Parallel Genotyping of Over 10,000 SNPs Using a One-Primer Assay on a High-Density Oligonucleotide Array (original) (raw)
Abstract
The analysis of single nucleotide polymorphisms (SNPs) is increasingly utilizedto investigate the genetic causes of complex human diseases. Here we present a high-throughput genotyping platform that uses a one-primer assay to genotype over 10,000 SNPs per individual on a single oligonucleotide array. This approach uses restriction digestion to fractionate the genome, followed by amplification of a specific fractionated subset of the genome. The resulting reduction in genome complexity enables allele-specific hybridization to the array. The selection of SNPs was primarily determined by computer-predicted lengths of restriction fragments containing the SNPs, andwas further driven by strict empirical measurements of accuracy, reproducibility, andaverage call rate, which we estimate to be >9.5%, >99.9%, and>95%, respectively. With average heterozygosity of 0.38 andgenome scan resolution of 0.31 cM, the SNP array is a viable alternative to panels of microsatellites (STRs). As a demonstration of the utility of the genotyping platform in whole-genome scans, we have replicated and refined a linkage region on chromosome 2p for chronic mucocutaneous candidiasis and thyroid disease, previously identified using a panel of microsatellite (STR) markers.
Single nucleotide polymorphisms (SNPs) are the most abundant form of genetic variation in the human genome (Brookes 1999). Recent estimates suggest that there may be ∼5 million SNPs with minor allele frequencies of at least 10%, and possibly as many as ∼11 million with minor allele frequencies of at least 1% (Kruglyak and Nickerson 2001). The Human Genome Project has provided a windfall of sequence polymorphism data, and because of collaborative SNP discovery initiatives such as the SNP Consortium (TSC), millions of human SNPs have been catalogued, many of which are publicly available in the TSC and NCBI dbSNP repositories (Thorisson and Stein 2003; http://snp.cshl.org/; http://www.ncbi.nlm.nih.gov/SNP/). The challenge is to develop a high-capacity SNP genotyping platform that is readily scalable yet highly accurate.
Almost all currently available SNP genotyping methods (for review, see Tsuchihashi and Dracopoli 2002; Kwok 2001) start with a locus-specific amplification step, followed by an allele discrimination step. At capacities of 1000 SNPs or less, locus-specific amplification is economically feasible in terms of oligonucleotide synthesis and other reagent costs. At capacities of 10,000 SNPs and greater, however, the costs of designing, synthesizing, and managing such an enormous number of oligonucleotides become prohibitive. Additionally, large amounts of starting sample DNA are required to genotype tens of thousands of SNPs in a locus-specific manner. Alternative nonlocus-specific approaches, such as degenerate oligonucleotide primer (DOP)-PCR, that genotype SNPs in a reduced complexity fraction of the genome have proven successful (Grant et al. 2002; Jordan et al. 2002). SNP genotyping has also been successfully demonstrated when reducing genome complexity by restriction digestion and either gel-based or PCR-based fragment size selection, followed by allele-specific hybridization to oligonucleotide arrays (Dong et al. 2001; Kennedy et al. 2003). High-density oligonucleotide arrays have been used to investigate polymorphisms (Chee et al. 1996), and have been applied to SNP genotyping (Wang et al. 1998; Fan et al. 2000; Carrasquillo et al. 2002).
Here we present a robust high-throughput SNP genotyping platform that is based on the approach proposed by Dong et al. (2001) and Kennedy et al. (2003). We rigorously optimized a nonlocus-specific amplification assay that generates a reduced fraction of the genome composed of restriction fragments in a size range that is highly reproducible across samples. Starting with computer-predicted lengths of restriction fragments containing SNPs drawn from the TSC repository, we applied strict acceptance criteria to large sets of empirical data in order to select a set of 11,555 TSC SNPs that have flanking sequences that are most amenable to allele-specific hybridization on high-density oligonucleotide arrays. A genotype-calling algorithm was developed to assign genotypes based on allele-specific hybridization intensities (Liu et al. 2003). We report here an evaluation of the genotyping accuracy and reproducibility of the genotyping platform, as well as a survey of SNP genotypes from 307 individuals across 13 ethnic groups that assesses the level of polymorphism represented in the 11,555 SNPs. Also reported are the genome-wide coverage and estimated genome scan resolution of the genotyped SNPs. As a demonstration of the utility of the genotyping platform in whole-genome scans, we have correctly replicated linkage on chromosome 2p for chronic mucocutaneous candidiasis and/or thyroid disease, previously identified using a panel of microsatellite (STR) markers (Atkinson et al. 2001).
RESULTS
Complexity Reduction Assay
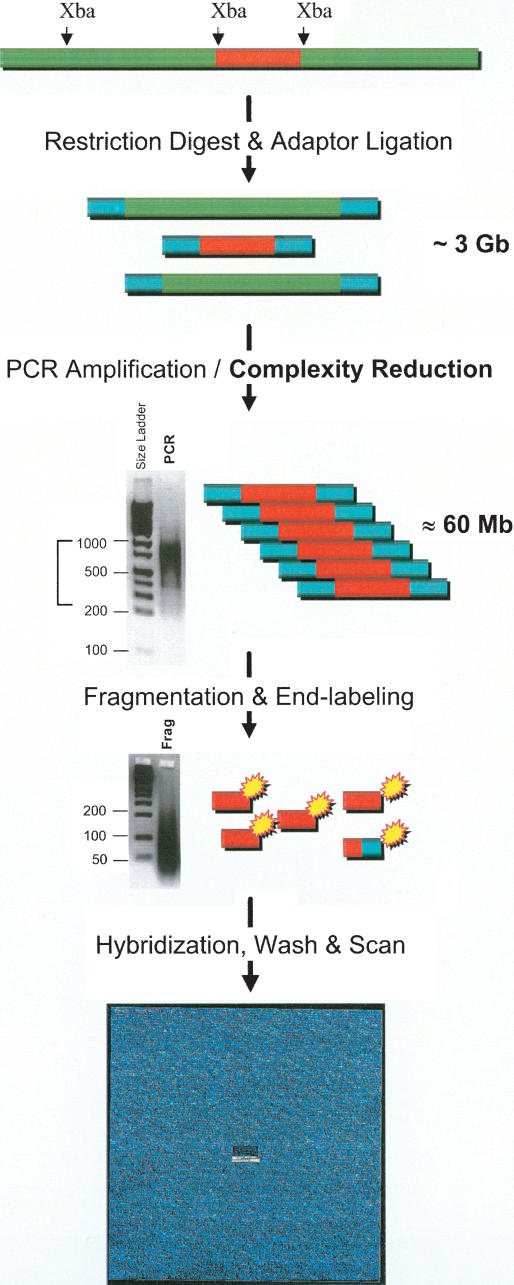
Figure 1 is a schematic of the one-primer amplification assay that reduces the complexity of the genome, and enables allele-specific hybridization. The assay involves five primary steps, starting with restriction digestion, ligation of adaptor, amplification, fragmentation, and labeling, prior to hybridization to the oligonucleotide array (Fig. 1). The complexity reduction occurs at the PCR step which preferentially amplifies restriction fragments that are between 250 and 1000 bp. The sequence complexity of the PCR products is estimated to be ∼60 Mbases, which represents a 50-fold reduction in genome complexity. The adaptor sequence was selected to have no homology with known genome sequences. The one primer used in the PCR is the forward strand of the adaptor; thus only two oligonucleotides are necessary for genotyping over 10,000 SNPs. In contrast, alternative genotyping methods, such as single-base extension (SBE; Nikiforov et al. 1994) and Invader (Hall et al. 2000), require three oligonucleotides to score each SNP. Because the adaptor and primer sequence is not locus-specific, the sequence can be interchanged with alternative sequences with no loss in PCR yields or changes in amplicon size distribution (data not shown). The interchange-ability of the adaptor and primer sequence is a safeguard against carryover contamination, which in contrast will severely compromise locus-specific amplification-based methods.
