Current opportunities and challenges in microbial metagenome analysis—a bioinformatic perspective (original) (raw)
Abstract
Metagenomics has become an indispensable tool for studying the diversity and metabolic potential of environmental microbes, whose bulk is as yet non-cultivable. Continual progress in next-generation sequencing allows for generating increasingly large metagenomes and studying multiple metagenomes over time or space. Recently, a new type of holistic ecosystem study has emerged that seeks to combine metagenomics with biodiversity, meta-expression and contextual data. Such ‘ecosystems biology’ approaches bear the potential to not only advance our understanding of environmental microbes to a new level but also impose challenges due to increasing data complexities, in particular with respect to bioinformatic post-processing. This mini review aims to address selected opportunities and challenges of modern metagenomics from a bioinformatics perspective and hopefully will serve as a useful resource for microbial ecologists and bioinformaticians alike.
Keywords: 16S rRNA biodiversity, binning, bioinformatics, Genomic Standards Consortium, metagenomics, next-generation sequencing
INTRODUCTION
The development of techniques for sequencing deoxyribonucleic acid (DNA) from environmental samples was a crucial factor for the discovery of the exceptional degree of diversity among prokaryotes. In particular, techniques to obtain 16S ribosomal ribonucleic acid (rRNA) sequences from the environment, such as the early reverse transcriptase-based approaches [1] and the later polymerase chain reaction-based methods have been cornerstones toward current large-scale studies of microbial biodiversity. More than 3 million 16S rRNA sequences of Bacteria and Archaea in the release 111 of the SILVA database [2] constitute an impressive hallmark of microbial versatility. This number is already in the order of magnitude of the estimated few million microbial species for the entire ocean [3], whereas on the other hand, it represents just a fraction of the diversity of soils where just a single ton is believed to potentially harbor millions of species [3, 4]. The extent of 16S rRNA gene variation recently discovered among lowly abundant species in the deep sea (‘rare biosphere’) [5–7] indicates that with respect to microbial diversity we so far have seen just the proverbial tip of the iceberg.
For a long time, microbial ecologists were mostly restricted to pure cultures of cultivable isolates to shed light on the diversity and functions of environmental microbes. Pure cultures allow the study of an isolate’s metabolism and of its gene repertoire by genome sequencing. Both provide valuable information for extrapolating on the isolate’s ecophysiological role. Cultivability of environmental microbes often ranges below 1% of the total bacteria [8], but depending on cultivation technique and habitat, much higher cultivation rates have been reported, for example up to 10% for a freshwater lake [9] and 23% for a marine tidal sediment [10]. Such successes notwithstanding, in almost all cases, a major fraction of Bacteria and Archaea evades current cultivation approaches and thus conventional whole genome shotgun sequencing.
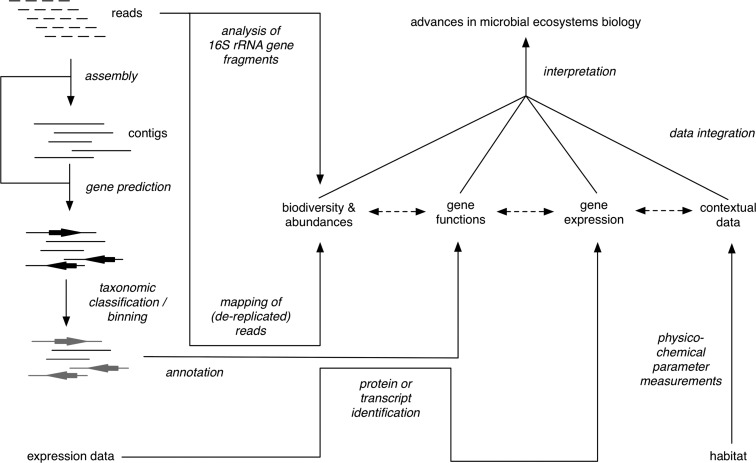
Solutions are to sequence either single microbial cells [11] or entire microbial communities—the latter is termed metagenomics [12, 13]. The classical metagenome approach involves cloning of environmental DNA into vectors with the help of ultra-competent bioengineered host strains. The resulting clone libraries are subsequently screened either for dedicated marker genes (sequence-driven approach) or metabolic functions (function-driven approach) [14]. The function-driven approach is still paramount for screening enzymes with prospects in biotechnology (see [15] for a recent mini review), whereas in microbial ecology, increasing throughput (i.e. base pairs per run) and diminishing costs for DNA sequencing have rendered the sequence-driven approach largely obsolete. Nowadays, direct sequencing of environmental DNA (aka shotgun metagenomics) is commonly used to study the gene inventories of microbial communities. By combining the resulting metagenomic data with biodiversity data (e.g. from 16S rRNA gene amplicon sequencing (A. Klindworth et al. submitted for publication), in situ expression data (metatranscriptomics and metaproteomics) and environmental parameters, a new type of holistic ecosystem studies has become feasible [16] (Figure 1). Similarly, metagenome data can be integrated with metabolome data [17]. Such integrative ‘ecosystems biology’ studies (e.g. [18, 19]) introduce a plethora of challenges with respect to experimental design and bioinformatic downstream processing. These involve considerations about the habitat, sampling strategy, sequencing technology, assembly, gene prediction, taxonomic classification and binning, biodiversity estimation, function predictions and analyses, data integration and subsequent interpretation and data deposition. This mini review aims to address some of these aspects and complement more elaborate full reviews of the matter (e.g. [20]).
Figure 1.
Scheme of the major stages of an integrative metagenomic ecosystems study on microbial ecology.
HABITAT
The biodiversity composition (richness and evenness) of a habitat has a profound impact on the quality of a metagenome. For metagenome analyses involving assembly (to generate longer genome fragments with multiple genes), habitats with few microbial species or an uneven population with few dominating species are more promising targets than habitats with many species of even abundance. However, more important than the absolute number of species is their level of genomic coherence. Even seemingly ideal habitats with a stable composition of few dominant species, for example microbial mats [21] or invertebrate bacterial symbioses [22], can be difficult to assemble when evolutionary micro-niche adaptations have led to large pan-genomes and thus to a low level of population clonality (see e.g. [23] for a discussion on sub-species fine-scale evolution and pan-genomes). In contrast, seemingly unsuitable habitats that harbor a multitude of species with dynamically changing compositions can yield long assemblies, when the species that thrive and dominate are largely clonal. This effect is observed when a second round of sequencing and reassembly of an environmental sample breaks rather than elongates assemblies from the previous round. The reason is that in habitats with little clonality, more sequencing covers more genomic heterogeneity. This increases incongruities in putative assemblies, which causes assemblers to generate smaller but congruent assemblies rather than long assemblies with high levels of positional variability.
These common issues in metagenomics might be overcome by switching either to a longer read sequencing technology or by more sequencing to increase coverage (i.e. the number of calls of a base in a given DNA sequence, typically attained by sequencing multiple molecules containing the respective sequence), but in any case they mean increasing costs, data volumes and complexity. Preceding biodiversity analysis can help to properly assess the required amount of sequencing, e.g. in form of full-length 16S rRNA clone libraries analysis (for fine-scale resolution) in conjunction with 16S rRNA gene tag analysis (for abundance estimations).
SAMPLING
When the study target is a specific uncultivable microbial species of low abundance, it is worthwhile to try to enrich the species after sampling. Sometimes favorable culture conditions can be found that result into co-cultures with substantially enriched target species or the species can be physically enriched, for example by methods such as fluorescence-activated cell sorting (e.g. [24, 25]) or by density gradient centrifugation (e.g. [26]). Subsequent multiple-displacement amplification and single cell sequencing might be a viable solution to obtain a draft genome. However, when the target is a habitat’s overall function, a representative sample must be studied, and data from single cells, enrichments or isolates—though valuable—are complementary. Sampling of microbes from environments usually involves a size selection (e.g. fractionating filtration) to minimize contaminations by viruses or eukaryotes. Such reduction of a sample’s complexity introduces a bias in the community composition, for example by under-sampling particle-associated, filamentous, aggregate-forming or very small microbes. This is the reason why even deeply covered metagenomes mostly represent only a select fraction of a habitat’s microbial gene inventory. It can be reasonable to reduce the complexity of an environmental sample by enrichment or size selection, in particular when multiple different enrichments or samples with different size fractions are taken and their results are combined, but such effects need to be taken into account in experiment design and interpretation of the final data.
