Differential Roles of Insulin Receptor Substrates in Brown Adipocyte Differentiation (original) (raw)
Abstract
Insulin promotes adipocyte differentiation via a complex signaling network involving multiple insulin receptor substrates (IRSs). In cultured brown preadipocytes, expression of IRS-1 and IRS-2 mRNAs and proteins was at relatively high levels before and after differentiation into mature fat cells, while IRS-3 transcript was not detectable in preadipocytes but increased during the course of differentiation, and IRS-4 mRNA was barely detected in both states. To determine more precisely the roles of various IRS proteins in adipogenesis, we established and characterized brown preadipocyte cell lines from wild-type and IRS knockout (KO) animals. While wild-type, IRS-2 KO, and IRS-4 KO cells fully differentiated into mature adipocytes, IRS-3 KO cells showed a moderate defect in differentiation and IRS-1 KO cells exhibited a severe defect in the process. Cells lacking both IRS-1 and IRS-3 completely failed to differentiate. Expression of the adipogenic markers peroxisome proliferator-activated receptor gamma (PPARγ), CCAAT/enhancer-binding protein alpha, fatty acid synthase, glucose transporter 4, and the transcription factor signal transducer and activator of transcription 5, as well as the brown-fat-specific markers PPARγ coactivator 1 alpha and uncoupling protein 1, mirrored the differentiation pattern. Reconstitution of the IRS-1 KO cells with IRS-1 and IRS-4, but not IRS-2 or IRS-3, compensated for the lack of differentiation in IRS-1 KO cells. A chimeric molecule containing the N terminus of IRS-1 and the C terminus of IRS-2, but not one with the N terminus of IRS-2 and the C terminus of IRS-1, also rescued differentiation. Expression of Wnt 10a, a molecule known to inhibit adipogenesis, was dramatically increased in the IRS-1 KO cells, and this could be reduced by overexpression of IRS-1 or IRS-4, which was correlated with restoration of differentiation. These data indicate that both IRS-1 and -3 play important roles in the differentiation of brown adipocytes and that the N terminus of IRS-1 is more important for this function of the molecule. Although IRS-4 is not essential for the process, overexpression of IRS-4 can compensate for the deficiency in differentiation in IRS-1 KO cells.
Adipose tissues play important roles in obesity, insulin resistance, and diabetes. Two functionally different types of fat are known in mammals. White adipose tissue is the primary site of deposit of triglycerides and release of fatty acids, and brown adipose tissue is specialized in thermogenic energy expenditure through the expression of uncoupling protein 1 (UCP-1) (6). The developmental patterns of the two tissues are quite distinct. Brown adipose tissue develops during fetal life and possesses all the features of mature tissue at birth, when the requirements for nonshivering thermogenesis are needed. In contrast, the development of white adipose tissue takes place after birth, and its mass increases during postnatal life (31, 32). Understanding the regulation of the differentiation of both types of adipose tissue is important for whole-body energy balance, since both obesity and lipoatrophy lead to profound metabolic diseases (21, 27).
Adipogenesis is a complex process that is tightly controlled by positive and negative stimuli, including a variety of hormones and nutrients (5, 17, 24, 36, 37). Recently, significant progress has been made in understanding the cellular and molecular mechanisms of white adipocyte differentiation; however, knowledge of brown adipocyte differentiation is still very limited. At the cellular and molecular levels, the program of white adipocyte differentiation can be divided into at least four stages: (i) preconfluent proliferation, (ii) confluence-growth arrest, (iii) hormonal induction-clonal expansion, and (iv) permanent growth arrest-terminal differentiation (7, 18). The later part of this process is under complex transcriptional control involving CCAAT/enhancer-binding protein β (C/EBPβ), -δ, and -α; peroxisome proliferator-activated receptor gamma (PPARγ); and other transcription factors that are induced in a specific sequence. This leads to the synthesis of proteins characteristic of a fully differentiated phenotype, including fatty acid synthase (FAS) and glucose transporter 4 (GLUT 4). In addition, the signal transducers and activators of transcription (STAT) family of transcription factors, especially STAT 1, 3, and 5, have recently been implicated in 3T3-L1 adipocyte differentiation (2, 20, 41). More recently, PPARγ coactivator-1 (PGC-1) has been identified as a potentially unique mechanism leading to brown fat differentiation (34, 35).
The upstream signaling pathways leading to the activation of these transcriptional events during adipocyte differentiation are still poorly understood. The phosphoinositide 3-kinase (PI3K) pathway appears to be required for differentiation of 3T3-L1 adipocytes (28, 16, 40). In contrast, mitogen-activated protein kinase (MAPK) pathways are inhibitory to adipogenesis (2, 15, 33). Insulin and insulin-like growth factor 1 (IGF-1) appear to play specific roles in adipocyte differentiation, presumably via activation of these pathways (18).
The insulin receptor substrates (IRSs) are a growing family of proteins that are phosphorylated by the activation of insulin, IGF-1, growth hormone, and other cytokine receptors (45). Although the four IRS proteins have similar overall architectures, disruption of each individual IRS gene causes distinct phenotypes in mice (1, 12, 26, 46), indicating that the four IRS proteins play different roles in the regulation of the pleiotropic effects of insulin, IGF-1, and other growth factors. Our laboratory has successfully established brown preadipocyte cell lines derived from different IRS knockout (KO) mice and has begun to use these cells as a model system to study the roles of various IRS proteins in the development of adipose cells (23). These cell lines retain the characteristics of brown fat, including the ability to accumulate lipids, express UCP-1, and respond to β-adrenergic stimulation (22). It has been found that differentiation of IRS-1 KO brown preadipocytes is impaired (13), whereas IRS-2 KO cells can be differentiated into mature adipocytes but have defects in insulin-induced glucose uptake (14). However, the complete extent to which the various IRS proteins play unique versus redundant or complementary roles in the regulation of adipogenesis remains to be determined.
In this study, we have investigated the roles of the IRS proteins in brown adipocyte differentiation by using cell lines derived from all four types of IRS KO mice. While wild-type, IRS-2 KO, and IRS-4 KO cells fully differentiate into mature adipocytes, IRS-3 KO cells show a moderate defect in differentiation, and IRS-1 KO cells exhibit a marked defect in the process. Reconstitution experiments indicate important roles for IRS-1, especially through its N-terminal domain, in the process of brown adipocyte differentiation, and this may involve the Wnt signaling pathway.
MATERIALS AND METHODS
Materials.
The antibodies used for immunoblotting included anti-IRS-1 (JD 287, which recognized amino acid residues 511 to 859), anti-IRS-2 (Upstate Biotechnology, Lake Placid, N.Y.), anti-phospho-p44/42 MAPK (Thr-202/Tyr-204), anti-p44/42 MAPK, anti-phospho-p38 MAPK (Thr-180/Tyr-182) and anti-p38 MAPK (Cell Signaling, Beverly, Mass.), anti-C/EBPδ, anti-PPARγ, and anti-C/EBPα (Santa Cruz Biotechnology, Santa Cruz, Calif.), anti-Glut4 (Chemicon International, Inc., Temecula, Calif.), and anti-STAT 5 (BD Biosciences, San Diego, Calif.). Immobilon P transfer membranes were from Millipore (Billerica, Mass.), and electrophoresis supplies were from Bio-Rad Laboratories (Hercules, Calif.). All other supplies were from Sigma Chemical Co. (St. Louis, Mo.) unless otherwise specified.
Cell isolation and culture.
