Identification of Cys255 in HIF-1α as a novel site for development of covalent inhibitors of HIF-1α/ARNT PasB domain protein–protein interaction (original) (raw)
Abstract
The heterodimer HIF-1α (hypoxia inducible factor)/HIF-β (also known as ARNT-aryl hydrocarbon nuclear translocator) is a key mediator of cellular response to hypoxia. The interaction between these monomer units can be modified by the action of small molecules in the binding interface between their C-terminal heterodimerization (PasB) domains. Taking advantage of the presence of several cysteine residues located in the allosteric cavity of HIF-1α PasB domain, we applied a cysteine-based reactomics “hotspot identification” strategy to locate regions of HIF-1α PasB domain critical for its interaction with ARNT. COMPOUND 5 was identified using a mass spectrometry-based primary screening strategy and was shown to react specifically with Cys255 of the HIF-1α PasB domain. Biophysical characterization of the interaction between PasB domains of HIF-1α and ARNT revealed that covalent binding of COMPOUND 5 to Cys255 reduced binding affinity between HIF-1α and ARNT PasB domains approximately 10-fold. Detailed NMR structural analysis of HIF-1α-PasB-COMPOUND 5 conjugate showed significant local conformation changes in the HIF-1α associated with key residues involved in the HIF-1α/ARNT PasB domain interaction as revealed by the crystal structure of the HIF-1α/ARNT PasB heterodimer. Our screening strategy could be applied to other targets to identify pockets surrounding reactive cysteines suitable for development of small molecule modulators of protein function.
Keywords: protein–protein interaction (PPI), crystal structure, covalent inhibitor, NMR spectroscopy, mass spectrometry, AlphaScreen, surface plasmon resonance (SPR), isothermal titration calorimetry (ITC)
Introduction
Hypoxia inducible factors (HIFs) are the key mediators of cellular response to hypoxia.1 HIF proteins are basic helix-loop-helix heterodimers composed of an oxygen-sensitive HIF-1α subunit and a constitutively expressed ARNT (also known as HIF-β subunit). Expression levels of HIF-1α are regulated by intracellular oxygen concentration. Under normoxic conditions, HIF-1α is continually degraded by ubiquitination and proteasomal degradation. Degradation occurs when HIF-1α is hydroxylated at residues Pro564 and Pro402, located in its oxygen-dependent degradation domain (ODDD). This process is facilitated by a family of prolyl hydroxylases (PHDs).2 Following hydroxylation, HIF-1α binds to the von Hippel–Lindau protein (pVHL) which is part of an E3-ubiquitin ligase complex that marks HIF-1α for proteasomal degradation through ubiquitination.3 Oxygen is required for the function of PHDs. Therefore, under hypoxic conditions HIF-1α is stabilized, accumulates, and translocates to the nucleus where it interacts with ARNT to form the active transcription factor, HIF-1. HIF-1 then specifically initiates expression of multiple genes that participate in angiogenesis, iron metabolism, glycolysis, glucose transport, cell proliferation, and survival. Increased levels of HIF-1α have also been associated with an aggressive phenotype and decreased patient survival in various types of cancer (breast, cervical, and endometrical cancers).4 A HIF-1α specific agent could also be used in combination therapy to overcome resistance to chemotherapy and radiation in cancer patients.5, 6
A number of strategies directed at modulation of HIF-1α function were undertaken previously to identify potential cancer therapeutics. Those strategies were mostly aimed at modulation of HIF-1α expression level by compounds affecting its transcription/translation,7 compounds activating PHD proteins which promote HIF-1α degradation in a VHL dependent manner,8 and compounds affecting HIF-1α/ARNT/p300/CBP complex formation by binding to p300/CBP.9 Additionally, an RNA antagonist, EZN-2968, composed of a third-generation oligonucleotide, locked nucleic acid technology that specifically binds and inhibits the expression of HIF-1α mRNA was developed and shown to inhibit tumor cell growth in glioblastoma and prostate cancer models.10
A second HIF isoform, HIF-2α, was identified that is regulated in a manner similar to HIF-1α.11, 12 Recent studies suggest that HIF-2α also regulates gene expression under physiological oxygen conditions and at prolonged hypoxia, whereas HIF-1α primarily governs acute hypoxic responses.13 Mutational analysis of the HIF-2α/ARNT PasB heterodimer interface based on the NMR structure of the HIF-2α PasB domain showed that mutations in the PasB domain of HIF-2α result in approximately a 50% reduction of transcriptional activity of the hypoxia response element (HRE). This suggests that direct modulation of the HIF-2α/ARNT PasB interface may reduce hypoxia-driven gene transcription.14, 15 While not as exhaustive as classical alanine scan mutagenesis studies for identification of “hot-spots” on protein–protein interfaces,16 these results suggest that the HIF-2α/ARNT PasB interface could be a potential target for therapeutic intervention.
