible on lane 13 may result from the non-specific amplification of some plasmid-encoded gene or/and sdiA rearrangement, since they were not obtained with DNA template from CTF073 strain (data not shown). Bands visible on lane 14 are non-specific
It is well known that uropathogenic strains possess genes encoding specific virulence factors that play an important role in the pathogenicity by overcoming host defence mechanisms and causing disease. The most common occurring are adhesins P fimbriae (pilus associated with pyelonephritis, pap), S fimbriae (sfa), afimbrial adhesin (afa) and curli fiber (csgA), siderophore aerobactin (aer), toxins hemolysin (hlyA) and cytotoxic necrotizing factor 1 (cnf1). As shown on Fig. 1b all of these genes are present in the genome of our UPEC strain except afa gene. The occurrence of some biofilm-related genes, namely ant43, luxS, sdiA, and mcbA suggests that analyzed E. coli strain has the ability to crate biofilm structure (Fig. 1c).
Antibacterial activity of plant extracts
Results obtained in the present study showed that the tested plant extracts possessed different antimicrobial activities (Fig. 2). The number of bacterial cells (c.f.u/mL) in control sample was 4.5 × 109. Increased concentrations of plant extracts caused decrease in survival of bacterial cells. The extracts of H. glabra and _V. vitis_-idaea showed the highest bactericidal activity (p < 0.05). The strongest inhibition of bacterial growth was observed at H. glabra extract concentrations of 1.0 5.0, 10.0, 15.0, 20.0 mg/mL. Very strong reduction of E. coli growth was also observed during incubation of bacteria in _V. vitis_-idaea extract concentrations of 10.0, 15.0, 20.0 mg/mL. Effect of B. pendula extract was slightly less efficient in comparison with H. glabra and V. vitis_-idaea extracts. The percentage viability of bacterial cells decreased from 51 % (at concentration of 0.125 mg/mL) to 6 % (at concentration of 20 mg/mL) of the control sample. This was statistically significant (p < 0.05). U. dioica and E. arvense extracts showed similar antimicrobial activity patterns. Exposure of rods to U. dioica extracts inhibited their growth from 72 % (at concentration of 0.125 mg/mL) to 59 % (at concentration of 20 mg/mL) of the control sample. The percent of survival of bacteria incubated in the presence of E. arvense extracts was reduced from 82 % (at concentration of 0.125 mg/mL) to 50 % (at concentration of 20 mg/mL) of the control. Figure 2 clearly shows that U. dioica and E. arvense extracts were more effective than G. odoratum extract and less effective than H. glabra, V. vitis_-idaea and B. pendula extracts. G. odoratum extract had the weakest antimicrobial activity with the exception of concentration of 20 mg/mL which represents 37 % of the control sample (p < 0.05).
Fig. 2.
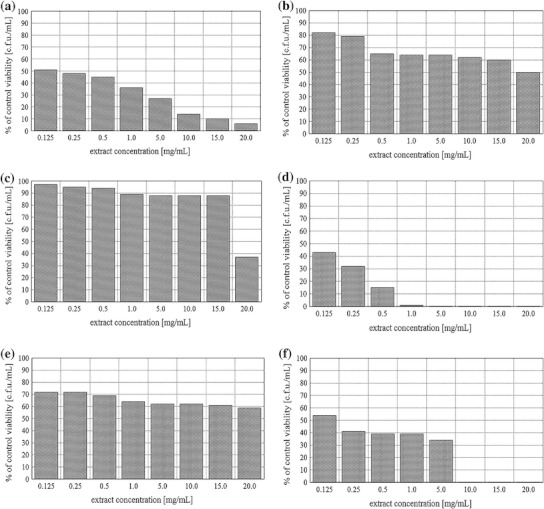
The percentage of E. coli strain survival after exposure to: a B. pendula, b E. arvense, c G. odoratum, d H. glabra, e U. dioica, f _V. vitis_-idaea extracts
Effect of plant extracts on hydrophobicity of bacterial cells
The results showing the effect of plant extracts on cell surface hydrophobicity are shown in Table 4. Surface hydrophobicity of autoaggregating E. coli strain changed after exposure to plant extracts, with the exception of _V. vitis_-idaea extract. The cell surface hydrophobicity changes were observed in bacteria treated with G. odoratum and U. dioica extracts at concentrations of 15.0 and 20.0 mg/mL. Cells surfaces were classified as hydrophilic because they aggregated in 3.2 M ammonium sulfate. This result was not statistically significant (p = 0.241). Other concentrations of G. odoratum and U. dioica extracts resulted in bacteria aggregation in the lower concentrations of ammonium sulfate (0.4–1.0 M) indicating strong hydrophobic bacterial cells surface. Rods incubated in all concentrations of E. arvense and H. glabra and in 5.0, 10.0, 15.0 and 20.0 mg/mL B. pendula extracts showed strong hydrophobic cells surface, because they aggregated in 0.4–1.0 M ammonium sulfate. The lower concentrations of B. pendula extracts resulted in bacteria aggregation in 0.1–0.2 M, hence their cell surface was considered to be very strong hydrophobic.