Figure 1.
Complexity reduction assay and array hybridization. Sample genomic DNAs are digested with Xba I, and adaptors are ligated to the ends of restriction fragments. The fragments are then amplified by using one of the strands of the adaptor as a primer. Restriction fragments in the size range 250–1000 bp are preferentially amplified as shown in the gel image of PCR products. The narrow size range of amplicons is estimated to represent ∼60 Mb of sequence complexity, which is a 50-fold reduction in genome complexity. To allow efficient hybridization to 25-mer oligonucleotide probes on the array, the PCR products are fragmented with DNAse I. The size range of the fragmented PCR products is shown in the second gel image. The fragmented products are biotinylated and then hybridized to the arrays. Following a series of stringent washing and signal generation steps, the arrays are scanned; genotypes are then determined based on hybridization signal intensities.
The choice of restriction enzyme determines the sequence content of the reduced fraction of the genome. The locations of restriction sites vary for each restriction enzyme, and sequence complexity is directly proportional to the frequency of restriction sites. We chose to use Xba I in the current implementation of the assay, and have also demonstrated the assay with Bgl II and EcoRI (Kennedy et al. 2003). In order to ensure that the sequence content of the reduced genome fractions is consistent across hundreds of samples, we optimized and determined robust operating windows of the critical assay steps, in particular the amplification, fragmentation, and labeling steps, as detailed in the Supplemental material (see Assay Optimization) available online at www.genome.org.
Oligonucleotide Array Design and SNP Content
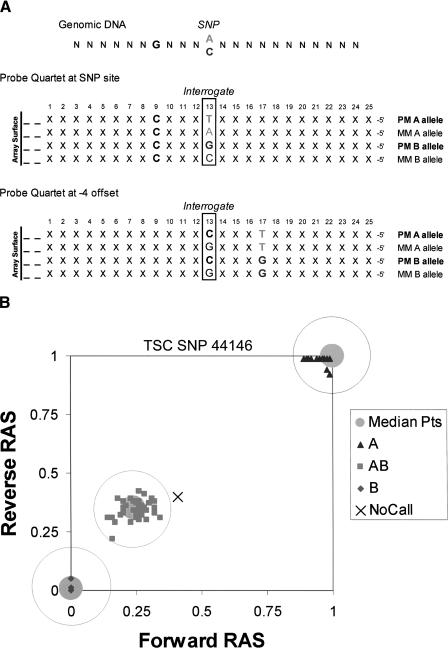
The array is composed of allele-specific hybridization probes that are complementary to SNP regions present in the reduced fraction of the genome amplified in the assay. Photolithography (Fodor et al. 1991) enables the synthesis of over 500,000 unique 25-mer oligonucleotide probe sequences each contained in an 18 μ2 feature. As shown in Figure 2A, the oligonucleotides are organized as pairs of perfect match (PM) and mismatch (MM) probes to allow discrimination between signal and noise. Probe pairs for the A allele and pairs for the B allele are grouped as probe quartets, and are the basis for allele discrimination. To provide data redundancy, each SNP region is oversampled with five probe quartets in both the forward and reverse orientations. By offsetting four of the five quartets from the SNP site by one to four nucleotides (Fig. 2A), each SNP is represented by 40 distinct probes sequences, which allows multiple but slightly different samplings of the same target SNP region in the hybridization. The probe pairs for a given SNP are geographically scattered throughout the array to mitigate the effects of array variations.
Figure 2.
(A) Sequence prototypes of the oligonucleotide probes. Twenty-five-mer oligonucleotides which are complementary to SNP sites and flanking sequences are synthesized on the surface of the array. The 13th nucleotide is the interrogative position where the probe sequences are either perfectly matched (PM) or mismatched (MM) to one of the two alleles of the SNP. The PM and MM probe pairs provide a basis for signal vs. noise measurements. The two probe pairs corresponding to the two alleles are grouped as probe quartets. Shown are the prototype sequences of the probe quartet at the SNP site, where the probe sequences differ only at the SNP site which is also the interrogative position. To provide data redundancy, four additional probe quartets are offset from the SNP site by one to four nucleotides in either direction. Also shown are prototype sequences for the probe quartet offset by –4. In this offset probe quartet, the SNP site has shifted to position 17 of the 25-mer. The probe sequences in this quartet are different at –4 (PM vs. MM) and at the SNP site (allele A vs. B). Each SNP is represented by five probe quartets (one at the SNP site and four offset) in both orientations, for a total of 40 oligonucleotide probes. (B) Genotype calling. The RAS is a measure of the signal intensities contributed from the A allele compared to signals from both alleles. Median RAS values from forward and reverse probe quartets define points in x_–_y coordinates. RAS points from the algorithm training set of 133 ethnically diverse individuals were clustered to determine the three median points, shown as large gray points. Genotypes are assigned for each SNP by determining the shortest distance between RAS points and one of the three median points. Call zones are drawn around the median points to increase the stringency of the genotype assignments. Shown are the RAS points from 99 individuals in 33 CEPH trios for TSC SNP 44146. One of the individuals was not assigned a genotype because the RAS point fell just outside the AB call zone.
The relative allele signal (RAS) is a measure of the signal intensities contributed from the A allele probes compared to signals from both A and B allele probes. In the ideal case, RAS values range from 1 for AA homozygotes to 0 for BB homozygotes, with AB heterozygotes in between at 0.5. For each SNP, two median RAS values are calculated separately for the five forward and five reverse probe quartets. The two median RAS values define points for each of the individuals assayed (Fig. 2B). A genotype-calling algorithm, described by Liu et al. (2003), clusters RAS points from a training set of 133 ethnically diverse individuals into three classes corresponding to the genotypes. For each SNP, the clustering aggregates the 133 points into three median points, each representing a group of individuals from the training set who share the same genotype. As illustrated in Figure 2B, genotype assignments are made for each SNP on the basis of the shortest Euclidean distance to one of three median points. Adjustable call zones are drawn around the median points to increase the stringency of the genotype assignment. In addition, signal-to-noise discrimination filters based on PM and MM probe pairs are applied to mitigate nonspecific hybridization. Signal intensities that fail to meet the discrimination filter criteria and RAS points that fall outside call zones are assigned as “no calls.”
The 11,555 SNPs are the result of a selection process that progressively imposed stricter criteria to cull SNPs from the TSC repository that were the most compatible with the one-primer amplification assay and allele-specific hybridization. SNP selection was primarily determined by computer-predicted fragment lengths based on restriction sites immediately upstream and downstream of the SNP sites. The restriction fragment predictions were initially done on BAC sequence records from GenBank, and later on contig sequence records from the UCSC Golden Path. Restriction fragment length predictions that spanned known contig gaps or other sequence gaps (>30 N's), particularly in draft sequence records were omitted. RepeatMasker (A.F.A. Smit and P. Green, unpubl., http://ftp.genome.washington.edu/RM/RepeatMasker.html) was run on the sequences flanking the SNP sites to check for proximity to known repeat regions. SNPs located inside or within 30 bp of known repeat regions were omitted. A total of 55,605 candidate SNPs drawn from the January 2001 and September 2001 releases of the TSC database were predicted to be on Xba I fragments in the size range of 250 to 1000 bp. Four primary selection criteria were applied to these SNPs: (1) clustering into the three expected genotype groups in 133 ethnically diverse individuals, (2) Mendelian inheritance across 33 families, (3) reproducibility across as many as 12 replicates, and (4) SNP call rates across more than 300 experiments. Additional criteria, including Hardy-Weinberg distribution, uniqueness of map positions, and cross-hybridization predictions, were applied to define the final set of 11,555 SNPs. A detailed accounting of the SNP selection is described in the Supplemental material (SNP Selection).
