Multiple sclerosis: Brain-infiltrating CD8+ T cells persist as clonal expansions in the cerebrospinal fluid and blood (original) (raw)
Abstract
We surveyed the T cell receptor repertoire in three separate compartments (brain, cerebrospinal fluid, and blood) of two multiple sclerosis patients who initially had diagnostic brain biopsies to clarify their unusual clinical presentation but were subsequently confirmed to have typical multiple sclerosis. One of the brain biopsy specimens had been previously investigated by microdissection and single-cell PCR to determine the clonal composition of brain-infiltrating T cells at the single-cell level. Using complementarity-determining region 3 spectratyping, we identified several identical, expanded CD8+ (but not CD4+) T cell clones in all three compartments. Some of the expanded CD8+ T cells also occurred in sorted CD38+ blood cells, suggesting that they were activated. Strikingly, some of the brain-infiltrating CD8+ T cell clones persisted for >5 years in the cerebrospinal fluid and/or blood and may thus contribute to the progression of the disease.
Classical experiments in an animal model of multiple sclerosis (MS), experimental autoimmune encephalomyelitis (EAE), suggested that myelin antigen-specific T cells play a crucial role because EAE could be transferred with CD4+ T cell lines specific for myelin basic protein (1). Obviously, such transfer experiments are impossible in human MS. Recently the possible contribution of CD8+ T cells has received increased attention, especially as effectors of the pathological immune reactions that damage the CNS of EAE animals and MS patients (2–6). Using molecular techniques, including single-cell PCR analysis of microdissected T cells, clonally expanded CD8+ (cytotoxic) T cells could be demonstrated in MS brain lesions at both perivascular and intraparenchymal sites (7). Another recent study found expanded CD8+ T cells in the cerebrospinal fluid (CSF) of MS patients (8). However, it is unclear whether the expanded CD8+ T cell populations observed at different tissue sites of MS patients are related and also relevant to the disease.
We had the opportunity to survey the T cell receptor (TCR) repertoire in three distinct tissue compartments (brain, CSF, and blood) of two MS patients who underwent a brain biopsy for diagnostic reasons. In one of these patients, the TCR repertoire of the putatively pathogenic T cell infiltrate had been previously studied by microdissection of individual brain-infiltrating CD8+ T cells and subsequently by single-cell PCR analysis of their rearranged TCR β-chain gene sequences (7). In the present study we applied complementarity-determining region 3 (CDR3) spectratyping, a screening technique for TCR repertoire analysis, to compare the TCR repertoires in the brain, CSF, and blood of these patients. Our findings demonstrate that CD8+ T cell clones that infiltrate the CNS may surprisingly persist in the CSF and blood for many years as clonal expansions. To our knowledge, these are among the first observations that directly relate the TCR repertoire in MS lesions with that in peripheral immune compartments of MS patients. The findings are consistent with the hypothesis that expanded clones of CD8+ T cells are involved in the pathogenesis of MS.
Materials and Methods
Patients and Clinical Samples. Patient A (F.E., male) presented in 1996 with recurrent episodes of left-sided hemianopia at the age of 49 years. Cranial MRI suggested the presence of a malignant glioma in the right temporooccipital lobe. Two weeks after the initial symptoms the lesion was completely removed. Part of the brain tissue was frozen in liquid nitrogen immediately after resection. Histopathological analysis showed a large area of inflammatory demyelination compatible with an MS lesion. CSF analysis revealed oligoclonal Ig bands. Within the subsequent 5 years, the patient experienced three relapses, which also supported the diagnosis of MS. Follow-up MRI confirmed the occurrence of new demyelinating lesions. The patient has been treated with IFN-β1a since the third relapse (1997) until submission of this paper. No further relapses have occurred during treatment. However, during the past years the clinical course has been slowly progressive. Patient A gave his informed consent to the use of brain material, CSF, and blood for research purposes. The blood and CSF samples analyzed in this study were taken 62 months after surgery. Another blood sample was analyzed in December 2003, i.e., 7 years after the brain biopsy. Patient A is seemingly homozygous for the haplotype HLA-A*0101, HLA-B*0801, HLA-Cw*0701, HLA-DRB1*0301, HLA-DRB3*0101, HLA-DQA1*05, HLA-DQB1*0201, because only one allele was detected for each of the loci analyzed.