Table 4.
Effect of plant extracts on hydrophobicity of E. coli bacterial cells
| Plants | Extract concentrations (mg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | 0.125 | 0.25 | 0.5 | 1.0 | 5.0 | 10.0 | 15.0 | 20.0 | |
| B. pendula | Autoaggregative# | Very strong hydrophobic (0.1)§ | Very strong hydrophobic (0.1) | Very strong hydrophobic (0.1) | Very strong hydrophobic (0.2) | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) |
| E. arvense | Autoaggregative | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) |
| G. odoratum | Autoaggregative | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Strong hydrophobic (0.8) | Hydrophilic (3.2) | Hydrophilic (3.2) |
| H. glabra | Autoaggregative | Strong hydrophobic (1.0) | Strong hydrophobic (1.0) | Strong hydrophobic (1.0) | Nt | Nt | Nt | Nt | Nt |
| U. dioica | Autoaggregative | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) | Strong hydrophobic (0.4) | Strong hydrophobic (1.0) | Strong hydrophobic (1.0) | Strong hydrophobic (1.0) | Hydrophilic (3.2) | Hydrophilic (3.2) |
| _V. vitis_-idaea | Autoaggregative | Autoaggregative | Autoaggregative | Autoaggregative | Autoaggregative | Autoaggregative | Nt | Nt | Nt |
Effect of plant extracts on swimming motility
The diverse effects of plant extracts on swimming motility of motile E. coli strain were recorded (Table 5). The largest concentrations of B. pendula (15.0, 20.0 mg/mL) and U. dioica extracts (20.0 mg/mL) significantly reduced the motility of the examined strain (p < 0.05). Swimming zone diameters ranged between 10.7 (±1.5) and 12.7 (±0.6) mm (Fig. 3b). Bacteria treated with B. pendula (5.0, 10.0 mg/mL) and U. dioica extracts (5.0–15.0 mg/mL) also showed decreased movement abilities. Motility of bacteria incubated in the presence of H. glabra and _V. vitis_-idaea extracts decreased slightly compared to the control sample (16.2 ± 2.0). Swimming zone diameters ranged from 14.0 (±2.0) to 16.0 (±2.6) mm. Low concentrations of B. pendula and U. dioica extracts (0.125–1.0 mg/mL) and all concentrations of E. arvense as well as G. odoratum extracts did not inhibit bacterial movement. On the contrary, they resulted in increasing of the swimming zone diameter [16.3 (±4.2) to 36.7 (±7.4) mm] (Fig. 3c).
Table 5.
Effect of plant extracts on E. coli swimming motility. Results showing the motility zone are the mean values from three experiments (±SD)
| Plants | Extract concentrations (mg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Control | 0.125 | 0.25 | 0.5 | 1.0 | 5.0 | 10.0 | 15.0 | 20.0 | |
| B. pendula | 16.2 (±2.0) | 18.7 (±3.1) | 18.3 (±1.5) | 19.3 (±4.2) | 18.7 (±5.5) | 15.0 (±1.7) | 12.7 (±2.1) | 11.7 (±3.1)* | 12.7 (±0.6)* |
| E. arvense | 16.2 (±2.0) | 18.7 (±2.1) | 20.3 (±0.6)* | 25.0 (±2.6)* | 34.3 (±7.5)* | 28.3 (±2.9)* | 36.7 (±7.4)* | 26.7 (±4.2)* | 23.0 (±1.7) |
| G. odoratum | 16.2 (±2.0) | 16.3 (±4.2) | 16.3 (±4.0) | 21.0 (±3.6)* | 17.0 (±3.5) | 23.3 (±1.5)* | 31.3 (±1.5)* | 32.7 (±5.5)* | 30.0 (±3.5)* |
| H. glabra | 16.2 (±2.0) | 15.7 (±4.0) | 15.7 (±3.2) | 16.0 (±2.6) | Nt | Nt | Nt | Nt | Nt |
| U. dioica | 16.2 (±2.0) | 21.3 (±3.1)* | 17.3 (±2.5) | 16.7 (±4.6) | 20.0 (±2.0)* | 12.7 (±4.2) | 15.3 (±2.1) | 14.0 (±2.0) | 10.7 (±1.5)* |
| _V. vitis_-idaea | 16.2 (±2.0) | 15.7 (±2.5) | 14.3 (±2.5) | 14.3 (±0.6) | 14.0 (±2.0) | 15.3 (±1.5) | Nt | Nt | Nt |
Fig. 3.