Genotyping Accuracy
Although concordance with reference genotypes is a measure of genotyping accuracy, the significance of the comparisons is dependent on the accuracy of the reference genotyping methods, and on sampling sizes, which are often constrained by the high costs and low throughputs of current genotyping methods. In order to conduct a representative assessment of genotyping accuracy, we compared genotype calls generated on the oligonucleotide array with reference genotypes generated by a variety of alternative genotyping methods. Further, Mendelian inheritance error analysis of genotypes from pedigrees provided another independent estimate of genotyping accuracy.
Concordance Analysis
Concordance was determined separately in three comparisons with different reference genotyping methods and sample sizes: (1) comparison with publicly available allele frequencies determined from genotypes generated by a variety of methods, (2) concordance with genotypes generated by single base extension (SBE), and finally (3) concordance with dideoxy sequencing.
The first concordance measure was a comparison with allele frequencies reported by the TSC. The TSC initiated the Allele Frequency Project to determine allele frequencies for ∼60,000 TSC SNPs in three major ethnic groups, Caucasian, African-American, and East Asian, each represented by 42 individuals from the Human Variation Panel and American Diabetes Association. TSC allele frequencies estimated from genotyping methods using pools of individual samples were excluded from the comparison. The overlap between the 11,555 SNPs represented on the array and TSC SNPs with reported allele frequencies based on nonpooled genotyping methods was 989 and 755 SNPs in Caucasians and African-Americans, respectively, with 741 SNPs in common between the two groups. Comparison of our allele frequencies in the same 42 individuals from the two groups revealed correlations of 0.992 and 0.993, respectively. The correlations are capped below 1.0 because of genotyping errors that are unaccounted for in the TSC data; also, TSC allele frequencies of many SNPs were based on only 30 or less individuals instead of 42. Despite these comparison limitations, the high correlations between allele frequencies confirmed overall agreement with data generated using a range of genotyping methods at the major sequencing centers, as well as at companies engaged in developing genotyping methods.
The second concordance measure was a comparison with genotypes generated by a proprietary high-throughput SBE platform. We compared genotype calls for 538 SNPs out of 11,560 SNPs across 40 individuals, and found 98 discordances in 21,191 comparisons giving a concordance of 99.5% (Table 1). A tallying of the discordances by SNP revealed that five of the 538 SNPs accounted for a disproportionate 59 of the 98 discordances in 37 of the individuals. That five SNPs representing less than 1% of the sampling set contributed to over 60% of the discordances is an indication of nonrandom and systematic error in either the reference SBE calls or the array-based calls; therefore, the five SNPs were excluded from the set of 11,555 SNPs. The remaining 39 of the 98 discordant calls were scattered among 28 SNPs across 25 individuals, which is a pattern more consistent with random and nonsystematic errors. We attempted to resolve these discordances by comparing both SBE-based and array-based calls with genotypes determined by dideoxy sequencing. Interestingly, 66% of array calls that were discordant with SBE calls were found to be concordant with sequencing calls. Assuming that genotypes concordant with sequencing are correct, the concordance with the SBE genotypes is an underestimate of the actual genotyping accuracy.
Table 1.
Genotyping Accuracy
| Sampled SNPs | Total SNPs | Individuals or families | Genotypes compared | Discordances or inheritance errors | Discordant SNPs | Concordance | |
|---|---|---|---|---|---|---|---|
| Single base extension | 543 | 11,560 | 40 | 21,191 | 98 | 33 | 99.5% ± 0.2% |
| Dideoxy sequencing | 60 | 11,555 | 6 | 341 | 1 | 1 | 99.7% ± 0.7% |
| Mendelian inheritance | 11,555 | 11,555 | 5 trios | 167,649 | 61 | 60 | 99.96% ± 0.01% |
The third concordance study is a comparison with a set of 60 SNPs genotyped by dideoxy sequencing in six individuals from the Human Variation Panel. To ensure equal representation of genotype calls in the comparison, the SNPs and individuals were chosen so that each SNP had two AA homozygotes, two AB heterozygotes, and two BB homozygotes. Sequencing-based genotypes were obtained for 341 of the 360 attempted calls. Of the 341 sequencing calls, there was one discordance with the array-based calls (Table 1). Based on trace data from three independent sequencing data sources, this one discordance with sequencing was due to an unexpected polymorphism immediately adjacent to the SNP site in one of the six individuals. Neither the TSC nor dbSNP had a record of a polymorphism at this adjacent position. The occurrence of unreported polymorphisms in close proximity to the interrogated SNP site can destabilize hybridization to probes for one or both alleles, and lead to erroneous genotype calls such as in this isolated instance.
Mendelian Inheritance Error Analysis
Genotype calls in pedigrees that do not adhere to Mendelian inheritance patterns are indicative of genotyping errors. PEDCHECK software (O'Connell and Weeks 1998) is a widely used genetic analysis tool for detecting the occurrence of inheritance errors in pedigree genotypes. PEDCHECK reports errors which include any inconsistencies between parents and offspring in nuclear families, as well as any male heterozygote calls in sex-linked chromosomes. A total of 38 family trios consisting of two parents and one child were genotyped. The families were from the Centre d'Etude du Polymorphisme Humain (CEPH) or the National Institute of General Medical Sciences (NIGMS) repositories. The majority of the trio genotyping data was used for SNP selection; however, five trios were reserved as a validation data set. There were 61 inheritance errors out of 167,649 genotype calls in the five trios, which is a 0.036% inheritance error rate and suggests accuracy as high as 99.96% (Table 1). Unlike discordances, which assume the accuracy of reference genotypes, inheritance errors are unequivocally indicative of genotyping errors. However, not all possible instances of genotyping errors are reported as inheritance errors. For example, if one parent is homozygous and the other is heterozygous, the child can either be called homozygous or heterozygous, and if both parents are called heterozygotes, the child can be called any of the three possible genotypes without generating an error. Therefore, the inheritance-based estimate of genotyping accuracy is an overestimate.
Reproducibility
Sets of replicate experiments demonstrated that the genotyping reproducibility in as many as nine replicates is 99.99% and can range as high as 99.999% (Table 2). Eight individuals from the Human Variation Panel were run in triplicate on three sets of arrays manufactured from three different wafer lots, for a total of nine replicates. The arrays are manufactured in sets of 49 arrays from one glass wafer. This set of experiments was designed to be a highly stringent test of assay reproducibility, as well as array manufacturing reproducibility. Genotype calls from each experiment were compared against a consensus set of genotype calls based on all nine replicates. Out of 820,235 genotypes in 72 experiments, there were 40 calls that were inconsistent with the consensus calls, which is an average reproducibility of 99.995% among the eight individuals. In one of the eight individuals, the reproducibility was 99.999%, where there was just one inconsistency out of 102,937 calls from nine replicates (Suppl. Table S-3). Of the 24 sets of triplicate experiments within arrays from the same wafer lot, 10 sets of triplicates had 100% reproducibility. A high level of reproducibility was also observed in replicate experiments run on peripheral blood samples. A peripheral blood sample was genotyped nine times. Out of 102,484 genotype calls in the nine replicates, there were only 12 calls that were in disagreement with consensus calls, which is a reproducibility of 99.988% (Table 2).
Table 2.