Brown adipocytes and their precursor cells were isolated from newborn wild-type and IRS KO mice by collagenase digestion as described previously (13, 14, 42). Preadipocytes were grown to confluence (day 0) in differentiation medium (Dulbecco's modified Eagle's medium containing 10% fetal bovine serum supplemented with 20 nM insulin and 1 nM 3,3′,5-triiodo-l-thyronine [T3]). Adipocyte differentiation was induced by treating confluent cells for 48 h in differentiation medium further supplemented with 0.5 mM isobutylmethylxanthine (IBMX), 0.5 μM dexamethasone, and 0.125 mM indomethacin. After this induction period (day 2), the cells were placed back in differentiation medium, which was then changed every second day. After four more days in differentiation medium (day 6), the cells exhibited a fully differentiated phenotype with massive accumulation of multilocular fat droplets.
Plasmids and retroviral infection of cells.
Full-length human IRS-1, mouse IRS-2, mouse IRS-3, and mouse IRS-4, as previously described, were cloned into a retroviral vector (13, 14, 42, 43). N1.C2 and N2.C1 chimeras were constructed by using _Afl_III sites of IRS-1 and IRS-2. The N1.C2 construct contains the N-terminal half of IRS-1 and the C-terminal region of IRS-2. Conversely, the N2.C1 chimera contains the N-terminal domains of IRS-2 and the C-terminal half of IRS-1 (42). Viral φ NX-packaging cells were transfected at 70% confluence by the calcium phosphate method (30), and the viral supernatants were harvested 48 h after transfection. IRS-1 KO cells were infected overnight at 60% confluence with Polybrene (4 μg/ml)-supplemented virus-containing supernatants. Selection with 250 μg of the bleomycin analogue Zeocin (Invitrogen, Carlsbad, Calif.)/ml was started 48 h after infection.
Quantitative RT-PCR analysis using Taqman procedure.
Total RNA isolated from cells was digested by DNase I and purified with RNeasy Mini Kit columns (Qiagen, Valencia, Calif.). The number of copies of transcript of each IRS were measured using the Applied Biosystems (ABI) Taqman procedure in an ABI Prism 7700 sequence detection system (PE Biosystems, Foster City, Calif.). This instrument uses a fluorescence detection system to follow PCR product formation. The cycle conditions were as follows: 1 cycle of 30 min at 50°C (reverse-transcription [RT] step), 1 cycle of 10 min at 95°C, and 40 cycles of 15 s at 95°C and 1 min at 60°C. Primers (90 nM) and probes (100 nM) specific for each IRS were added to the reaction mixture containing reverse transcriptase and RNA templates. Plasmids with cDNA for each IRS were used to prepare calibration curves for the IRS (H. J. Goren, R. N. Kulkarni, and C. R. Kahn, submitted for publication). The sequences of the primers and probes used in this study are as follows: 5′IRS-1/2, CTGGAGTATTATGAGTCGAGAAGAAGTGG; 5′IRS-2/3, CTGGAGTTCTACGAGACGAGAAGAAGT; 5′IRS-4, CTGTACCAATGCTTCTCCGTGA; 3′IRS-1, GTAGAGAGCCACCAGGTGCTTGT; 3′IRS-2, TGTAGAGGGCGATCAGGTACTTGT; 3′IRS-3, ACGATCAGGTGGCGCTGAC; 3′IRS-4, AGGGCAATGAGGTGTCGGT; IRS-1 probe, 6FAM-CATCAACAAGCGGGCTGACTCCAAGA-TAMRA; IRS-2 probe, TET-TCAACAAGCGCGCGGACGC-TAMRA; IRS-3 probe, VIC-AGCAAGCGCGCGGATGCC-TAMRA; and IRS-4 probe, 6FAM-CCAGCGCGCCGATGCCA-TAMRA.
Quantitative RT-PCR analysis using SYBR Green procedure.
cDNA was prepared from 1 μg of RNA using the Advantage RT-PCR kit (BD Biosciences) according to the manufacturer's instructions. Five microliters of cDNA was used in a 25-μl PCR (SYBR Green; PE Biosystems) containing primers at a concentration of 300 nM each. PCRs were run in triplicate and quantitated in the ABI Prism 7700 sequence detection system. The results were expressed as arbitrary mRNA units. The sequences of the Wnt 10a and PGC-1α primers are as follows: 5′Wnt 10a, CACCCGGCCATACTTCCT; 3′Wnt 10a, CACTTACGCCGCATGTTCT; 5′PGC-1α, GTCAACAGCAAAAGCCACAA; 3′PGC-1α, TCTGGGGTCAGAGGAAGAGA.
Oil red O staining.
Dishes were washed twice with phosphate-buffered saline and fixed with 10% buffered formalin for at least 1 h at room temperature. Cells were then stained for 2 h at room temperature with a filtered oil red O solution (0.5% oil red O in isopropyl alcohol), washed twice with distilled water, and visualized.
Western blot analysis.
Cells were harvested in lysis buffer (50 mM HEPES, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 10 mM Na2P2O7, 10 mM NaF, 2 mM EDTA, 10% glycerol, 1% Igepal CA-630, 2 mM vanadate, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, 2 mM phenylmethylsulfonyl fluoride, pH 7.4). After lysis, the lysates were clarified by centrifugation at 12,000 × g for 10 min at 4°C, and the amounts of protein in the supernatants were determined by the Bradford protein assay (Bio-Rad). Equal amounts of protein (100 μg) were directly solubilized in Laemmli sample buffer. The lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes. The membranes were blocked for 30 min and incubated with the appropriate antibody for 2 h at room temperature. Specifically bound primary antibodies were detected with peroxidase-coupled secondary antibody and enhanced chemiluminescence (Amersham Biosciences, Piscataway, N.J.).
Northern blot analysis.
Total cellular RNA was isolated using the ULTRASPEC RNA isolation system (Biotecx Laboratories, Inc., Houston, Tex.) following the manufacturer's instructions. Twenty micrograms of RNA was then subjected to Northern analysis with various probes as described previously (22), exposed to a PhosphorImager screen, and quantitated with a Molecular Dynamics densitometer.
RESULTS
IRS-1 and IRS-3 play important roles in brown adipocyte differentiation.
Brown adipocyte precursor cells were induced to differentiate into adipocytes using insulin, T3, dexamethasone, IBMX, and indomethacin as described in Materials and Methods. A fat-specific dye, oil red O, was used to monitor lipid accumulation. On day 6 of differentiation, adipocytes from wild-type, IRS-2 KO, and IRS-4 KO mice differentiated into mature brown adipocytes as shown by uniform oil red O staining. Cells from IRS-3 KO mice differentiated to a lesser extent, and cells from IRS-1 KO mice showed a marked defect in differentiation (Fig. 1A) (13). This defect in differentiation was maintained even when the cells were allowed to accumulate lipid through day 13 (data not shown). The roles of IRS-1 and IRS-3 in brown adipocyte differentiation were further addressed by using cell lines lacking both proteins (25). Consistent with the lipoatrophic phenotype found in the double-KO animals, brown precursor cells lacking both IRS-1 and IRS-3 totally failed to differentiate into adipocytes (Fig. 1B). Thus, these data suggest that IRS-4 and IRS-2 are not required for brown adipocyte differentiation but IRS-1 and IRS-3 are essential and might play complementary roles in adipogenesis.
FIG. 1.
IRS-1 and IRS-3 play important roles in brown adipocyte differentiation. (A) Differentiation of wild-type and all four IRS KO brown adipocytes. (B) Comparison of differentiation of wild-type, IRS-1 KO, IRS-3 KO, and IRS-1-IRS-3 double-KO adipocytes. Brown adipose precursor cells isolated from newborn wild-type and different IRS KO mice were grown to confluence. Differentiation was induced as described in Materials and Methods. On day 6, the cells were fixed and stained with oil red O. The results are representative of at least four independent experiments.