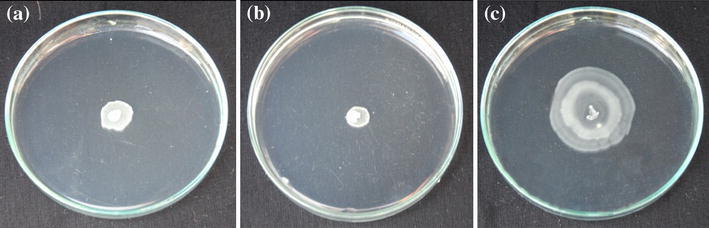
Representative images of E. coli swimming motility under control conditions (a); in the presence of _U. dioica_—20.0 mg/mL (b) and _E. arvense_—10.0 mg/mL (c)
Effect of plant extracts on hemagglutination and expression of P fimbriae
The effects of plant extracts on hemagglutination and expression of P fimbriae are shown in Table 6. They depended on the type and concentration of the plant extract used. The lowest extract concentration causing no agglutination was 1.0 mg/mL and it was observed in case of _V. vitis_-idaea extract. Bacteria incubated with G. odoratum lost their hemagglutination ability at extract concentrations of 5.0 mg/mL and higher. In case of B. pendula and U. dioica, examined rods failed to agglutinate erythrocytes after exposure to extract concentrations of 10.0, 15.0 and 20.0 mg/mL. Bacteria growing in the presence of E. arvense and H. glabra extracts did not show any alteration of the analyzed properties.
Table 6.
Effect of plant extracts on P fimbriae (P) and curli fibers (C) synthesis by E. coli
| Plants | Extract concentrations (mg/mL) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | 0.125 | 0.25 | 0.5 | 1.0 | 5.0 | 10.0 | 15.0 | 20.0 | |||||||||
| P | C | P | C | P | C | P | C | P | C | P | C | P | C | P | C | ||
| B. pendula | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | + |
| E. arvense | + | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − |
| G. odoratum | + | + | + | + | + | + | + | + | + | − | + | − | + | − | + | − | + |
| H. glabra | + | + | + | + | + | + | + | Nt | Nt | Nt | Nt | Nt | Nt | Nt | Nt | Nt | Nt |
| U. dioica | + | + | + | + | + | + | + | + | + | + | + | − | + | − | + | − | + |
| _V. vitis_-idaea | + | + | − | + | − | + | − | − | − | − | − | Nt | Nt | Nt | Nt | Nt | Nt |
Effect of plant extracts on curli expression
The impact of plant extracts on the occurrence of the curli fibers is shown in Table 6. Only bacteria incubated in the presence of all concentrations of _V. vitis_-idaea extract and 10.0, 15.0 and 20.0 mg/mL of E. arvense extracts formed white colonies which indicated the loss of curli fimbriae. Other extracts did not inhibit the synthesis of curli fibers. Bacterial colonies had red color.
Effect of plant extracts on biofilm formation
The activities of plant extracts at concentration 0.125 mg/mL against total biomass of E. coli biofilm are shown in Table 7. In all cases, the strongest and statistically significant biofilm reduction was noticed after 4, 5, and 6 days of bacterial incubation (p < 0.05). The amount of biofilm ranged from 1.4 to 2.0 % in comparison to control samples (ODs = 0.0001). Equally strong inhibition of biofilm mass production was observed after 9- and 10-day incubation in E. arvense and H. glabra extracts and after 10-day treatment in case of _V. vitis_-idaea extract (ODs = 0.0001).
Table 7.
