Influence of Salmonella enterica Serovar Enteritidis Infection on the Development of the Cecum Microbiota in Newly Hatched Chicks (original) (raw)
Abstract
Terminal restriction fragment length polymorphism and quantitative PCR showed that the cecal microbiota of chicks up to the age of 21 days was dominated by representatives of the orders Enterobacteriales, Clostridiales, and Lactobacillales. Salmonella enterica serovar Enteritidis infection caused the greatest changes in the gut microbiota when 1-day-old chicks were infected, compared with the infection of 4- and 16-day-old chicks.
TEXT
Unlike all other farm animals, chicks are hatched in a clean hatchery environment without any contact with adult chickens and colonization of the intestine is therefore dependent only on environmental sources. If a pathogen appears in the environment, the sterile intestinal tract of the newly hatched chick represents an empty ecological niche enabling such a pathogen essentially unrestricted multiplication.
Infection of chicks with Salmonella enterica is manifested as a transient inflammation of the intestinal tract, especially the cecum (1, 2). The induction of inflammation may be one of S. enterica's evolutionary adaptations that provide S. enterica a growth advantage over the resident microbiota (3–5). In this study, we were therefore interested in the development of the cecal microbiota of newly hatched chicks and also the effect of S. enterica serovar Enteritidis (S. Enteritidis) infection on the composition of the gut microbiota.
Male ISA Brown chicks were used in all experiments. Three chicks each were sacrificed on days 1, 2, 3, 4, 7, 11, 14, 19, and 26 of life. In addition, 1-, 4-, and 16-day-old chicks (six birds in each group) were infected orally with 1 × 107 CFU of S. Enteritidis 147 and sacrificed at 3 days (three birds) and 10 days (the remaining three birds) postinfection. This experiment was repeated on two independent occasions. During postmortem analysis, the cecal contents were removed and homogenized and DNA was extracted with the QIAamp DNA stool minikit (Qiagen). The purified DNA was used as a template in a PCR with fluorescently labeled primers specific for the conserved regions of bacterial 16S rRNA genes (27F, 6-carboxyfluorescein–5′ AGA GTT TGA TCM TGG CTC AG 3′; 1492R, 5′ GGY TAC CTT GTT ACG ACT T 3′). Following PCR, the amplification products were digested with HaeIII and the resulting fragments were separated by capillary electrophoresis with an ABI 310 Genetic Analyzer (Applied Biosystems). The data were processed as described previously (6).
In addition, a set of seven primer pairs (Table 1) used to detect representatives of higher taxonomic levels were designed from the variable regions of 16S rRNA genes by using PRIMROSE software (http://www.cardiff.ac.uk/biosi/research/biosoft/). Real-time PCR was carried out by using the QuantiTect SYBR green PCR kit (Qiagen) and a LightCycler LC480 thermocycler (Roche). After PCR, the cycle threshold (CT) values were normalized to an average CT value of amplifications (Δ_CT_) performed with 2 different universal primer pairs for the domain Bacteria (7, 8). The relative amount of each taxon was finally calculated as 2−Δ_CT_.
Table 1.
Taxon-specific primers used in this study
| Primer | Sequence (5′–3′) | Amplicon size (bp) | Target organisms |
|---|---|---|---|
| 16S_Bacteroid-F | CGC ACA AGC GGA GGA AC | 155 | Order Bacteroidales |
| 16S_Bacteroid-R | CGA CAC CTC ACG GCA CG | ||
