The phage-related chromosomal islands of Gram-positive bacteria (original) (raw)
. Author manuscript; available in PMC: 2012 Dec 15.
Published in final edited form as: Nat Rev Microbiol. 2010 Aug;8(8):541–551. doi: 10.1038/nrmicro2393
Abstract
The phage-related chromosomal islands (PRCIs) were first identified in Staphylococcus aureus as highly mobile, superantigen-encoding genetic elements known as the S. aureus pathogenicity islands (SaPIs). These elements are characterized by a specific set of phage-related functions that enable them to use the phage reproduction cycle for their own transduction and inhibit phage reproduction in the process. SaPIs produce many phage-like infectious particles; their streptococcal counterparts have a role in gene regulation but may not be infectious. These elements therefore represent phage satellites or parasites, not defective phages. In this Review, we discuss the shared genetic content of PRCIs, their life cycle and their ability to be transferred across large phylogenetic distances.
Horizontal gene transfer has an extremely important role in bacterial evolution. It has been estimated that some 20% of the extant genetic content of any given bacterial species has been acquired from other organisms1. Perhaps half of this 20% consists of mobile genetic elements (MGEs) that have moved freely within and between species and have occasionally crossed boundaries between genera.
In facultative pathogens, MGEs are largely responsible for antibiotic resistance, environmental adaptations and the wide range of adaptations to life in host tissues that we perceive as pathogenesis. Among these pathogens is Staphylococcus aureus, which is a major scourge of the hospital environment and has become increasingly important of late as a cause of infections among otherwise healthy individuals. Not only are some strains resistant to methicillin and oxacillin owing to the acquisition of the staphylococcal cassette chromosome mec (SCC_mec_) element2, but also the species as a whole has increased its virulence and transmissibility. A possible contributor to this increased virulence is Panton-Valentine leukocidin (PVL), the prophage-encoded toxin responsible for necrotizing pneumonitis, which can be fatal within 24–48 hours, as recently described3. Superantigens are among the other important staphylococcal virulence factors; they are responsible for food poisoning, toxic shock syndrome (TSS) and necrotizing fasciitis. Toxic shock syndrome toxin (TSST1) is particularly insidious, especially in surgical wounds, because it inhibits the production of most other virulence factors and, consequently, causes little inflammation4.
All known classes of MGEs in S. aureus and other pathogenic bacteria, including temperate phages, plasmids, transposons and other horizontally acquired units, may contribute to pathogenesis; it is particularly striking that nearly all of the bacterial toxins that cause specific toxin-mediated diseases, such as PVL pneumonia, TSS, necrotizing fasciitis and food poisoning — known as toxinoses — are encoded by MGEs5. Although there are well-defined classes of MGEs, there are also transitional forms that can be classified as chromosomal islands; these are discrete chromosomal segments that have been acquired by horizontal transfer, that are usually flanked by direct repeats and that lack essential genes. Chromosomal islands range in size from two genes (called ‘islets’) to several hundred genes. Most do not seem to be mobile anymore, probably owing to the loss of mobilization functions. Those chromosomal islands that are mobile use either phage-mediated transfer or self-coded conjugative transfer machinery to move between cells (DNA-mediated transformation is not known to be an MGE-specific mode of transfer). The discovery of the highly mobile S. aureus pathogenicity islands (SaPIs), which carry genes encoding TSST1, staphylococcal enterotoxin B (SEB; also known as EntB) and other superantigens6, was an important advance in our understanding of the mobile character of pathogenicity islands (BOX 1). These elements do not encode any machinery for their horizontal transfer and, instead, hijack the capsids of phages (known as helper phages) for their transduction. SaPI gene expression is carefully regulated to take advantage of the lytic cycle of the helper phage and to maximize the transfer of progeny. Many S. aureus genomes contain one or more SaPIs, including several that do not carry any genes encoding superantigens or any other discernable accessory genes. Virtually all clinical toxic shock-causing isolates contain two or more SaPIs with various combinations of genes, suggesting that there has been extensive recombination between SaPIs7.
Box 1| History of Staphylococcus aureus pathogenicity islands.
Following the identification of toxic shock syndrome toxin 1 (TSST1)32, it was observed that the toxin was produced by only 10–20% of natural staphylococcal isolates33 and that non-producers lacked the gene encoding the toxin (tst) plus some 15 kb of additional DNA34, suggesting that the gene was carried by a mobile genetic element. As chromosomal mapping revealed two different locations for tst35, the mobile element was assumed to be a transposon, and it was designated Tn_557_. However, Southern blotting revealed that the two _tst_-carrying elements were different, leading to their designation as a family of Staphylococcus aureus pathogenicity islands (SaPIs)6.
Using a tetracycline resistance marker in the tst gene, the SaPI transduction frequency with staphylococcal phage 80α (but not with the related phage ϕ11) was found to be around 107-fold higher than that for the same marker integrated elsewhere in the chromosome. This high frequency was phage specific and recombinase A (RecA) independent. Further, SaPI1 induction interfered strongly with multiplication of the inducing (or helper) phage, phage 80α, blocking plaque formation and reducing the phage burst size by 10-fold to 100-fold, but had no effect on the non-helper phage ϕ11. Finally, SaPI1 DNA was found to be encapsidated in small-headed infective phage-like particles that accommodated the smaller SaPI genome20 and were composed exclusively of phage virion proteins17,28.
The recent identification of closely related elements in other staphylococcal species8,9 and in other genera10 (BOX 2) has prompted us to suggest a general designation for all these elements of ‘phage-related chromosomal islands’ (PrCIs), owing to the similarity of these elements to phages (BOX 1). In Staphylococcus epidermidis the PrCI SePI-I11 encodes the toxins staphylococcal enterotoxin C3 (SEC3; also known as EntC) and staphylococcal enterotoxin L (SEL); this is the first example of super antigen production by staphylococci other than S. aureus. SePI-I was probably acquired from S. aureus, but it lacks many of the genes that are found in SaPIs and is probably non-transmissible. PCrIs have also been identified in recently sequenced isolates of Lactococcus lactis (r.P.N., unpublished observations), Streptococcus spp.10 and Enterococcus faecalis (J.r.P., unpublished observations).
Box 2| Phage-related chromosomal island nomenclature.
The finding of phage-related chromosomal islands (PRCIs) in other genera suggests that such islands are widespread and possibly co-ancestral, and that they require a well-defined nomenclature. We propose that the individual PRCIs are designated with reference to their species — thus, SaPIn, SeCIn, ShCIn, SsCIn, LlCIn, SsuCIn and SpyCIn would be used for Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus saprophyticus, Lactococcus lactis, Streptococcus suis and Streptococcus pyogenes, respectively, where ‘n’ refers to a specific island (for example, SsCI15305 for the PRCI identified in the genome sequence of S. saprophyticus str. 15305). We further propose to retain the term SaPI rather than change it to SaCI, because of the carriage of virulence factors in this case. It is used in this Review in reference to the S. aureus elements. Note that the ‘n’ in SaPIn1 as previously published36 refers to S. aureus str. n315.