Effect of plant extracts on E. coli biofilm formation
| Time of incubation (days) | Biofilm formation | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| control | B. pendula | E. arvense | G. odoratum | H. glabra | U. dioica | _V. vitis_-idaea | ||||||||
| OD (±SD) | (%) | OD (±SD) | (%) | OD (±SD) | (%) | OD (±SD) | (%) | OD (±SD) | (%) | OD (±SD) | (%) | OD (±SD) | (%) | |
| 1 | 0.004 (±0.001) | 100.0 | 0.002* (±0.002) | 50.0 | 0.0001* (±0.000) | 2.5 | 0.0001* (±0.000) | 2.5 | 0.001* (±0.0005) | 25.0 | 0.0001* (±0.000) | 2.5 | 0.0001* (±0.000) | 2.5 |
| 2 | 0.005 (±0.002) | 100.0 | 0.004 (±0.003) | 80.0 | 0.0001* (±0.000) | 2.0 | 0.004 (±0.002) | 80.0 | 0.002* (±0.001) | 40.0 | 0.002* (±0.001) | 40.0 | 0.003* (±0.002) | 60.0 |
| 3 | 0.01 (±0.002) | 100.0 | 0.01 (±0.003) | 100.0 | 0.002* (±0.0002) | 20.0 | 0.007 (±0.003) | 70.0 | 0.003* (±0.001) | 30.0 | 0.007 (±0.003) | 70.0 | 0.01* (±0.003) | 100.0 |
| 4 | 0.005 (±0.002) | 100.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 |
| 5 | 0.005 (±0.001) | 100.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 | 0.0001* (±0.000) | 2.0 |
| 6 | 0.007 (±0.001) | 100.0 | 0.003* (±0.001) | 42.8 | 0.0001* (±0.000) | 1.4 | 0.0001* (±0.000) | 1.4 | 0.0001* (±0.000) | 1.4 | 0.0001* (±0.000) | 1.4 | 0.0001* (±0.000) | 1.4 |
| 7 | 0.01 (±0.002) | 100.0 | 0.006* (±0.002) | 60.0 | 0.002* (±0.001) | 20.0 | 0.005* (±0.002) | 50.0 | 0.003* (±0.001) | 30.0 | 0.004* (±0.001) | 40.0 | 0.0001* (±0.000) | 1.0 |
| 8 | 0.014 (±0.002) | 100.0 | 0.009* (±0.003) | 64.3 | 0.003* (±0.001) | 21.4 | 0.008* (±0.002) | 57.1 | 0.004* (±0.002) | 28.6 | 0.008* (±0.002) | 57.1 | 0.01 (±0.003) | 71.4 |
| 9 | 0.008 (±0.002) | 100.0 | 0.008 (±0.003) | 100.0 | 0.0001* (±0.000) | 1.3 | 0.003* (±0.001) | 37.5 | 0.0001* (±0.000) | 1.3 | 0.003* (±0.001) | 37.5 | 0.007 (±0.0025) | 87.5 |
| 10 | 0.005 (±0.001) | 100.0 | 0.005 (±0.003) | 100.0 | 0.0001* (±0.000) | 2.0 | 0.001* (±0.0004) | 20.0 | 0.0001* (±0.000) | 2.0 | 0.002* (±0.001) | 40.0 | 0.0001* (±0.000) | 2.0 |
The detailed analysis of the obtained results showed the most effective inhibitory effect of E. arvense extract. This plant significantly (p < 0.05) reduced the optical density (ODs ≤0.003). The maximum amount of biofilm mass was only 21.4 % of the control sample (8th day). Similar strong inhibitory effect was shown for H. glabra extract. The largest amount of biofilm mass represented only 40 % of the control.
The weakest effect was demonstrated for B. pendula extract. After 3, 9 and 10 days, it did not inhibit biofilm formation. In the remaining days, the amount of biofilm was reduced from 42.8 to 80 %, with the exception of 4 and 5 days, when the synthesis of biofilm mass was strongly inhibited (2 % of the control).
Discussion
It is known that in the prevention and treatment of urinary tract infections, one should use medicinal herbs as supplement of the daily diet. Cranberry is one of the most recommended plants by both, doctors and pharmacists. Due to its properties, this fruit prevents adhesion of pathogenic bacteria to uroepithelial tissue, what results in inhibition of their growth and multiplication. Apart from cranberry; however, many other plants are used in folk medicine to prevent or to treat bacterial infections. As an example of such plants may serve B. pendula, E. arvense, G. odoratum, H. glabra, U. dioica and _V. vitis_-idaea, which are used in traditional medicine. Many reports describe their medical properties (diuretic, diastolic, diaphoretic activities and anti-inflammatory effect) which are caused by their chemical composition typical for each species. Phytochemical investigations have shown that these plants contain mainly flavonoids, glycosides, saponins, tannins and terpene derivatives [18, 19]. According to the results presented in this paper, B. pendula, E. arvense, G. odoratum, H. glabra, U. dioica and _V. vitis_-idaea extracts should also be remembered and added to the list of herbs which can be used in UTIs. Unfortunately, only a few research groups described the antibacterial activity of birch, horsetail, woodruff, rupturewort, nettle and lingonberry extracts [20–22]. For this reason, the purpose of our study was to determine the effect of these extracts on bacterial survival, virulence factors involved in tissue colonization and biofilm formation.