REPLICATION
It is good scientific practice to analyze true replicates of a sample and to assess whether observed differences within one sample are statistically meaningful. However, this is rarely done in microbial ecology [27]. One reason is that in many habitats it is almost impossible to take true replicates. For example, sediment cores that have been taken only centimeters away from each other might host slightly different microbial communities due to environmental patchiness. Similarly, water samples that were taken within few minutes might differ because the sampled water was moving and not perfectly homogeneous. Hence, comparing such alleged replicates reveal little information on methodological reproducibility. Our own comparisons of true 454/Roche pyrosequencing replicates have shown that library preparation and sequencing are highly reproducible, which is corroborated by a recent comparison of 454/Roche pyrosequencing and Illumina sequencing [28]. Thus it is understandable that environmental biologists prefer to analyze more samples rather than to invest in replication, in particular in expensive large-scale projects. This, however, does not release scientists from assessing the reproducibility of their methods. Part of this can be addressed by pseudo-replication [29], such as sub-sample analysis and comparison of samples within time series [30], and by independent assessments of methodological reproducibility using representative test data sets.
SEQUENCING
Next-generation sequencing (NGS) has been nothing less than a paradigm shift for metagenomics. Not long ago, the classical clone-based metagenome approach in combination with Sanger sequencing usually allowed for obtaining only few selected inserts, as sequencing was the limiting factor. NGS has obliterated the cloning step and its inherent problems and enabled to sequence environmental DNA directly. Initially, 454/Roche pyrosequencing was most widely used, because it generated substantially longer reads than competing platforms. Meanwhile, in particular large-scale metagenome projects make increasing use of the Illumina and, to a lesser extent, SOLiD platforms. Although the latter two still provide shorter reads than pyrosequencing, they offer a much higher throughput and hence coverage for the same price. The read length of 2 × 150 bp provided by the current Illumina GA IIx line of instruments basically matches that of the first generation 454 Life Sciences GS20 instrument and high coverage in conjunction with mate-pair libraries facilitate assembly and can compensate for the lack of read length. A recent comparative study on a freshwater lake planktonic community has shown that Illumina and 454 pyrosequencing lead to similar results with respect to assemblies and the covered taxonomic and functional repertoires [28]. In addition, protocols have been proposed that allow for obtaining longer ‘composite reads’ from short read platforms [31].
It remains to be seen what impact newer sequencing platforms will have on the metagenomic field. The lately announced Ion Torrent Proton fits in between the 454/Roche and Illumina platforms in terms of read length and throughput at a seemingly competitive price. At the same time, single-molecule detection methods such as Pacific Bioscience’s PacBio RS and the recently announced Oxford Nanopore Technologies (ONT) GridION and MinION systems offer much longer read lengths, albeit at the expense of higher sequencing errors (PacBio RS: ∼10–15% and ONT GridION: ∼4%). However, in contrast to the inherent systematic errors of other platforms, these errors are mostly random, and once these platforms improve, they could be reduced in an almost linear fashion by increased multifold sequencing of the exact same DNA molecule and increased coverage. The specific value of PacBio and ONT sequencing is that they provide read lengths that are long enough to span multiple prokaryote genes and thus are able to provide reliable genetic contexts. Currently, a combination of long- and short-read technologies constitutes a particularly promising approach in future metagenomics that bears the potential to significantly advance the field.
Read length, error rate and throughput/coverage of NGS technologies determine the resolution at which we can investigate gene inventories of natural microbial communities in a very similar way as the magnification and aberrations of optical microscopes determine the resolution at which microbes can be directly seen with the human eye. In this respect, advances in sequencing technologies will continue to shape the field of metagenomics and extend our possibilities to address habitats of increasing complexity.
ASSEMBLY
It is a non-trivial question, whether to assemble a metagenome. An assembly yields larger genomic fragments that allow for the study of gene arrangements. Valuable functional knowledge can be deduced from gene neighborhoods, e.g. when a gene of unknown function always appears together with a gene whose function is well known [32, 33]. Large-scale systematic investigations of such gene syntenies across metagenomes have the potential to uncover as yet unknown functional couplings.
Assembly of sequences from metagenomic libraries can result in good draft or even complete genomes when the target species shows little intraspecies variation, but this usually requires a substantial amount of sequencing. For example, massive sequencing allowed Pelletier et al. [34] to obtain a good draft genome of ‘Candidatus Cloacamonas acidaminovorans’ from a wastewater anaerobic digester. Similarly, Hess et al. [35] were able to reconstruct 15 draft genomes by direct assembly of a cow rumen metagenome. Erkel et al. [36] could even obtain a complete genome of a methanogenic archaeon from the Rice Cluster I clade by direct assembly of rice soil-derived metagenome, albeit not from a sample with natural diversity but from an enrichment. Similarly, Iverson et al. [37] succeeded in retrieving a complete genome of a marine group II euryarchaeon from an extensive sea surface water metagenome, despite the genome was represented by less than 2% of the reads. Spiking experiments of metagenomes with a pure culture isolate have suggested that a genome with little intraspecies variation can be retrieved from a metagenome when it is covered at least 20-fold [38].
Although assembly does yield longer sequences, it also bears the risk of creating chimeric contigs, in particular in habitats with closely related species or highly conserved sequences that occur across species (for example as a result from high transposase, phage and lateral gene transfer activities). Furthermore, assembly distorts abundance information, as overlapping sequences from an abundant species will be identified as belonging to the same genome and consequently joined. This leads to a relative underrepresentation of sequences of abundant species. Hence, gene frequencies are better compared based on read representation rather than on the basis of assemblages. An alternative is to back-trace all reads that constitute a given contig (or gene), either by direct mapping of the reads on the assemblies (Figure 1) or by extracting the respective information from the assembly ACE file.
Assemblers yield similar results when the coverage is high, but our own experience indicates that at low coverage, the assembler and its settings can have a notable effect. Furthermore, assemblers are mainly built for assembling all reads into a single sequence, which is exactly the opposite of the separation of sequences of different organisms, which metagenomics strives for. Furthermore, metagenome assemblers need to be more fault tolerant than genome assemblers to account for strain-level genomic heterogeneity, which on the other hand elevates the risk for chimeric assemblies. Dedicated metagenome assemblers that try to address these problems are Genovo [39], Meta-IDBA [40], MetaVelvet [41] and MAP [42]. The first three of these are intended for short-read data, whereas MAP also handles longer reads as they are produced by current 454 FLX+ pyrosequencers. All four assemblers are claimed to yield longer assemblies and more representative taxonomic representations than conventional assemblers. A dedicated stand-alone metagenome scaffolder that can be used to post-process the unitig graphs of other assemblers is BAMBUS2 [43].
One particular problem is that the increasing throughput of NGS platforms imposes challenges on assembly, in particular with respect to memory requirements. A current Illumina HiSeq2000 sequencer can generate 600 Gb in a single run, and higher throughput technologies are almost given in the nearer future. As a result, metagenomics is currently experiencing a split between smaller, more targeted projects with assemblies and large-scale projects without assemblies. The trend in metagenomics for tremendous data scales has been anticipated even before second- and third-generation NGS platforms became available and has been termed ‘megagenomics’ [44]. Such megagenome projects, as for example the Human Microbiome [45] and Earth Microbiome [46, 47] Projects, require dedicated bioinformatic post-processing and data integration pipelines, some of which have yet to be developed.