However, the nomenclature of the SaPIs is somewhat controversial. Lindsay et al.6 proposed the SaPIn designation, in which ‘n’ represents a numerical series. This was adhered to in the description of the first S. aureus genome36 but not in the description of the second one12. In this case, a complex classification scheme was proposed instead, using ‘ν’ as the prefix for most of the chromosomal islands, renaming the SaPIs along with all other putative islands (except for the staphylococcal cassette chromosome mec elements (SCC_mec_s), which continued to be listed separately). Lindsay and Holden37 retained the SaPI designation but changed the specific (numerical) designations to include only those from strains that had been sequenced at the time. They proposed that all SaPI-like elements located at a single chromosomal site be assigned to a single class. We agree with this, but we must point out that the SaPIs have recombined promiscuously7, resulting in the presence of several diverse elements at single sites. These are listed in TABLE 1 and illustrated in Supplementary information S7 (figure), which should be compared with FIG. 1. A SaPI-like PRCI in the well-known fusidic acid-resistant strain of S. aureus was designated SaRIfusB38 (for S. aureus resistance island fusB) and has not been renamed.
All of the sequenced staphylococcal genomes contain two other pathogenicity islands, νSaα and νSaβ, which contain sets of serine protease-like and superantigen-like genes, respectively, but lack flanking direct repeats and mobilization functions12, and it is unclear whether these elements are acquired through horizontal transfer.
In this review, we address the basic molecular genetics of SaPIs, which are prototypical PrCIs, focusing on their genomic organization and regulation, their mobility (including the potential to disseminate superantigens and other virulence factors), and their interaction with and use of helper phages. we also comment briefly on the trans-genera transfer of SaPIs and on the existence of similar elements in other bacteria.
Genome organization
Conserved core genes
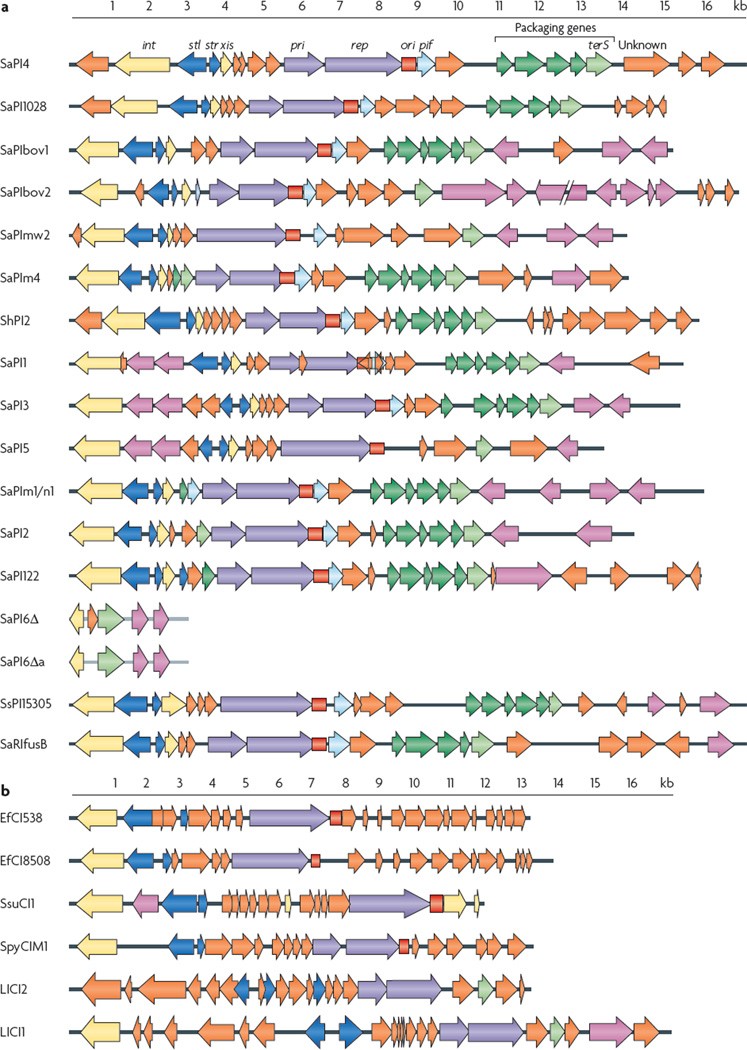
All SaPIs share a core set of genes that regulate their life cycle (FIG. 1; TABLE 1). At the left end of the SaPI genome is a homologue of the typical phage integrase gene (int), which is required for the integration of the element. Adjacent to int, sometimes with one or more intervening accessory genes, are two divergently oriented promoters that regulate two major transcription units: stl on the left, and str on the right. These two genes resemble the divergent cI and cro genes, respectively, of temperate coliphages, which encode regulator proteins. Stl is a master repressor for the SaPI excision–replication–encapsidation cycle13 and is de-activated during phage induction; the main function of Stl is to repress transcription of str. To the right of str is xis (which encodes excisionase), followed by the replication module, consisting of pri, which encodes a primase, rep, which encodes a replication initiator, and a SaPI-specific origin. In some cases the primase is fused to the replication initiator; it enhances replication but is not absolutely required13. rep is similar to replicon-specific replication initiators of other types of replicons and has a helicase activity13 that is similar to that of α-protein of phage P4 (REF. 14). The replication origin consists of two sets of inversely oriented hexanucleotide-to-octanucleotide iterons flanking an AT-rich region (see Supplementary information S1 (figure)). In SaPIbov1, gene 12, called pif, is to the right of the replication origin and is responsible for interference with phage growth15. This gene is followed by gene 11, which encodes a protein of unknown function and an operon that is involved in genome packaging. The operon is controlled by the SOS response-specific repressor LexA13,15–17 and includes terS, a close homologue of the terminase small subunit of several phages from Gram-positive bacteria. Interestingly, most of the predicted SaPI-specific genes encoding proteins with unknown functions do not have close orthologues. These gene identifications discussed above are based mostly on experiments performed with SaPIbov1, but many have been confirmed in other SaPIs, primarily SaPI1 (REF. 7), SaPI2 (REF. 7) and SaPIn1 (J.r.P., unpublished observations).
Figure 1. comparison of phage-related chromosomal island genomes.
Genomes are aligned according to the prophage convention, with the integrase gene (int) at the left end. Genes are coloured according to their sequence and function: int and xis (excisionase) are yellow; transcription regulators are dark blue; replication genes (including the primase gene (pri) and the replication initiator gene (rep)) are purple; the replication origin (ori) is red; encapsidation genes are green, with the terminase small subunit gene (terS) in light green; superantigen and other accessory genes are pink; and pif (which functions in phage interference) is light blue. Genes encoding hypothetical proteins are orange. a | The gene organization of Staphylococcus aureus pathogenicity islands (SaPIs). b | Putative phage-related chromosomal islands from genera other than Staphylococcus. SaPIm1/n1 indicates SaPIn1 (from S. aureus str. n315) and SaPIm1 (from S. aureus str. mu50), which are essentially identical.
Table 1.