The findings of the present study clearly indicate that the tested extracts exhibit significant differences in their antimicrobial activities against E. coli rods. The susceptibility order was as follows: H. glabra > _V. vitis_-idaea > B. pendula > E. arvense > U. dioica > G. odoratum. Based on the qualitative and quantitative analysis of plant extracts (Tables 2, 3), it can be concluded that the differences in susceptibility of bacterial strains to these extracts do not depend on the content of phenolic compounds. It can, therefore, be suspected that other compounds such as saponins, tannins and terpenes are responsible for the bacterial growth inhibition. Khanna and Kannabiran [23] reported that saponin fractions purified from leaves of some plants show antimicrobial activity against Gram-negative strains: Pseudomonas aeruginosa, E. coli, Salmonella typhi, Klebsiella pneumoniae, and Proteus mirabilis. Research carried out by Ho et al. [20] and Barile et al. [24] revealed that saponins, terpenes and tannins found in Vernonia amygdalina, Allium minutiflorum and _V. vitis_-idaea constitute the antimicrobial components of these plants. In our experiments, we established that high concentrations of saponin-rich H. glabra and _V. vitis_-idaea extracts show the strongest bactericidal activity among the rest tested plants. The growth of bacteria was totally inhibited by extracts concentrations ranging from 1.0 to 20.0 mg/mL for H. glabra and 10.0–20.0 m/mL in case of _V. vitis_-idaea.
In contrast to our above-presented results, Kylli et al. [21] found that phenolic proanthocyanidins-rich extracts of lingonberries had antimicrobial effect only on Gram-positive, but not on Gram-negative bacteria: Salmonella enterica sv. Typhimurium, Lactobacillus rhamnosus and E. coli. This result shows that separately isolated compounds do not always have to be as effective as multi-component extracts. Saric et al. [25] examined the influence of B. pendula ethanolic extract on the growth of different bacterial species and found that the concentration 10 mg/mL of this extract possesses the strongest antimicrobial effect against Bacillus cereus. This plant exhibited moderate antimicrobial activity against the other investigated bacteria. Njume et al. [26] examined several different extracts of Combretum molle against Helicobacter pylori. Their results have shown the significant correlation between bacterial survival and the extracts concentration and the type of solvent used in the extraction process.
Looking at the results obtained in our investigation (Fig. 2), B. pendula extract should be forcefully considered as an efficient E. coli multiplication inhibitor and hence as agent reducing the bacterial survival. All tested concentrations of this extract significantly decreased bacterial growth. The least effective extracts were those derived from U. dioica and G. odoratum leaves. Even their highest used concentrations (20.0 mg/mL) in this study did not fully inhibit bacterial growth but only reduced their survival. The viability level was 59 % for bacteria treated with U. dioica and 37 % in case of G. odoratum compared to the control samples. Similar to the above-presented results, Singh et al. [21] established no activity either of U. dioica aqueous or methanol and ethyl-acetate extracts against Gram-positive Staphylococcus aureus, as well as Gram-negative Shigella flexneri, P. aeruginosa, K. pneumoniae and Salmonella typhi. The chloroform extract of common nettle showed only moderate action against tested microorganisms, while good antibacterial properties against all studied bacterial species were found for hexane extract of U. dioica. These results clearly show that the antibacterial abilities of the extract strongly depend on the reagents used during its extraction.
The limited number of publications describing the influence of plant extracts on bacterial virulence factors prompted us to perform research in this area. The most important pathogenic factors involved in bacterial adhesion to uroepithelial cells are hydrophobic surface, the ability to movement and synthesis of adhesins.