Reproducibility
| Replicates | Individuals | Genotypes | Discordances | Call rate | Reproducibility | |
|---|---|---|---|---|---|---|
| Human variation panel | 9 | 8 | 820,235 | 40 | 98.59% ± 0.42% | 99.995% ± 0.003% |
| Peripheral blood | 9 | 1 | 102,484 | 12 | 98.55% ± 0.81% | 99.988% |
Detection of Sample Contamination and Degradation
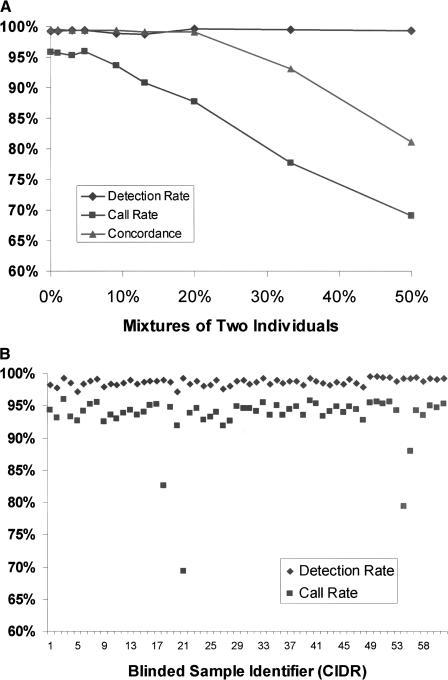
A potential problem, particularly in high sample-throughput situations, is inadvertent mixing of DNA from different individuals. To assess the ability of the platform to identify cases of mixed samples, DNA from two individuals were combined in various amounts. The two individuals were from the Human Variation Panel, and reference genotypes based on SBE were available for both. Figure 3A shows that call rates, which are the percentage of SNPs assigned a genotype instead of a no call, decreased as the proportion of the second individual was increased, whereas the detection rate, a measure of the number of SNPs passing the signal versus noise discrimination filter, remained constant. As the two individuals were increasingly mixed together, the RAS values were gradually shifted in any SNP where the genotypes differed. For example, in SNPs that were homozygous in one individual but heterozygous in the second, the RAS points would gradually shift toward a midpoint between the two genotypes. The occurrence of no calls steadily increased as more RAS values shifted outside the call zones drawn around the median points (described above). Interestingly, the concordance with reference genotypes remained high whereas call rates fell much more rapidly, demonstrating that the genotyping algorithm gives priority to high accuracy over call rates by assigning no calls rather than muddled and incorrect genotypes.
Figure 3.
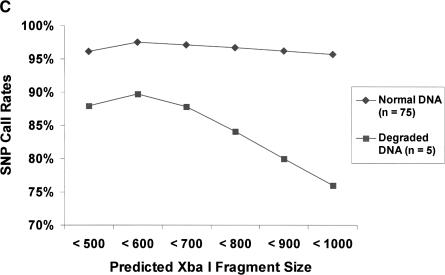
(A) Detection of sample contamination. Sample DNAs from two individuals were progressively mixed together and genotyped as mixtures. The detection rate is a measure of the number of SNPs passing the signal vs. noise discrimination filter. The call rate is the percentage of SNPs that were assigned a genotype instead of a no call. The concordance rate is based on comparison of genotype calls with SBE reference genotypes for one of the two individuals. At 50% mixture, the concordance rate was nearly identical compared to SBE reference genotypes from either individual (data not shown). (B) Blinded samples from the CIDR. Detection and call rates from 61 blinded samples from the CIDR. Blinded samples containing mixtures of two individuals were identified as having high detection rates, but low call rates. Other blinded samples included eight samples that were previously problematic when genotyped at STR loci. The samples also included 18 members of a family affected with chronic mucocutaneous candidiasis and thyroid disease, as well as CEPH pedigree samples. (C) Degraded sample DNA and SNP call rates. Call rates were calculated for each SNP across multiple experiments. There were five experiments in the case of degraded sample DNAs, and for comparison, 75 experiments in the case of normal DNAs. SNPs were binned into six groups on the basis of the predicted lengths of the restriction fragments containing the SNPs. Each bin represented restriction fragment lengths in increments of 100 bp, and SNP call rates were averaged within each of the bins.
A blinded set of 61 samples from the Center for Inherited Disease Research (CIDR) was genotyped, among which were mixtures containing contamination of a second individual. Blinded samples with detection rates >98% and usually low call rates of 87.3%, 83.2%, 79.4%, and 69.3% (Fig. 3B) were successfully detected as mixed samples of 20%, 40%, 50%, and 50%, respectively. Eight samples in the blinded set that previously had low and intermediate call rates with STR genotyping, all had >90% call rates by our SNP genotyping method (Fig. 3B).
Another potential problem with starting samples is sheared or degraded DNA. Five degraded DNA samples, as judged by gel electrophoresis, were genotyped, but resulted in low call rates, which ranged from 82.8% to 86.7%. SNP call rates were plotted against the predicted lengths of Xba I restriction fragments containing the SNPs (Fig. 3C). For comparison, SNP call rates from 75 nondegraded DNA samples were plotted, showing that the SNP call rates in nondegraded samples were for the most part independent of the predicted amplicon length. In contrast, SNP call rates in the five degraded samples were lower across the size range, and as expected, particularly lower for SNPs predicted to be on longer amplicons.
Call Rates and Marker Heterozygosities
Individuals belonging to 13 ethnic groups from various geographic locations and demographic histories were genotyped. The ethnic groups were each represented by at least 19 individuals, and in the case of groups from the Human Variation Panel, there were up to 42 individuals. In total there were over 3.4 million genotype calls from 307 individuals, giving an overall call rate of 95.9% and an average of 11,075 genotypes per individual (Table 3A). Differences in the average call rates between groups are due to variations in the condition of the sample DNAs, and are not due to any biases introduced from the ethnicities of the samples used to train the genotyping algorithm. Of the 133 individuals in the genotyping algorithm training set, Caucasians represented ∼40%, African-Americans represented ∼30%, and East Asians ∼15%, with ethnicity-blinded Polymorphism Discovery samples making up the remainder.
Table 3A.
Call Rates and Heterozygosities in Thirteen Ethnic Groups
| Individuals | Mean call rate | Median heterozygosity | Mean heterozygosity | Nonpolymorphic | |
|---|---|---|---|---|---|
| African-American | 42 | 96.5% ± 1.2% | 0.39 | 0.35 ± 0.14 | 109 |
| Caucasian | 42 | 98.5% ± 0.6% | 0.40 | 0.35 ± 0.15 | 402 |
| Mende | 22 | 95.0% ± 1.6% | 0.35 | 0.31 ± 0.16 | 868 |
| South Asian | 22 | 96.6% ± 0.9% | 0.39 | 0.35 ± 0.15 | 555 |
| Mbuti Pygmy | 20 | 96.3% ± 1.3% | 0.29 | 0.27 ± 0.17 | 1518 |
| East Asian | 20 | 97.3% ± 1.2% | 0.38 | 0.32 ± 0.17 | 1168 |
| Nahua | 20 | 93.1% ± 1.5% | 0.32 | 0.28 ± 0.18 | 1865 |
| Puerto Rican | 20 | 92.7% ± 3.0% | 0.40 | 0.35 ± 0.14 | 362 |
| Quechua | 20 | 95.1% ± 0.9% | 0.32 | 0.29 ± 0.18 | 1566 |
| Altaian | 20 | 97.5% ± 2.1% | 0.39 | 0.34 ± 0.15 | 601 |
| Spanish | 20 | 93.8% ± 2.3% | 0.38 | 0.33 ± 0.16 | 710 |
| Burunge | 20 | 92.9% ± 4.8% | 0.38 | 0.33 ± 0.15 | 599 |
| Nasioi | 19 | 97.1% ± 1.8% | 0.30 | 0.27 ± 0.18 | 2145 |
| *Across all groups | 307 | 95.9% ± 2.6% | 0.41 | 0.38 ± 0.12 | 4 |
To discount the possibility of biases introduced from the ethnicities in the training data set, a second algorithm training set was constructed using 103 individuals from five other ethnic groups. Genotypes were determined based on this alternative training set, and the overall call rate across 307 individuals was 96.5%. To assess the accuracy of these new genotypes, calls were compared with SBE genotypes in 40 individuals. The concordance was 99.5%, where there were 105 discordances in 20,953 comparisons, which was essentially the same as the 99.5% concordance based on the original training set (Table 1). Similarly, the occurrence of inheritance errors in the five CEPH trios (described above) was fairly consistent between the two algorithm training sets, at 0.083% compared with 0.036% previously. Thus, call rates as well as measures of genotyping accuracy are not dependent on a particular algorithm training data set.