Patient B (P.I., female) presented at the age of 32 with right-sided hemianopia in April 2001. Four weeks later she had a single generalized epileptic seizure, severe left-frontal headache, and nausea. MRI revealed four small lesions in the subcortical white matter and a large (4 × 2 × 2 cm) T2w-hyperintense region in the left occipitotemporal lobe, which suggested a low-grade glioma. Subsequently, the patient underwent diagnostic stereotactic brain biopsy. Histology showed inflammatory demyelination compatible with MS, and CSF analysis revealed oligoclonal Ig bands. Within the subsequent 11 months she suffered four additional relapses, which supported the diagnosis of MS. Blood and CSF samples analyzed in this study were taken in November 2001, 6 months after the brain biopsy. Patient B was typed as HLA-A*0301, -; B*0702, 4002; Cw*0202, 0702; DRB1*1101, 1501; DRB3*0202, -; DRB5*0101, -; DQB1*0301, 0602.
Isolation of Peripheral Blood Mononuclear Cells (PBMC) and T Cell Subsets. PBMC were isolated with standard-density centrifugation. CD8+, CD4+, and CD38+ T cells were isolated from the PBMC with antibody-coated magnetic beads according to the manufacturer's recommendations (Dynabeads, Dynal, Hamburg, Germany).
Isolation of RNA and cDNA Synthesis. Total RNA was extracted from brain, CSF, and blood cells with TRIzol-LS reagent (GIBCO/BRL). For cDNA synthesis, oligo(dT) and reverse transcriptase (SuperScript II, GIBCO/BRL) were used according to the recommendations of the suppliers.
CDR3 Spectratyping. CDR3 spectratype (“immunoscope”) analysis was performed essentially as described (9). In brief, cDNA was subjected to TCR-Vβ (TCR-BV) gene family-specific PCR in 26 separate reactions, each containing 1 of 26 BV family-specific and 1 Cβ (BC)-specific primer (10). With 1 of 13 fluorescence-labeled Jβ (BJ)-specific oligonucleotides (11), single-stranded runoff products were generated from each BV-specific PCR product. Transcripts were resolved on a sequencing gel, and fluorescence intensities were measured with an automated DNA sequencer (ABI377, Applied Biosystems). Clonal expansions appear as distinct peaks of a given CDR3 length above the Gaussian-like background of polyclonal cells. Expanded candidate BV–BJ subpopulations were subamplified from the initial BV–BC amplification product by using BV- and BJ-specific primers and were directly sequenced (9). If the population of TCR-β-chain mRNA molecules amplified by the BV primer combination was dominated by a single clonal transcript, the CDR3 sequence could be read by direct sequencing. For the detection of selected clones (as indicated) on an oligoclonal background, clone-specific BJ primers, complementary to part of the J and part of the N-D-N region, were used. The sequence identity of the amplification product was determined by direct sequencing as above. The sequences of the clone-specific BJ primers were as follows: BV1–BJ2.3, 5′-AAAATACTGCGTATCCTTCCT-3′; BV9–BJ2.7, 5′-CCCGAAGTACTGCTCGCCGAC-3′; BV22–BJ2.1, 5′-GAACTGCTCATTGTGTTCTCC-3′.
Immunohistochemistry, Microdissection, and Single-Cell PCR Analysis of Brain Tissue from Patient A. T cell infiltrates in the brain tissue of patient A had been previously analyzed by microdissection and single-cell PCR (7). In that study, TCR-BV sequences were amplified from single TCR-encoding, rearranged DNA molecules of brain-infiltrating T cells from perivascular or intraparenchymal locations (7). CD8+ T cells outnumbered CD4+ T cells by 5- to 6-fold in the parenchyma and by ≈3-fold at perivascular locations.
CD4+ and CD8+ T cells were identified by staining with monoclonal antibodies (7). The cells were then mobilized from the surrounding tissue and transferred into PCR tubes by means of hydraulic micromanipulators. TCR-BV gene rearrangements were amplified by seminested PCR. Briefly, a first round of amplification was performed by using a mix of 24 BV gene family-specific and 8 external BJ-specific primers. In a second round of PCR, two to three of the same BV primers were used together with a mix of seven internal BJ-specific primers in separate reactions (7).
Of 24 rearrangements obtained from single parenchymal CD8+ cells of patient A, 17 (71%) originated from 9 different clones; only 7 sequences were unique (7). Forty-six of the 52 parenchymal CD4+ T cells could not be assigned to any clonal expansion, whereas six (12%) belonged to three clones. Of the 46 sequences derived from perivascular cells, 26% could be assigned to clonal expansions. Except for clone BV13.2-2.2, all clones could be assigned to the CD8 or CD4 subset, because the clonal V region sequence was obtained at least once from a single parenchymal CD8+ or CD4+ cell. Only potentially functional rearrangements were used for the present study.