In our study, the changes in the cell surface properties were observed for E. coli incubated in G. odoratum and U. dioica extracts. Very strong hydrophobic cells’ surface of autoaggregative E. coli strain used in our experiments became hydrophilic after exposure only to the highest concentrations (15.0 and 20.0 mg/mL) of both extracts. Hydrophilic cell surface nature impedes the colonization of the host tissues. Therefore, despite the weak growth-inhibiting properties, G. odoratum and U. dioica can be applied in UTIs prevention. Similar results of bacterial hydrophobicity changes have been noticed by Razak et al. [27]. The authors established that the hydrophobicity of Streptococcus mitis, Streptococcus sanguinis and Actinomyces sp. incubated in the presence of Psidium guajava extract was reduced.
The analysis of the swimming zone diameters obtained in our research showed that some of the extracts have reduced, and other have increased the bacterial motility. This phenomenon is difficult to explain, because there are no reports describing similar experiments. Probably, this effect is associated with different pH values of the plant extracts. Hattermann and Ries [28] found that bacteria growing at pH 6 and 7 were more motile than those grown at pH 5.8–10.0. Hidalgo et al. [29] showed that swimming and swarming motilities were hindered when E. coli CFT073 strain was grown in the presence of the cranberry compounds. This result was due to inhibition of flagellin gene (fliC) expression. Transmission electron microscopy imaging of bacteria exposed to cranberry materials revealed fewer flagella than in control bacteria.
The presence of fimbrial adhesins promotes the attachment of the bacterial cells to the host tissues and protects them against removing from the urinary tract with urine. We established that extracts of silver birch, sweet woodruff, common nettle and lingonberry inhibited erythrocyte hemagglutination by uropathogenic E. coli strain, which indicates the dysfunction of P fimbriae. It is known that these plants are rich in tannins—compounds with the structure very similar to receptors found on bladder and kidney cells [30]. Therefore, these compounds act by binding to fimbriae and thereby preventing their attachment to the host tissue. Ahuja et al. [31] found that E. coli rods growing in the presence of the cranberry juice lost the expression of P fimbriae leading to a loss of the ability to epithelial cells colonization. Proanthocyanidins present in cranberry fruits are responsible for this phenomenon. Strong inhibition of adherence of multi-drug resistant E. coli strains treated with proanthocyanidins to uroepithelial cells was also observed by Gupta et al. [32]. Curli fibers play an important role in biofilm formation by rods belonging to Enterobacteriaceae family [33]. It has been also shown that the reduction of pili correlated with the loss of the ability of uropathogenic E. coli strains to colonize bladder cells and to form biofilm [34]. In our study, bacteria growing in all tested concentrations of _V. vitis_-idaea extracts and the highest concentrations of E. arvense extracts (10.0–20.0 mg/mL) showed no expression of curli fimbriae.
According to some researchers, recurrent UTIs are caused by microorganisms that invade the urinary tract and form a biofilm structures [35]. The results of our study indicate that the exposure of E. coli rods to plant extracts significantly reduced or inhibited the biofilm production. Such activity of these plant extracts can be explained by the presence of flavonoids. It is known that flavonoids such as quercetin, kaempherol, naringenin and apigenin reduce biofilm synthesis because they can suppress autoinducer-2 activity which is responsible for cell-to-cell communication [36]. Lee et al. [37] confirmed that phloretin belonging to flavonoids suppressed autoinducer-2 importer genes of E. coli O157:H7 biofilm cells.
The results obtained in our study suggest that anti-biofilm effect of plant extracts can be caused by modifications in the bacterial surface structures responsible for binding to the occupied surface.
Conclusion
All plant extracts used in our study showed antibacterial activity and/or reduction of the biofilm mass. Only some extracts altered virulence factors in examined rods. Therefore, our results should be confirmed in clinical trials to be able to recommend tested plant extracts in prevention of and treatment of UTIs. Moreover, considering the wide variety of the antibacterial activities of plant extracts, depending on the solvent, it would be worthwhile in the future studies to compare our current results with, e.g., methanol- or ethanol-derived extracts.
Acknowledgments
Authors are grateful to Professor Andrzej Hendrich for reading the manuscript and helpful discussions. This research was supported by a grant of the Wrocław Medical University, Wrocław, Poland (protocol number: 1906).