| 16S_Bifido-F | GGT GTG AAA GTC CAT CG | 85 | Order Bifidobacteriales |
| 16S_Bifido-R | ACC GGG AAT TCC AGT CT | ||
| 16S_Clostrid-F | GCG TTA TCC GGA TTT AC | 286 | Order Clostridiales |
| 16S_Clostrid-R | ACA CCT AGT ATT CAT CG | ||
| 16S_Enterobac-F | STG AGA CAG GTG CTG CA | 85 | Order Enterobacteriales |
| 16S_Enterobac-R | AAA GGA TAA GGG TTG CG | ||
| 16S_Fusobac-F | CGG CNA CAA GGG RAC TG | 136 | Phylum Fusobacteria |
| 16S_Fusobac-R | CTG AAA GMA CTT TAC AW | ||
| 16S_Lactobac-F | CTT GAG TGC AGA AGA GG | 74 | Order Lactobacillales |
| 16S_Lactobac-R | CAC TGG TGT TCT TCC AT | ||
| 16S_Verruco-F | CAG TAT GGC CCT TAY GC | 103 | Order Verrucomicrobiales |
| 16S_Verruco-R | GAA CTG RGC CCA GTT TT |
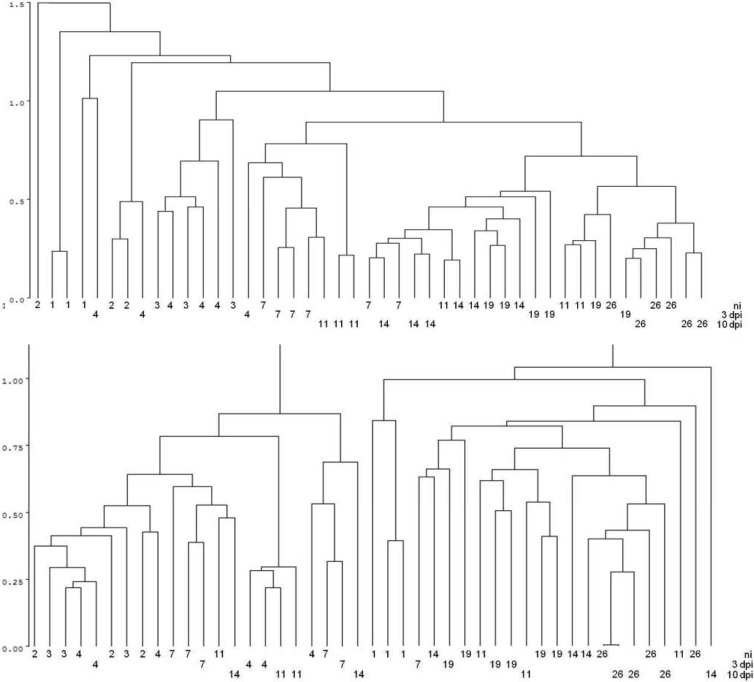
In healthy chicks, the complexity of the microbiota, expressed as the number of terminal restriction fragments (TRF), increased from day 1 until day 26 of the chick's life, with the most dynamic development within the first 4 days of life. Cluster analysis of the TRF profiles revealed separate clusters of samples from 1-, 2-, 3- and 4-day-old chicks. On the other hand, samples from chicks 1 to 3 weeks old did not form a well-defined cluster (Fig. 1). These findings could be explained by yolk sac absorption, which is completed between days 4 and 7 of the chick's life (9, 10) and makes the microbiota of the young bird different from that which develops later in life (11–13). Cloning and sequencing of 16S rRNA PCR products obtained by amplification of cecal DNA from 1- and 14-day-old chicks showed that the microbiota of chicks commonly included members of the families Enterobacteriaceae, Lachnospiraceae, Clostridiaceae, Eubacteriaceae, Peptostreptococcaceae, and Pseudomonadaceae.
Fig 1.
Cluster analysis of TRF data originating from cecal samples from individual chicks. Each number indicates the age of a particular chick. Line ni, noninfected chicks of the ages indicated; line 3 dpi, ages of chicks at 3 days postinfection with S. Enteritidis; line 10 dpi, ages of chicks at 10 days postinfection with S. Enteritidis. Upper panel, results of the first experiment; lower panel, results of the repeat experiment.
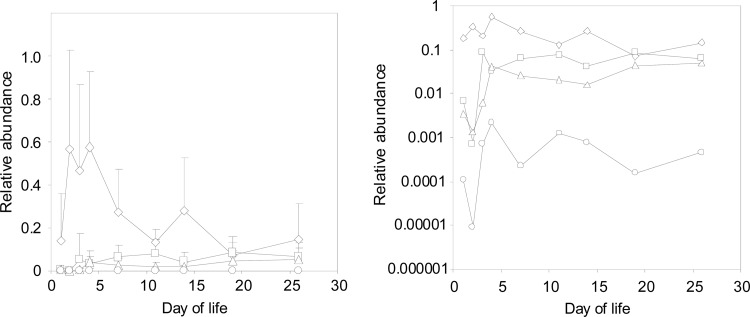
Real-time PCR data yielded negative results for members of the phylum Fusobacteria and the orders Verrucomicrobiales and Bacteroidales. The cecal microbiota of chicks up to 1 week old was dominated by Enterobacteriales. Clostridiales and Lactobacillales were present at a prevalence 10 times lower than that of Enterobacteriales, and Bifidobacteriales members were the least predominant component of the cecal microbiota, similar to previous reports (12–14). With increasing chick age, the presence of Enterobacteriales bacteria decreased while that of Clostridiales and Lactobacillales gradually increased so that nearly the same prevalence was detected in the ceca of 3-week-old chicks (Fig. 2).
Fig 2.
Chick cecum colonization as determined by quantitative PCR analysis with taxon-specific primers. Diamonds, Enterobacteriales; squares, Clostridiales; triangles, Lactobacillales; circles, Bifidobacteriales. The two panels were created by using the same data. The only difference is the y axis scaling, which is linear in the left panel and logarithmic in the right panel. Data in the left panel are the averages ± standard deviations combined from both experiments. Data in the right panel are only the average values combined from both experiments.