Genes of three Stapylococcus aureus pathogenicity island prototypes
| ORF number (gene name)* | orientation | Annotation or function | ||
|---|---|---|---|---|
| SaPi1 | SaPibov1 | SaPi2 | ||
| attL | attL | attL | NA | Repeat region |
| 26 (int) | 21 (int) | 1 (int) | − | Integrase |
| 25 | Absent | Absent | − | Hypothetical protein |
| 24 (sek) | Absent | Absent | − | Enterotoxin K |
| 23 (seq) | Absent | Absent | − | Enterotoxin Q |
| 22 (stl) | 20 (stl) | 2 (stl) | − | SaPI master repressor |
| 21 (str) | 19 (str) | 3 (str) | + | Regulatory protein |
| 20 | 18 (xis) | 4 | + | Excisionase in SaPIbov1 |
| 19 | 17 | 5 | + | Hypothetical protein |
| Absent | Absent | 6 | + | Hypothetical protein |
| 18 | 16 | 7 | + | Hypothetical protein |
| 17 (pri) | 15 (pri) | 8 (pri) | + | Similar to DNA primase |
| 15 (rep) | 13 (rep) | 9 (rep) | + | Replication initiator with helicase activity |
| ori | ori | ori | + | Replication origin |
| 11 (pif) | 12 (pif) | 10 (pif) | + | Phage interference |
| 10 | 11 | Absent | + | Hypothetical protein |
| 9 | Absent | 11 | + | Hypothetical protein |
| Absent | Absent | 12 | + | Hypothetical protein |
| 8 | 10 | 13 | + | Hypothetical protein |
| 7 (cp3) | 9 (cp3) | 14 (cp3) | + | Capsid size determinant |
| 6 (cp2) | 8 (cp2) | 15 (cp2) | + | Capsid size determinant |
| 5 (cp1) | 7 (cp1) | 16 (cp1) | + | Capsid size determinant |
| 4 | 6 | 17 | + | Hypothetical protein |
| 3 (terS) | 5 (terS) | 18 (terS) | + | Terminase small subunit |
| 2 (tst) | 4 (tst) | 19 (tst) | − | Toxic shock syndrome toxin 1 |
| 1 (ear) | Absent | Absent | + | Homologous to penicillin binding protein |
| Absent | 3 | Absent | + | Hypothetical protein |
| Absent | 2 (sec) | Absent | + | Enterotoxin type C |
| Absent | 1 (sel) | Absent | − | Enterotoxin type L |
| Absent | Absent | 20 (eta) | − | Similar to Staphylococcus hyicus exfoliatin A |
| attR | attR | attR | NA | Repeat region |
PRCI-specific accessory genes
The accessory genes can be located at either or both ends of the PrCI. The putative functions of many of these genes have not been analysed and are assigned on the basis of homology, location or both. In addition to superantigen genes, PrCIs carry genes for antibiotic and phage resistance as well as genes encoding biofilm-inducing proteins, a putative ferrichrome ATP-binding cassette (ABC) transporter, a membrane protein (Mmp) and a putative acetyl transporter (Act) (TABLE 2), although several PrCIs lack any identifiable accessory genes. orthologues of these genes can be found in other genomes, suggesting that some random genetic exchange may be responsible for their acquisition by PrCIs, although several of the most important staphylococcal superantigen genes, including tst (encoding TSST1), seb and the sec genes, are found only in SaPI genomes.
Table 2.
Accessory genes in phage-related chromosomal islands
| gene | Product | Function | carried by |
|---|---|---|---|
| tst | Toxic shock syndrometoxin 1 (TSST1) | Superantigen | SaPI1, SaPI2,SaPIbov1, SaPIn1and SaPIm1 |
| seb | Enterotoxin B | Superantigen | SaPI3 |
| sec | Enterotoxin C | Superantigen | SaPIbov1, SaPIn1,SaPIm1, SaPImw2and SePI1 |
| ear | Penicillin-biding proteinfragment | Penicillin resistance(in Escherichia coli) | SaPI1, SaPI3 andSaPI5 |
| sek | Enterotoxin K | Superantigen | SaPI1 and SaPI3 |
| sel | Enterotoxin L | Superantigen | SaPIbov1, SaPI3 andSePI1 |
| seq | Enterotoxin Q | Superantigen | SaPI1 and SaPI5 |
| eta | Exfoliatin A | Epidermolytic toxin | SaPI2 |
| bap | Biofilm-associatedprotein (BAP) | Biofilm formation inthe bovine udder | SaPIbov2 |
| fhuD | Ferrichrome ABCtransporter homologue | Iron transport | SaPIm4 |
| mdr | Multidrug resistanceprotein | Multidrug exporter | SaPI122 |
| aad | Aminoglycoside adenyltransferase | Aminoglycosideresistance | SsPI15305 |
| fosB | Glutathione thionylphosphatase | Fosfomycinresistance | SsPI15305 |
| fusB | FusB | Fusidic acidresistance | SaRIfusB |
| ermA | Ribosome methylase | MLS resistance | SpPI1 |
| Unnamed | Unnamed | Phage resistance | LlCI1 |
| actA | GNAT familyhomologue | Acetyl transferprotein | SaPI6Δ |
| None | NA | NA | SaPI4, SaPI1028,ShPI2 and LlCI2 |
PRCIs of other Gram-positive bacteria
Several putative PrCIs have been identified in other Gram-positive bacterial genera, either by searching the published genome sequences or by tracking phenotypes. These share the basic features of the staphylococcal pathogenicity islands but differ in certain details (FIG. 1b). The two PCrIs identified in L. lactis most closely resemble the SaPIs; the other non-staphylococcal PrCIs usually lack an identifiable terS homologue. It is possible that this function is carried out by a protein that does not resemble TerS or that these PrCIs use alternative packaging strategies. A genetic element in Streptococcus pyogenes, SF370.4, which was described by McShan and colleagues as a prophage remnant10, has all of the genomic features of a PrCI. we suggest, with the authors’ concurrence (w. McShan, personal communication), that it be re-designated as PrCI SpyCIM1. unlike most PrCIs, however, SpyCIM1 excises and reintegrates during growth10. E. faecalis carries the PrCI EfCI, which can be transferred to and replicate in S. aureus (J.r.P., unpublished observations).