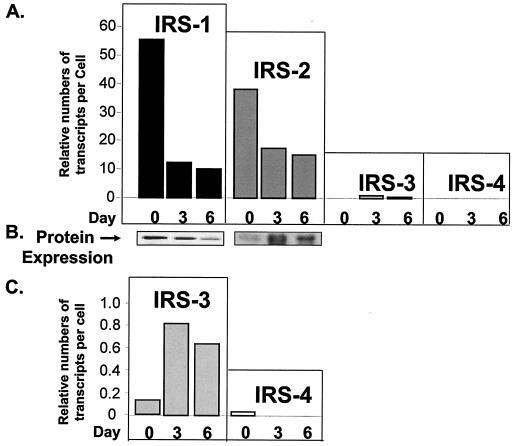
Using quantitative RT-PCR, we estimated the copy numbers per cell of each IRS transcript in wild-type cells during the course of differentiation. The levels of IRS-1 and IRS-2 transcripts were high at the preadipocyte stage and then decreased during differentiation (Fig. 2A). This expression pattern of IRS-1 mRNA perfectly correlated with the levels of protein during differentiation, whereas IRS-2 protein was very low on day 0, peaked on day 3, and then decreased somewhat by day 6 (Fig. 2B). The discrepancy between the expression patterns of IRS-2 mRNA and protein might be due to additional regulation of IRS-2 protein via the proteasome pathway, as recently described by Rui et al. (39). By comparison, the expression level of IRS-3 was much lower than those of IRS-1 and IRS-2 but increased on day 3 and then slightly decreased by day 6 of differentiation. The expression of IRS-4 was barely detected in both preadipocytes and adipocytes, consistent with previous findings (11, 26) (Fig. 2C).
FIG. 2.
Expression of IRS mRNAs and proteins in wild-type brown adipocytes during differentiation. (A) Quantitative real-time PCR by the ABI Taqman procedure was used to measure the transcript numbers for each IRS in total RNA extracted from wild-type cells on days 0, 3, and 6 of differentiation. Details of RNA preparation, primers, probes, and the method of quantification are described in Materials and Methods. The data are presented as relative numbers of transcripts per cell using calibration curves prepared for each IRS. (B) Protein expression of IRS-1 and IRS-2 in wild-type brown adipocyte differentiation. Specific antibodies against IRS-1 or IRS-2 were used in Western blot analysis as described in Materials and Methods. (C) Blow-up graph of panel A to better illustrate the expression levels of IRS-3 and IRS-4 mRNAs during differentiation. The results are means from two to four experiments.
Patterns of differentiation correlate with expression of adipogenic markers.
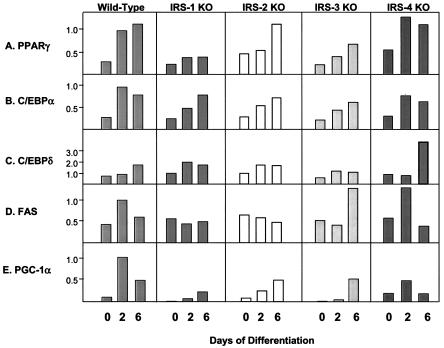
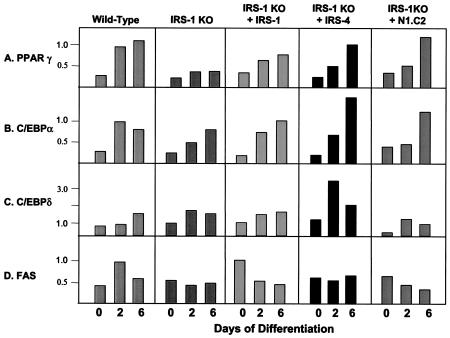
To further investigate the roles of the IRS proteins in differentiation, we monitored the expression of the adipogenic markers C/EBPδ, C/EBPα, PPARγ, GLUT 4, FAS, STAT 5, PGC-1α, and UCP-1 at the mRNA and protein levels during the differentiation process. PPARγ showed a progressive increase in expression between days 0 and 6 of differentiation in wild-type cells. IRS-2 and IRS-4 KO cells showed a similar pattern, while IRS-3 KO cells showed somewhat decreased expression, and the IRS-1 KO cells, which failed to differentiate, showed the lowest expression of PPARγ (Fig. 3A). C/EBPα showed a pattern similar to that of PPARγ in all cell types. The transcripts of C/EBPα peaked on day 2 in wild-type and IRS-4 KO cells, whereas IRS-1 KO, IRS-2 KO, and IRS-3 KO cells showed delayed responses (Fig. 3B). The expression levels of C/EBPδ were generally low in all of the cell lines with the exception of the IRS-4 KO cells, which expressed an ∼3-fold-higher level of C/EBPδ than wild-type cells by day 6 (Fig. 3C). For FAS, both wild-type and IRS-4 KO cells showed peak expression on day 2; however, this response was blunted in IRS-1 KO and IRS-2 KO cells and delayed in IRS-3 KO cells (Fig. 3D). The expression levels of PGC-1 peaked on day 2 and remained at a high level on day 6 in wild-type cells, whereas this expression was markedly blunted in IRS-1 KO cells (Fig. 3E). Thus, by both lipid accumulation and mRNA expression pattern, IRS-4 KO cells most closely mirror wild-type cells, confirming that IRS-4 is not required for differentiation in brown fat cells.
FIG. 3.
mRNA expression of adipocyte differentiation markers in wild-type, IRS-1 KO, IRS-2 KO, IRS-3 KO, and IRS-4 KO cells on days 0, 2, and 6 of differentiation. Total RNA was isolated from the cells, and 20 μg of RNA was subjected to Northern analysis with specific probes for the adipogenic markers PPARγ, C/EBPα, C/EBPδ, and FAS. Expression of PGC-1 was measured by quantitative RT-PCR analysis using the SYBR Green protocol as described in Materials and Methods. The data are presented as arbitrary units after normalization to day 2 wild-type levels for each experiment. The results are means of three independent experiments. The average standard error of the mean was 20.3% at all data points.
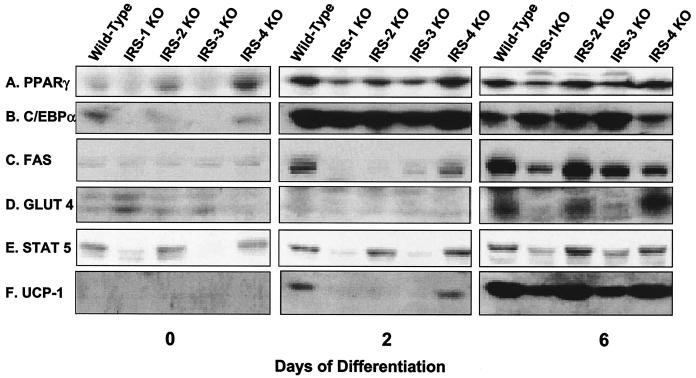
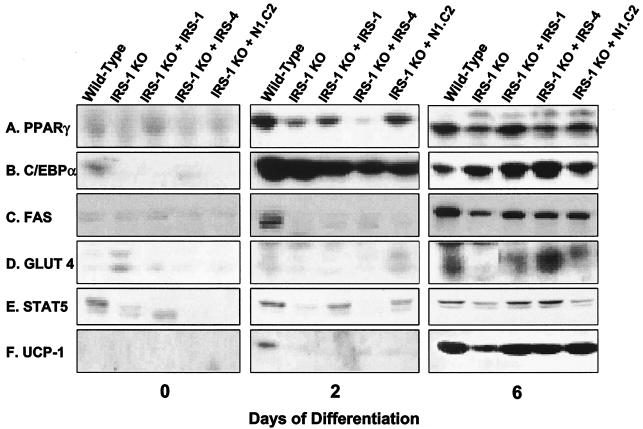
At the protein level, PPARγ showed a pattern of expression comparable to mRNA levels. On day 0, PPARγ was already detectable in wild-type, IRS-2 KO, and IRS-4 KO cells but was at much lower levels in IRS-1 and IRS-3 KO cells. As differentiation progressed, so did the expression of PPARγ, with wild-type, IRS-2 KO, and IRS-4 KO cells expressing the highest levels, whereas IRS-1 KO and IRS-3 KO cells lagged behind (Fig. 4A). The pattern of protein expression for C/EBPα was similar to that of PPARγ. C/EBPα was expressed at low but detectable levels in preadipocytes (day 0) and rose dramatically in all cells undergoing differentiation (Fig. 4B). Expression of C/EBPδ protein was barely detected in all cell lines despite mRNA expression (data not shown). FAS was detected on day 2 of differentiation in wild-type and IRS-4 KO cells and reached maximum levels in all cell lines on day 6, with the lowest level of expression in the IRS-1 null cells (Fig. 4C). Similarly, GLUT 4 was most highly expressed in wild-type, IRS-2 KO, and IRS-4 KO cells on day 6. The slight increase in GLUT 4 protein in IRS-1 KO cells on day 0 was not consistently observed in cell lines derived from different mice (13) and probably reflected the variance of different cell lines derived from mice of mixed genetic background, such as the IRS-1 KO mice (Fig. 4D).