Heterozygosities of the 11,555 genotyped SNPs were calculated across the 13 ethnic groups (Table 3A). When all 307 individuals were aggregated together, the overall median and mean heterozygosity values were 0.41 and 0.38, respectively. For comparison, commonly used panels of ∼400 STRs have average heterozygosities of ∼0.8 (Dubovsky et al. 1995). The distributions of heterozygosity values and minor allele frequencies are shown in Table 3B. Around 8% (966) of the SNPs had minor allele frequencies of 10% or lower, and only 0.2% (23) had minor allele frequencies of 1% or lower (Table 3B). Heterozygosity values varied between the ethnic groups (Table 3A), and reflect the degree of isolation of certain populations, such as the Mbuti Pygmy (Ituri Forest) and Nasioi (Bougainville) groups which had the lowest values. Although nonpolymorphic SNPs were found within each ethnic group, there were only four SNPs that were not polymorphic when data were combined from all 13 ethnic groups. These four SNPs were polymorphic within the Polymorphism Discovery panel samples that were used for SNP discovery by the TSC (Altshuler et al. 2000; Mullikin et al. 2000) and for algorithm training (data not shown). Interestingly, the African-American group had the lowest number of nonpolymorphic SNPs (Table 3A), which is consistent with this group's diverse origins in Africa and history in North America.
Table 3B.
Histograms of Heterozygosities and Minor Allele Frequencies Determined in 307 Individuals From 13 Ethnic Groups
| Heterozygosity | Number of SNPs | % of SNPs | Minor allele frequency | Number of SNPs | % of SNPs |
|---|---|---|---|---|---|
| 0 | 4 | .03% | 0% | 4 | .03% |
| ≤0.05 | 63 | .6% | ≤1% | 19 | .2% |
| ≤0.10 | 208 | 2.4% | ≤5% | 220 | 2.1% |
| ≤0.15 | 379 | 5.7% | ≤10% | 723 | 8.4% |
| ≤0.20 | 544 | 10.4% | ≤15% | 1050 | 17.4% |
| ≤0.25 | 737 | 16.7% | ≤20% | 1311 | 28.8% |
| ≤0.30 | 930 | 24.8% | ≤25% | 1416 | 41.0% |
| ≤0.35 | 1202 | 35.2% | ≤30% | 1421 | 53.3% |
| ≤0.40 | 1412 | 47.4% | ≤35% | 1362 | 65.1% |
| ≤0.45 | 1826 | 63.2% | ≤40% | 1376 | 77.0% |
| ≤0.50 | 4250 | 100.0% | ≤45% | 1352 | 88.7% |
| ≤50% | 1301 | 100.0% |
Genome-Wide Coverage
Of the 11,555 genotyped SNPs, 11,384 (98.5%) are currently mapped to unique positions in the UCSC Golden Path (release hg13, November 2002). The remaining unmapped SNPs are on sequence records that allowed restriction fragment predictions, but could not be assigned to unique positions in this build of the Golden Path. To visualize the genome-wide coverage of the genotyped SNPs, physical maps of the chromosomes were plotted with red vertical bars representing the presence of at least one SNP in 100-kb regions, and black vertical bars representing large contig gaps that are 100,000 N's or longer (Fig. 4A). Large contig gaps are gaps between map contigs, and also represent large regions of heterochromatin, including centromeres and telomeres. The SNPs are well distributed across the genome, but coverage is not absolutely uniform, with some regions containing fewer markers than other regions. The distribution of the genotyped SNPs is determined by the occurrence of Xba I sites in the genome, which is essentially random, but certain regions have fewer sites. Also, regions of the genome heavily represented by BAC and contig records in draft stages containing many clone gaps will have given fewer predictions of restriction fragments and contributed proportionately fewer candidate SNPs from which the 11,555 were selected. SNPs with physical map positions were assigned genetic distances by interpolating against a high-resolution genetic map based on 5136 STR markers, made available by deCODE (Kong et al. 2002). Genetic views of the chromosomes were plotted with red bars representing the presence of at least one genotyped SNP in 0.1 cM intervals (Fig. 4B). The distribution of interpolated genetic distances shows broad coverage across the chromosomes, but reveals a number of underrepresented intervals, particularly in chromosome 19 and the X chromosome.
Figure 4.
(A) Distribution of SNPs across the genome. Physical map assignments of 11,384 SNPs are based on the November 2002 release of the UCSC Golden Path (http://genome.ucsc.edu/). Red vertical bars represent the presence of at least one SNP in 100-kb regions. Black vertical bars represent large contig gaps that are 100,000 N's or longer. The Y chromosome is not shown because none of the 11,555 SNPs map to this chromosome. (B) SNPs distributed on the basis of genetic distances across the genome. Red bars represent the presence of at least one genotyped SNP in 0.1-cM intervals. Genetic distance assignments were based on interpolations against the deCODE STR genetic map (Kong et al. 2002). STR markers that had discrepant chromosome assignments and ordering between the deCODE map and Golden Path were omitted from the interpolation. Pairs of deCODE STRs with unique physical positions were used as local landmarks to interpolate genetic distances for SNPs located between the STRs. The interpolation makes the simplifying assumption that physical and genetic distances correlate linearly over the short localized regions of the chromosomes defined by pairs of STRs drawn from the 5136 STRs on the deCODE map. Physical map positions as well as the interpolated genetic distances are reported for each SNP in the Supplemental material (SNP Information).
Inter-SNP distances provide an estimate of SNP coverage across the genome and are a useful measure of marker utility. Physical distances between the 11,384 mapped SNPs were calculated with and without accounting for large contig gaps. There were 927 contig gaps of size 10,000 N's or longer which all together totaled over 209 Mb (roughly 9% of the genome). In addition, 568 of the 11,384 SNPs had at least one contig gap in between. The median and mean inter-SNP distances were 104.0 kb and 209.8 kb, respectively, when the large contig gaps (10,000 N's or longer) were excluded from the genome. The longest inter-SNP distance was a 4-Mbase stretch in chromosome 7. We found that 49% of the genotyped SNPs are less than 100 kb apart, and 97% of the SNPs are less than 1 Mb apart (Suppl. Table S-4B). The median and mean when including the contig gaps were artificially higher at 116.2 kb and 254.1 kb, respectively (Suppl. Table S-4A), and the longest inter-SNP distance was the 24-Mbase centromere in chromosome 1. Inter-SNP genetic distances were estimated based on interpolated genetic distances (Suppl. Table S-4A). The median inter-SNP distance was 0.10 cM, and the mean was 0.31 cM. The longest distance was a 9.98-cM span in chromosome 19. Fifty percent of the genotyped SNPs had distances less than 0.1 cM, and over 92% had distances less than 1 cM (Suppl. Table S-4B). Because of cases where multiple SNPs were located between pairs of unresolved STRs, the interpolated genetic distances of these genotyped SNPs were not unique, and account for the 684 SNPs, or 6%, with zero inter-SNP distances. For comparison, panels of ∼400 STRs that are commonly used for genome-wide scans in linkage analysis have average intermarker distances of ∼10 cM (Dubovsky et al. 1995).