The SaPI lifestyle
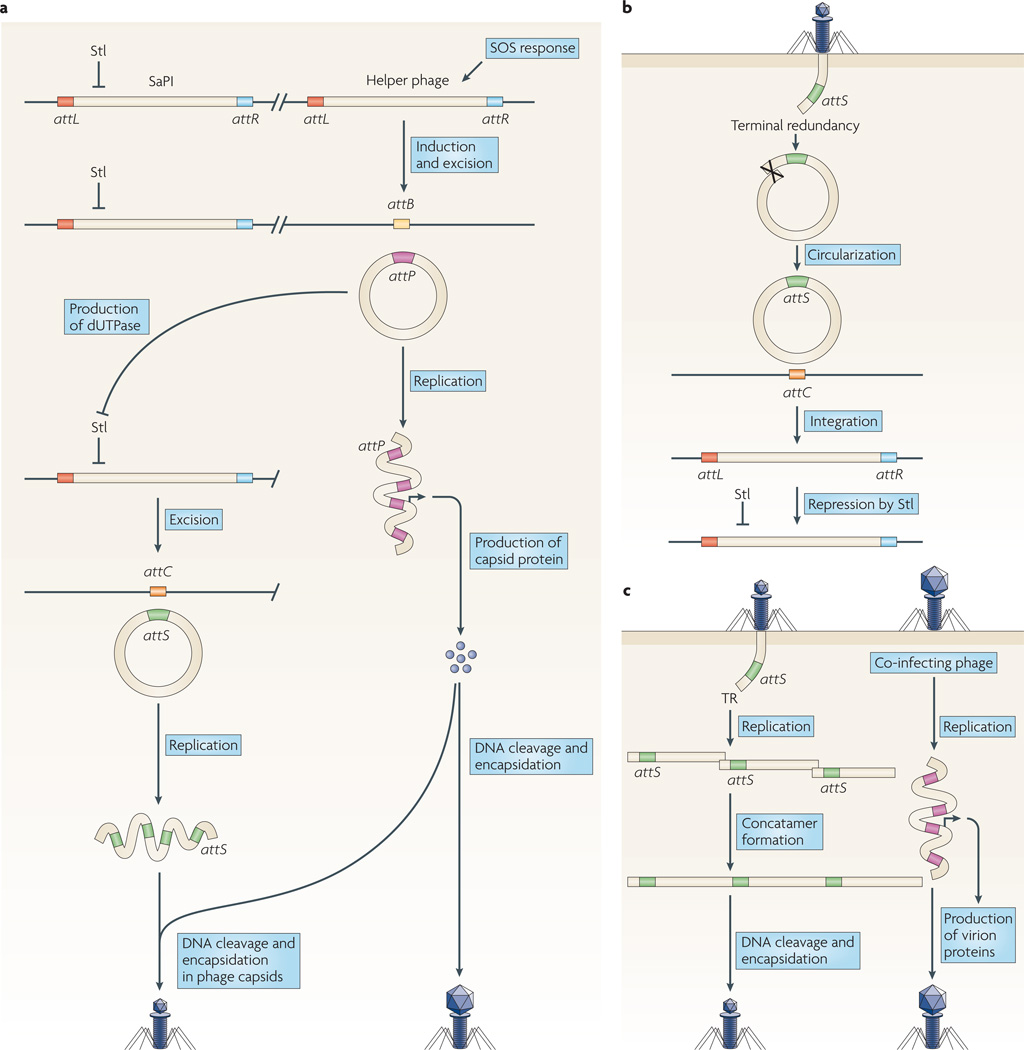
SaPIs reside stably in the host chromosome, like prophages, but their excision and replication are not induced by the SoS response; instead, they require a helper phage. Following superinfection by a helper phage or induction of a resident helper prophage by the SoS response, the SaPI genome is excised by Int and Xis, presumably using the Campbell mechanism19, and replicates independently to form a concatamer. Alongside the production of phage virions, smaller SaPI capsids are produced from phage proteins. Packaging of SaPI DNA into these phage capsids is initiated by cleavage of the DNA at SaPI pac sites (terminase recognition sites) by TerS. Phage-induced lysis releases both mature phages and SaPI particles (FIG. 2a). on entry into a new host cell, SaPI DNA follows a replicative pathway if the DNA is accompanied by an incoming phage genome and an integrative pathway if not (FIG. 2b,c).
Figure 2. Staphylococcal pathogenicity island replication scenarios.
a | Staphylococcus aureus pathogenicity island (SaPI) induction by an SOS-induced helper prophage. attP and attS are the prophage and SaPI core attachment sequences, respectively; attB and attC are the prophage and SaPI core chromosomal attachment sequences, respectively. After induction and excision of the helper phage, phage dUTPase relieves the Stl-mediated repression of the SaPI, allowing production of SaPI proteins. SaPI excisionase (Xis) subsequently promotes the excision of the SaPI through a Campbell mechanism, restoring the attC and attS sites. Subsequent SaPI replication produces hundreds of copies in the form of a concatemer, which is cleaved by the terminase complex into individual copies and packed into phage particles consisting entirely of phage proteins but with a smaller head than the phage capsids, owing to the action of SaPI proteins. b | SaPI infection. Terminal redundancy allows the incoming SaPI DNA to circularize, after which the circular DNA is integrated at the chromosomal attC site by crossover with the SaPI attS site. This process requires the SaPI integrase. Stl silences the expression of SaPI genes, keeping the element integrated. c | Co-infection of a SaPI and a helper phage allows the SaPI DNA to be replicated prior to integration. The synthesis of phage proteins leads to the production of phage capsids, which are used to package the replicated SaPI DNA. As in part a, SaPI proteins affect the size of the capsids that are produced.
Integration
Chromosomally located MGEs — such as PrCIs, SCC_mec_s, ICEs (integrative and conjugative elements) and prophages — that encode and use sites-pecific integrases integrate at a site (known as the attC site) that is determined by the specificity of the integrase, and these elements are flanked by directly repeated sequences that comprise the core integrase recognition site. SaPIs require only integrase for insertion, although integration seems to be inefficient, as incoming SaPI DNA persists for several hours in the autonomous (but non-replicating) state20, perhaps because integrase is expressed poorly under these conditions.
The known SaPIs occupy six different attC sites in the staphylococcal genome21 (TABLE 3; see Supplementary information S2 (figure)); these sites correspond to the attB sites for bacteriophage integration. The SaPIs each contain a corresponding insertion site sequence, attS, which corresponds to the classical attP integration site of bacteriophages. The six different attC sequences are unrelated to each other and occur only once in each of the sequenced S. aureus genomes. when S. aureus carries more than one SaPI, as is common among the sequenced strains (TABLE 3), the SaPIs occupy different chromosomal attC sites; no strain has been found to contain two SaPIs at the same attC site to date.
Table 3.