FIG. 4.
Protein expression of adipocyte differentiation markers in wild-type, IRS-1 KO, IRS-2 KO, IRS-3 KO, and IRS-4 KO cells on days 0, 2, and 6 of differentiation. Protein lysates were prepared from the cells and analyzed for the adipogenic markers PPARγ, C/EBPα, FAS, GLUT 4, STAT 5, and UCP-1 by Western blot analysis as described in Materials and Methods. The results are representative of at least two independent experiments.
It has been shown that expression of STAT 5 is tightly correlated with the adipocyte phenotype in 3T3-L1 cells (41). Consistent with this finding, levels of STAT 5 were lower in cells with a deficiency in differentiation than in those cells that could differentiate into mature adipocytes (Fig. 4E). On day 2, expression of the thermogenic protein UCP-1 was detected only in wild-type and IRS-4 KO cells, and the levels had increased in all cell lines by day 6, with the lowest expression in IRS-1 KO and IRS-3 KO cells (Fig. 4F). Thus, for each IRS KO cell line, protein expression parallels mRNA expression, and this also matches differentiation as monitored by oil red O staining.
Reconstitution of IRS-1 KO and IRS-1-IRS-3 double-KO cells and regulation of adipocyte differentiation.
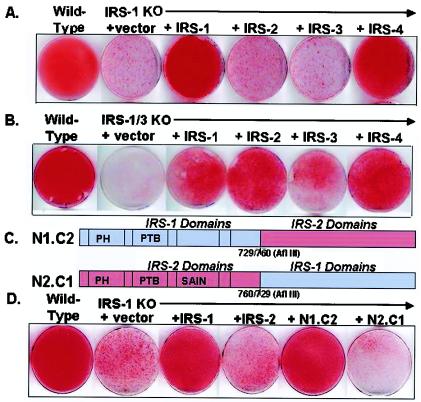
To examine the possible complementary versus redundant roles among IRS family members, we used retrovirus-mediated gene transfer to reconstitute the IRS-1 KO cells with full-length IRS-1, IRS-2, IRS-3, and IRS-4. As noted above, using oil red O staining, wild-type cells were fully differentiated by day 6 while IRS-1 KO cells did not undergo differentiation. Reexpression of IRS-1 in the IRS-1 null cells compensated for this defect, whereas overexpression of IRS-2 or IRS-3 did not compensate. Surprisingly, although the IRS-4 KO cells had no defect in differentiation, overexpression of IRS-4 fully compensated for the lack of differentiation in IRS-1 KO cells (Fig. 5A). Similar reconstitution experiments were also performed with the IRS-3 KO and IRS-1-IRS-3 double-KO cells. As expected, IRS-3 was able to completely restore the moderate decrease in differentiation of IRS-3 KO cells (data not shown), and either IRS-1 or IRS-3 partially compensated for the severe defect in the IRS-1-IRS-3 double-KO cells. Overexpression of IRS-4 in the cells lacking both IRS-1 and IRS-3 almost fully restored differentiation. Interestingly, the double-KO cells overexpressing IRS-2 showed partial rescue of adipocyte differentiation (Fig. 5B). This might be due to a notable supraphysiological level of IRS-2 protein expression in these cells (data not shown). The structure-function relationships of IRS proteins were further assessed using overexpression of IRS-1- IRS-2 chimeric proteins in the IRS-1 KO cells. An N1.C2 construct containing the N-terminal half of IRS-1 and the C-terminal half of IRS-2 (Fig. 5C) was able to fully rescue differentiation, as determined by oil red O staining of accumulated lipid (Fig. 5D). By contrast, the N2.C1 chimera, with the N-terminal half of IRS-2 and the C-terminal half of IRS-1, did not support differentiation to the same extent (Fig. 5D).
FIG. 5.
Reconstitution of the IRS-1 KO and IRS-1-IRS-3 double-KO cells with various IRS proteins or IRS-1-IRS-2 chimeras. IRS-1 KO cells (A) or IRS-1-IRS-3 double-KO cells (B) were reconstituted with (+) IRS-1, IRS-2, IRS-3, or IRS-4 as described in Materials and Methods. Differentiation was induced as described in the legend to Fig. 1. On day 6, the cells were fixed and stained with oil red O. (C) Schematic diagram showing the structures of IRS-1-IRS-2 chimeras. (D) On day 6 of differentiation, wild-type and IRS-1 KO cells and IRS-1 KO cells reconstituted with IRS-1, IRS-2, N1.C2, and N2.C1 were fixed and stained with oil red O.
Next, we examined mRNA and protein expression of adipogenic markers in these cells. As in the previous experiment, the mRNA expression of the adipocyte differentiation marker PPARγ and C/EBPα increased steadily in wild-type cells during differentiation. IRS-1 KO cells had a reduced level of PPARγ that was partially restored by IRS-1 and IRS-4 reconstitution, as well as by the N1.C2 chimera (Fig. 6A and B). The expression levels of C/EBPδ were low in all of the cell lines, with the exception that overexpression of IRS-4 in the IRS-1 KO cells promoted C/EBPδ expression on day 2 of differentiation (Fig. 6C). FAS mRNA expression was elevated on day 2 in wild-type cells, and this response was blunted in IRS-1-deficient cells. Overexpression of IRS-1, IRS-4, or N1.C2 did not restore the peak response on day 2, but IRS-1-reconstituted cells showed increased expression of FAS on day 0 (Fig. 6D). At the protein level, PPARγ, C/EBPα, FAS, GLUT 4, STAT 5, and UCP-1 were reduced in IRS-1 KO cells and were partially or completely restored by reconstitution of the cells with IRS-1, IRS-4, or the N1.C2 chimeric protein (Fig. 7). These markers also demonstrated protein expression comparable to mRNA expression. Thus, these data confirm that IRS-4, as well as the N1.C2 chimera, can replace IRS-1 in brown fat cells in the differentiation process.
FIG. 6.
mRNA expression of adipocyte differentiation markers in wild-type and IRS-1 KO cells and IRS-1 KO cells reconstituted with (+) IRS-1, IRS-4, or the N1.C2 chimera on days 0, 2, and 6 of differentiation. Total RNA was isolated from the cells, and 20 μg was subjected to Northern analysis with specific probes for the adipogenic markers PPARγ, C/EBPα, C/EBPδ, and FAS. The data are presented as arbitrary units after normalization to day 2 wild-type levels for each experiment. The results are means of three independent experiments. The average standard error of the mean was 21.4% at all data points.