GENE PREDICTION
Many conventional gene finders require longer stretches of sequence to discriminate coding from non-coding sequences. Furthermore, many gene finders require training sequences from a single species that is subsequently used to build a species-specific gene prediction model. This is unsuitable for metagenomes that are constituted as a mixture of sequences from different organisms and often comprise only a limited number of long contigs but mainly short assemblies and unassembled reads. Furthermore, partial genes must be predicted missing proper gene starts, stops or even both. In addition, metagenomes (in particular at low coverage) are often riddled with frame shifts. This makes gene prediction for metagenomes a non-trivial task [48].
Dedicated gene prediction programs have been developed for metagenomes, such as MetaGene [49], MetaGeneAnnotator [50], Orphelia [51, 52] and FragGeneScan [53]. All these programs have been built for short reads, but they follow different approaches (such as machine learning techniques and Markov models), differ in the precision of ribosomal binding site and thus correct start prediction and in their tolerance for sequencing errors.
No matter how you look at it, the quality of gene predictions in microbial metagenome data sets is inferior to those of sequenced genomes. Combining multiple gene finders, screening intergenic regions for overlooked genes and using dedicated frameshift detectors [54, 55] are common strategies to overcome at least some of these limitations.
TAXONOMIC CLASSIFICATION AND BINNING
One of the key problems of current metagenomics is to assign the obtained sequences and their gene functions to dedicated taxa in the habitat. Phylogenetic marker genes are sparse and thus allow only taxonomic assignment of a minor portion of sequences. Hence, other approaches are needed that can partition metagenomes into taxonomically distinct bins (taxobins) that provide taxon-specific gene inventories with ecologically indicative functions.
A number of such approaches have been developed that can be categorized into classification and binning approaches. Classification approaches assign taxonomies based on similarities between metagenomic sequences and sequences of known taxonomy. Binning approaches work intrinsically (i.e. without reference sequences) and cluster sequences based on compositional characteristics. In general, one can discriminate methods that operate on the level of protein sequences (gene-based classification), on the level of intrinsic DNA characteristics (signature-based binning/classification) and those that map DNA reads to reference sequences (mapping-based classification).
Gene-based classification
Gene-based classification requires all the metagenomic sequences’ potentially full and partial protein-coding regions to be translated into their corresponding protein sequences. There are two main approaches.
The first is to use conventional basic local alignment search tool (BLAST) searches [56] against protein databases such as the non-redundant NCBI database or UniProt [57] and to derive taxonomic information from the resulting hits. This can be done either by constructing a multiple sequence alignment from the best matching hits with subsequent phylogenetic reconstruction, as implemented in Phylogena [58], or it can be done directly based on the BLAST results. Of course, as BLAST is a heuristic for fast sequence database searches and not a phylogenetic algorithm per se, the top BLAST hit does not necessarily agree with the taxonomic affiliation of the gene in question [59]. However, it has been shown that post-processing a larger number of BLAST hits can reveal useful taxonomic assignments, for example by using consensus information as implemented in the lowest common ancestor algorithm of MEGAN [60, 61] or the Darkhorse [62] and Kirsten [19] algorithms.
A second possibility is to infer taxonomic information from HMMer searches against Pfam models [63] as implemented in CARMA [64, 65] or Treephyler [66]. The principle of CARMA is to align a sequence hitting a Pfam model with the model’s curated seed alignment, construct a neighbor-joining tree from the alignment and use this tree to infer the sequence’s taxonomy. Treephyler follows a similar approach but uses speed-optimized Pfam domain prediction and treeing methods. Both approaches provide more accurate classifications than those based on BLAST, but they work for fewer sequences, as Pfam hits are less frequent than BLAST hits (typically ∼20% of the genes).
Signature-based binning/classification
DNA base compositional asymmetries carry a weak but detectable phylogenetic signal [67] that is most pronounced within the patterns of statistical over- and underrepresentation of tetra- to hexanucleotides [68]. Various algorithms have been used to discriminate this signal from the DNA-compositional background noise and to use it for taxonomic inference, e.g. simple [67, 69–71] and advanced Markov models such as interpolated context models (ICMs) [72], Bayesian classifiers [73] and machine-learning algorithms such as support vector machines (SVMs) [68], kernelized nearest-neighbor approaches [74] and self-organizing maps (SOMs) [75–81]. Also, weighted PCA-based [82], Spearman distance-based [83, 84] and Markov Chain Monte Carlo-based [85] assessments of oligomer counts have been used. As the information that DNA composition-based methods rely on is a function of sequence lengths, most of these methods deteriorate below 3–5 kb and perform poorly on sequences shorter than 1 kb. Nonetheless, methods have been developed for successful binning of short reads as they are produced by Illumina machines, e.g. AbundanceBin, which is an unsupervised l-tuple-abundance-based clustering method [86]. Recently, a signature-based method has been developed for fast taxonomic profiling of metagenomes that is independent of length and can be used with very short reads [87].
Mapping-based classification
Sequenced genomes have been used as references with known taxonomies for read recruitment in metagenome studies [18]. This approach is particularly useful for habitats with species that have closely related sequenced relatives. A variant of this approach is to use habitat-specific sets of reference genomes for a competitive metagenome read mapping [19, 88]. Such sets can be compiled using the EnvO-lite environmental ontology [89]. Low-quality repetitive reads should be excluded from the mapping using tools such as mreps [90], and phages should be masked to minimize misclassifications. The mapping itself can be done with tools such as SSAHA2 [91] or its successor SMALT (http://www.sanger.ac.uk/resources/software/smalt). Combined mapping information of the reads constituting a contig can be subsequently combined into a taxonomy consensus.
Combinatory classification
All aforementioned methods have specific advantages and disadvantages, and all are limited by the amount of information that can be retrieved from a sequence at all. Although protein-based methods tend to be more accurate than DNA-based methods, especially on shorter sequences or even reads, they can only classify sequences with existing homologues in public databases. Unfortunately, this is not the case for a large fraction of the genes within environmental microorganisms that are typically the focus of metagenomic studies. At least half of the genes of novel sequenced environmental microbes lack dedicated known functions, and a large proportion of these genes are hypothetical or conserved hypothetical genes that have either no or insufficient homologues with known taxonomic affiliation. This limitation does not apply to DNA-based methods, which, for their part, have other limitations. For example, methods such as ICMs, SVMs or SOMs need to be pre-trained, which is computationally expensive and must be continually done to keep pace with the fast-growing amount of new sequences. On the other hand, pre-training might lead to better prediction accuracy, especially when there is prior knowledge about a habitat’s biodiversity that allows restriction to a dedicated set of training sequences.
In general, DNA-based methods suffer much more from a decrease in prediction accuracy when sequences get shorter than protein-based methods, even though good classification accuracies have been reported for sequences ∼100 bp [72, 86]. One must, however, critically reflect that these results have been obtained either with simulated metagenomes or with real metagenomes of rather low complexity that are not representative for many environmental settings. Signature-based classifications hence work best with sequences from low- to medium diverse habitats where ideally longer assemblies can be obtained or with habitats that feature species with a pronounced DNA composition bias.
Mapping-based classification is the most precise but is often hampered by the availability of suitable reference sequences to map to. As of this writing, 3171 genomes have been completed and 10 536 are ongoing according to the Genomes OnLine Database [92, 93]. Although this number seems impressive, entire clades of the microbial tree are not represented and others only poorly. However, this issue is becoming less and less limiting, as targeted sequencing of as yet unsequenced taxa like in the GEBA project [94] and large-scale metagenome projects like the Earth Microbiome Project [46, 47] start to deliver large quantities of microbial genomes at an increasing pace. Hence, read mapping to closely related reference genomes might become the main method for metagenome taxonomic classifications in the not too distant future.