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
References
- 1.Ejrnaes K, Stegger M, Reisner A, Ferry S, Monsen T, Holm SE, Lundgren B, Frimodt-Moller N. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence. 2011;2:528–537. doi: 10.4161/viru.2.6.18189. [DOI] [PubMed] [Google Scholar]
- 2.Emody L, Kerenyi M, Nagy G. Virulence factors of uropathogenic Escherichia coli. Int J Antimicrob Agents. 2003;22:S29–S33. doi: 10.1016/S0924-8579(03)00236-X. [DOI] [PubMed] [Google Scholar]
- 3.Norinder BS, Koves B, Yadav M, Brauner A, Svanborg C. Do Escherichia coli strains causing acute cystitis have a distinct virulence repertoire? Microb Pathog. 2012;52:10–16. doi: 10.1016/j.micpath.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Wojnicz D, Sycz Z, Walkowski S, Gabrielska J, Włoch A, Kucharska A, Sokół-Łętowska A, Hendrich AB. Study on the influence of cranberry extract Żuravit S·O·S(®) on the properties of uropathogenic Escherichia coli strains, their ability to form biofilm and its antioxidant properties. Phytomedicine. 2012;19:506–514. doi: 10.1016/j.phymed.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 5.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arisoy M, Aysev D, Ekim M, Ozel D, Kose SK, Ozsoy ED, Akar N. Detection of virulence factors of Escherichia coli from children by multiplex polymerase chain reaction. Int J Clin Pract. 2006;60:170–173. doi: 10.1111/j.1742-1241.2005.00668.x. [DOI] [PubMed] [Google Scholar]
- 7.Burgos Y, Beutin L. Common origin of plasmid encoded alpha-haemolysin genes in Escherichia coli. BMC Microbiol. 2010;10:193. doi: 10.1186/1471-2180-10-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Liu B, Chen M, Guo D, Guo X, Liu F, Feng L, Wang L. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods. 2010;82:71–77. doi: 10.1016/j.mimet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Lindahl M, Faris A, Wadstrom T, Hjerten S. A new test based on “salting out” to measure relative surface hydrophobicity of bacterial cells. Biochim Biophys Acta. 1981;77:471–476. doi: 10.1016/0304-4165(81)90261-0. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez-Torrez V, Hu H, Wood TK. GGDEF proteins YeaI, YedQ, and YfiN reduce early biofilm formation and swimming motility in Escherichia coli. Appl Microbiol Biotechnol. 2011;90:651–658. doi: 10.1007/s00253-010-3074-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Latham RH, Stamm WE. Role of fimbriated Escherichia coli in urinary tract infections in adult women: correlation and localization studies. J Infect Dis. 1984;149:835–840. doi: 10.1093/infdis/149.6.835. [DOI] [PubMed] [Google Scholar]
- 12.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x. [DOI] [PubMed] [Google Scholar]
- 13.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 14.Stepanovic S, Cirkovic I, Ranin L, Svabic-Vlahovic M. Biofilm formation by Salmonela spp. and Listeria monocytogenes on plastic surface. Lett Appl Microbiol. 2004;38:428–432. doi: 10.1111/j.1472-765X.2004.01513.x. [DOI] [PubMed] [Google Scholar]
- 15.Di Bonaventura G, Spedicato I, D’Antonio D, Robuffo I, Piccolomini R. Biofilm formation by Stenotrophomonas maltophilia modulation by quinolones, trimethoprim-sulfamethoxazole, and ceftazidime. Agents Chemother. 2004;48:151–160. doi: 10.1128/AAC.48.1.151-160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herzer PJ, Inouye S, Inouye M, Whittam TS. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J Bacteriol. 1990;172:6175–6181. doi: 10.1128/jb.172.11.6175-6181.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J Clin Microbiol. 2002;40:3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freiler M, Resnicek G, Jurenitsch J, Kubelka W. New triterpene saponins from Herniaria glabra. Helv Chim Acta. 1996;79:385–390. doi: 10.1002/hlca.19960790207. [DOI] [Google Scholar]
- 19.Galashkina NG, Vedernikov DN, Roschin VI. Flavonoids of Betula pendula Roth buds. Rastitelnye Resursy. 2004;40:62–68. [Google Scholar]
- 20.Ho KY, Tsai CC, Huang JS, Chen CP, Lin TC, Lin CC. Antimicrobial activity of tannin components from Vaccinium vitis-idaea L. J Pharm Pharmacol. 2001;53:187–791. doi: 10.1211/0022357011775389. [DOI] [PubMed] [Google Scholar]