FIG. 7.
Protein expression of adipocyte differentiation markers in wild-type and IRS-1 KO cells and IRS-1 KO cells reconstituted with (+) IRS-1, IRS-4, or the N1.C2 chimera on days 0, 2, and 6 of differentiation. Protein lysates were prepared from the cells and analyzed for the adipogenic markers PPARγ, C/EBPα, FAS, GLUT 4, STAT 5, and UCP-1 by Western blot analysis as described in Materials and Methods. The results are representative of at least two independent experiments.
Wnt signaling is involved in IRS-1-mediated brown adipocyte differentiation.
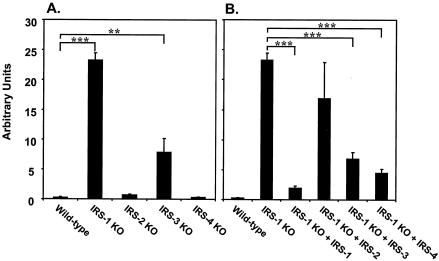
To address the potential mechanisms by which IRS-1 regulated adipogenesis of brown fat, we investigated molecules that have been described as playing a role in adipogenesis. The Wnt signaling pathway has been shown to exert an inhibitory function in the differentiation of 3T3-L1 preadipocytes through down-regulation of the adipogenic transcription factors C/EBPα and PPARγ (4, 38). Using quantitative RT-PCR, we examined the expression of different components of the Wnt signaling pathway and found a dramatic increase in the expression of Wnt 10a in IRS-1 KO cells. More interestingly, the levels of Wnt 10a expression in different IRS KO cells were inversely correlated with their abilities to differentiate (Fig. 8A), suggesting dose-dependent regulation of adipocyte differentiation by Wnt 10a. To confirm that Wnt 10a expression was mediated through an IRS-1-dependent pathway, we examined its expression in IRS-1-reconstituted cells and found that IRS-1 reexpression significantly reduced the elevated levels of Wnt 10a transcript in IRS-1 KO cells. Overexpression of IRS-2 in the IRS-1-deficient cells had no effect on Wnt 10a expression, whereas IRS-3 or IRS-4 reconstitution partially decreased the levels of Wnt 10a mRNA.
FIG. 8.
Expression of Wnt 10a mRNA in wild-type and different IRS KO cells (A) and IRS-1 KO cells reconstituted with (+) various IRS proteins (B) at the preadipocyte stage. Quantitative RT-PCR analysis using the SYBR Green protocol was used to measure the expression levels of Wnt 10a in various cell lines as described in Materials and Methods. The data are from three independent experiments and are expressed as the mean plus standard error of the mean. Significance is determined relative to wild-type (A) or IRS-1 KO (B) cells by Student's t test. **, P < 0.01; ***, P < 0.0001.
Tyrosine phosphorylation and differentiation.
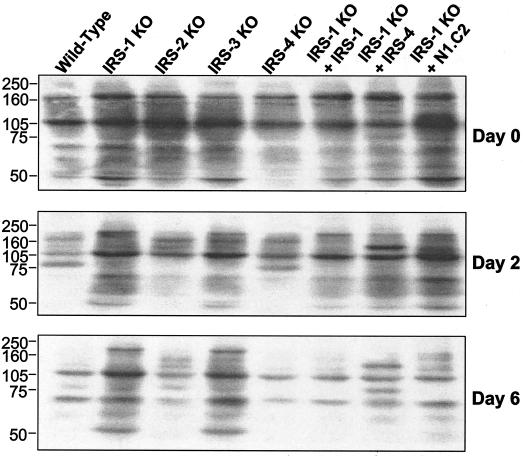
The phosphorylation of signaling proteins on tyrosine residues leads to downstream effects of many growth factors and hormones, including insulin and IGF-1. To assess the overall tyrosine phosphorylation of proteins during differentiation in IRS KO cells and cells reconstituted with IRS-1, IRS-4, or the N1.C2 chimera, Western blot analysis was performed on direct lysates from these cells (Fig. 9). As differentiation progressed, overall tyrosine phosphorylation of proteins decreased in cells that differentiated, including wild-type, IRS-2 KO, IRS-4 KO, and IRS-1 null cells reconstituted with IRS-1, IRS-4, and the N1.C2 chimera. Conversely, in cells that did not differentiate, i.e., the IRS-1 KO and IRS-3 KO cells, total tyrosine phosphorylation of proteins decreased to a lesser extent. This was also the case for the IRS-1 null cells reconstituted with the N2.C1 construct (data not shown). These data suggest that the degree of overall tyrosine phosphorylation of cellular proteins inversely correlates with adipocyte differentiation. This may reflect a change in growth factor function or the action of phosphotyrosine phosphatases during adipogenesis.
FIG. 9.
Overall tyrosine phosphorylation of proteins decreases during differentiation. Wild-type, IRS-1 KO, IRS-2 KO, IRS-3 KO, and IRS-4 KO cells and IRS-1 KO cells reconstituted with (+) IRS-1, IRS-4, or the N1.C2 chimera were differentiated, and protein lysates were prepared on days 0, 2, and 6 of differentiation. The lysates were separated by sodium dodecyl sulfate-6% polyacrylamide gel electrophoresis and analyzed by Western blotting using antiphosphotyrosine antibody (4G10). Representative blots are shown. Numbers on the left are protein molecular weight markers.
MAPK pathways may be differentially regulated during the differentiation process.
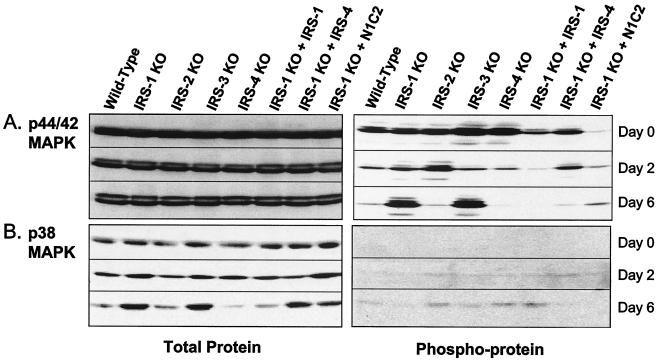
An important role of PI3K and its downstream effector, Akt, has been demonstrated in the differentiation of brown fat (13). Thus, both PI3K activity and Akt phosphorylation are impaired in the IRS-1 KO cells, and these defects can be restored by reconstitution with IRS-1 in the IRS-1 null cells. For MAPK pathways, however, the literature is controversial. In the present study, we found that protein levels of p44/42 MAPK were unchanged during adipogenesis, but phosphorylation of this protein decreased as differentiation progressed (Fig. 10A). In cells that had reduced differentiation (IRS-1 KO and IRS-3 KO), phosphorylation of p44/42 MAPK remained elevated even on day 6. Reconstitution of the IRS-1 null cells with IRS-1, IRS-4, or the N1.C2 chimera rescued this phenomenon (Fig. 10A).
FIG. 10.
Expression of total proteins and phosphoproteins of p42/44 MAPK (A) and p38 MAPK (B). Wild-type, IRS-1 KO, IRS-2 KO, IRS-3 KO, and IRS-4 KO cells and IRS-1 KO cells reconstituted with (+) IRS-1, IRS-4, or the N1.C2 chimera were differentiated, and protein lysates were prepared on days 0, 2, and 6 of differentiation. The lysates were separated by sodium dodecyl sulfate-0% polyacrylamide gel electrophoresis and analyzed by Western blotting using antibodies against p44/42 MAPK and p38 MAPK (left) or phospho-p44/42 MAPK and phospho-p38 MAPK (right).