The staphylococcal pathogenicity island family
| Element | Staphylococcal genome | Baba* | LindsayandHolden‡ | Size(kb) | Inducingphages | att site core (location, att/_int_group) | Refs |
|---|---|---|---|---|---|---|---|
| SaPI4 | S. aureus str. MRSA252 | NA | SaPI4 | 15.1 | Endogenousprophage | AAAGAAGAACAATAATAT (~8′, I) | 7,39 |
| SaPI1028 | S. aureus str. NY940 | NA | NA | 15.6 | Endogenousprophage | AAAGAAGAACAATAATAT (~8′, I) | 7,40 |
| SaPIbov1 | S. aureus str. RF122 | νSa2 | NA | 15.8 | φ11 and 80α | TAATTATTCCCACTCAAT (~9′, II) | 25,41 |
| SaPIbov2 | S. aureus str. V329 | NA | NA | 27 | 80α | TAATTATTCCCACTCGAT (~9′, II) | 25 |
| SaPIm4 | S. aureus str. mu50 | νSa3type I | NA | 14.4 | Endogenousprophage | TCCCGCCGTCTCCAT (~18′, III) | 7,12 |
| SaPImw2 | S. aureus str. mw2 | νSa3type II | SaPI3 | 14.4 | Endogenousprophage | TCCCGCCGTCTCCAT (~18′, III) | 7,12 |
| SePI1 | S. aureus str. FRI909 | NA | NA | 9.9 | Not known | TCCCGCCGTCTCCAT (location unknown§, III) | 11 |
| ShPI2 | S. haemolyticus | νSh2 | NA | 16.6 | Not known | TCCCGCCGTCTCCAT (48′, III)‖ | 8 |
| SaPI1 | S. aureus str. RN4282 | νSa1 | NA | 15.2 | 80α and φ13 | TTATTTAGCAGGAATAA (~19′, IV) | 6 |
| SaPI3 | S. aureus str. COL | νSa1 | SaPI1 | 15.6 | Not known | TTATTTAGCAGGAATAA (~19′, IV) | 42 |
| SaPI5 | S. aureus str. USA300 | NA | NA | 14.0 | Not known | TTATTTAGCAGGAATAA (~19′, IV) | 43 |
| SaPIn1andSaPIm1 | S. aureus str. n315 and_S. aureus_ str. mu50,respectively | νSa4type I | SaPI2 | 15 | 80α | GTTTTACCATCATTCCCGGCAT (~44′, V) | 36 and J.R.P., unpublished observations |
| SaPI2 | S. aureus str. RN3984 | NA | NA | 14.7 | 80 and 80α | ATTTTACATCATTCCTGGCAT (~44′, V) | 7,20 |
| SaRIfusB | S. aureus European fusidicacid-resistant impetigo cloneCS6 | NA | NA | 20.7 | Not known | ATGCCAGGTATGATGTAAAAC (~44′, V) | 38 |
| SaPI122 | S. aureus str. RF122 | NA | NA | 17.9 | Endogenousprophage | GTTTTACATCATTCCTGGCAT (~44′, V) | NA¶ |
| SaPI6Δ | S. aureus strains 8325, COL,USA300, MSSA476, Newmanand mw2 | νSa4type II | NA | 3.14 | Not known | GTTTTACCATCATTCCCGGCAT,GTTTTACATCATTCCTGGCAT (~44′, V) | 12 |
| SsPI15305 | S. saprophyticus str. 15305 | νSs15305 | NA | 16.7 | Not known | Unknown sequence (~48′, VI) | 9 |
When the SaPI1 attC site is deleted22, SaPI1 inserts into secondary att sites at nearly the same collective frequency as it inserts into its primary site22 (see Supplementary information S3 (table)). The sequence conservation at the core of the secondary att sites is low, and the sequences flanking the core are not at all conserved. The specificity of the SaPI1 Int therefore seems to be between that of a classical integrase, which has a very high specificity, and a transposase, which has a much lower specificity.
Regulation
After integration of the SaPI into the host chromosome, SaPI gene expression is repressed by the master regulator Stl (FIG. 1,2a). Mutational inactivation of the Stl of SaPIbov1 (REF. 13) and SaPI1 (A. Matthews and r.P.N., unpublished observations) leads to SaPI excision and replication in the absence of a helper phage. This is probably due to reversible integration–excision, because the autonomous (that is, unintegrated) form is extremely unstable in the absence of a functional Int (see below). Therefore, the primary, if not the only, regulatory function of the helper phage is to relieve Stl-mediated repression. However, phages differ in their ability to induce SaPIs; in one study, phage 80α could induce all of the five SaPIs tested, whereas phage ϕ11 induced only SaPIbov1 (REF. 23) and phage 85 could not induce any.
The site of action of Stl is in the intergenic region between the stl and str promoters, suggesting that it regulates transcription from both promoters23. In the integrated state, none of the rightward genes are expressed, whereas int, which always lies to the left, is expressed constitutively but at a low level from the stl promoter (r. Adhikari and r.P.N., unpublished observations). Although str is negatively regulated by Stl24, the regulatory function of Str is unclear, as an str mutation in SaPIbov1 has no obvious effect on the SaPI replication cycle13.
Relief of repression by helper phages
Phage 80α mutants that were selected for their ability to form plaques on SaPIbov1-containing staphylococcal strains were all found to contain mutations in dut, the phage gene encoding duTPase. These mutations eliminated SaPIbov1 induction. Furthermore, the addition of duTPases from either phage ϕ11 or phage 80α enabled non-helper phages 147 and 85 to induce SaPIbov1 (REF. 24). Interestingly, duTPase is a bifunctional ‘moon-lighting’ protein, as mutations affecting duTPase activity but not SaPIbov1 induction, and vice-versa, have been isolated24. Although duTPase is highly conserved among staphylococcal and other phages, there is a sharply divergent central region of about 40 amino acids (see Supplementary information S4 (figure)) that is involved in SaPI mobilization. This region is absent from non-inducing phages, such as the S. epidermidis_-infecting phage PH15, and from chromosomally encoded duTPases, such as that of Mycobacterium tuberculosis25. duTPase relieves SaPIbov1 repression by binding to Stl, thus activating transcription from both the stl and str promoters24. Remarkably, phage 80α Δ_dut does not induce SaPI2 but does induce SaPI1 and SaPIbov2 (see Supplementary information S5 (table)), suggesting that different phage functions are required for the induction of different SaPIs. These were identified by selection for phage 80α mutants that were able to form plaques on staphylococcal strains containing the other SaPIs24. The gene responsible for SaPI1 induction is sri, which encodes a DnaI-binding protein that blocks bacterial DNA replication, and the gene responsible for SaPIbov2 induction is orf15, which encodes a small protein of unknown function. Both of these genes are absent from the phage ϕ11 genome but present in the genomes of many other staphylococcal phages. Phage 80α with mutations in sri are defective in the induction of SaPI1 but not of SaPIbov1 or SaPI2; mutations in orf15 eliminate SaPIbov2 induction but have no effect on the other pathogenicity islands (see Supplementary information S5 (table)). In each case, expression of the cloned gene induces excision and replication of the responsive SaPI24. It will be interesting to unravel the evolutionary process underlying the development of the SaPI–phage interactions, including the induction of SaPIs by SaPI-specific phage proteins and the interference of specific SaPI-encoded proteins with phage reproduction.
Excision
Excision requires an Xis function13,26. As xis is repressed by Stl, the general SaPI repressor, excision occurs only following induction; excision of the uninduced SaPI genome is very rare or undetectable. This serves the needs of the island: spontaneous excision in the presence of a functional repressor would generally cause loss of the island if cell division were to occur before it could reintegrate. Indeed, at least two of the known SaPIs undergo rare spontaneous excision and are slightly unstable, whereas others do not and are very stable20,26. Constitutive expression of int also serves the needs of the island, as it must integrate following cell entry or it will be lost.
Replication
After excision, SaPIs are replicated extensively, ensuring that many particles will be released from the cell. SaPI replication is similar to that of other small bacterial replicons. SaPIs encode replicon-specific rep proteins with matched replication origins (ori sites). origin specificity is determined by the sequence and arrangement of iterons in the ori (see Supplementary information S1 (figure)) and a short carboxy-terminal region of rep27. Initiation requires the AT-rich region between the iterons and the inversely oriented iterons themselves. we propose that rep binds to the iterons, inducing melting of the AT-rich region and initiating replication through the helicase activity of the protein. A host DNA polymerase then replicates the DNA, leading to concatemer formation by an unidentified mechanism. During replication several hundred copies of SaPI DNA are produced. In the absence of a replicating phage, the incoming SaPI DNA circularizes before integration20. This circularization presumably occurs through recombination between the redundant termini, and it must involve an unknown recombinase, as SaPI transfer is recombinase A (recA) independent6.