Results
The brain tissue from patient A (the lesion had been surgically removed in 1996) was first analyzed by spectratype analysis of the TCR-BV CDR3 regions. Several peaks indicating clonally expanded T cell populations were observed. The peaks were compared to the TCR-BV sequences of expanded T cell clones previously amplified by single-cell PCR from microdissected brain-infiltrating CD8+ T cells (ref. 7; GenBank accession nos. AJ405646–AJ405745) and to clonal expansions detected by CDR3 spectratyping in CSF cells and PBMC subpopulations obtained 5 years after the brain biopsy. Corresponding peaks that occurred in two or more different samples were sequenced. Altogether, 13 TCR sequences were identified in this way (Tables 1 and 2).
Table 1. TCR-BV sequences identified in brain cells, CSF cells, and sorted peripheral blood cells (PBL) from Patient A by single-cell PCR and CDR3 spectratyping.
| Single-cell PCR of brain cells, 1996* | CDR3 spectratyping | ||||||
|---|---|---|---|---|---|---|---|
| Frequency of parenchymal CD8+ T cells | Frequency of perivascular lymphocytes | TCR-BV CDR3 sequence | Brain 1996 | CSF 2001 | CD4+ PBL 2001 | CD8+ PBL 2001 | CD38+ PBL 2001 |
| 1/24 | 1/46 | BV1 - C A S-T P E R D P S - N E Q - BJ2.1 | + | - | - | - | - |
| 1/24 | 2/46 | BV1 - C A S S - I S G R K - D T Q - BJ2.3 | + | (+)# | - | + | + |
| 4/24 | 2/46 | BV4 - C S - V W E V - S G A-BJ2.6 | + | + | - | + | - |
| 2/24 | BV13.1-C A S S - L G A - D T Q - BJ2.3 | + | - | - | - | - | |
| 2/24 | 1/46 | BV13.2 - C A S - R A L V A T - Y N E - BJ2.1 | + | - | - | - | - |
| 2/46 | BV13.2 - CASS-YP - GEL - BJ2.2 | + | - | - | - | - | |
| 1/24 | 3/46 | BV13.3 - C A S S - P G D R A Q - Y N E - BJ2.1 | + | + | - | (+) | - |
| 1/24 | 1/46 | BV14-C A S S - P L W E G G I G - N T E - BJ1.1 | + | + | - | + | - |
| 3/24 | BV22 - C A S S - E G A G E H - N E Q - BJ2.1 | + | (+)# | - | - | - |
Table 2. TCR-BV sequences identified in brain tissue, CSF cells, and sorted peripheral blood cells (PBL) from Patient A by CDR3 spectratyping.
| TCR-BV CDR3 sequence | Brain 1996 | CSF 2001 | CD4+ PBL 2001 | CD8+ PBL 2001 | CD38+ PBL 2001 |
|---|---|---|---|---|---|
| BV9 - C A S S - P W E E G - S G N - BJ1.3 | + | + | - | - | - |
| BV9 - C A S S - Q I R G V G - E Q Y - BJ2.7 | + | (+)# | - | - | + |
| BV16 - C A S - G G T S G K - D T Q - BJ2.3 | + | (+) | - | - | - |
| BV17 - C A S S - W A I S D R - Q P Q - BJ1.5 | + | (+) | - | - | - |
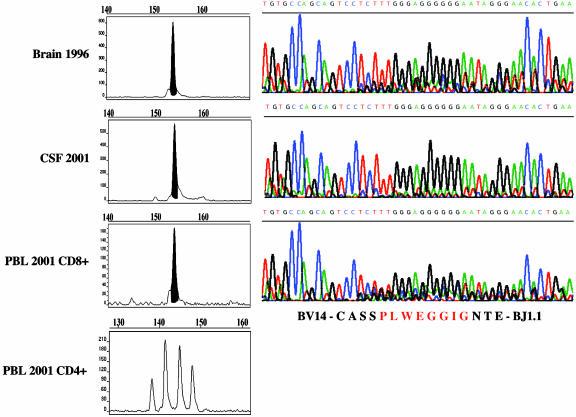
Nine of the 13 sequences derived by CDR3 spectratyping from brain tissue corresponded to one of the TCR sequence(s) previously identified by single-cell PCR (Table 1). Five of these nine sequences were also detected as clonal peaks in the CSF or blood of the same patient (Table 1). Specifically, the sequence BV1-CASS-ISGRK-DTQ-BJ2.3 was found in CD8+ and CD38+ (activated) blood cells and in the CSF; the sequence BV4-CS-VWEV-SGA-BJ2.6 was found in CD8+ blood cells and in the CSF; the sequence BV13.3-CASS-PGDRAQ-YNE-BJ2.1 was found in CD8+ blood cells and CSF; the sequence BV14-CASS-PLWEGGIG-NTE-BJ1.1 was found in CD8+ cells and CSF; and the sequence BV22-CASS-EGAGEH-NEQ-BJ2.1 was found in the CSF (Table 1). Fig. 1 shows an example of CDR3 spectratype peaks in different samples and the corresponding TCR-BV CDR3 sequence (BV14-CASS-PLWEGGIG-NTE-BJ1.1). This sequence was undetectable in CD4+ T cells (Fig. 1).