In cells that differentiated (wild type, IRS-2 KO, IRS-4 KO, and IRS-1 KO reconstituted with IRS-1), p38 MAPK protein levels decreased during differentiation, while in cells that did not differentiate (IRS-1 KO and IRS-3 KO), p38 MAPK protein increased by day 6. Interestingly, in the IRS-1 KO cells reconstituted with IRS-4 and the N1.C2 chimera, although the cells did differentiate into mature adipocytes, total p38 MAPK protein increased to almost the same levels seen in the cells with deficiencies in differentiation. Phosphorylation of p38 MAPK was barely detectable on day 0 and day 2 of differentiation but showed a pattern inverse to that of total protein on day 6 (Fig. 10B).
DISCUSSION
Adipose tissue not only serves as a fat depot but also acts as a secretory or endocrine organ and plays a central role in the regulation of energy balance and thermoregulation by virtue of the mitochondrial protein UCP-1 expressed in brown fat. Understanding the regulation of white adipocyte differentiation has been one of the main focuses of cell biology and obesity-related research, while knowledge about brown adipocyte differentiation is more limited. In vitro, a combination of dexamethasone, IBMX, and insulin is commonly used to induce differentiation of both white and brown preadipocytes; however, exactly what signaling pathways are used by these factors to induce adipogenesis is still incompletely understood.
The IRS proteins are important mediators in insulin signaling and thus may play a crucial role in insulin-regulated biological effects, including adipogenesis. Using brown preadipocytes isolated from wild-type and IRS KO mice, important roles of IRS-1 in both the adipogenic (13) and antiapoptotic (42) functions of insulin and IGF-1 have been demonstrated. By contrast, IRS-2 KO cells can differentiate into mature adipocytes but have impaired insulin-stimulated glucose uptake (14). This function of IRS-2 correlates with a higher level of protein expression in mature wild-type adipocytes than in preadipocytes. In this study, we have further extended these observations and defined special roles for IRS-3 and IRS-4 in this process. Thus, IRS-3 KO cells have a partial defect in differentiation, and although deletion of the IRS-4 gene has no effect on brown preadipocyte differentiation, overexpression of this protein in IRS-1 null cells completely restores the deficiency in differentiation. Taken together, these data suggest that the IRS proteins may play unique, as well as complementary, roles in adipogenesis.
Consistent with these findings, IRS-1- IRS-3 double-KO mice exhibit marked generalized lipoatrophy with hyperglycemia, hyperinsulinemia, and severe insulin resistance (25). Using fibroblasts derived from IRS-deficient embryos, Miki et al. have also demonstrated a critical role of IRS-1 in adipocyte differentiation (29). Furthermore, embryonic fibroblasts lacking both IRS-1 and IRS-2 completely fail in differentiation. Since double KO of IRS-1 and IRS-2 genes causes embryonic lethality, we are unable to generate brown preadipocyte cell lines lacking both proteins. Consistent with the recent findings by Valverde et al. (44), we also find that expression of the UCP-1 protein is significantly reduced in IRS-1 KO cells after differentiation. This may be due to a decrease of PGC-1α expression in the IRS-1 KO preadipocytes. Taking these data together, it is evident that IRS-1 plays a critical role in both white and brown adipocyte differentiation. IRS-2, on the other hand, is not required for brown fat differentiation but may play an important role in the differentiation of white fat, as suggested by Miki et al. (29). In this report, we have shown for the first time that IRS-3 has a moderate effect on brown adipocyte differentiation. Furthermore, mice with a combined deficiency of IRS-1 and IRS-3 are severely lipoatrophic, indicating that in vivo IRS-3 may also play an important role, along with IRS-1, in the development of white fat.
Although IRS-4 does not appear to be required for adipogenesis in vitro, overexpression of IRS-4 in IRS-1 KO or IRS-1-IRS-3 double-KO cells results in a fully differentiated phenotype. In addition, the ability of IRS-4 to restore the defect in differentiation in cells lacking IRS-1 is well demonstrated by induction of expression of the adipogenic markers PPARγ, C/EBPα, and C/EBPδ, indicating an adipogenic potential for IRS-4 when overexpressed in cells with deficiencies in differentiation. This occurs despite the fact that knocking out IRS-4 alone has no effect on adipogenesis in whole animals (12) or on the differentiation of cultured brown adipocytes in this study. An increase has been observed in the basal activities of PI3K and Akt in IRS-1 KO brown preadipocytes reconstituted with IRS-4 (42). Since both kinases have been inferred by other studies to play an important role in adipocyte differentiation (16, 28, 40), this may also provide potential pathways which IRS-4 utilizes to compensate for the adipogenic function of IRS-1. These findings present additional lines of evidence for redundant versus complementary functions in the IRS protein family.
The Wnt signaling pathway (specifically, Wnt 1 and Wnt 10b) has been shown to inhibit 3T3-L1 adipocyte differentiation (4, 38). In this study, we found that expression of the other member of the Wnt family, Wnt 10a, is markedly elevated in the IRS-1 KO preadipocytes and can be reduced by reconstitution of the IRS-1 KO cells with either IRS-1 or IRS-4, or IRS-3 to a lesser extent. The distinction between Wnt 10b in 3T3-L1 adipocytes and Wnt 10a in brown preadipocytes may provide the first evidence for the involvement of different members of the Wnt family of proteins in the regulation of white versus brown fat differentiation. We have also begun to search for other potential mechanisms utilized by various IRS proteins to regulate adipogenesis by using expression-profiling approaches. Preliminary data suggest that many of the extracellular matrix components, as well as cell cycle regulators, may play important roles in these processes (Y. H. Tseng, A. Butte, E., V. K. Yechoor, and C. R. Kahn, unpublished observations).
Although IRS-1 and IRS-2 are well conserved in most regions of known functional importance and are both highly expressed in brown preadipocytes, only IRS-1 is required for differentiation. Interestingly, the chimeric protein N1.C2, which contains the N-terminal half of IRS-1 and the C-terminal half of IRS-2, but not the reversed construct N2.C1, is able to rescue differentiation, implying that the PH and PTB domains or other surrounding sequence motifs located in the amino terminus of IRS-1 possess some information that is important in adipocyte differentiation. This region of IRS-1 is known to play a role in its interaction with the plasma membrane and the juxtamembrane region of the insulin receptor (3, 8), suggesting that some aspect of these interactions is crucial for adipocyte differentiation.
The signaling pathways mediating white adipocyte differentiation have been intensively studied; however, there is still some confusion as to the “essential” pathways for this process, in part due to the different systems utilized to study the process. It is generally believed that activation of the PI3K pathway is required for differentiation of 3T3-L1 adipocytes (16, 28, 40). It has been shown that PI3K activity is also necessary for brown adipocyte differentiation (13). The role of the MAPK pathway in adipogenesis is less clear (2, 15, 33). In this study, we found that the levels of phosphorylation of p44/42 MAPK decreased as differentiation proceeded in wild-type brown preadipocytes, while in cells with a deficiency in differentiation, the levels of phospho-p44/42 MAPK remained high (Fig. 10). On the other hand, treatment of the wild-type cells with the MEK inhibitor PD98059 had no effect on adipogenesis (13), indicating that activation of the p44/42 MAPK pathway is not essential for brown adipocyte differentiation. Furthermore, it has been shown that the levels of p44/42 MAPK phosphorylation in response to acute IGF-1 stimulation are not altered in IRS-1-deficient brown preadipocytes compared to those in the wild-type cells (42). Therefore, the increased levels of phosphorylation of p44/42 MAPK on day 6 in IRS-1 KO and IRS-3 KO cells may be a result of the defect in differentiation rather than the cause of this process.