Replication of Linkage at a Disease Locus
Atkinson et al. (2001) identified a candidate linkage region on chromosome 2p using a 10-cM STR genome scan in a family with a combination of chronic mucocutaneous candidiasis and thyroid disease. Detailed information on this large nonconsanguineous family is given in that study (Atkinson et al. 2001). We attempted to replicate this linkage result using our SNP genotyping method, and generated genotypes from 18 of the individuals in this family. The overall call rate for the family samples was 97.98%, and the Mendelian inheritance error rate was 0.04%. Nonparametric linkage analyses were performed using the software package Merlin 0.9.3 and GENEHUNTER version 2.1 (Kruglyak et al. 1996; Abecasis et al. 2002; see Methods for details). A total of 961 SNPs on chromosome 2 were used in the two-point analysis. Using 525 informative SNPs on chromosome 2, multipoint nonparametric LOD (NPL) scores reached 8.39 (P = 0.0078) in the 38.28 to 56.01 cM region (Table 4). Nonparametric linkage analyses were also performed on 10-cM genome scan data from 28 STR markers on chromosome 2. The STR marker genotyping on the same samples gave a maximum multipoint NPL score of 4.21 (P = 0.0039) in the region between 38.33 and 73.61 cM (Table 4). In conclusion, we were able to replicate the linkage region and refine the linkage interval into a 17.73-cM region on chromosome 2.
Table 4.
Replication of Linkage Region in a Family With a Combination of Chronic Mucocutaneous–Candidiasis and Thyroid Disease, Using SNPs
| Informative markers on Chr 2 | LOD score | _P_-value | Interval | |
|---|---|---|---|---|
| 10 cM STR genome scan | 28 | 4.21 | 0.0039 | 35.28 cM |
| SNP array (11,555 SNPs) | 525 | 8.39 | 0.0078 | 17.73 cM |
DISCUSSION
We have shown that the complexity reduction and parallel genotyping platform is a highly accurate method for high-throughput genome-wide SNP genotyping. Based on concordance measures with current genotyping methods, and analysis of inheritance, the genotyping accuracy is conservatively estimated to be >99.5%. The reproducibility in as many as nine replicate experiments was 99.99%. The assay procedures have been rigorously optimized to achieve robustness.
As demonstrated by the replication of linkage of chronic mucocutaneous candidiasis and/or thyroid disease on chromosome 2p, the set of 11,555 SNPs genotyping on the array presents a highly attractive alternative to panels of STR markers for whole-genome scans. The average heterozygosity of the 11,555 SNPs in 307 individuals from 13 ethnic groups was 0.38. The call rate across the 307 individuals, representing DNA isolated by a variety of methods, was 95.9%. The genotyped SNPs are spaced on average every 210 kb across the genome, and based on interpolated genetic distances are spaced every 0.31 cM. The average inter-SNP distance of 0.31 cM suggests that the genome scan resolution of the 11,555 SNPs on the array may be as much as 30-fold higher than currently used panels of ∼400 STR markers. Although STRs have more allelic variation than bi-allelic SNPs, when genotyped in greater numbers, SNPs may provide higher power and accuracy in disease mapping linkage studies (Xiong and Jin 1999). In addition, the SNP array can be used to investigate loss of heterozygosity (LOH) in paired sets of tumor and control samples. A caveat is that samples, particularly from paraffin-fixed tissues, cannot be degraded. Furthermore, our survey of SNP genotypes across 13 ethnic groups is representative of how the genotyping platform can advance studies that investigate genetic variations across populations and across human history (for review, see Miller and Kwok 2001).
In study designs that involve isolated populations, a proportion of the 11,555 SNPs are likely to be noninformative. The utility of any given SNP is highly dependent on the ethnic context of the individuals that are genotyped. Moreover, very rare polymorphisms and mutations may disrupt the ability to genotype particular SNPs in certain individuals. The occurrence of an unexpected polymorphism immediately adjacent to an SNP site resulted in an erroneous genotype call. Similarly, rare polymorphisms or mutations that disrupt Xba I restriction sites can result in no calls or possibly incorrect genotype calls.
The genotype-calling algorithm prioritizes accuracy over call rates. The current implementation of call zones drawn around median points assumes that the scatter of RAS points about a median point is (1) circular and normally distributed, and (2) equal for all three genotypes. Neither of these simplifying assumptions, however, is true. There are cases where there is orientation-specific hybridization asymmetry that appears as RAS points scattered in one axis but not the other; and, there are instances of allele-specific hybridization asymmetry that appears as RAS points scattered for heterozygotes and one of the homozygotes, but not for the opposite homozygotes. The noncircular and unequal distributions of RAS points observed in the training data, however, are lost when the clustering process aggregates over 100 points down to three median points. Model-based algorithms that retain the RAS point distributions could capture more genotype calls from RAS points scattered outside call zones, while correctly filtering out spurious RAS points as no calls. However, such model-based algorithms have been difficult to generalize across all SNPs, particularly in low-heterozygosity SNPs where there are very few minor allele homozygotes from which to construct meaningful distribution models. Improvements to the genotype-calling algorithm and alternative approaches, exemplified by Cutler et al. (2001), have been in development, and when implemented should increase call rates, while maintaining the high levels of accuracy.
The scalability of the genotyping platform is driven by two underlying trends: (1) continuous SNP discovery, and (2) increasing density of oligonucleotide arrays. The November 2002 release of the TSC database contains over 1.8 million SNPs, which are a subset of the over 4 million human reference SNP (RS) cluster records contained in the current build of the dbSNP (build 114). In the near future, these public SNP repositories combined with large private repositories will undoubtedly contain a complete catalog of SNPs in the genome. To access greater numbers of SNPs, the complexity reduction assay can be run in parallel on different fractions of the genome defined by more than one restriction enzyme. The parallel use of multiple restriction enzymes should also result in a more uniform distribution of SNPs across the genome by compensating for the scarcity of particular restriction sites in certain regions. Multiple genome fractions coupled with very-high-density oligonucleotide arrays containing several million probes will enable the parallel genotyping of hundreds of thousands of SNPs. Ultimately, scaling to upwards of half a million SNPs should enable whole-genome case-control association studies that may help identify the causative genes and mechanisms at work in complex diseases (for review, see Jorde 2000).
In conclusion, we have developed a genotyping platform that represents a new approach to genome-wide SNP genotyping. The platform extracts information value from publicly available data, in the form of SNP content provided by the TSC and sequences from Human Genome Project, by combining a simple complexity reduction assay with the enormous capacity and allele-specific sensitivity of high-density oligonucleotide arrays. The high levels of throughput, genotyping accuracy, marker heterozygosity, and genome-wide coverage each contribute to the functionality of the genotyping platform to greatly broaden the scope of previous studies, and accelerate advancements across a range of applications, starting with linkage analysis, LOH, population genetics, and ultimately whole-genome association studies.
METHODS
Preparation of Reduced Complexity Samples
To increase sample throughputs, procedures were carried out in 96-well plates. For each individual assayed, 250 ng of genomic DNA was digested with 10 U of Xba I (New England BioLabs) in a volume of 15 μL for 2 h at 37°C. Following heat inactivation at 70°C for 20 min, 0.25 μM of adaptor (5′phosphate-CTAGAGATCAGGCGTCTGTCGTGCTCATAA-3′, and 5′-ATTATGAGCACGACAGACGCCTGATCT-3′ synthesized by QIAGEN) was ligated to the digested DNA with T4 DNA Ligase (New England BioLabs) in 25 μL for 2 h at 16°C. The ligation was stopped by heating to 70°C for 20 min, and then diluted fourfold with water. For each sample, four PCRs were run using 10 μL of the diluted ligation reaction (25 ng of starting DNA) in 100 μL volumes containing 0.75 μM of primer (5′phosphate-CTAGAGATCAGGCGTCTGTCGTGCTCATAA-3′), 0.25 mM dNTPs, 2.5 mM MgCl2, 10 U AmpliTaq Gold (Applied Biosystems), and PCR Buffer (Applied Biosystems). Thirty-five cycles of PCRs were done in either MJ DNA Engine Tetrad (MJ Research) or GeneAmp PCR System 9700 (Applied Biosystems) cyclers. The cycling program in the MJ Tetrads was 95°C denaturation for 20 sec, 59°C annealing for 15 sec, and 72°C extension for 15 sec. The denaturation, annealing, and extension times were each increased to 30 sec when using the GeneAmp cycler. As a check, 3 μL of PCR products were visualized on 2% TBE agarose gels to confirm the size range of amplicons. PCR products from the four reactions were combined and purified over MinElute 96 UF PCR Purification plates (QIAGEN). PCR amplicons from the four 100 μL reactions were recovered in 40 μL of EB buffer (QIAGEN). PCR yields, based on absorbance readings at 260 nm, were typically ∼30 μg. To allow efficient hybridization to the 25-mer oligonucleotides on the array, PCR amplicons were fragmented with DNAse I (Amersham Biosciences). Here, 0.24 U of DNAse I was added to 20 μg of purified PCR amplicons in a 55 μL volume containing 50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol for 30 min at 37°C, followed by heat inactivation at 95°C for 15 min. Fragmentation products were visualized on 4% TBE agarose gels. The 3′ ends of the fragmented amplicons were biotinlyated by adding 143 μM of a proprietary DNA labeling reagent (Affymetrix) using Terminal Deoxynucleotidyl Transferase (Promega) in a 70 μL volume containing 100 mM cacodylic acid (pH 6.8), 0.1 mM dithiothreitol, and 1 mM CoCl2 for 2 h at 37°C, followed by heat inactivation at 95°C for 15 min.
