The Crystal Structure of Human Argonaute2 (original) (raw)
. Author manuscript; available in PMC: 2013 May 25.
Published in final edited form as: Science. 2012 Apr 26;336(6084):1037–1040. doi: 10.1126/science.1221551
Abstract
Argonaute proteins form the functional core of the RNA-induced silencing complexes (RISCs) that mediate RNA silencing in eukaryotes. The 2.3 Å resolution crystal structure of human Argonaute2 (Ago2) reveals a bi-lobed molecule with a central cleft for binding guide and target RNAs. Nucleotides 2 to 6 of a heterogeneous mixture of guide RNAs are positioned in an A-form conformation for base pairing with mRNA targets. Between nucleotides 6 and 7 there is a kink, which may function in miRNA target recognition or release of sliced RNA products. Tandem tryptophan binding pockets in the PIWI domain define a likely interaction surface for recruitment of GW182 or other tryptophan-rich cofactors. These results will enable structure-based approaches for harnessing the untapped therapeutic potential of RNA silencing in humans.
RNA silencing processes, such as the RNA interference (RNAi) and microRNA (miRNA) pathways, are mediated by a specialized family of RNA-binding proteins named Argonaute. Argonaute proteins bind small regulatory RNAs (21–23 nt) and use the encoded sequence information to locate and silence complementary target RNAs. Targeted RNAs are silenced either by direct cleavage via the endonucleolytic “slicing” reaction catalyzed by some Argonaute proteins (1, 2), or by Argonaute-mediated recruitment of additional silencing factors (3–5). Structural studies of prokaryotic homologs, which use DNA guides to recognize and cleave target oligonucleotides, revealed a bi-lobed architecture composed of four globular domains (N, PAZ, MID and PIWI) connected through two structured linker domains (L1 and L2) (6). The two lobes form the walls of a central cleft that cradles guide DNAs and complementary targets (7–9). An RNase H-like active site in the PIWI domain catalyzes the cleavage of target nucleic acids (6, 10, 11). Although structures of isolated PAZ, MID and MID-PIWI domains from several eukaryotic Argonaute proteins have been reported (12–18), the extent to which the structures and mechanisms of full-length Argonautes resemble those of their prokaryotic cousins is not known.
We determined the crystal structure of full-length human Ago2 to a resolution of 2.3 Å (Table S1). Ago2 has a bi-lobed structure reminiscent of that seen in prokaryotes (Fig. 1 and S1). However, the lobes of Ago2 do no align with the corresponding lobes derived from prokaryotic structures, revealing large structural differences between Argonautes from different kingdoms of life (Fig. 2A and S2). In contrast, the individual domains of Ago2 superimpose reasonably well with their prokaryotic counterparts (Fig. 2B). Therefore, the major architectural differences between prokaryotic and eukaryotic Argonautes appear mainly in the relative positions of well-conserved core domain structures (Fig. S3 and S4). The core domains in Ago2 also have extended loops and additional secondary structures, not present in bacteria, that are likely to play roles in guide binding, target RNA recognition and recruitment of Ago2-assoicated protein factors (Fig. S5 and S6).
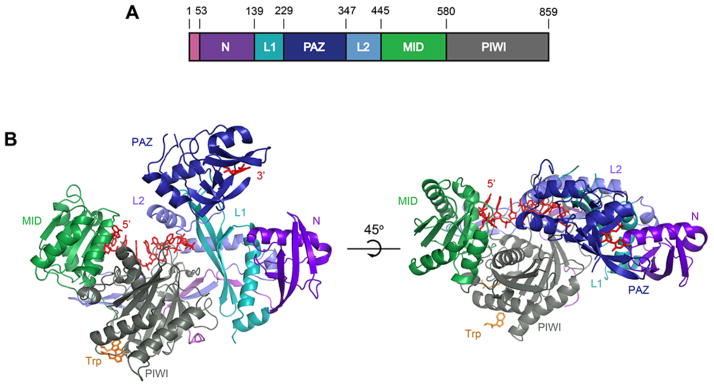
Figure 1. Structure of human Ago2.
A. Schematic of the Ago2 primary sequence. B. Front and top views of Ago2 with the N (purple), PAZ (navy), MID (green), PIWI (grey) domains and linkers L1 (teal) and L2 (blue). A generic guide RNA (red) can be traced for nucleotides 1–8 and 21. Tryptophan molecules (orange) bind to tandem hydrophobic pockets in the PIWI domain.