- 21.Kylli P, Nohynek L, Puupponen-Pimia R, Westerlund-Wikstrom B, Leppanen T, Welling J, Moilanen E, Heinonen M. Lingonberry (Vaccinium vitis-idaea) and European cranberry (Vaccinium microcarpon) proanthocyanidins: isolation, identification, and bioactivities. J Agric Food Chem. 2011;59:3373–3384. doi: 10.1021/jf104621e. [DOI] [PubMed] [Google Scholar]
- 22.Singh R, Dar SA, Sharma P (2012) Antibacterial activity and toxicological evaluation of semi purified hexane extract of Urtica dioica leaves. Res J Med Plant. doi:10.3923/rjmp.2012
- 23.Khanna VG, Kannabiran K. Antimicrobial activity of saponin fractions of leaves of Gymnema sylvestre and Eclipta prostrata. World J Microbiol Biotech. 2008;4:2737–2740. doi: 10.1007/s11274-008-9758-7. [DOI] [Google Scholar]
- 24.Barile E, Bonanomi G, Antignani V, Zolfaghari B, Sajjadi SE, Scala F, et al. Saponins from Allium minutiflorum with antifungal activity. Phytochemistry. 2007;68:596–603. doi: 10.1016/j.phytochem.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Saric CL, Cabarkapa SI, Beljkas MB, Misan CA, Sakac BM, Plavsic VD. Antimicrobial activity of plant extracts from Serbia. Food Process Qual Saf. 2009;1–2:1–5. [Google Scholar]
- 26.Njume C, Afolayan AJ, Samie A, Ndip RN. Inhibitory and bactericidal potential of crude acetone extracts of Combretum molle (Combretaceae) on drug-resistant strains of Helicobacter pylori. J Health Popul Nutr. 2011;29:438–445. doi: 10.3329/jhpn.v29i5.8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razak FA, Othman RY, Rahim ZH. The effect of Piper betle and Psidium guajava extracts on the cell-surface hydrophobicity of selected early settlers of dental plaque. J Oral Sci. 2006;48:71–75. doi: 10.2334/josnusd.48.71. [DOI] [PubMed] [Google Scholar]
- 28.Hattermann DR, Ries SM. Motility of Pseudomonas syringae pv. glycinea and its role in infection. Phytopathology. 1989;79:284–289. doi: 10.1094/Phyto-79-284. [DOI] [Google Scholar]
- 29.Hidalgo G, Chan M, Tufenkji N. Inhibition of Escherichia coli CFT073 fliC expression and motility by cranberry materials. Appl Environ Microbiol. 2011;77:6852–6857. doi: 10.1128/AEM.05561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Howell AB, Vorsa N, Der Marderosian A, Foo LY. Inhibition of the adherence of P-fimbriated Escherichia coli to uroepithelial-cell surfaces by proanthocyanidin extracts from cranberries. N Engl J Med. 1998;339:1085–1086. doi: 10.1056/NEJM199810083391516. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja S, Kaack B, Roberts J. Loss of fimbrial adhesion with the addition of Vaccinium macrocarpon to the growth medium of P-fimbriated Escherichia coli. J Urol. 1998;159:559–562. doi: 10.1016/S0022-5347(01)63983-1. [DOI] [PubMed] [Google Scholar]
- 32.Gupta A, Dwivedi M, Mahdi AA, Nagana Gowda GA, Khetrapal CL, Bhandari M. Inhibition of adherence of multi-drug resistant E. coli by proanthocyanidin. Urol Res. 2012;40:143–150. doi: 10.1007/s00240-011-0398-2. [DOI] [PubMed] [Google Scholar]
- 33.Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aberg V, Almqvist F. Pilicides-small molecules targeting bacterial virulence. Org Biomol Chem. 2007;5:1827–1834. doi: 10.1039/b702397a. [DOI] [PubMed] [Google Scholar]
- 35.Anderson GG, Dodson KW, Hooton TM, Hultgren SJ. Intracellular bacterial communities of uropathogenic Escherichia coli in urinary tract pathogenesis. Trends Microbiol. 2004;12:424–430. doi: 10.1016/j.tim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 36.Vikram A, Jayaprakasha GK, Jesudhasan PR, Pillai SD, Patil BS. Suppression of bacterial cell–cell signalling, biofilm formation and type III secretion system by citrus flavonoids. J Appl Microbiol. 2010;109:515–527. doi: 10.1111/j.1365-2672.2010.04677.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee JH, Regmi SC, Kim JA, Cho MH, Yun H, Lee CS, Lee J. Apple flavonoid phloretin inhibits Escherichia coli O157:H7 biofilm formation and ameliorates colon inflammation in rats. Infect Immun. 2011;79:4819–4827. doi: 10.1128/IAI.05580-11. [DOI] [PMC free article] [PubMed] [Google Scholar]