As of today, there is no standard for the taxonomic classification of metagenome sequences. Also, taxonomic sequence classification can be error prone, in particular for habitats with a complex diversity or high proportions of as yet barely characterized taxa (e.g. [88]). Rather than using a single method, a combination of individual methods is currently the most reasonable approach to partition metagenomes into taxobins. Such combinations have for example been implemented in PhymmBL that combines ICMs and BLAST [72] and in CARMA3 [95] that combines the original CARMA-approach with BLAST. In both cases the combination has already been shown to lead to increased classification accuracy. A combination of BLAST-, CARMA-, SOM- and 16S rRNA gene fragment-based classification termed ‘Taxometer’ was used in recent metagenome studies [19, 88]. Also, different binning methods have been successfully combined to improve accuracy [96]. Besides combining different methods, it has recently been shown that combining multiple related metagenomes in a joint analysis is a way to improve binning accuracy [97].
One interesting aspect of taxonomic sequence classification is that it allows extrapolations onto relative taxon abundances. Although abundance information is lost in the assembly process due to the merger of similar sequences, abundance information can be obtained either from taxonomically classified reads or by back mapping of reads onto taxonomically classified assemblies (Figure 1). It has been shown that relative abundances obtained this way can be close to quantitative cell-based abundances assessments by CARD-FISH [19].
Pre-assembly taxonomic classification and binning
Binning and taxonomic classification methods are typically applied after the assembly. However, these methods can also be used prior to assembly to partition reads into taxonomic bins, which has the potential to substantially reduce the complexity of metagenome assemblies. This strategy might be particularly useful, when sequences from the habitat are already available (e.g. fosmids) that can serve as seeds in an iterative binning-assembly procedure.
BIODIVERSITY ESTIMATION BY 16S rRNA GENE ANALYSIS
About one in every few thousand genes in a metagenome data set is a 16S rRNA gene. With 454 pyrosequencing, this typically translates to ∼1000 reads per picotiter plate (∼1 million reads) that harbor partial 16S rRNA genes with sufficient lengths and quality for phylogenetic analysis. Depending on the length and region of the retrieved partial 16S rRNA gene sequence, phylogenetic analysis can result into varying taxonomic depths. However, since the introduction of 454+ a substantial fraction of the respective reads allows for a genus level assignment, and this situation is expected to even improve with future increases in pyrosequencing read length. A limitation with pyrosequencing is that the number of obtained high-quality 16S rRNA genes might not be sufficient for a representative biodiversity estimation, particular not for lowly abundant taxa. Illumina does not have this problem due to its much higher throughput but on the other hand is plagued by its comparatively short reads that can compromise the depth and quality of the taxonomic assignments.
Dedicated analysis frameworks [98] have been proposed for clustering such data into operational taxonomic units (OTUs). Representative sequences for OTUs can subsequently be mapped against a 16S rRNA reference tree for classification [2, 99]. The advantage of this method over 16S rRNA gene clone libraries is that no primers are involved and hence no primer bias exists (A. Klindworth et al. submitted for publication). The disadvantage, besides not obtaining full-length high-quality 16S rRNA gene sequences, is that different taxa harbor different numbers of rRNA operons, which can distort metagenomic 16S rRNA gene abundances. For example, some Planctomycetes feature large genomes but only a single disjoint rRNA operon [100], which would lead to an underestimation of their abundance in relation to average-sized genomes with more rRNA operons. These limitations notwithstanding, analysis of metagenomic partial 16S rRNA genes provides a direct way to assess a habitat’s biodiversity that in the case of 454+ often provides a resolution down to the genus level. The resulting information is essential for identifying misclassifications in the taxonomic classification of other sequences (as outlined earlier) and identifying taxa that were missed in the taxonomic classification process.
FUNCTIONAL ANALYSIS
Analysis of metagenomes involves functional annotation of the predicted genes by database comparison searches. This typically includes protein BLAST searches against databases such as SWISSPROT, NCBI nr or KEGG [101], HMMer searches against the Pfam [102] and TIGRfam [103] databases, as well as predictions of tRNA [104] and rRNA [104] genes, signal peptides [105], transmembrane regions [106, 107], CRISPR repeats [108] and sub-cellular localization (e.g. using CoBaltDB [109], GNBSL [110], PSLpred [111], CELLO [112] or PSORT-B [113]). Also, dedicated databases are available for special functions, for example the CAZY [111, 114] and dbCAN (http://csbl.bmb.uga.edu/dbCAN/) databases for carbohydrate-active enzymes, the TSdb [115] and TCDB [116, 117] databases for transporters and the MetaBioMe [115] database for enzymes with biotechnological prospects. The resulting annotations are then used as a basis for functional data mining including metabolic reconstruction. Dedicated metagenome annotation systems have been developed to aid these tasks, e.g. WebMGA [118], IMG/M [119–121] and MG-RAST [61, 122, 123]. All three have expanded beyond mere annotation systems and continue to add useful features such as biodiversity analysis, taxonomic classification and metagenome comparisons.
For the latter, a number of dedicated comparison tools have been developed as well, including METAREP [124], STAMP [125], CoMet [126] and RAMMCAP [127]. METAREP and STAMP do not take sequence but already pre-processed data as inputs—tabulated annotations (such as gene ontology (GO) terms, enzyme commission numbers, Pfam hits and BLAST hits) in the case of METAREP and a contingency table of properties (for example exports from Metagenomics Rapid Annotation using Subsystems Technology (MG-RAST), Integrated Microbial Genomes (IMG)/M or CoMet) in the case of STAMP. Both tools feature various statistical tests and visualizations. METAREP is a web service developed by the J. Craig Venter Institute that can compare up to 20 or more metagenomes, whereas STAMP is a stand-alone software. The CoMet and RAMNCAP web servers in contrast do not require pre-computed data. CoMet takes sequence files as an input, does an Orphelia gene prediction, subsequently runs HMMer against the Pfam database followed by multi-dimensional scaling and hierarchical clustering analysis on the Pfam hits and associated GO terms plus visualization of the data. RAMMCAP takes raw reads as an input, does a six-reading frame open reading frame (ORF) prediction, clusters reads and ORFs, does a HMMer and BLAST-based annotation and allows comparison of the data, e.g. by similarity matrices. RAMMCAP is part of the CAMERA data portal [128, 129], which currently comes closest to an integrative processing pipeline for metagenomes with various tools for data retrieval, upload, querying and analysis.
Although automatic in silico annotation is essential for metagenome analysis, one should not forget that a substantial proportion of such annotations are erroneous or even incorrect. Aside from well-studied pathways of the core metabolism, automatic annotations are also often unspecific, i.e. restricted to assigning general functions (e.g. lipase, oxidoreductase, alcohol dehydrogenase) without resolving the involved specific substrates and products. This reflects a fundamental lack of knowledge rather than a limitation of bioinformatic methods per se and can only be addressed by future high-throughput functional screening pipelines.
One of the intriguing aspects of metagenomics is that typically about half of the genes in a metagenome have as yet unknown functions. Hence, restricting metagenome analyses to genes with functional annotations equals to ignoring large proportions of the genes. As a solution, it has been proposed to cluster and analyze metagenomic ORFs in a similar way as OTUs in biodiversity analyses. Such clusters have been termed operational protein families and can be analyzed, for example with MG-DOTUR [130].