Fig. 1.
Example of an expanded CD8+ T cell clone that persists in brain, CSF, and blood of an MS patient (patient A). (Left) TCR CDR3 spectragrams of the BV14–BJ1.1 PCR runoff (see Materials and Methods for details). Polyclonal CDR3 spectragrams reveal a Gaussian-like length distribution. As shown in the bottom spectragram, this was the case for CD4+ blood T cells. In contrast, a clonal peak was observed in the brain biopsy specimen of 1996 (top spectragram) and in CSF and CD8+ blood T cells sampled 5 years later (middle spectragrams). The clonal peaks could be directly sequenced on an automated sequencer. (Right) Sequencing data. The nucleotide sequences are encoded in color as indicated in small letters above each curve. The amino acid sequence is shown at the bottom.
Four of the TCR sequences that had been identified in the 1996 brain biopsy by CDR3 spectratyping did not correspond to any of the single-cell PCR-derived sequences (ref. 7 and Table 2). However, they could be traced in CSF cells obtained 5 years after the brain biopsy. One TCR sequence (BV9-CASS-QIRGVG-EQY-BJ2.7) also appeared in activated CD38+ blood T cells (Table 2).
Altogether, nine expanded CD8+ T cell clones from the MS brain lesion were tracked in the CSF, blood, or both (Tables 1 and 2). Five of these T cell clones were found in the CSF and blood, and four were found only in the CSF (Tables 1 and 2). None of the clones was detected in the CD4+ PBMC population.
Additional blood samples from patient A were analyzed in December 2003, i.e., 7 years after the initial brain biopsy. CDR3 spectratyping of purified CD8+ T cells revealed a conspicuous TCR-BV1-BJ2.3 peak. The cDNA sequence of this peak was identical to the cDNA sequence coding for the BV1-CASS-ISGRK-DTQ-BJ2.3 sequence, which had been previously identified in CD8+ blood and CSF T cells in 2001, and in the brain biopsy specimen obtained in 1996.
Similar results were obtained for patient B (Table 3). This patient had had a stereotactic brain biopsy in May 2001. CDR3 spectratyping analysis of brain tissue revealed several clonal peaks that could be traced in CSF as well as in CD8+ and CD38+ blood cells obtained 6 months after the brain biopsy (Table 3). In this patient, one clonal peak appeared in the CD4+ fraction of blood cells and in the CSF but not in the brain (Table 3).
Table 3. TCR-BV sequences identified in brain tissue, CSF cells, and sorted peripheral blood cells (PBL) from patient B by CDR3 spectratyping.
| TCR-BV CDR3 sequence | Brain 05/2001 | CSF 11/2001 | CD4+ PBL 12/2001 | CD8+ PBL 12/2001 | CD38+ PBL 12/2001 |
|---|---|---|---|---|---|
| BV16 - C A S S - R L T D S T - T D T - BJ2.3 | (+) | - | - | + | - |
| BV19 - C A S S - H P G T G S N - E Q Y - BJ2.7 | + | + | - | (+) | - |
| BV20 - C A W S - A P D - N T E - BJ1.1 | (+) | - | - | (+) | - |
| BV23 - C A S S - P G R A G - S P L - BJ1.6 | - | + | + | - | - |
| BV24 - C A T S - R L Q G G - T G E - BJ2.2 | + | + | - | - | + |
Discussion
Our present results shed new light on the possible pathogenic importance of CD8+ T cells in MS. First, several expanded clones of CD8+ T cells were present in MS brain lesions (as demonstrated by single-cell PCR and/or CDR3 spectratyping) and also in the CSF or blood (as demonstrated by CDR3 spectratyping). Second, the shared expansions were observed only in the CD8+ but not in the CD4+ T cell population. The only exception was an expanded CD4+ T cell clone that was present in both the CSF and the CD4+ blood cells. However, this CD4+ T cell clone was not detectably expanded in brain tissue (Table 3). Third, some of the expanded CD8+ T cells identified in brain tissue were also detected in sorted CD38+ blood cells, suggesting that they were activated at the time of sampling. Finally, several brain-infiltrating CD8+ T cell clones had persisted for many years in the blood or CSF, suggesting a strong, persisting memory response and/or ongoing exposure to an unknown antigen. All this is consistent with an important role of CD8+ T cells in MS pathogenesis.