Infection with S. Enteritidis delayed microbiota development mainly when 1- or 4-day-old chicks were infected. The terminal restriction fragment length polymorphism profiles of the cecal contents of 4-, 7-, 11-, and 14-day-old chicks, i.e., chicks infected with S. Enteritidis on day 1 and day 4 of life and sacrificed 3 and 10 days later, clustered with those of younger, noninfected chicks. Infection of 16-day-old chicks did not affect the clustering of such cecal samples (Fig. 1).
The number of Enterobacteriales bacteria in the ceca of chicks infected with S. Enteritidis at 1 day of age and sacrificed 3 and 10 days later was greater than that in noninfected controls. This increase corresponded to a decrease in the numbers of Clostridiales, Lactobacillales, and Bifidobacteriales at 3 days postinfection and a decrease in the numbers of Lactobacillales and Bifidobacteriales bacteria at 10 days postinfection. None of the taxa differed significantly when 4-day-old chicks were infected with S. Enteritidis; however, the same general trend as in the 1-day-old birds was observed. S. Enteritidis infection of 16-day-old chicks was associated with an increase in the number of Enterobacteriales bacteria at 3 days postinfection and a decrease in the numbers of Clostridiales bacteria at both 3 and 10 days postinfection, Lactobacillales bacteria at 3 days postinfection, and Bifidobacteriales bacteria at 10 days postinfection; however, these differences did not reach statistical significance.
In this study, we have shown that despite the absence of any clinical signs of infection, infection of chicks with S. Enteritidis caused changes in the cecal microbiota. However, the results are best described as a trend because the differences were repeatable but minor. One of the possible explanations for the trend is the nature of the samples that were analyzed. Inflammation induced by S. Enteritidis in chicks is restricted to the epithelial surface and does not result in electrolyte efflux, tissue damage, and diarrhea as in humans. This may mean that the luminal microbiota present in the whole cecal contents, which were collected and analyzed, could be only marginally affected by S. Enteritidis infection, while more significant changes in the microbiota composition can be observed at the epithelium and gut surface, a hypothesis which we are currently testing.
ACKNOWLEDGMENTS
This work was supported by EMIDA project HealthyGut and projects AdmireVet CZ.1.05/2.1.00/01.0006 and ED0006/01/01 from the Ministry of Education of the Czech Republic and project 0002716202 from the Ministry of Agriculture of the Czech Republic.
We thank Peter Eggenhuizen for his English language corrections.
Footnotes
Published ahead of print 9 November 2012
REFERENCES
- 1.Berndt A, Wilhelm A, Jugert C, Pieper J, Sachse K, Methner U. 2007. Chicken cecum immune response to Salmonella enterica serovars of different levels of invasiveness. Infect. Immun. 75:5993–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crhanova M, Hradecka H, Faldynova M, Matulova M, Havlickova H, Sisak F, Rychlik I. 2011. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar Enteritidis infection. Infect. Immun. 79:2755–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, Mak TW, Tsolis RM, Bevins CL, Solnick JV, Dandekar S, Baumler AJ. 2009. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5:476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, Dougan G, von Mering C, Hardt WD. 2007. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 5:2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, Roth JR, Baumler AJ. 2010. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467:426–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938 [DOI] [PubMed] [Google Scholar]
- 7.Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, Arai H, Tanimoto I, Nishimura F, Takashiba S. 2003. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39:81–86 [DOI] [PubMed] [Google Scholar]
- 8.Tseng CP, Cheng JC, Tseng CC, Wang C, Chen YL, Chiu DT, Liao HC, Chang SS. 2003. Broad-range ribosomal RNA real-time PCR after removal of DNA from reagents: melting profiles for clinically important bacteria. Clin. Chem. 49:306–309 [DOI] [PubMed] [Google Scholar]
- 9.Moran ET., Jr 2007. Nutrition of the developing embryo and hatchling. Poult. Sci. 86:1043–1049 [DOI] [PubMed] [Google Scholar]
- 10.Noy Y, Sklan D. 1998. Yolk utilisation in the newly hatched poult. Br. Poult. Sci. 39:446–451 [DOI] [PubMed] [Google Scholar]
- 11.Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD. 2003. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69:6816–6824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mead GC, Adams BW. 1975. Some observations on the caecal microflora of the chick during the first two weeks of life. Br. Poult. Sci. 16:169–176 [DOI] [PubMed] [Google Scholar]
- 13.Wise MG, Siragusa GR. 2007. Quantitative analysis of the intestinal bacterial community in one- to three-week-old commercially reared broiler chickens fed conventional or antibiotic-free vegetable-based diets. J. Appl. Microbiol. 102:1138–1149 [DOI] [PubMed] [Google Scholar]
- 14.Zhu XY, Zhong T, Pandya Y, Joerger RD. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl. Environ. Microbiol. 68:124–137 [DOI] [PMC free article] [PubMed] [Google Scholar]