In this report, we have documented that the levels of overall tyrosine phosphorylation are decreased as brown fat differentiation progresses, while in cells that do not differentiate, total tyrosine phosphorylation decreases to a lesser extent. Related to these findings, a recent study by Harmon et al. has demonstrated that the tyrosine kinase inhibitor genistein inhibits 3T3-L1 adipocyte differentiation by blocking C/EBPβ activity (19). Whether this is specific to 3T3-L1 adipogenesis or a general event in both white and brown fat differentiation is still not clear. Taking these data together, we speculate that IRS-1 and IRS-3 may play a critical role in tyrosine phosphorylation of certain key proteins in the regulation of brown fat differentiation.
Recently, p38 MAPK, a stress-activated MAPK, has been reported to play a role in the differentiation of 3T3-L1 cells (9, 10). Consistent with these findings, we find that cells with defects in differentiation display decreased levels of phospho-p38 MAPK on day 6 compared to those cells that differentiate. Interestingly, the p38 MAPK protein was also increased in both IRS-1 KO and IRS-3 KO cells. This may represent a failed “compensatory” mechanism which is not sufficient to overcome the defect in differentiation. Moreover, the patterns of expression of p38 MAPK protein, as well as its phosphorylation, in wild-type cells during adipogenesis are somewhat distinct from those found in 3T3-L1 cells, likely reflecting intrinsic differences between white and brown adipocytes.
In summary, IRS-1 and IRS-3 play important roles in brown adipocyte differentiation. Defects in differentiation in the IRS-1 KO cells can be restored by reconstitution of these cells with IRS-1 itself and IRS-4, as well as a chimeric protein containing the N terminus of IRS-1 and the C terminus of IRS-2, but not with IRS-2 or IRS-3. Expression of the adipogenic markers PPARγ, C/EBPα, FAS, GLUT 4, and STAT 5, as well as the brown fat-specific markers PGC-1α and UCP-1, mirrored the differentiation pattern. These data suggest that the IRS proteins may play unique, as well as complementary, roles in the regulation of adipogenesis, and this may be due to activation of distinct downstream effectors and/or target genes.
Acknowledgments
We thank M. Fasshauer, J. Klein, and K. Ueki for preparation of cell lines. We gratefully acknowledge A. Entingh for help with the kinase assay, H. J. Goren for valuable advice on quantitative RT-PCR analysis, and M. Benito for introduction of the brown adipocyte cell lines to our laboratory. We are also grateful to J. Marr for excellent secretarial assistance.
This work was supported in part by the National Institutes of Health grants DK33201 and DK55545 (to C.R.K.) and DK101183 (to Y.-H.T.).
REFERENCES
- 1.Araki, E., M. A. Lipes, M. E. Patti, J. C. Brüning, B. L. Haag III, R. S. Johnson, and C. R. Kahn. 1994. Alternative pathway of insulin signaling in mice with targeted disruption of the IRS-1 gene. Nature 372**:**186-190. [DOI] [PubMed] [Google Scholar]
- 2.Aubert, J., N. Belmonte, and C. Dani. 1999. Role of pathways for signal transducers and activators of transcription, and mitogen-activated protein kinase in adipocyte differentiation. Cell. Mol. Life Sci. 56**:**538-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backer, J. M., C. Wjasow, and Y. Zhang. 1997. In vitro binding and phosphorylation of insulin receptor substrate 1 by the insulin receptor. Role of interactions mediated by the phosphotyrosine-binding domain and the pleckstrin-homology domain. Eur. J. Biochem. 245**:**91-96. [DOI] [PubMed] [Google Scholar]
- 4.Bennett, C. N., S. E. Ross, K. A. Longo, L. Bajnok, N. Hemati, K. W. Johnson, S. D. Harrison, and O. A. MacDougald. 2002. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 277**:**30998-31004. [DOI] [PubMed] [Google Scholar]
- 5.Camp, H. S., D. Ren, and T. Leff. 2002. Adipogenesis and fat-cell function in obesity and diabetes. Trends Mol. Med. 8**:**442-447. [DOI] [PubMed] [Google Scholar]
- 6.Collins, S., W. Cao, K. W. Daniel, T. M. Dixon, A. V. Medvedev, H. Onuma, and R. Surwit. 2001. Adrenoceptors, uncoupling proteins, and energy expenditure. Exp. Biol. Med. 226**:**982-990. [DOI] [PubMed] [Google Scholar]
- 7.Cowherd, R. M., R. E. Lyle, and R. E. J. McGehee. 1999. Molecular regulation of adipocyte differentiation. Semin. Cell Dev. Biol. 10**:**3-10. [DOI] [PubMed] [Google Scholar]
- 8.Eck, M. J., S. Dhe-Paganon, T. Trub, R. T. Nolte, and S. E. Shoelson. 1996. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell 85**:**695-705. [DOI] [PubMed] [Google Scholar]
- 9.Engelman, J. A., A. H. Berg, R. Y. Lewis, A. Lin, M. P. Lisanti, and P. E. Scherer. 1999. Constitutively active mitogen-activated protein kinase kinase 6 (MKK6) or salicylate induces spontaneous 3T3-L1 adipogenesis. J. Biol. Chem. 274**:**35630-35638. [DOI] [PubMed] [Google Scholar]
- 10.Engelman, J. A., M. P. Lisanti, and P. E. Scherer. 1998. Specific inhibitors of p38 mitogen-activated protein kinase block 3T3-L1 adipogenesis. J. Biol. Chem. 273**:**32111-32120. [DOI] [PubMed] [Google Scholar]
- 11.Fantin, V. R., B. E. Lavan, Q. Wang, N. A. Jenkins, D. J. Gilbert, N. G. Copeland, S. R. Keller, and G. E. Lienhard. 1999. Cloning, tissue expression, and chromosomal location of the mouse insulin receptor substrate 4 gene. Endocrinology 140**:**1329-1337. [DOI] [PubMed] [Google Scholar]
- 12.Fantin, V. R., Q. Wang, G. E. Lienhard, and S. R. Keller. 2000. Mice lacking insulin receptor substrate 4 exhibit mild defects in growth, reproduction, and glucose homeostasis. Am. J. Physiol. Endocrinol. Metab. 278**:**E127-E133. [DOI] [PubMed] [Google Scholar]
- 13.Fasshauer, M., J. Klein, K. M. Kriauciunas, K. Ueki, M. Benito, and C. R. Kahn. 2001. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol. Cell. Biol. 21**:**319-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fasshauer, M., J. Klein, K. Ueki, K. M. Kriauciunas, M. Benito, M. F. White, and C. R. Kahn. 2000. Essential role of insulin receptor substrate-2 in insulin stimulation of glut4 translocation and glucose uptake in brown adipocytes. J. Biol. Chem. 275**:**25494-25501. [DOI] [PubMed] [Google Scholar]
- 15.Font, D. M., A. Porras, N. Ahn, and E. Santos. 1997. Mitogen-activated protein kinase activation is not necessary for, but antagonizes, 3T3-L1 adipocytic differentiation. Mol. Cell. Biol. 17**:**6068-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gagnon, A., C. S. Chen, and A. Sorisky. 1999. Activation of protein kinase B and induction of adipogenesis by insulin in 3T3-L1 preadipocytes: contribution of phosphoinositide-3,4,5-trisphosphate versus phosphoinositide-3,4-bisphosphate. Diabetes 48**:**691-698. [DOI] [PubMed] [Google Scholar]