Genotyping by Allele-Specific Hybridization
The fragmented and biotinylated PCR amplicons were combined with 11.5 μg/mL human Cot-1 (Invitrogen) and 115 μg/mL herring sperm (Promega) DNAs. The DNAs were added to a hybridization solution containing 2.69 M tetramethylamonium chloride (TMACl), 56 mM MES, 5% DMSO, 2.5 X Denhardt's solution, and 0.0115% Tween-20 in a final volume of 260 μL. The hybridization solution was heated to 95°C for 10 min, then placed on ice. Next, 200 μL of the hybridization solution was injected into cartridges housing the oligonucleotide arrays (Affymetrix GeneChip 10K Mapping Array version Xba131, version Xba130 for early access, or version XbaDev2 for R&D). Hybridization was carried out at 48°C for 16–18 h in a rotisserie rotating at 60 rpm. Following the overnight hybridization, the arrays were washed with 6X SSPE and 0.01% Tween-20 at 25°C, then more stringently washed with 0.6X SSPE and 0.01% Tween-20 at 45°C. Hybridization signals were generated in a three-step signal amplification process: 10 μg/mL streptavidin (Pierce) was added to the biotinylated targets hybridized to the oligonucleotide probes, and washed with 6X SSPE and 0.01% Tween-20 at 25°C, followed by the addition of 5 μg/mL biotinylated goat anti-streptavidin (Vector) to increase the effective number of biotin molecules on the target; finally, streptavidin R-phycoerythrin (SAPE) conjugate (Molecular Probes) was added and washed extensively with 6X SSPE and 0.01% Tween-20 at 30°C. The Streptavidin, Antibody, and SAPE were added to arrays in 6X SSPE, 1X Denhardt's solution, and 0.01% Tween-20 at 25°C for 10 min. The washing and staining procedures were performed using Affymetrix fluidics stations. Arrays were scanned using either the GeneArray (Agilent) or GCS3000 (Affymetrix) scanners. Scan images were processed to get hybridization signal intensity values using either Micro Array Suite (MAS) v5 software (Affymetrix) or GCOS v1 software (Affymetrix). The genotype-calling algorithm as described in Liu et al. (2003) was implemented in GenoTyping Tools (GTT; Affymetrix) and GDAS v2 (Affymetrix) analysis software. Default algorithm parameters, that is, a discrimination score cutoff of 0.08 and call zones of 0.8, were used to make all of the genotype calls.
Sources of Samples and Reference Genotypes
The 133 individuals in the algorithm training set consisted of 24 from the Polymorphism Discovery Panel, and 42 Caucasians, 42 African-Americans, and 20 East Asians (10 Chinese, 10 Japanese) from the Human Variation Panel, as well as five Caucasian individuals from CEPH families (Utah pedigrees). Sample DNAs from the Polymorphism Discovery and Human Variation Panels as well as CEPH and NIGMS families were purchased from the Coriell Institute for Medical Research. The alternative algorithm training set of 103 individuals consisted of 22 South Asians (Southern India), 20 Altaians (Siberia), 19 Nasioi (Bougainville), 20 Mbuti Pygmies (Ituri Forest), and 22 Mende (Sierra Leone). DNA from individuals belonging to the ethnic groups Puerto Rican, Spanish (Valencia), Nahua (Mexico), and Quechua (Cusco, Peru), and Altaian (Siberia) were from the Department of Anthropology, Pennsylvania State University. DNA from individuals in the Mbuti Pygmy (Ituri Forest) and South Asian (Southern India) groups were from the Department of Human Genetics, University of Utah. DNA samples from the Burunge (Tanzania) and Mende (Sierra Leone) were from the Department of Biology, University of Maryland. DNAs from the Nasioi (Bougainville) were from the Department of Anthropology, Temple University. DNA from members of a family affected with chronic mucocutaneous candidiasis and thyroid disease were from the University of Alabama Medical Center.
TSC allele frequency data from the TSC Allele Frequency Project were downloaded from the FTP site: ftp://snp.cshl.org/pub/SNP/frequency/. Allele frequency contributors included the Whitehead Institute, Sanger Center, Washington University, Orchid Biosciences, Celera, and Motorola. For SNPs that had frequencies reported by more than one contributor, the frequency value based on the higher number of individuals was used in the allele frequency comparison. Genotypes based on single base extension (SBE) were obtained from one of the allele frequency contributors. Dideoxy sequencing was performed by Qiagen Genomics, SeqWright, and Lark Technologies.
Restriction Fragment Predictions and SNP Mapping
TSC SNP map positions and flanking sequences were downloaded from the TSC FTP site: ftp://snp.cshl.org/pub/SNP/. BAC and other sequence records containing SNP sites were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/index.html). Assembled contigs and physical map positions of SNPs on the UCSC Golden Path were downloaded from http://genome.ucsc.edu/. SNPs that did not have positions reported in the UCSC data were mapped with BLAT (Kent 2002). Restriction site searches on sequence records were done using BioPerl modules (http://bioperl.org/). Predicted restriction fragment lengths were based on restriction sites immediately upstream and downstream of the SNP site; in cases where there were >30 N's between the SNP site and either restriction site, a fragment prediction was not made for that particular SNP. The deCODE STR genetic map (Kong et al. 2002) is available at http://www.nature.com/ng/journal/v31/n3/suppinfo/ng917_S1.html.
Linkage Analysis
Nonparametric linkage analyses were performed using the software package Merlin 0.9.3 and GENEHUNTER version 2.1 (Kruglyak et al. 1996; Abecasis et al. 2002). Due to the constraint of the software, a trimmed version of the pedigree containing 20 individuals with chronic mucocutaneous candidiasis and/or thyroid disease with 11 genotyped individuals was used for the analyses. A two-point LOD score for every SNP on chromosome 2 was calculated. Multipoint LOD scores were calculated after excluding markers having two-point NPL scores between –.05 and 0.05.
Acknowledgments
We thank Kimberly F. Doheny, Elizabeth W. Pugh, and Paul Boyce for facilitating our linkage study and providing critical comments; Richard Chiles, Fred Christians, Carsten Rosenow, Teresa Webster and David Kulp for helpful discussions, and Sarah A. Tishkoff, Jonathan S. Friedlaender, Theodore G. Schurr, and W. Scott Watkins for contributing valuable DNA samples.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2014904.
Footnotes
[Supplemental material is available online at www.genome.org. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: S.A. Tishkoff, J.S. Friedlaender, T.G. Schurr, and W.S. Watkins.]