AUTOMATIZATION, STANDARDIZATION AND CONTEXTUAL DATA
Until recently, the capacity to sequence has been the limiting factor for metagenome analysis. However, the continual increase in sequencing capacity and decline of costs meanwhile have turned post-metagenomic data analysis into the main bottleneck. Although progress in sequencing technologies still continues at an exponential pace, individuals who analyze the data do not scale equally well. As a consequence, the cost of sequencing drops continuously, whereas the costs for bioinformatic data analysis go up [131]. This is still not recognized widely enough, as metagenome projects tend to suffer from insufficient resource allocation for data post-processing. The latter stresses the needs for further development of semi-automated metagenome analysis tools that allow scientist to handle the wealth of data from recent metagenomics. Steps in metagenome analyses that can be automated should be automated to ensure quality, but this requires the establishment of commonly accepted data formats for metagenome sequences and their associated contextual (meta)data, as well as defined interfaces for data exchange and integration—a task that is tackled by the Genomic Standards Consortium (GSC, http://gensc.org) [132]. Contextual data are among the key factors for successful metagenomes analyses, in particular when it comes to interpretation of time series or biogeographic data. Contextual data are all the data that are associated with a metagenome, such as habitat description (including geographic location and common physicochemical parameters) and sampling procedure (including sampling time). The GSC has published standards for the minimum information about a metagenome sequence (MIMS) [133] as part of the minimum information about any sequence (MIxS) standards and checklist [134], which are supported by the International Nucleotide Sequence Databases Collaboration (INSDC). Similarly, standards have been devised in terms of data formats to ensure data inter-operability, such as the genomic contextual data markup language [135]. It is important that contextual data are collected and integrated into databases, because in the long run these data will allow to extract correlations between geography, time, prevailing environmental conditions and functions from metagenomic data that otherwise never would be uncovered [136].
As of today, there is no comprehensive tool for metagenome analysis that incorporates all types of analysis (biodiversity analysis, taxobinning, functional annotation, metabolic reconstruction and sophisticated statistical comparisons). Consequently, scientists/bioinformaticians in this field need to operate, merge and interpret results from various tools, which for larger data sets can be a daunting task. In terms of pipelines, MetaAMOS [137] provides an integrated solution for the initial post-processing of mated read metagenome data that supports different assemblers, the BAMBUS 2 scaffolder and various gene prediction, annotation and taxonomic classification tools. In terms of data integration, CAMERA has so far developed the most comprehensive infrastructure for holistic metagenome analyses, and further tools and pipelines are currently developed in the GSC and Micro B3 project (http://www.microb3.eu/) frameworks.
DATA SUBMISSION
Progress in sequencing allows for metagenomes with increasing sizes. A full run on an Illumina HiSeq2000 sequencer does not only produce 600 Gb of sequences but also the FASTQ raw data files are multiple times as large. Although sequencing facilities send these data to their customers on Terabyte-scale hard disc drives, such data volumes are certainly not suitable anymore for upload to data analysis servers or INSDC databases for submission, even not with fast user datagram protocoll (UDP) protocols such as Aspera Connect [138]. This is a problem that is as yet unsolved. Also, the INSDC databases are currently not prepared for handling the quality of many metagenomes (pervasiveness of frameshifts, automatically generated non-standard annotations and large amounts of partial genes) and their accessory data (such as lists of metagenomic 16S rRNA gene fragments including taxonomic classifications). It is clear that currently sequencing technologies evolve faster than bioinformatic infrastructures for post-genomic analysis are built. As mentioned before, this has been recognized and efforts such as those of the GSC, Micro B3, CAMERA, MG-RAST and IMG/M are on the way to define standards and develop pipelines for future metagenome data handling. However, it is the authors’ conviction that ultimately the INSDC databases have the mandate and should maintain such tools and the associated infrastructure in the long run. The European Bioinformatics Institute has recognized this and recently has made a submission and analysis pipeline for 454/pyrosequencing metagenome data available (https://www.ebi.ac.uk/metagenomics).
FUTURE PERSPECTIVES
The newest generation of sequencers, such as the PacBio RS, the Ion Torrent Proton or the ONT GRIDIon/MINIon, will continue to propel the field of metagenomics, and who knows whether at some point in the future technologies such as the conceptual IBM/Roche DNA transistor [139] will revolutionize the field again. On the one hand, development of affordable bench-top devices (454 Junior, Illumina MiSeq, Ion Torrent PGM and Proton) has led to a democratization of sequencing, and future devices such as the ONT MINIon could even be used for metagenomic analyses directly in the field. On the other hand, the ever-growing throughput of NGS sequencers is making data analysis increasingly complex.
Although smaller and medium-sized metagenomes can be analyzed with the resources described so far, different infrastructures and bioinformatic pipelines are necessary for future large-scale projects. ‘Megagenome’ projects reach the size of many terabytes of sequences (and beyond), and instead of moving these data around, it is reasonable that they reside at the sequencing institution and that these institutions provide pipelines for remote data analyses. This implies that large-scale sequencing and large-scale computing have become inseparable. For example, the BGI (formerly Beijing Genomics Institute, http://en.genomics.cn) has projected an integrated national center for sample storage, sequencing, data storage and analysis. Monolithic data centers are one way to address this, but also cloud computing such as Amazon’s EC2/S3 can be used as a viable and scalable alternative for large-scale metagenome data analysis [140], provided that the data can be transferred to the cloud, and appropriate data security is guaranteed.
Metagenomics constitutes an invaluable tool for investigating complete microbial communities in situ, in particular when integrated with biodiversity, expression and contextual data (metadata). Continuous advancements in sequencing technologies not only allow for addressing more and more complex habitats but also impose growing demands on bioinformatic data post-processing. Not long ago, sampling and associated logistics, clone library construction and Sanger sequencing of a couple of inserts were the time-consuming steps in metagenomics. Nowadays, analyzing the wealth of data has become the bottleneck, in particular for larger metagenome projects. This stresses the importance of integrative bioinformatic software pipelines for metagenomics/megagenomics, something that we as scientists must support with all efforts to get the most out of metagenome data.
Key Points.
- Metagenomics has become an indispensable and widely affordable tool for studying as yet uncultivable microbes (Bacteria, Archaea and viruses).
- Progress in NGS allows for larger metagenomes, for studying series of metagenomes over time and space and for addressing increasingly complex habitats.
- A new type of integrative ecosystems biology study seeks to combine metagenomics with metatranscriptome, metaproteome, metabolome and biodiversity and contextual (meta)data analyses.
- There are several bioinformatic tools and pipelines for different aspects of metagenome analysis, but there is no standardized, comprehensive pipeline covering all aspects. Large-scale ‘megagenome’ projects are particularly affected and hence face challenges with respect to data handling, data integration and data analysis.
- Ongoing international efforts strive to establish standards and tools for future large-scale metagenome analysis that are necessary to turn the proverbial metagenomic data deluge into knowledge.
Funding
Funding was provided by the Max Planck Society.
ACKNOWLEDGEMENTS
The authors acknowledge Johannes Werner and Pelin Yilmaz for critical reading of the manuscript. Observations from the following publically funded projects have been incorporated in this mini review: MIMAS (Microbial Interactions in Marine Systems; http://mimas-project.de) funded by the German Federal Ministry of Education and Research (BMBF), MAMBA (Marine Metagenomics for New Biotechnological Applications; http://mamba.bangor.ac.uk/) and Micro B3 (Biodiversity, Bioinformatics, Biotechnology; http://www.microb3.eu/), both funded by the FP7 of the European Union.
Biographies
Hanno Teeling followed independent educations as a chemist and biologist before specializing on bioinformatics for microbial genomics. He has worked for 12 years in this field and is currently a scientist at the Max Planck Institute for Marine Microbiology in Bremen.
F rank Oliver Glöckner entered the field of bioinformatics more than 15 years ago. He specialized on tools and databases for microbial biodiversity analysis and microbial genomics. He is the head of the Microbial Genomics and Bioinformatics Research Group at the Max Planck Institute for Marine Microbiology and Professor of Bioinformatics at the Jacobs University Bremen.