The two MS patients from whom we obtained brain biopsies, blood, and CSF samples had had an unusual initial clinical presentation, necessitating a diagnostic brain biopsy. However, in both cases, the histological findings plus the subsequent clinical course and laboratory findings proved to be typical of MS and met the current diagnostic criteria (12). Obviously, our findings cannot be generalized to all MS patients, especially in view of the growing evidence for the pathological heterogeneity of MS (13). Furthermore, imaging studies suggest that the mechanisms of lesion genesis might be diverse (14). As regards the possible role of CD8+ T cells, it is noteworthy that a recently published study of 36 MS patients showed an overrepresentation of CD8+ T cells expressing particular TCR-BV chains in the CSF compared with blood cells in the majority of patients (8). Although TCR sequences were obtained in only a small number of these patients, the data indicate that CD8+ T cell expansions occur in the CSF of most MS patients (8).
CD8+ T cells are the predominating type of T cell in MS lesions (5, 15). It has also been known for some time that the T cell infiltrates of MS lesions express a limited repertoire of antigen receptors (TCR) (16). However, only very recently has it become technically possible to link the immunocytochemical localization of CD8 with PCR analysis of the TCR expressed in individual, morphologically characterized CD8+ T cells (7, 17, 18). In MS patient A, the TCR repertoire had been previously studied with unprecedented precision by microdissection and single-cell PCR analysis of brain-infiltrating CD8+ T cells (7). When we compared the previously identified TCR sequences with the TCR sequences obtained 5 years later by CDR3 spectratyping analysis of CSF and blood cells, five of nine previously observed sequences recurred (Table 1). One of these T cell clones, which had been present in brain, CSF, and activated CD8+ blood T cell preparations, was again detectable as a conspicuous clonal expansion in CD8+ blood T cells 7 years after the biopsy.
It appears that the TCR repertoire expressed in the brain is similar to but not identical with the repertoire in the peripheral immune compartments of this patient. The differences could have statistical reasons (sampling fluctuations) or could reflect systematic changes of the TCR repertoire over time (e.g., because of epitope spreading). In addition, the CSF and blood might contain clonally expanded bystander cells (see below). This must be taken into consideration when CSF or blood samples are used for immunological studies in MS patients.
It is noteworthy that patient A had been treated for many years with IFN-β when the follow-up CSF and blood samples were obtained. So far he has had no further clinical exacerbations during the treatment period. Our observation that more than half of the initially identified brain-infiltrating, expanded CD8+ T cell clones persisted during many years of treatment would be consistent with the assumed mechanism of action of IFN-β, which is thought to act as an antigen-nonspecific immunomodulatory agent (19). It often dampens, but does not completely halt, disease activity and would not be expected to eradicate (auto)-antigen-specific T cells.
Our observation that the TCR repertoire of CD8+ T cells in the CSF is similar to the TCR repertoire of brain-infiltrating T cells may seem surprising at first glance. It has been well established that leukocyte entry into the CNS is restricted because of the blood brain barrier and that the trafficking of immunocompetent cells into the CNS is tightly regulated (20, 21). At least three distinct routes of leukocyte entry into the CNS have been defined: (i) from blood to CSF across the choroid plexus, (ii) from blood to subarachnoid space, and (iii) from blood to parenchymal perivascular space (20). Migration along these routes is guided by special adhesion and chemokine systems. This helps to explain why the distribution of leukocyte subsets differs markedly among the brain parenchyma, meninges, CSF, and blood. For example, the vast majority of leukocytes present in the CSF of normal subjects and MS patients belong to the CD4+ subset of T cells (22), making it all the more remarkable that we observed T cell expansions shared among brain, CSF, and blood mainly in the CD8+ compartment.