The pri–rep–ori region of SaPIbov1 can promote the replication of an Escherichia coli vector in S. aureus27. However, the plasmid is extremely unstable, with a segregation probability of about 0.4; this is probably because the replication product, like that of the intact SaPI, is a linear concatemer and so is likely to have a very low copy number and to be unable to segregate normally to daughter cells.
Initiation of the SaPI replication cycle can be induced in three ways: following SoS response-mediated induction of a helper prophage present in the same cell as the SaPI (FIG. 2a); following superinfection of a SaPI-containing non-lysogen by a helper phage; or following the joint entry of SaPI and helper phage DNAs into a cell lacking both5 (FIG. 2c). SpyCIM1 is an example of a SaPI that is induced after activation of the SoS system, but the possible participation of endogenous helper prophages has not yet been addressed. After insertion into the chromosome, SpyCIM1 interrupts the DNA mismatch repair (mutS–mutL) operon, blocking expression of mutL10. It can excise spontaneously during exponential growth, thereby transiently activating mutL expression. It then replicates transiently and is reintegrated, shutting off mutL, as soon as the bacterium exits the exponential phase.
when the SaPI replication cycle is initiated by SoS response-mediated induction of a helper phage or by superinfection with a helper phage, the initial event is relief of SaPI repression, but in the case of joint entry of the SaPI and the helper phage, repression is not established. In the absence of a helper phage, incoming SaPI1 DNA does not detectably replicate, almost certainly because _stl_-mediated repression is rapidly established. In all three scenarios, linear monomeric SaPI DNA is produced; it appears immediately following infection but much later following induction of an endogenous SaPI, when it results from the disruption of intracellular SaPI particles16. After SoS response-mediated induction of a helper phage or superinfection by a helper phage, it is assumed that phage-induced SaPI excision occurs by the Campbell mechanism, but the expected circular excision product is not detected. Instead, replicating SaPI DNA migrates with the bulk DNA in agarose gels, suggesting that the SaPI is present as concatenated, linear copies. This indicates that SaPI replication may be initiated on either linear or circular DNA. Following joint entry of a SaPI and a helper phage, the amount of linear DNA increases, possibly reflecting SaPI replication. Late in infection (at 50–60 minutes after infection), supercoiled SaPI monomers appear and SaPI DNA can be detected in the bulk DNA, indicating that SaPI integration has occurred.
Packaging
SaPIs are packaged in particles composed exclusively of phage proteins17,28. Packaging requires the conserved SaPI-encoded TerS homologue, presumably complexed with the phage-encoded large terminase subunit, which cleaves the multimeric DNA at the SaPI-specific pac site and conducts it to the phage-encoded portal protein for threading into capsids. Filling of the heads is followed by cleavage of the DNA by the large terminase subunit, generating terminally redundant monomeric SaPI DNA, analogous to the DNA of a typical pac phage15.
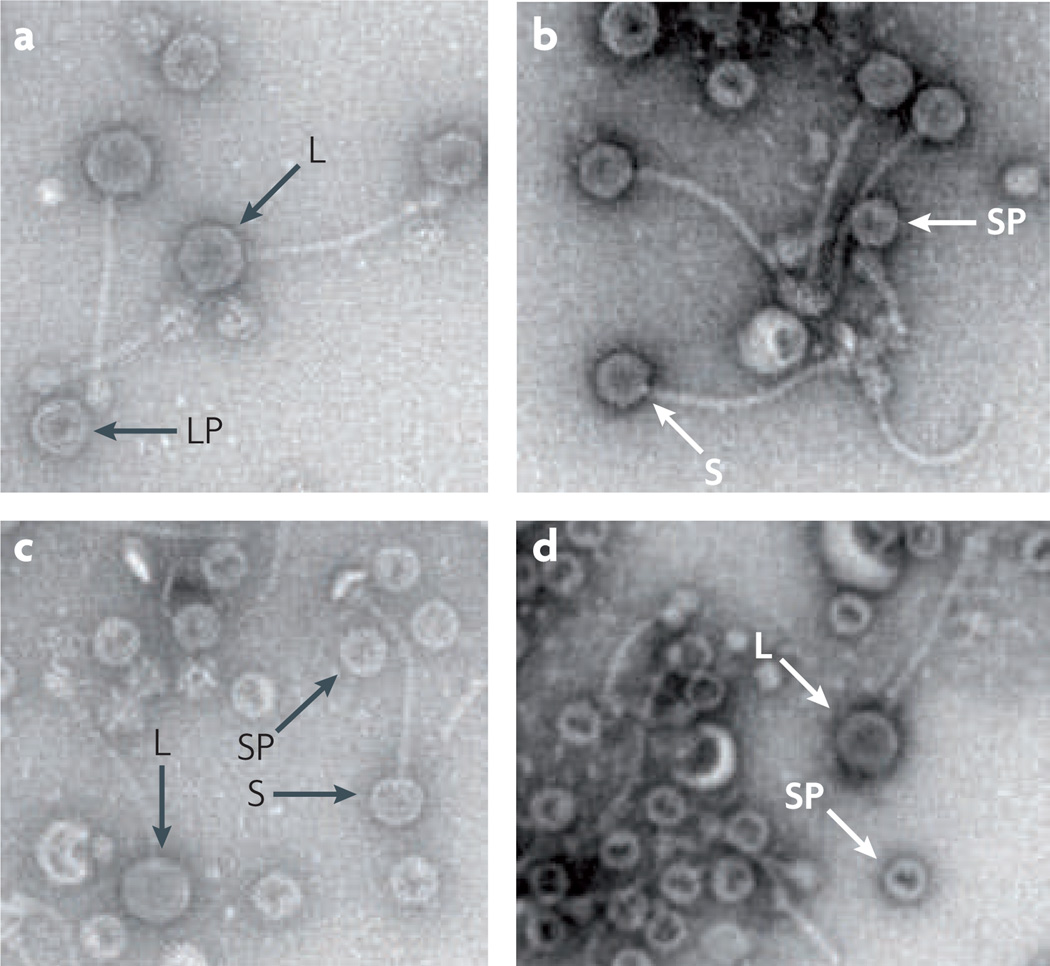
SaPI1 and SaPIbov1 remodel the phage capsid proteins to generate capsids that are one-third of the size of the helper phage capsids, to accommodate the smaller SaPI genomes but exclude complete helper phage genomes. This requires two SaPI genes, cp1 and cp2, which are adjacent in a LexA-regulated operon called operon I that contains six genes encoding proteins involved in packaging16 as well as terS. The product of cp2 is present in purified procapsids and is predicted to form a complex with the helper phage scaffolding protein to regulate the size of the capsid29. The product of the operon I gene cp3 seems to affect the amount of SaPI-specific DNA found in small capsids, but whether this protein alters the number of small capsids or affects DNA packaging remains to be determined. The geometry of the typical staphylococcal phage capsomere can accommodate these two capsid sizes; whether other sizes are possible is unknown. of the 16 known SaPIs, 13 are predicted to contain such capsid-remodelling genes (FIG. 1a). one of the exceptions, SaPIbov2, does not produce small capsids23, in keeping with its much larger genome (~27 kb). The capsid sizes of the other two exceptions, SaPI5 and SaPImw2, have not been examined, but their genomes are in the typical SaPI size range. SaPI DNA packaging is nonspecific; SaPI DNA can be packaged in large and small phage heads, but there is an interesting difference in the relative proportions of these between SaPI and SaPIbov1. SaPI1 DNA is found primarily in small capsids (FIG. 3), whereas SaPIbov1 is packaged into capsids of both sizes with approximately equal efficiency15. Furthermore, fragments of helper phage DNA are readily packaged into small capsids during mobilization of SaPI1. SaPIbov1, however, specifically excludes phage DNA from its small capsids15. This exclusion depends on a single SaPIbov1 gene, gene 12 (see below).