References
- 1.Lane DJ, Pace B, Olsen GJ, et al. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82(20):6955–9. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruesse E, Quast C, Knittel K, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–96. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99(16):10494–9. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309(5739):1387–90. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 5.Sogin ML, Morrison HG, Huber JA, et al. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA. 2006;103(32):12115–20. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quince C, Curtis TP, Sloan WT. The rational exploration of microbial diversity. ISME J. 2008;2(10):997–1006. doi: 10.1038/ismej.2008.69. [DOI] [PubMed] [Google Scholar]
- 7.Huse SM, Welch DM, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12(7):1889–98. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–69. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruns A, Nubel U, Cypionka H, Overmann J. Effect of signal compounds and incubation conditions on the culturability of freshwater bacterioplankton. Appl Environ Microbiol. 2003;69(4):1980–89. doi: 10.1128/AEM.69.4.1980-1989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Köpke B, Wilms R, Engelen B, et al. Microbial diversity in coastal subsurface sediments: a cultivation approach using various electron acceptors and substrate gradients. Appl Environ Microbiol. 2005;71(12):7819–30. doi: 10.1128/AEM.71.12.7819-7830.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalisky T, Quake SR. Single-cell genomics. Nat Methods. 2011;8(4):311–14. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 12.Handelsman J, Rondon MR, Brady SF, et al. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol S. 1998;5(10):R245–9. doi: 10.1016/s1074-5521(98)90108-9. [DOI] [PubMed] [Google Scholar]
- 13.Schloss PD, Handelsman J. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol. 2005;6(8):229. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riesenfeld CS, Schloss PD, Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–52. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- 15.Ekkers DM, Cretoiu MS, Kielak AM, Elsas JD. The great screen anomaly—a new frontier in product discovery through functional metagenomics. Appl Microbiol Biotechnol. 2012;93(3):1005–20. doi: 10.1007/s00253-011-3804-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simon C, Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77(4):1153–61. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Gordon JI. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134(5):708–13. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5(6):999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teeling H, Fuchs BM, Becher D, et al. Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science. 2012;336(6081):608–11. doi: 10.1126/science.1218344. [DOI] [PubMed] [Google Scholar]
- 20.Kunin V, Copeland A, Lapidus A, et al. A bioinformatician's guide to metagenomics. Microbiol Mol Biol Rev. 2008;72(4):557–78. doi: 10.1128/MMBR.00009-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyerdierks A, Kube M, Kostadinov I, et al. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol. 2010;12(2):422–39. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 22.Woyke T, Teeling H, Ivanova NN, et al. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature. 2006;443(7114):950–5. doi: 10.1038/nature05192. [DOI] [PubMed] [Google Scholar]
- 23.Pena A, Teeling H, Huerta-Cepas J, et al. Fine-scale evolution: genomic, phenotypic and ecological differentiation in two coexisting Salinibacter ruber strains. ISME J. 2010;4(7):882–95. doi: 10.1038/ismej.2010.6. [DOI] [PubMed] [Google Scholar]
- 24.Woyke T, Xie G, Copeland A, et al. Assembling the marine metagenome, one cell at a time. PLoS One. 2009;4(4):e5299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Garcia M, Brazel DM, Swan BK, et al. Capturing single cell genomes of active polysaccharide degraders: an unexpected contribution of verrucomicrobia. PLoS One. 2012;7(4):e35314. doi: 10.1371/journal.pone.0035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleiner M, Wentrup C, Lott C, et al. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci USA. 2012;109(19):E1173–82. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prosser JI. Replicate or lie. Environ Microbiol. 2010;12(7):1806–10. doi: 10.1111/j.1462-2920.2010.02201.x. [DOI] [PubMed] [Google Scholar]
- 28.Luo C, Tsementzi D, Kyrpides N, et al. Direct comparisons of illumina vs. Roche 454 sequencing technologies on the same microbial community DNA sample. PLoS One. 2012;7(2):e30087. doi: 10.1371/journal.pone.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurlbert SH. Pseudoreplication and the design of ecological field experiments. Ecol Monogr. 1984;54(2):187–211. [Google Scholar]
- 30.Lennon JT. Replication, lies and lesser-known truths regarding experimental design in environmental microbiology. Environ Microbiol. 2011;13(6):1383–6. doi: 10.1111/j.1462-2920.2011.02445.x. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigue S, Materna AC, Timberlake SC, et al. Unlocking short read sequencing for metagenomics. PLoS One. 2010;5(7):e11840. doi: 10.1371/journal.pone.0011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Overbeek R, Fonstein M, D'Souza M, et al. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96(6):2896–901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osterman A, Overbeek R. Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol. 2003;7(2):238–51. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier E, Kreimeyer A, Bocs S, et al. “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J Bacteriol. 2008;190(7):2572–9. doi: 10.1128/JB.01248-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hess M, Sczyrba A, Egan R, et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–7. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 36.Erkel C, Kube M, Reinhardt R, Liesack W. Genome of Rice Cluster I archaea—the key methane producers in the rice rhizosphere. Science. 2006;313(5785):370–2. doi: 10.1126/science.1127062. [DOI] [PubMed] [Google Scholar]
- 37.Iverson V, Morris RM, Frazar CD, et al. Untangling genomes from metagenomes: revealing an uncultured class of marine Euryarchaeota. Science. 2012;335(6068):587–90. doi: 10.1126/science.1212665. [DOI] [PubMed] [Google Scholar]
- 38.Luo C, Tsementzi D, Kyrpides NC, Konstantinidis KT. Individual genome assembly from complex community short-read metagenomic datasets. ISME J. 2012;6(4):898–901. doi: 10.1038/ismej.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laserson J, Jojic V, Koller D. Genovo: de novo assembly for metagenomes. J Comput Biol. 2011;18(3):429–43. doi: 10.1089/cmb.2010.0244. [DOI] [PubMed] [Google Scholar]
- 40.Peng Y, Leung HC, Yiu SM, Chin FY. Meta-IDBA: a de novo assembler for metagenomic data. Bioinformatics. 2011;27(13):i94–101. doi: 10.1093/bioinformatics/btr216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Namiki T, Hachiya T, Tanaka H, Sakakibara Y. In: Proceedings of the 2nd ACM Conference on Bioinformatics, Computational Biology and Biomedicine 2011. New York, NY, USA: ACM; MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai B, Ding R, Li Y, et al. A de novo metagenomic assembly program for shotgun DNA reads. Bioinformatics. 2012;28:1455–62. doi: 10.1093/bioinformatics/bts162. [DOI] [PubMed] [Google Scholar]
- 43.Koren S, Treangen TJ, Pop M. Bambus 2: scaffolding metagenomes. Bioinformatics. 2011;27(21):2964–71. doi: 10.1093/bioinformatics/btr520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Handelsman J. Metagenomics or megagenomics? Nat Rev Microbiol. 2005;3(6):457–8. [Google Scholar]
- 45.Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project. Nature. 2007;449(7164):804–10. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert JA, Meyer F, Antonopoulos D, et al. Meeting report: the terabase metagenomics workshop and the vision of an earth microbiome project. Stand Genomic Sci. 2010;3(3):243–8. doi: 10.4056/sigs.1433550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert JA, Meyer F, Jansson J, et al. The earth microbiome project: meeting report of the “1 EMP meeting on sample selection and acquisition” at Argonne National Laboratory October 6 2010. Stand Genomic Sci. 2010;3(3):249–53. doi: 10.4056/aigs.1443528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoff KJ. The effect of sequencing errors on metagenomic gene prediction. BMC Genomics. 2009;10(1):520. doi: 10.1186/1471-2164-10-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34(19):5623–30. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Noguchi H, Taniguchi T, Itoh T. MetaGeneAnnotator: detecting species-specific patterns of ribosomal binding site for precise gene prediction in anonymous prokaryotic and phage genomes. DNA Res. 2008;15(6):387–96. doi: 10.1093/dnares/dsn027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoff KJ, Tech M, Lingner T, et al. Gene prediction in metagenomic fragments: a large scale machine learning approach. BMC Bioinformatics. 2008;9(1):217. doi: 10.1186/1471-2105-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoff KJ, Lingner T, Meinicke P, Tech M. Orphelia: predicting genes in metagenomic sequencing reads. Nucleic Acids Res. 2009;37(Web Server issue):W101–5. doi: 10.1093/nar/gkp327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38(20):e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kislyuk A, Lomsadze A, Lapidus AL, Borodovsky M. Frameshift detection in prokaryotic genomic sequences. Int J Bioinform Res Appl. 2009;5(4):458–77. doi: 10.1504/IJBRA.2009.027519. [DOI] [PubMed] [Google Scholar]