The slight pleocytosis that is often observed in the CSF of MS patients could be related to perivascular, meningeal, and/or parenchymal inflammation (23). The data obtained by microdissection and single-cell PCR of individual brain-infiltrating T cells clearly establish that some of the expanded CD8+ T cell clones occur in both brain lesions and CSF. Even T cell clones that deeply infiltrated the brain parenchyma could be found in the CSF (Table 1). Thus, the extent to which the CSF and blood mirror the immune cell repertoire of MS lesions seems to be greater than previously appreciated. This finding raises important questions. There could be a bidirectional traffic of CD8+ T cells between the CNS and peripheral compartments, or there could be unidirectional migration of CD8+ T cells from a constantly supplying peripheral source into the CNS. In the latter case, the CNS might act as a “sink” where the infiltrating T cells eventually die by apoptosis. Presently we cannot distinguish between these possibilities.
In principle, oligoclonal expansions of CD8+ can also occur in the normal immune system. For example, they can be sometimes observed in the blood of healthy elderly subjects (24), or they could nonspecifically infiltrate tissue lesions (25). However, their occurrence at different sites, striking persistence, and signs of recent activation indicate that at least some of the expanded CD8+ T cell clones observed in our study have a more relevant role. Assuming that this is the case, it is intriguing to speculate about their function in MS lesions. In principle, they could be involved in regulatory networks (26–28), or they might act as cytotoxic effector cells. Consistent with the latter possibility, the clinical course of patient A has been slowly progressive, despite therapy with IFN-β. Furthermore, it has been demonstrated that some MS lesions contain CD8+ T cells that directly contact demyelinated axons and polarize their cytotoxic granules to the contact zone (5). These CD8+ T cells seem to attack neurons and axons, which can express MHC class I antigens and therefore are susceptible to antigen-specific lysis by CD8+ T cells in vitro (5). In addition, the CD8+ T cells might interact with MHC-class I-expressing glia cells (5). Quantitative histological analyses of MS lesions indeed revealed a positive correlation between acute axonal damage and the number of infiltrating CD8+ T cells (29, 30). Furthermore, there is a positive correlation between activated cytokine-producing CD8+ T cells detectable in the blood of MS patients and MRI features of tissue destruction (31).
It is not yet known which antigen(s) are recognized by these CD8+ T cells. Viral antigens would be the obvious candidates. So far, however, all attempts to pinpoint a virus as the cause of MS have yielded inconclusive results. Alternatively, the CD8+ T cells might recognize genuine CNS autoantigen(s). Myelin antigens can indeed be recognized by human MHC class I-restricted CD8+ T cells (32, 33), and myelin-specific CD8+ T cells are encephalitogenic in mice (2, 3).
Our findings by no means rule out that CD4+ T cells have an equally important role. Indeed, experimental autoimmune encephalomyelitis can be induced in both CD4 and CD8 genedeleted mice, indicating that both T cell subsets play a role in pathogenesis (34). It is likely that CD8+ T cells depend on “help” from CD4+ regulatory T cells. In addition, CD4+ effector cells might directly contribute to inflammatory tissue injury. Our failure to identify CD4+ clonal expansions shared between the different tissue compartments could indicate that the sizes of CD4+ expansions are smaller than those of CD8+ expansions. Perhaps the CD4+ T cell response is more diverse than the CD8+ response and thus it is more difficult to detect clonal expansions with the CDR3 spectratyping method. Alternatively, CD4+ T cell responses might be fluctuating (35), or they might preferentially occur at tissue sites not sampled in our experiments.
The CD8+ T cell expansions should be useful in future studies for monitoring disease activity and assessing responses to immunosuppressive and immunomodulatory therapies. They might also serve as targets for T cell-directed immunotherapies, including T cell vaccination strategies (19, 36–38). Furthermore, they might provide clues to the identity of the enigmatic antigenic trigger of MS, whether it is an unidentified virus or autoantigen. It may be possible to clone expanded CD8+ T cells from the CSF or blood and test them with large arrays of candidate antigens, including synthetic peptide libraries (39). To study antigen recognition by these T cells in vivo, it might be possible to construct transgenic mice that express a human TCR together with an appropriate MHC class I molecule, as has been elegantly shown for MHC class II-restricted TCRs from myelin basic protein-specific CD4+ human T cell clones (40). Finally, our findings reemphasize the importance of cytotoxic mechanisms in myelin and axonal injury, calling for neuroprotective and repair strategies.
Acknowledgments
We thank Ms. Ingrid Eiglmeier for excellent technical assistance and Ms. Judy Benson for help with editing the manuscript. This study is part of the M.D. thesis of C.S. This work was supported by the Deutsche Forschungsgemeinschaft (SFB 571, A3 and A1).