Figure 3. Electron microscopy of phage and Staphylococcus aureus pathogenicity island particles.
Lysates prepared from Staphylococcus aureus strains carrying the relevant S. aureus pathogenicity island (SaPI) and/or helper phage were centrifuged, and the pellets were resuspended in buffer, applied to copper grids and negatively stained with uranyl acetate. a | Lysate from bacteria carrying phage 80α alone, containing a normal-sized capsids. b | Lysate from cells carrying phage 80α and SaPI1. c | Lysate from cells carrying phage 80α and SaPI1, showing both the large phage capsids and the small SaPI capsids. d | Lysate from cells carrying phage 80α and SaPIbov1Δ_terS_. Despite the absence of TerS, SaPIbov1 induces the formation of small capsids. L, large capsid; LP, large procapsid; S, small capsid; SP, small procapsid.
Interestingly, phage ϕ13, a cos phage with a virion genome that contains cohesive termini, can induce excision, circularization and replication of SaPI1 but cannot produce infective SaPI1 particles20. This is presumably due to the inability of the ϕ13 cos packaging machinery to package SaPI1 DNA. However, phage ϕ11 can package SaPI1 DNA after its excision and replication have been induced by phage ϕ13 (A. Mathews and r.P.N., unpublished observations). Furthermore, phages that cannot induce the SaPI1 excision–replication–packaging cycle, such as phage ϕ147 and phage ϕ85, can package SaPI DNA if excision and replication have been activated by an stl mutation13.
Packaging and transfer of the PrCIs LlCI1 and LlCI2 in L. lactis, EfCI538 in E. faecalis, and SsuC1 and SpyCIM1 in streptococci have yet to be studied. Some of these lack an identifiable terS, and it is not yet known whether they are packaged.
SaPI–phage interactions
In general, SaPI induction results in a substantial decrease of phage reproduction, usually blocking plaque formation. SaPIbov1 gene 12 (pif), and, presumably, SaPI1 gene 11 are responsible for this interference and are expressed following induction of the SaPI excision–replication–packaging cycle. Expression of pif alone substantially inhibits phage maturation15. There are two versions of pif among the known SaPI genomes, as exemplified by SaPIbov1 and SaPI1, which contain pif genes with around 30% similarity (see Supplementary information S6 (table)). As noted above, pif affects packaging specificity as well as phage reproduction. SaPI1 does not exclude phage DNA from its small capsids (although it interferes with phage maturation more strongly than SaPIbov1), nor does it affect phage DNA packaging; in fact, most of the SaPI1 and phage DNAs in mature particles are in small capsids15.
Trans-species and trans-genera transfer
SaPIbov1 can be transduced to several staphylococci other than S. aureus, including Staphylococcus xylosus, Staphylococcus chromogenes, S. epidermidis, and Staphylococcus intermedius29. recent reports have indicated that SaPI1 and SaPIbov1 can be transduced to Listeria monocytogenes (see Supplementary information S7 (table)) but not to streptococci, lactobacilli or Bacillus subtilis22.
A very high transfer frequency (>108 per ml of phage lysate) was observed for SaPI1 in some L. monocytogenes strains, and analysis of the resulting transductants suggested that both integrated and autonomous forms were present, indicating that SaPI1 can exist as a plasmid as well as in an integrated form in L. monocytogenes. The transfer frequency of a SaPI1 int mutant was the same as that of the wild type, and the stability of the transferred SaPI was similar to that of the autonomously replicating SaPI plasmid in S. aureus (J. Chen and r.P.N., unpublished observations), indicating that the element can be maintained as a plasmid. Mapping of the integration sites revealed a set of insertion sites that are similar to the secondary attS sites in S. aureus22. (see Supplementary information S3 (table)) Although they could clearly infect L. monocytogenes, phage 80α, phage ϕ11 and several other staphylococcal phages did not form plaques on any of the L. monocytogenes strains tested22.
The dramatic finding of SaPI transfer to L. monocytogenes has raised the interesting question of whether superantigens expressed by an intracellular pathogen within a eukaryotic host cell are toxic, and it has also been suggested that a novel biohazard will emerge: superantigen-expressing L. monocytogenes. Although no naturally-occurring strain of Listeria containing any PrCI-like island that produces staphylococcal super-antigens has been identified to date, we consider it highly likely that SaPI transfer to L. monocytogenes has already occurred or will soon occur, because both S. aureus and L. monocytogenes are commonly found in the bovine udder, most bovine and ovine S. aureus isolates carry genes for one or more superantigens, and phage therapy of bovine mastitis is increasing30.
The staphylococcal phages that induce SaPI transfer to L. monocytogenes do not form plaques, so silent, phage-mediated transfer may represent a new and important mechanism of horizontal gene transfer among bacteria. Furthermore, given the very high transfer frequency that is observed, this may not be confined to highly mobile units such a SaPIs.
Concluding comments
Studies of the staphylococcal PrCIs — the SaPIs in particular — have revealed the very intimate adaptation of a MGE to the phage life cycle, encoding just those genetic functions that enable the element to use the phage to activate its own replicative machinery and to provide the structural components of its own specific capsids. we suggest that PrCIs are not defective prophages but, rather, that they have evolved from prophages in a highly specific manner and that they represent a branch of the pathway leading from fully lytic to temperate phages. At the same time, their high mobility places them at one end of the MGE mobility spectrum, and they are a connecting link between prophages and the other MGEs. In the staphylococci, they are the only known repository of several superantigen genes, including tst, seb and the sec genes, and they therefore have a major role in the dissemination of genes that impact both human health and adaptation of the organism to the animal host. They are very common in staphylococci and are probably widespread among other Gram-positive bacteria. Although they have not been described in Gram-negative bacteria or Archaea, they bear some similarity to other MGEs that exploit helper phages, such as satellite phage P4 and Sulfolobus islandicus plasmid pSSVx31, and it is probably only a matter of time until they are recognized in these groups of organisms as well.
The remaining experimental challenges are, among others: to determine the mechanism of SaPI inhibition of helper phage reproduction; to clarify the relationship between relief of SaPI repression and packaging specificity; to elucidate the nature of the interactions between co-resident SaPIs; to define the molecular genetics and physiology of SaPIs infecting other genera; to determine the bacterial spectrum of PrCIs and their functionality; and to determine the prevalence of PrCI-mediated and phage-mediated silent trans-genera gene transfer.