References
- Abecasis, G.R., Cherny, S.S., Cookson, W.O., and Cardon, L.R. 2002. Merlin—Rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30**:** 97–101. [DOI] [PubMed] [Google Scholar]
- Altshuler, D., Pollara, V.J., Cowles, C.R., Van Etten, W.J., Baldwin, J., Linton, L., and Lander, E.S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407**:** 513–516. [DOI] [PubMed] [Google Scholar]
- Atkinson, T.P., Schaffer, A.A., Grimbacher, B., Schroeder Jr., H.W., Woellner, C., Zerbe, C.S., and Puck, J.M. 2001. An immune defect causing dominant chronic mucocutaneous candidiasis and thyroid disease maps to chromosome 2p in a single family. Am. J. Hum. Genet. 69**:** 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes, A.J. 1999. The essence of SNPs. Gene 234**:** 177–186. [DOI] [PubMed] [Google Scholar]
- Carrasquillo, M.M., McCallion, A.S., Puffenberger, E.G., Kashuk, C.S., Nouri, N., and Chakravarti, A. 2002. Genome-wide association study and mouse model identify interaction between RET and EDNRB pathways in Hirschsprung disease. Nat. Genet. 32**:** 237–244. [DOI] [PubMed] [Google Scholar]
- Chee, M., Yang, R., Hubbell, E., Berno, A., Huang, X.C., Stern, D., Winkler, J., Lockhart, D.J., Morris, M.S., and Fodor, S.P. 1996. Accessing genetic information with high-density DNA arrays. Science 274**:** 610–614. [DOI] [PubMed] [Google Scholar]
- Cutler, D.J., Zwick, M.E., Carrasquillo, M.M., Yohn, C.T., Tobin, K.P., Kashuk, C., Mathews, D.J., Shah, N.A., Eichler, E.E., Warrington, J.A., et al. 2001. High-throughput variation detection and genotyping using microarrays. Genome Res. 11**:** 1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, S., Wang, E., Hsie, L., Cao, Y., Chen, X., and Gingeras, T.R. 2001. Flexible use of high-density oligonucleotide arrays for single-nucleotide polymorphism discovery and validation. Genome Res. 11**:** 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubovsky, J., Sheffield, V.C., Duyk, G.M., and Weber, J.L. 1995. Sets of short tandem repeat polymorphisms for efficient linkage screening of the human genome. Hum. Mol. Genet. 4**:** 449–452. [DOI] [PubMed] [Google Scholar]
- Fan, J.B., Chen, X., Halushka, M.K., Berno, A., Huang, X., Ryder, T., Lipshutz, R.J., Lockhart, D.J., and Chakravarti, A. 2000. Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 10**:** 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor, S.P., Read, J.L., Pirrung, M.C., Stryer, L., Lu, A.T., and Solas, D. 1991. Light-directed, spatially addressable parallel chemical synthesis. Science 251**:** 767–773. [DOI] [PubMed] [Google Scholar]
- Grant, S.F., Steinlicht, S., Nentwich, U., Kern, R., Burwinkel, B., and Tolle, R. 2002. SNP genotyping on a genome-wide amplified DOP-PCR template. Nucleic Acids Res. 30**:** e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, J.G., Eis, P.S., Law, S.M., Reynaldo, L.P., Prudent, J.R., Marshall, D.J., Allawi, H.T., Mast, A.L., Dahlberg, J.E., Kwiatkowski, R.W., et al. 2000. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc. Natl. Acad. Sci. 97**:** 8272–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, B., Charest, A., Dowd, J.F., Blumenstiel, J.P., Yeh Rf, R.F., Osman, A., Housman, D.E., and Landers, J.E. 2002. Genome complexity reduction for SNP genotyping analysis. Proc. Natl. Acad. Sci. 99**:** 2942–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde, L.B. 2000. Linkage disequilibrium and the search for complex disease genes. Genome Res. 10**:** 1435–1444. [DOI] [PubMed] [Google Scholar]
- Kennedy, G.C., Matsuzaki, H., Dong, S., Liu, W.M., Huang, J., Liu, G., Su, X., Cao, M., Chen, W., Zhang, J., et al. 2003. Large-scale genotyping of complex DNA. Nat. Biotechnol. 21**:** 1233–1237. [DOI] [PubMed] [Google Scholar]
- Kent, W.J. 2002. BLAT—The BLAST-like alignment tool. Genome Res. 12**:** 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., Gudbjartsson, D.F., Sainz, J., Jonsdottir, G.M., Gudjonsson, S.A., Richardsson, B., Sigurdardottir, S., Barnard, J., Hallbeck, B., Masson, G., et al. 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31**:** 241–247. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L. and Nickerson, D.A. 2001. Variation is the spice of life. Nat. Genet. 27**:** 234–236. [DOI] [PubMed] [Google Scholar]
- Kruglyak, L., Daly, M.J., Reeve-Daly, M.P., and Lander, E.S. 1996. Parametric and nonparametric linkage analysis: A unified multipoint approach. Am. J. Hum. Genet. 58**:** 1347–1363. [PMC free article] [PubMed] [Google Scholar]
- Kwok, P.Y. 2001. Methods for genotyping single nucleotide polymorphisms. Annu. Rev. Genomics Hum. Genet. 2**:** 235–258. [DOI] [PubMed] [Google Scholar]
- Liu, W.-m., Di, X., Yang, G., Matsuzaki, H., Huang, J., Mei, R., Ryder, T.B., Webster, T.A., Dong, S., Liu, G., et al. 2003. Algorithms for large scale genotyping microarrays. Bioinformatics 19**:** 2397–2403. [DOI] [PubMed] [Google Scholar]
- Miller, R.D. and Kwok, P.Y. 2001. The birth and death of human single-nucleotide polymorphisms: New experimental evidence and implications for human history and medicine. Hum. Mol. Genet. 10**:** 2195–2198. [DOI] [PubMed] [Google Scholar]
- Mullikin, J.C., Hunt, S.E., Cole, C.G., Mortimore, B.J., Rice, C.M., Burton, J., Matthews, L.H., Pavitt, R., Plumb, R.W., Sims, S.K., et al. 2000. An SNP map of human chromosome 22. Nature 407**:** 516–520. [DOI] [PubMed] [Google Scholar]
- Nikiforov, T.T., Rendle, R.B., Goelet, P., Rogers, Y.H., Kotewicz, M.L., Anderson, S., Trainor, G.L., and Knapp, M.R. 1994. Genetic Bit Analysis: A solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 22**:** 4167–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell, J.R. and Weeks, D.E. 1998. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63**:** 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson, G.A. and Stein, L.D. 2003. The SNP Consortium website: Past, present and future. Nucleic Acids Res. 31**:** 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihashi, Z. and Dracopoli, N.C. 2002. Progress in high throughput SNP genotyping methods. Pharmacogenomics J. 2**:** 103–110. [DOI] [PubMed] [Google Scholar]
- Wang, D.G., Fan, J.B., Siao, C.J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E., Spencer, J., et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280**:** 1077–1082. [DOI] [PubMed] [Google Scholar]
- Xiong, M. and Jin, L. 1999. Comparison of the power and accuracy of biallelic and microsatellite markers in population-based gene-mapping methods. Am. J. Hum. Genet. 64**:** 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://snp.cshl.org/; SNP Consortium (TSC) home page.
- http://www.ncbi.nlm.nih.gov/SNP/; dbSNP (NCBI) home page.
- http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker documentation.
- http://www.ncbi.nlm.nih.gov/Genbank/index.html; GenBank (NCBI) home page.
- http://genome.ucsc.edu/; UCSC Genome Bioinformatics home page.
- http://bioperl.org/; BioPerl home page.
- http://www.nature.com/ng/journal/v31/n3/suppinfo/ng917_S1.html; Web supplement to Kong et al. (2002).