- 55.Antonov I, Borodovsky M. Genetack: frameshift identification in protein-coding sequences by the Viterbi algorithm. J Bioinform Comput Biol. 2010;8(3):535–51. doi: 10.1142/s0219720010004847. [DOI] [PubMed] [Google Scholar]
- 56.Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 57.Apweiler R, Bairoch A, Wu CH, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32(Database issue):D115–9. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanekamp K, Bohnebeck U, Beszteri B, Valentin K. PhyloGena—a user-friendly system for automated phylogenetic annotation of unknown sequences. Bioinformatics. 2007;23(7):793–801. doi: 10.1093/bioinformatics/btm016. [DOI] [PubMed] [Google Scholar]
- 59.Koski LB, Golding GB. The closest BLAST hit is often not the nearest neighbor. J Mol Evol. 2001;52(6):540–2. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- 60.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–86. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitra S, Rupek P, Richter DC, et al. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinformatics. 2011;12(Suppl. 1):S21. doi: 10.1186/1471-2105-12-S1-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podell S, Gaasterland T. DarkHorse: a method for genome-wide prediction of horizontal gene transfer. Genome Biol S. 2007;8(2):R16. doi: 10.1186/gb-2007-8-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28(3):405–20. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 64.Krause L, Diaz NN, Goesmann A, et al. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 2008;36(7):2230–9. doi: 10.1093/nar/gkn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerlach W, Junemann S, Tille F, et al. WebCARMA: a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinformatics. 2009;10:430. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schreiber F, Gumrich P, Daniel R, Meinicke P. Treephyler: fast taxonomic profiling of metagenomes. Bioinformatics. 2010;26(7):960–1. doi: 10.1093/bioinformatics/btq070. [DOI] [PubMed] [Google Scholar]
- 67.Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13(2):145–58. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McHardy AC, Martin HG, Tsirigos A, et al. Accurate phylogenetic classification of variable-length DNA fragments. Nat Methods. 2007;4(1):63–72. doi: 10.1038/nmeth976. [DOI] [PubMed] [Google Scholar]
- 69.Rocha EP, Viari A, Danchin A. Oligonucleotide bias in Bacillus subtilis: general trends and taxonomic comparisons. Nucleic Acids Res. 1998;26(12):2971–80. doi: 10.1093/nar/26.12.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Teeling H, Meyerdierks A, Bauer M, et al. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ Microbiol. 2004;6(9):938–47. doi: 10.1111/j.1462-2920.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- 71.Reva ON, Tümmler B. Global features of sequences of bacterial chromosomes, plasmids and phages revealed by analysis of oligonucleotide usage patterns. BMC Bioinformatics. 2004;5:90. doi: 10.1186/1471-2105-5-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6(9):673–6. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sandberg R, Winberg G, Branden CI, et al. Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome Res. 2001;11(8):1404–9. doi: 10.1101/gr.186401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Diaz NN, Krause L, Goesmann A, et al. TACOA: taxonomic classification of environmental genomic fragments using a kernelized nearest neighbor approach. BMC Bioinformatics. 2009;10:56. doi: 10.1186/1471-2105-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abe T, Kanaya S, Kinouchi M, et al. Informatics for unveiling hidden genome signatures. Genome Res. 2003;13(4):693–702. doi: 10.1101/gr.634603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Abe T, Sugawara H, Kinouchi M, et al. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 2005;12(5):281–90. doi: 10.1093/dnares/dsi015. [DOI] [PubMed] [Google Scholar]
- 77.Chan C-KK, Hsu AL, Tang S-L, Halgamuge SK. Using growing self-organising maps to improve the binning process in environmental whole-genome shotgun sequencing. J Biomed Biotechnol. 2008;2008:513701. doi: 10.1155/2008/513701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chan CK, Hsu AL, Halgamuge SK, Tang SL. Binning sequences using very sparse labels within a metagenome. BMC Bioinformatics. 2008;9(1):215. doi: 10.1186/1471-2105-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin C, Diaz NN, Ontrup J, Nattkemper TW. Hyperbolic SOM-based clustering of DNA fragment features for taxonomic visualization and classification. Bioinformatics. 2008;24(14):1568–74. doi: 10.1093/bioinformatics/btn257. [DOI] [PubMed] [Google Scholar]
- 80.Dick GJ, Andersson AF, Baker BJ, et al. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009;10(8):R85. doi: 10.1186/gb-2009-10-8-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber M, Teeling H, Huang S, et al. Practical application of self-organizing maps to interrelate biodiversity and functional data in NGS-based metagenomics. ISME J. 2011;5(5):918–28. doi: 10.1038/ismej.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chatterji S, Yamazaki I, Bai Z, Eisen JA. In: Proceedings of the 12th Annual International Conference on Research in Computational Molecular Biology 2008. Heidelberg: Springer-Verlag Berlin; CompostBin: a DNA composition-based algorithm for binning environmental shotgun reads; pp. 17–28. [Google Scholar]
- 83.Yang B, Peng Y, Leung H, et al. In: Proceedings of the First ACM International Conference on Bioinformatics and Computational Biology 2010. New York, NY, USA: ACM; MetaCluster: unsupervised binning of environmental genomic fragments and taxonomic annotation; pp. 170–9. [Google Scholar]
- 84.Leung HCM, Yiu SM, Yang B, et al. A robust and accurate binning algorithm for metagenomic sequences with arbitrary species abundance ratio. Bioinformatics. 2011;27(11):1489–95. doi: 10.1093/bioinformatics/btr186. [DOI] [PubMed] [Google Scholar]
- 85.Kislyuk A, Bhatnagar S, Dushoff J, Weitz JS. Unsupervised statistical clustering of environmental shotgun sequences. BMC Bioinformatics. 2009;10:316. doi: 10.1186/1471-2105-10-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu YW, Ye Y. A novel abundance-based algorithm for binning metagenomic sequences using l-tuples. J Comput Biol. 2011;18(3):523–34. doi: 10.1089/cmb.2010.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meinicke P, Asshauer KP, Lingner T. Mixture models for analysis of the taxonomic composition of metagenomes. Bioinformatics. 2011;27(12):1618–24. doi: 10.1093/bioinformatics/btr266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferrer M, Werner J, Chernikova TN, et al. Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ Microbiol. 2011;14(1):268–81. doi: 10.1111/j.1462-2920.2011.02634.x. [DOI] [PubMed] [Google Scholar]
- 89.Hirschman L, Clark C, Cohen KB, et al. Habitat-lite: a GSC case study based on free text terms for environmental metadata. OMICS. 2008;12(2):129–36. doi: 10.1089/omi.2008.0016. [DOI] [PubMed] [Google Scholar]
- 90.Kolpakov R, Bana G, Kucherov G. mreps: efficient and flexible detection of tandem repeats in DNA. Nucleic Acids Res. 2003;31(13):3672–8. doi: 10.1093/nar/gkg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ning Z, Cox AJ, Mullikin JC. SSAHA: a fast search method for large DNA databases. Genome Res. 2001;11(10):1725–9. doi: 10.1101/gr.194201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kyrpides NC. Genomes OnLine Database (GOLD 1.0): a monitor of complete and ongoing genome projects world-wide. Bioinformatics. 1999;15(9):773–4. doi: 10.1093/bioinformatics/15.9.773. [DOI] [PubMed] [Google Scholar]