Abbreviations: TCR, T cell receptor; BV, Vβ; BJ, Jβ; BC, Cβ; CSF, cerebrospinal fluid; CDR3, complementarity-determining region 3; MS, multiple sclerosis; PBMC, peripheral blood mononuclear cell.
Data deposition: Sequence data for TCR β-chains in Tables 2 and 3 have been deposited in the GenBank database (accession nos. AY534307–AY534315).
References
- 1.Ben-Nun, A., Wekerle, H. & Cohen, I. R. (1981) Eur. J. Immunol. 11**,** 195–199. [DOI] [PubMed] [Google Scholar]
- 2.Sun, D. M., Whitaker, J. N., Huang, Z. G., Liu, D., Coleclough, C., Wekerle, H. & Raine, C. S. (2001) J. Immunol. 166**,** 7579–7587. [DOI] [PubMed] [Google Scholar]
- 3.Huseby, E. S., Liggitt, D., Brabb, T., Schnabel, B., Öhlén, C. & Goverman, J. (2001) J. Exp. Med. 194**,** 669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steinman, L. (2001) J. Exp. Med. 194**,** F27–F30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann, H., Medana, I. M., Bauer, J. & Lassmann, H. (2002) Trends Neurosci. 25**,** 313–319. [DOI] [PubMed] [Google Scholar]
- 6.Liblau, R. S., Wong, F. S., Mars, L. T. & Santamaria, P. (2002) Immunity 17**,** 1–6. [DOI] [PubMed] [Google Scholar]
- 7.Babbe, H., Roers, A., Waisman, A., Lassmann, H., Goebels, N., Hohlfeld, R., Friese, M., Schroder, R., Deckert, M., Schmidt, S., et al. (2000) J. Exp. Med. 192**,** 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacobsen, M., Cepok, S., Quak, E., Happel, M., Gaber, R., Ziegler, A., Schock, S., Oertel, W. H., Sommer, N. & Hemmer, B. (2002) Brain 125**,** 538–550. [DOI] [PubMed] [Google Scholar]
- 9.Goebels, N., Hofstetter, H., Schmidt, S., Brunner, C., Wekerle, H. & Hohlfeld, R. (2000) Brain 123**,** 508–518. [DOI] [PubMed] [Google Scholar]
- 10.Monteiro, J., Hingorani, R., Peroglizzi, R., Apatoff, B. & Gregersen, P. K. (1996) Autoimmunity 23**,** 127–138. [DOI] [PubMed] [Google Scholar]
- 11.Puisieux, I., Even, J., Pannetier, C., Jotereau, F., Favrot, M. & Kourilsky, P. (1994) J. Immunol. 153**,** 2807–2818. [PubMed] [Google Scholar]
- 12.McDonald, W. I., Compston, A., Edan, G., Goodkin, D., Hartung, H. P., Lublin, F. D., McFarland, H. F., Paty, D. W., Polman, C. H., Reingold, S. C., et al. (2001) Ann. Neurol. 50**,** 121–127. [DOI] [PubMed] [Google Scholar]
- 13.Lassmann, H., Brück, W. & Lucchinetti, C. (2001) Trends Mol. Med. 7**,** 115–121. [DOI] [PubMed] [Google Scholar]
- 14.Wolinsky, J. S. (1999) Brain 122**,** 1211–1212. [DOI] [PubMed] [Google Scholar]
- 15.Hauser, S. L., Bhan, A. K., Gilles, F., Kemp, M., Kerr, C. & Weiner, H. L. (1986) Ann. Neurol. 19**,** 578–587. [DOI] [PubMed] [Google Scholar]
- 16.Oksenberg, J. R., Stuart, S., Begovich, A. B., Bell, R. B., Erlich, H. A., Steinman, L. & Bernard, C. C. A. (1990) Nature 345**,** 344–346. [DOI] [PubMed] [Google Scholar]
- 17.Hofbauer, M., Wiesener, S., Babbe, H., Roers, A., Wekerle, H., Dornmair, K., Hohlfeld, R. & Goebels, N. (2003) Proc. Natl. Acad. Sci. USA 100**,** 4090–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornmair, K., Goebels, N., Weltzien, H. U., Wekerle, H. & Hohlfeld, R. (2003) Am. J. Pathol. 163**,** 1215–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohlfeld, R. (1997) Brain 120**,** 865–916. [DOI] [PubMed] [Google Scholar]