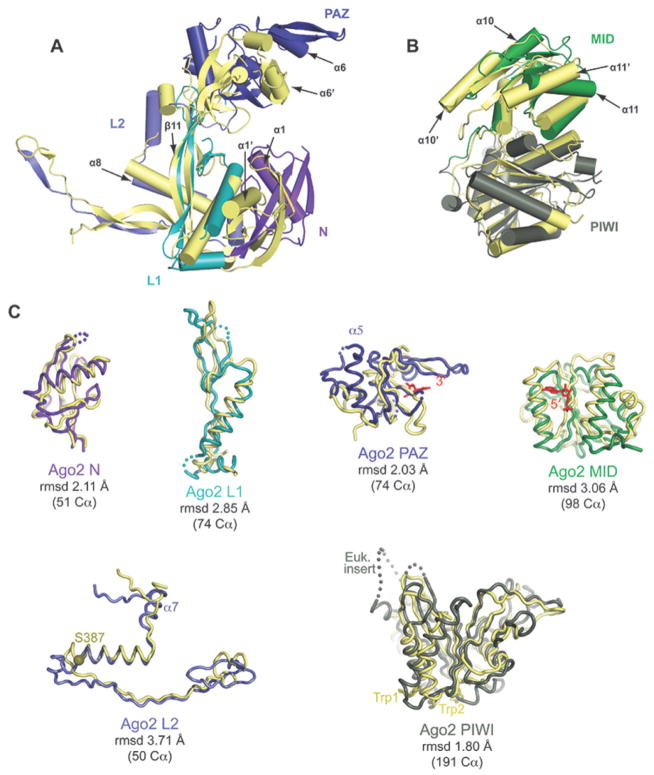
Figure 2. Comparison of bacterial and human Argonaute structures.
A. Superposition of N-PAZ and MID-PIWI lobes of Ago2 (colored as in Fig. 1) onto corresponding lobes from T. thermophiles (yellow). B. Individual domains of Ago2 superimposed on the corresponding domains from T. thermophilus with root-mean-square deviation (rmsd) values for equivalent alpha-carbons indicated. Functional points of interest in Ago2 are labeled.
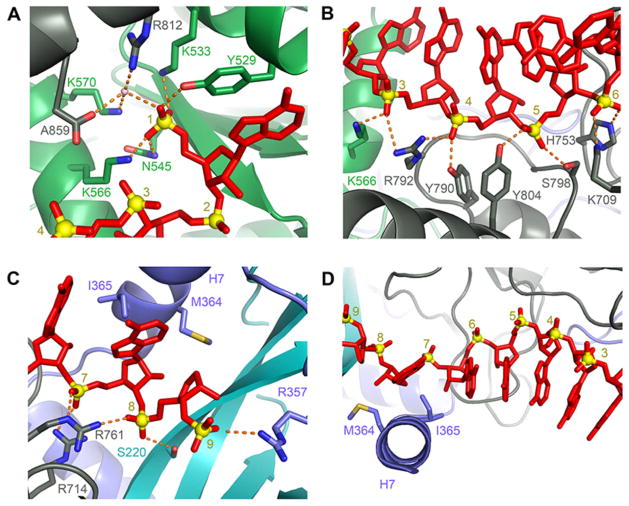
We observed electron density for an eight-nucleotide stretch of single stranded RNA extending across the MID/PIWI/L2/L1 interface (Fig. 3). This density likely arises from heterogeneous, small cellular RNAs (~20–10 nt in length) that accompany Ago2 in our preparations (Fig. S7). The RNA is bound in a conformation similar of guide DNAs in bacterial structures. The electron density for nucleotides 1–7 is well defined, indicating that Ago2 positions this segment of guide RNAs in a uniform conformation that is largely sequence independent. We modeled the guide RNA as poly adenosine because the experimental electron density accommodated purine bases (Fig. S8). The 5′ base of the RNA stacks against Y529, which also forms a hydrogen bond to the 5′ phosphate along with side chains of Y529, K533, N545, and K566 (Fig. 3A), as seen in the structure of the isolated MID domain (18). Additional water-mediated contacts to the 5′ phosphate are made by K570, R812, and the carboxyl terminus (A859). In contrast to prokaryotic structures, we did not observe a magnesium ion in the 5′ phosphate binding site. Ago2 contacts the guide RNA primarily through hydrogen bonds and salt linkages to the phosphate backbone and Van der Waals interactions with the body of the ribose sugar. Residues K566, K709, H753, Y790, R792, S798, and Y804 in the MID and PIWI domains contact phosphates 3–6 of the guide (Fig. 3B). Residues S220, R357, R714, and R761 of L1, L2, and the PIWI domain contact phosphates 7–9 (Fig. 3C). The 3′ binding pocket of the PAZ domain contained some weak electron density, which we modeled as a single nucleotide with a refined occupancy of 0.75.