- 17.Gregoire, F. M. 2001. Adipocyte differentiation: from fibroblast to endocrine cell. Exp. Biol. Med. 226**:**997-1002. [DOI] [PubMed] [Google Scholar]
- 18.Gregoire, F. M., C. M. Smas, and H. S. Sul. 1998. Understanding adipocyte differentiation. Physiol. Rev. 78**:**783-809. [DOI] [PubMed] [Google Scholar]
- 19.Harmon, A. W., Y. M. Patel, and J. B. Harp. 2002. Genistein inhibits CCAAT/enhancer-binding protein beta (C/EBPβ) activity and 3T3-L1 adipogenesis by increasing C/EBP homologous protein expression. Biochem. J. 367**:**203-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harp, J. B., D. Franklin, A. A. Vanderpuije, and J. M. Gimble. 2001. Differential expression of signal transducers and activators of transcription during human adipogenesis. Biochem. Biophys. Res. Commun. 281**:**907-912. [DOI] [PubMed] [Google Scholar]
- 21.Klaus, S. 1997. Functional differentiation of white and brown adipocytes. BioEssays 19**:**215-223. [DOI] [PubMed] [Google Scholar]
- 22.Klein, J., M. Fasshauer, M. Ito, B. B. Lowell, M. Benito, and C. R. Kahn. 1999. Beta(3)-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin induced glucose uptake in brown adipocytes. J. Biol. Chem. 274**:**34795-34802. [DOI] [PubMed] [Google Scholar]
- 23.Klein, J., M. Fasshauer, H. H. Klein, M. Benito, and C. R. Kahn. 2002. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays 24**:**382-388. [DOI] [PubMed] [Google Scholar]
- 24.Koutnikova, H., and J. Auwerx. 2001. Regulation of adipocyte differentiation. Ann. Med. 33**:**556-561. [DOI] [PubMed] [Google Scholar]
- 25.Laustsen, P. G., M. D. Michael, B. E. Crute, S. E. Cohen, K. Ueki, R. N. Kulkarni, S. R. Keller, G. E. Lienhard, and C. R. Kahn. 2002. Lipoatrophic diabetes in Irs1(−/−)/Irs3(−/−) double knockout mice. Genes Dev. 16**:**3213-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu, S. C., Q. Wang, G. E. Lienhard, and S. R. Keller. 1999. Insulin receptor substrate 3 is not essential for growth or glucose homeostasis. J. Biol. Chem. 274**:**18093-18099. [DOI] [PubMed] [Google Scholar]
- 27.MacDougald, O. A., and S. Mandrup. 2002. Adipogenesis: forces that tip the scales. Trends Endocrinol. Metab. 13**:**5-11. [DOI] [PubMed] [Google Scholar]
- 28.Magun, R., B. M. Burgering, P. J. Coffer, D. Pardasani, Y. Lin, J. Chabot, and A. Sorisky. 1996. Expression of a constitutively activated form of protein kinase B (c-Akt) in 3T3-L1 preadipose cells causes spontaneous differentiation. Endocrinology 137**:**3590-3593. [DOI] [PubMed] [Google Scholar]
- 29.Miki, H., T. Yamauchi, R. Suzuki, K. Komeda, A. Tsuchida, N. Kubota, Y. Terauchi, J. Kamon, Y. Kaburagi, J. Matsui, Y. Akanuma, R. Nagai, S. Kimura, K. Tobe, and T. Kadowaki. 2001. Essential role of insulin receptor substrate 1 (IRS-1) and IRS-2 in adipocyte differentiation. Mol. Cell. Biol. 21**:**2521-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18**:**3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moulin, K., N. Truel, M. Andre, E. Arnauld, M. Nibbelink, B. Cousin, C. Dani, L. Penicaud, and L. Casteilla. 2001. Emergence during development of the white-adipocyte cell phenotype is independent of the brown-adipocyte cell phenotype. Biochem. J. 356**:**659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nedergaard, J., E. Connally, and B. Cannon. 1986. Brown adipose tissue in the mammalian neonate, p. 152-213. In P. Trayhurn and D. G. Nicholls (ed.), Brown adipose tissue. Edward Arnold, Baltimore, Md.
- 33.Prusty, D., B. H. Park, K. E. Davis, and S. R. Farmer. 2002. Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPARγ) and C/EBPα gene expression during the differentiation of 3T3-L1 preadipocytes. J. Biol. Chem. 277**:**46226-46232. [DOI] [PubMed] [Google Scholar]
- 34.Puigserver, P., and B. M. Spiegelman. 2003. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 24**:**78-90. [DOI] [PubMed] [Google Scholar]
- 35.Puigserver, P., A. Wu, C. W. Park, R. Graves, M. Wright, and B. R. Spiegelman. 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 92**:**829-839. [DOI] [PubMed] [Google Scholar]
- 36.Rangwala, S. M., and M. A. Lazar. 2000. Transcriptional control of adipogenesis. Annu. Rev. Nutr. 20**:**535-559. [DOI] [PubMed] [Google Scholar]
- 37.Rosen, E. D., C. J. Walkey, P. Puigserver, and B. M. Spiegelman. 2000. Transcriptional regulation of adipogenesis. Genes Dev. 14**:**1293-1307. [PubMed] [Google Scholar]
- 38.Ross, S. E., N. Hemati, K. A. Longo, C. N. Bennett, P. C. Lucas, R. L. Erickson, and O. A. MacDougald. 2000. Inhibition of adipogenesis by Wnt signaling. Science 289**:**950-953. [DOI] [PubMed] [Google Scholar]
- 39.Rui, L., T. L. Fisher, J. Thomas, and M. F. White. 2001. Regulation of insulin/insulin-like growth factor-1 signaling by proteasome-mediated degradation of insulin receptor substrate-2. J. Biol. Chem. 276**:**40362-40367. [DOI] [PubMed] [Google Scholar]
- 40.Sakaue, H., W. Ogawa, M. Matsumoto, S. Kuroda, M. Takata, T. Sugimoto, B. M. Spiegelman, and M. Kasuga. 1998. Posttranscriptional control of adipocyte differentiation through activation of phosphoinositide 3-kinase. J. Biol. Chem. 273**:**28945-28952. [DOI] [PubMed] [Google Scholar]
- 41.Stephens, J. M., R. F. Morrison, and P. F. Pilch. 1996. The expression and regulation of STATs during 3T3-L1 adipocyte differentiation. J. Biol. Chem. 271**:**10441-10444. [DOI] [PubMed] [Google Scholar]
- 42.Tseng, Y. H., K. Ueki, K. M. Kriauciunas, and C. R. Kahn. 2002. Differential roles of insulin receptor substrates in the anti-apoptotic function of insulin-like growth factor-1 and insulin. J. Biol. Chem. 277**:**31601-31611. [DOI] [PubMed] [Google Scholar]
- 43.Tsuruzoe, K., R. Emkey, K. M. Kriauciunas, K. Ueki, and C. R. Kahn. 2001. Insulin receptor substrate 3 (IRS-3) and IRS-4 impair IRS-1- and IRS-2-mediated signaling. Mol. Cell. Biol. 21**:**26-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valverde, A. M., M. Arribas, C. Mur, P. Navarro, S. Pons, A. M. Cassard-Doulcier, C. R. Kahn, and M. Benito. 2003. Insulin-induced up-regulated uncoupling protein-1 expression is mediated by insulin receptor substrate 1 through the phosphatidylinositol 3-kinase/Akt signaling pathway in fetal brown adipocytes. J. Biol. Chem. 278**:**10221-10231. [DOI] [PubMed] [Google Scholar]
- 45.White, M. F. 2002. IRS proteins and the common path to diabetes. Am. J. Physiol. Endocrinol. Metab. 283**:**E413-E422. [DOI] [PubMed] [Google Scholar]
- 46.Withers, D. J., J. S. Gutierrez, H. Towery, D. J. Burks, J. M. Ren, S. Previs, Y. Zhang, D. Bernal, S. Pons, G. I. Shulman, S. Bonner-Weir, and M. F. White. 1998. Disruption of IRS-2 causes type 2 diabetes in mice. Nature 391**:**900-904. [DOI] [PubMed] [Google Scholar]