- 20.Ransohoff, R. M., Kivisäkk, P. & Kidd, G. (2003) Nat. Rev. Immunol. 3**,** 569–581. [DOI] [PubMed] [Google Scholar]
- 21.Pachter, J. S., de Vries, H. E. & Fabry, Z. (2003) J. Neuropathol. Exp. Neurol. 62**,** 593–604. [DOI] [PubMed] [Google Scholar]
- 22.Panitch, H. S. & Francis, G. S. (1982) N. Engl. J. Med. 307**,** 560–561. [DOI] [PubMed] [Google Scholar]
- 23.Kivisakk, P., Mahad, D. J., Callahan, M. K., Trebst, C., Tucky, B., Wei, T., Wu, L., Baekkevold, E. S., Lassmann, H., Staugaitis, S. M., et al. (2003) Proc. Natl. Acad. Sci. USA 100**,** 8389–8394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posnett, D. N., Sinha, R., Kabak, S. & Russo, C. (1994) J. Exp. Med. 179**,** 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotet, E., Peyrat, M.-A., Saulquin, X., Retière, C., Couedel, C., Davodeau, F., Dulphy, N., Toubert, A., Bignon, J. D., Lim, A., et al. (1999) Eur. J. Immunol. 29**,** 973–985. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, V. & Sercarz, E. (2001) Immunol. Rev. 182**,** 113–121. [DOI] [PubMed] [Google Scholar]
- 27.Najafian, N., Chitnis, T., Salama, A. D., Zhu, B., Benou, C., Yuan, X., Clarkson, M. R., Sayegh, M. H. & Khoury, S. J. (2003) J. Clin. Invest. 112**,** 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, H., Curran, S., Ruiz-Vazquez, E., Liang, B., Winchester, R. & Chess, L. (2003) Proc. Natl. Acad. Sci. USA 100**,** 8378–8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bitsch, A., Schuchardt, A., Bunkowski, S., Kuhlmann, T. & Brück, W. (2000) Brain 123**,** 1174–1183. [DOI] [PubMed] [Google Scholar]
- 30.Kuhlmann, T., Lingfeld, T., Bitsch, A., Schuchardt, J. & Brück, W. (2002) Brain 125**,** 2202–2212. [DOI] [PubMed] [Google Scholar]
- 31.Killestein, J., Eikelenboom, M. J., Izeboud, T., Kalkers, N. F., Ader, H. J., Barkhof, F., Van Lier, R. A., Uitdehaag, B. M. & Polman, C. H. (2003) J. Neuroimmunol. 142**,** 141–148. [DOI] [PubMed] [Google Scholar]
- 32.Tsuchida, T., Parker, K. C., Turner, R. V., McFarland, H. F., Coligan, J. E. & Biddison, W. E. (1995) Proc. Natl. Acad. Sci. USA 91**,** 10859–10863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jurewicz, A., Biddison, W. E. & Antel, J. P. (1998) J. Immunol. 160**,** 3056–3059. [PubMed] [Google Scholar]
- 34.Abdul-Majid, K. B., Wefer, J., Stadelmann, C., Stefferl, A., Lassmann, H., Olsson, T. & Harris, R. A. (2003) J. Neuroimmunol. 141**,** 10–19. [DOI] [PubMed] [Google Scholar]
- 35.Muraro, P. A., Bonanni, L., Mazzanti, B., Pantalone, A., Traggiai, E., Massacesi, L., Vergelli, M. & Gambi, D. (2002) J. Neuroimmunol. 127**,** 149–159. [DOI] [PubMed] [Google Scholar]
- 36.Cohen, I. R. (1992) Immunol. Today 13**,** 490–494. [DOI] [PubMed] [Google Scholar]
- 37.Lohse, A. W. & Cohen, I. R. (1991) Autoimmunity 9**,** 119–121. [DOI] [PubMed] [Google Scholar]
- 38.Martin, R., Sturzebecher, C. S. & McFarland, H. F. (2001) Nat. Immunol. 2**,** 785–788. [DOI] [PubMed] [Google Scholar]
- 39.Hemmer, B., Vergelli, M., Pinilla, C., Houghten, R. & Martin, R. (1998) Immunol. Today 19**,** 163–168. [DOI] [PubMed] [Google Scholar]
- 40.Madsen, L. S., Andersson, E. C., Jansson, L., Krogsgaard, M., Andersen, C. B., Engsberg, J., Strominger, J. L., Svejgaard, A., Holmdahl, R., Wucherpfennig, K. W., et al. (1999) Nat. Genet. 23**,** 343–347. [DOI] [PubMed] [Google Scholar]