Figure 3. Conformation of bound guide RNAs.
A. The 5′ nucleotides of guide RNAs are recognized by extensive interactions with the MID and PIWI domains. An ordered water molecule is shown as a pink sphere. Hydrogen bonds are shown as dashed orange lines B, C. Ago2 organizes the seed region in an A-form helix by extensive interactions with the phosphate backbone. D. Helix-7 (α7) introduces a kink in the guide RNA between bases 6 and 7 that disrupts helical stacking.
Ago2 does not appear to rely heavily on direct hydrogen bonds to the 2′ hydroxyls of the guide for RNA recognition. We observe only two hydrogen bonds between the protein and 2′ hydroxyls of the guide: Nucleotide 5 hydrogen bonds to the main chain amide of I756, and nucleotide 7 hydrogen bonds to the main chain carbonyl of A221 (Fig. S9). The 2′ hydroxyl of nucleotide 2 makes a water-mediated contact to main chain carbonyls of N562 and R792. However, we observe no hydrogen bond acceptors or donors less than 3.8 Å away from the 2′ hydroxyls of nucleotides 1, 3, 4, 6 and 8. This may explain why DNA bases and 2′ fluoro substitutions are tolerated in the antisense strand of small interfering RNAs (siRNAs) (19, 20). We also note that the guide-binding pocket of the bacterial Argonaute is more hydrophobic than the same region of human Ago2, and may not accommodate several 2′ hydroxyls (Fig. S10). These observations may explain why the prokaryotic enzyme prefers guide DNAs over RNAs (10).
Nucleotides 2–6 of the guide RNA are splayed out, with Watson-Crick faces exposed to the bulk solvent, in an A-form conformation (Fig. 3D). This observation supports the “seed-pairing” model of miRNA targeting, in which Argonaute is proposed to pre-arrange miRNA nucleotides 2–7 in an A-from configuration, thereby reducing the entropic cost associated with forming a stable duplex with target RNAs (21, 22). However, there is a distinct kink between nucleotides 6 and 7 of the RNA that breaks the A-form structure in this region of the guide. The kink appears to be introduced by Ile-365, which is inserted between the faces of bases 6 and 7. The position of nucleotide 7 is further stabilized by Met-364, which interacts with the minor-grove edge of the base. Met-364 and Ile-365 reside on α-helix-7 in a region of L2 that is conserved in eukaryotic Argonautes (Fig. 3D and S11). The archeal Argonaute from the Pyrococcus furiosus has a similar helix (Fig. S1). In contrast, T. thermophilus Argonaute, the best structurally characterized member of the super-family, lacks an analogous helix.
Docking an A-form duplex onto the guide RNA reveals that helix-7 would have to shift to accommodate target pairing to nucleotides 6 and 7 of the guide (Fig. S12). A shift in helix-7 would also likely release the constraints on nucleotide 7, allowing to the guide to form a contiguous A-form helix. These types of conformational changes may relate to the importance of pairing to nucleotide 7 for effective miRNA-targeting (23) and could possibly be used as a readout for recognition of miRNA target sites. The ability of Ago2 to kink guide RNAs may also facilitate release of sliced RNA products by disrupting guide-target pairing in this region.