- 93.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29(1):126–7. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu D, Hugenholtz P, Mavromatis K, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462(7276):1056–60. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gerlach W, Stoye J. Taxonomic classification of metagenomic shotgun sequences with CARMA3. Nucleic Acids Res. 2011;39(14):e91. doi: 10.1093/nar/gkr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Saeed I, Tang SL, Halgamuge SK. Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acids Res. 2012;40(5):e34. doi: 10.1093/nar/gkr1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baran Y, Halperin E. Joint analysis of multiple metagenomic samples. PLoS Comput Biol. 2012;8(2):e1002373. doi: 10.1371/journal.pcbi.1002373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS One. 2012;7(2):e30126. doi: 10.1371/journal.pone.0030126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yilmaz P, Iversen MH, Hankeln W, et al. Ecological structuring of bacterial and archaeal taxa in surface ocean waters. FEMS Microbiol Ecol. doi: 10.1111/j.1574-6941.2012.01357.x. In press. Doi: 10.1111/j.1574-6941.2012.01357.x. [DOI] [PubMed] [Google Scholar]
- 100.Glöckner FO, Kube M, Bauer M, et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100(14):8298–303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kotera M, Hirakawa M, Tokimatsu T, et al. The KEGG databases and tools facilitating omics analysis: latest developments involving human diseases and pharmaceuticals. Methods Mol Biol. 2012;802:19–39. doi: 10.1007/978-1-61779-400-1_2. [DOI] [PubMed] [Google Scholar]
- 102.Punta M, Coggill PC, Eberhardt RY, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Selengut JD, Haft DH, Davidsen T, et al. TIGRFAMs and genome properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35(Database issue):D260–64. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–64. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2(4):953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 106.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol S. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 107.Liakopoulos TD, Pasquier C, Hamodrakas SJ. A novel tool for the prediction of transmembrane protein topology based on a statistical analysis of the SwissProt database: the OrienTM algorithm. Protein Eng. 2001;14(6):387–90. doi: 10.1093/protein/14.6.387. [DOI] [PubMed] [Google Scholar]
- 108.Bland C, Ramsey TL, Sabree F, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goudenege D, Avner S, Lucchetti-Miganeh C, Barloy-Hubler F. CoBaltDB: complete bacterial and archaeal orfeomes subcellular localization database and associated resources. BMC Microbiol. 2010;10:88. doi: 10.1186/1471-2180-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Guo J, Lin Y, Liu X. GNBSL: a new integrative system to predict the subcellular location for Gram-negative bacteria proteins. Proteomics. 2006;6(19):5099–105. doi: 10.1002/pmic.200600064. [DOI] [PubMed] [Google Scholar]
- 111.Bhasin M, Garg A, Raghava GP. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics. 2005;21(10):2522–4. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- 112.Yu CS, Lin CJ, Hwang JK. Predicting subcellular localization of proteins for Gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004;13(5):1402–6. doi: 10.1110/ps.03479604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gardy JL, Spencer C, Wang K, et al. PSORT-B: improving protein subcellular localization prediction for Gram-negative bacteria. Nucleic Acids Res. 2003;31(13):3613–7. doi: 10.1093/nar/gkg602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park BH, Karpinets TV, Syed MH, et al. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010;20(12):1574–84. doi: 10.1093/glycob/cwq106. [DOI] [PubMed] [Google Scholar]
- 115.Zhao M, Chen Y, Qu D, Qu H. TSdb: a database of transporter substrates linking metabolic pathways and transporter systems on a genome scale via their shared substrates. Sci China Life Sci. 2011;54(1):60–4. doi: 10.1007/s11427-010-4125-y. [DOI] [PubMed] [Google Scholar]
- 116.Saier MHJ, Tran CV, Barabote RD. TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34(Database issue):D181–6. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Saier MHJ, Yen MR, Noto K, et al. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37(Database issue):D274–8. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wu S, Zhu Z, Fu L, et al. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12(1):444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Markowitz VM, Ivanova N, Palaniappan K, et al. An experimental metagenome data management and analysis system. Bioinformatics. 2006;22(14):e359–67. doi: 10.1093/bioinformatics/btl217. [DOI] [PubMed] [Google Scholar]
- 120.Markowitz VM, Ivanova NN, Szeto E, et al. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 2008;36(Database issue):D534–8. doi: 10.1093/nar/gkm869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Markowitz VM, Chen IM, Chu K, et al. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 2012;40(Database issue):D123–9. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meyer F, Paarmann D, D'Souza M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9(1):386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Glass EM, Wilkening J, Wilke A, et al. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5368. pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 124.Goll J, Rusch DB, Tanenbaum DM, et al. METAREP: JCVI metagenomics reports—an open source tool for high-performance comparative metagenomics. Bioinformatics. 2010;26(20):2631–2. doi: 10.1093/bioinformatics/btq455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Parks DH, Beiko RG. Identifying biologically relevant differences between metagenomic communities. Bioinformatics. 2010;26(6):715–21. doi: 10.1093/bioinformatics/btq041. [DOI] [PubMed] [Google Scholar]
- 126.Lingner T, Asshauer KP, Schreiber F, Meinicke P. CoMet—a web server for comparative functional profiling of metagenomes. Nucleic Acids Res. 2011;39(Web Server issue):W518–23. doi: 10.1093/nar/gkr388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li W. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics. 2009;10:359. doi: 10.1186/1471-2105-10-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Seshadri R, Kravitz SA, Smarr L, et al. CAMERA: a community resource for metagenomics. PLoS Biol. 2007;5(3):e75. doi: 10.1371/journal.pbio.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun S, Chen J, Li W, et al. Community cyberinfrastructure for Advanced Microbial Ecology Research and Analysis: the CAMERA resource. Nucleic Acids Res. 2011;39(Database issue):D546–51. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schloss P, Handelsman J. A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics. 2008;9(1):34. doi: 10.1186/1471-2105-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sboner A, Mu XJ, Greenbaum D, et al. The real cost of sequencing: higher than you think! Genome Biol. 2011;12(8):125. doi: 10.1186/gb-2011-12-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Field D, Amaral-Zettler L, Cochrane G, et al. The Genomic Standards Consortium. PLoS Biol. 2011;9(6):e1001088. doi: 10.1371/journal.pbio.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Field D, Garrity G, Gray T, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–7. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yilmaz P, Kottmann R, Field D, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat Biotechnol. 2011;29(5):415–20. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kottmann R, Gray T, Murphy S, et al. A standard MIGS/MIMS compliant XML schema: toward the development of the genomic contextual data markup language (GCDML) OMICS J Integr Biol. 2008;12(2):115–New Orleans, LA22. doi: 10.1089/omi.2008.0A10. [DOI] [PubMed] [Google Scholar]
- 136.Yilmaz P, Gilbert JA, Knight R, et al. The genomic standards consortium: bringing standards to life for microbial ecology. ISME J. 2011;5(10):1565–7. doi: 10.1038/ismej.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Treangen TJ, Koren S, Astrovskaya I, et al. MetAMOS: a metagenomic assembly and analysis pipeline for AMOS. Genome Biol. 2011;12(Suppl 1):25. doi: 10.1186/gb-2013-14-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Beloslyudtsev D. Sequence Read Archive Handbook [Internet] Bethesda (MD): National Center for Biotechnology Information (US); 2010. Aspera transfer guide. [Google Scholar]
- 139.Luan B, Martyna G, Stolovitzky G. Characterizing and controlling the motion of ssDNA in a solid-state nanopore. Biophys J. 2011;101(9):2214–22. doi: 10.1016/j.bpj.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wilkening J, Wilke A, Desai N, Meyer F. Cluster Computing and Workshops, 2009 CLUSTER'09 IEEE International Conference. New Orleans, LA: IEEE; 2009. Using clouds for metagenomics: a case study; pp. 1–6. [Google Scholar]