Supplementary Material
S1
S2
S3
S4
S5
S6
S7
Acknowledgements
We acknowledge, with gratitude, the scientific and intellectual contributions of the many members of our respective laboratories.
Abbreviations
Prophage
A quiescent form of a bacteriophage (usually inserted into the chromosome of its host), in which the lytic functions of the phage are repressed.
Iteron
One member of a set of short repeated DNA sequences that are located at a bacterial origin of replication and are required for the initiation of replication.
SOS response
A global stress response to DNA-damaging agents such as ultraviolet light or mitomycin C.
Campbell mechanism
A recombinational mechanism for the insertion of a genetic element, such as a phage genome or a plasmid, into the bacterial chromosome, involving circularization of the element followed by a single crossover with a chromosomal target site.
Lysogen
A bacterium containing an inducible prophage.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
SUPPLEMENTARY INFORMATION
See online article: S1 (figure) | S2 (figure) | S3 (table) |S4 (figure) | S5 (table) | S6 (table) | S7 (table)
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 2002;19:2226–2238. doi: 10.1093/oxfordjournals.molbev.a004046. [DOI] [PubMed] [Google Scholar]
- 2.Katayama Y, Ito T, Hiramatsu K. A new class of genetic element, staphylococcus cassette chromosome mec encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gillet Y, et al. Association between Staphylococcus aureus strains carrying gene for Panton-Valentine leukocidin and highly lethal necrotising pneumonia in young immunocompetent patients. Lancet. 2002;359:753–759. doi: 10.1016/S0140-6736(02)07877-7. [DOI] [PubMed] [Google Scholar]
- 4.Vojtov N, Ross HF, Novick RP. Global repression of exotoxin synthesis by staphylococcal superantigens. Proc. Natl Acad. Sci. USA. 2002;99:10102–10107. doi: 10.1073/pnas.152152499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/s0147-619x(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 6.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 7.Subedi A, Ubeda C, Adhikari RP, Penades JR, Novick RP. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiology. 2007;153:3235–3245. doi: 10.1099/mic.0.2007/006932-0. [DOI] [PubMed] [Google Scholar]
- 8.Takeuchi F, et al. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of humancolonizing staphylococcal species. J. Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuroda M, et al. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl Acad. Sci. USA. 2005;102:13272–13277. doi: 10.1073/pnas.0502950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott J, Thompson-Mayberry P, Lahmamsi S, King CJ, McShan WM. Phage-associated mutator phenotype in group A streptococcus. J. Bacteriol. 2008;190:6290–6301. doi: 10.1128/JB.01569-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madhusoodanan J, et al. Abstracts of the 107th General Meeting of the American Society for Microbiology. Washington DC: ASM; 2007. [Google Scholar]
- 12.Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 13.Ubeda C, Barry P, Novick RP, Penades JR. Characterization of mutations defining SaPI functions and enabling autonomous replication in the absence of helper phage. Mol. Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 14.Ziegelin G, Linderoth NA, Calendar R, Lanka E. Domain structure of phage P4 α protein deduced by mutational analysis. J. Bacteriol. 1995;177:4333–4341. doi: 10.1128/jb.177.15.4333-4341.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ubeda C, et al. Specificity of staphylococcal phage and SaPI DNA packaging as revealed by integrase and terminase mutations. Mol. Microbiol. 2009;72:98–108. doi: 10.1111/j.1365-2958.2009.06634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ubeda C, et al. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol. Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 17.Tormo MA, et al. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry P. Microbiology. New York: New York Univ. Press; 2006. [Google Scholar]
- 19.Campbell AM. In: Modern Prespectives in Biology. Halvorson HO, Roman HL, Bell E, editors. New York: Harper & Row; 1969. [Google Scholar]
- 20.Ruzin A, Lindsay J, Novick RP. Molecular genetics of SaPI1 – a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 21.Novick RP, Subedi A. The SaPIs: mobile pathogenicity islands of Staphylococcus. Chem. Immunol. Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 23.Maiques E, et al. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. J. Bacteriol. 2007;189:5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tormo-Mas MA, et al. Moonlighting bacteriophage proteins derepress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan S, et al. Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism. J. Mol. Biol. 2004;341:503–517. doi: 10.1016/j.jmb.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 26.Ubeda C, et al. Sip, an integrase protein with excision, circularization and integration activities, defines a new family of mobile Staphylococcus aureus pathogenicity islands. Mol. Microbiol. 2003;49:193–210. doi: 10.1046/j.1365-2958.2003.03577.x. [DOI] [PubMed] [Google Scholar]
- 27.Ubeda C, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc. Natl Acad. Sci. USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tallent SM, Langston TB, Moran RG, Christie GE. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poliakov A, et al. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J. Mol. Biol. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gill JJ, et al. Efficacy and pharmacokinetics of bacteriophage therapy in treatment of subclinical Staphylococcus aureus mastitis in lactating dairy cattle. Antimicrob. Agents Chemother. 2006;50:2912–2918. doi: 10.1128/AAC.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold HP, et al. The genetic element pSSVx of the extremely thermophilic crenarchaeon Sulfolobus is a hybrid between a plasmid and a virus. Mol. Microbiol. 1999;34:217–226. doi: 10.1046/j.1365-2958.1999.01573.x. [DOI] [PubMed] [Google Scholar]
- 32.Schlievert PM, Shands KN, Dan BB, Schmid GP, Nishimura RD. Identification and characterization of an exotoxin from Staphylococcus aureus associated with toxic-shock syndrome. J. Infect. Dis. 1981;143:509–516. doi: 10.1093/infdis/143.4.509. [DOI] [PubMed] [Google Scholar]
- 33.Altemeier WA, et al. Staphylococcus aureus associated with toxic shock syndrome. Ann. Int. Med. 1982;96:978–982. doi: 10.7326/0003-4819-96-6-978. [DOI] [PubMed] [Google Scholar]
- 34.Kreiswirth BN, Projan SJ, Schlievert PM, Novick RP. Toxic shock syndrome toxin 1 is encoded by a variable genetic element. Rev. Infect. Dis. 1989;11(Suppl. 1):S83–S88. doi: 10.1093/clinids/11.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 35.Chu MC, et al. Association of toxic shock toxin-1 determinant with a heterologous insertion at multiple loci in the Staphylococcus aureus chromosome. Infect. Immun. 1988;56:2702–2708. doi: 10.1128/iai.56.10.2702-2708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuroda M, et al. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay JA, Holden MT. Staphylococcus aureus : superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 38.O’Neill AJ, Larsen AR, Skov R, Henriksen AS, Chopra I. Characterization of the epidemic European fusidic acid-resistant impetigo clone of Staphylococcus aureus. J. Clin. Microbiol. 2007;45:1505–1510. doi: 10.1128/JCM.01984-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holden MT, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl Acad. Sci. USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwan T, Liu J, DuBow M, Gros P, Pelletier J. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl Acad. Sci. USA. 2005;102:5174–5179. doi: 10.1073/pnas.0501140102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fitzgerald JR, et al. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3:585–594. doi: 10.1016/s1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 43.Diep BA, et al. The complete genome sequence of USA300, an epidemic community-acquired methicillin-resistant Staphylococcus aureus strain. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1
S2
S3
S4
S5
S6
S7