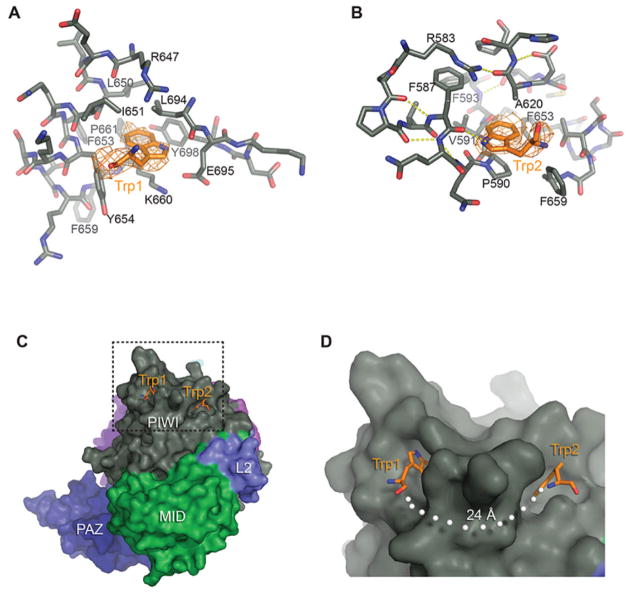
Argonaute-associated proteins often contain glycine-tryptophan (GW) rich regions, which are believed to interact with the Argonaute PIWI domain (3, 17, 24–27). The tryptophan residues in GW proteins are essential for mediating protein-Argonaute interactions (25, 28, 29). To identify possible GW interaction sites, we determined the structure of Ago2 crystallized in the presence of free tryptophan (Table S2). Unambiguous electron density for tryptophan molecules was observed in two adjacent hydrophobic pockets in the PIWI domain (Fig. 4). Tryptophan 1 stacks over the aliphatic segment of K660 and packs against the side chains of L650, I651, Y654, L694 and Y698. Tryptophan 2 stacks against P590, forms a hydrogen bond to the main chain carbonyl of F587 through the amine group in its indole ring, and packs against the side chains of F659, F587, V591, A620 and F653. In the absence of tryptophan, phenol (present at 100 mM in the –tryptophan crystallization conditions) was observed in tryptophan-binding site 1 (Fig. S13).
Figure 4. Tandem tryptophan-binding pockets in the PIWI domain.
A, B. Residues forming the binding pockets of tryptophan 1 and 2 shown, with hydrogen bonds indicated (dashed, yellow lines). Tryptophan molecules are shown with surrounding unbiased Fo-Fc maps contoured at two sigma (orange). C. Surface representation showing the tryptophan binding pockets. D. Close up view of boxed area in panel C. White dots indicate the shortest direct path connecting the two sites along the surface of Ago2.
Both tryptophan molecules bound with indole side-chains inserted into Ago2 and main chain atoms extended out towards the bulk solvent, as would be expected for binding tryptophans attached to a polypeptide. We therefore suggest that the observed tryptophan binding pockets are interaction sites for binding to GW motifs in Argonaute-associated proteins. Consistent with this idea, mutations that disrupt these binding sites in Ago2 and Drosophila Ago1 specifically reduce binding to GW182 without disrupting miRNA binding (Fig. S14) (17, 25)
We note that tryptophan residues in various GW proteins often occur in pairs, separated by a flexible linker of 8–14 amino acid residues (25, 28, 29). An extended 8-amino acid peptide spans a distance of about 24 Å, which closely matches the distance between the two tryptophan-binding sites, when measuring along the surface of Ago2 (Fig. 4C). Based on these observations, we hypothesize that Argonaute may specifically recognize GW proteins by binding to tandem tryptophan residues that are separated by an appropriately sized flexible linker. Consistent with this notion, the carboxylic acid of tryptophan 1 is oriented towards the amino group of tryptophan 2, as would be expected for tryptophan residues aligned on a single polypeptide chain (Fig. 4C).
A major motivation for previous studies of prokaryotic forms of Argonaute was that understanding their structures might provide mechanistic insights relevant to Argonaute in humans (1, 6, 9). The data presented here validate this assumption, showing that the active site structure is conserved (Fig S15), and seed region of the guide is pre-ordered for pairing to targets. The structure of Ago2 also reveals new features not present in bacteria. The rearrangement of domains within the bi-lobed structure likely influences how the protein interacts with guide and target molecules. Indeed, helix-7 influences the conformation of guide RNAs in a manner that is not seen for guide DNAs in bacteria. Moreover, the tryptophan binding sites in the PIWI domain form a likely interaction surface for additional RISC components for which no known homologs exist in the prokaryotic kingdom. The structures presented here extend studies of the prokaryotic into understanding Argonaute in humans. Bridging this gap is an essential step towards leveraging structural information for design and delivery strategies for silencing human disease factors using RNAi.
Supplementary Material
Supplement 1
Acknowledgments
We thank the laboratories of D. Stout, E. O. Saphire, and I. Wilson for sharing synchrotron time and for helpful discussions. Crystallization screens were carried out at the Joint Center for Structural Genomics, supported by the National Institute of General Medical Sciences (NIGMS) Protein Structure Initiative (U54 GM074898). Diffraction data were collected on beamlines 24-ID-E at the Advanced Photon Source and 11-1 at the Stanford Synchrotron Radiation Lightsource. This work was supported by NIGMS grant R01 GM086701 to I.J.M. I.J.M. is a Pew Scholar in the Biomedical Sciences. Coordinates of Ago2 and Ago2 bound to tryptophan have been deposited in the Protein Data Bank (4EI1 and 4EI3).
References
- 1.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004 Sep 3;305:1437. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 2.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004 Jul 23;15:185. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 3.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006 Jul 15;20:1885. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011 Oct 7;44:120. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Fabian MR, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011 Nov;18:1211. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- 6.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004 Sep 3;305:1434. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, et al. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008 Dec 18;456:921. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009 Oct 8;461:754. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008 Nov 13;456:209. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan YR, et al. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005 Aug 5;19:405. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma JB, et al. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005 Mar 31;434:666. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004 May 20;429:318. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003 Nov 27;426:465. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 14.Song JJ, et al. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003 Dec;10:1026. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 15.Yan KS, et al. Structure and conserved RNA binding of the PAZ domain. Nature. 2003 Nov 27;426:468. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- 16.Boland A, Tritschler F, Heimstadt S, Izaurralde E, Weichenrieder O. Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 2010 Jul;11:522. doi: 10.1038/embor.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proc Natl Acad Sci U S A. 2011 Jun 28;108:10466. doi: 10.1073/pnas.1103946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010 Jun 10;465:818. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 19.Chiu YL, Rana TM. siRNA function in RNAi: a chemical modification analysis. RNA. 2003 Sep;9:1034. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deleavey GF, et al. Synergistic effects between analogs of DNA and RNA improve the potency of siRNA-mediated gene silencing. Nucleic Acids Res. 2010 Jul;38:4547. doi: 10.1093/nar/gkq181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004 Jan 23;116:281. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005 Jan 14;120:15. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan 23;136:215. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lian SL, et al. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009 May;15:804. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eulalio A, Helms S, Fritzsch C, Fauser M, Izaurralde E. A C-terminal silencing domain in GW182 is essential for miRNA function. RNA. 2009 Jun;15:1067. doi: 10.1261/rna.1605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.El-Shami M, et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 2007 Oct 15;21:2539. doi: 10.1101/gad.451207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bies-Etheve N, et al. RNA-directed DNA methylation requires an AGO4-interacting member of the SPT5 elongation factor family. EMBO Rep. 2009 Jun;10:649. doi: 10.1038/embor.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol. 2009 Aug;29:4144. doi: 10.1128/MCB.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Till S, et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat Struct Mol Biol. 2007 Oct;14:897. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 30.Boland A, Huntzinger E, Schmidt S, Izaurralde E, Weichenrieder O. Crystal structure of the MID-PIWI lobe of a eukaryotic Argonaute protein. Proceedings of the National Academy of Sciences of the United States of America; Jun 6, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Till S, et al. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nature structural & molecular biology. 2007 Oct;14:897. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- 32.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nature structural & molecular biology. 2008 Apr;15:346. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 33.De N, Macrae IJ. Purification and assembly of human Argonaute, Dicer, and TRBP complexes. Methods Mol Biol. 2011;725:107. doi: 10.1007/978-1-61779-046-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minor ZOaW. In: Methods in Enzymology. CWC Jr, Sweet RM, editors. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 35.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66:125. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger TC. Reciprocal-space solvent flattening. Acta Crystallogr D Biol Crystallogr. 1999 Nov;55:1863. doi: 10.1107/S0907444999010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011 Apr;67:355. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1