Structural analysis of the HIF-2α/ARNT PasB domain interface revealed that both proteins interact through their beta-sheet surfaces, burying a total surface area of 2050 Å2 upon complex formation (PDB ID 3F1N).17 Additionally, structural analysis revealed the presence of an irregularly shaped 290 Å3 internal cavity within the HIF-2α domain. Gardner and coworkers identified a series of HIF-2α selective low molecular weight compounds that bind to the HIF-2α PasB internal cavity modulating the interaction between the HIF-2α and ARNT PasB domains (PDB ID 3H7W and 3H82).18 Their work represents the first example of specific inhibitors targeting the protein–protein interaction (PPI) of HIF-2α/ARNT and makes this particular system suitable for fragment-based lead discovery. Fragment-based screening represents a lead generation approach in drug discovery that identifies low molecular weight compounds (∼150–300 Da) with low/modest affinity to the target protein.19, 20 Fragment hits can then be further optimized to increase affinity for the target. This fragment approach often generates molecules in a novel chemical space not previously explored by more classical lead generation strategies, such as high through-put screening.21
Because the acute response to hypoxia relies on HIF-1α rather than HIF-2α, we focused on developing a fragment-based approach targeted to this isoform. The X-ray crystal structure of the HIF-1α/ARNT PasB heterodimer presented here reveals the presence of a homologous PPI interface and internal cavity in the HIF-1α PasB domain, opening a possibility to identify compounds that bind to PasB allosteric binding pocket similar to the ones described for HIF-2α.18 Since the HIF-1α cavity contains several Cys residues, we explored the use of irreversible fragments to fill the internal cavity of HIF-1α PasB domain and potentially disrupt HIF-1α/ARNT. This approach was pioneered by Wells and coworkers22 with thiol-containing fragments. Recently, there is a renewed interest to target cysteine-containing proteins with irreversible inhibitors.23
Considering the compact nature of the HIF-1α internal cavity and studies that indicate weak reactivity of thiol-based probes to cysteine residues buried in HIF-1α (unpublished results), we chose a “reactive electrophile library” for identification of covalent fragments that may play a role in modulation of HIF-1α/ARNT PPI in an allosteric manner similar to the ones previously described for HIF-2α. Here, we present the identification of COMPOUND 5 from a prototypical reactomics library containing 177 compounds armed with acrylamide or other reactive groups. COMPOUND 5 modifies Cys255 of HIF-1α PasB domain and modulates the PPI between HIF-1α and ARNT PasB domains by inducing local conformational change in the HIF-1α PasB domain.
Results
Structural and biophysical characterization of the HIF-1α/ARNT heterodimer
Analogous to previously described mutations in the HIF-2α/ARNT complex,17 we made the mutation R245E in the HIF-1α PasB domain to reverse a salt bridge between ARNT residue 362 (E362R mutation) resulting in stabilization of the HIF-1α/ARNT PasB complex.24 This mutation significantly improved solution behavior compared to the wild type HIF-1α PasB domain and allowed detailed biophysical characterization of the HIF-1α/ARNT interaction. The E362R mutation of ARNT was generated to complement R245E of HIF-1α. AlphaScreen protein competition studies show that the wild-type HIF-1α, construct 900 is ineffective as a competitive protein while the salt bridge mutant 947 has an IC50 of 43 n_M_ (Table III). Isothermal titration calorimetry (ITC) studies were carried out to determine the solution equilibrium dissociation constant (K_D) of the HIF-1α/ARNT PasB complex. In addition to K_D, ITC also provides a direct measure of the enthalpy of binding (Δ_H). For the 947 complex with ARNT a K_D of 125 n_M was observed at 10°C with Δ_H of +2.4 kcal mol−1. The free energy change (Δ_G_) of −8.9 kcal mol−1 is due to a favorable entropy change (T.Δ_S_) of +11.3 kcal mol−1 (Table I, Supporting Information Fig. S2). Thermodynamic data for the HIF-2α/ARNT interaction revealed a similar Δ_S_ driven binding event consistent with the high structural homology between the two complexes. However, the HIF-2α/ARNT complex showed more than an order of magnitude higher K_D (lower affinity) compared to HIF-1α. The lower binding affinity of HIF-2α PasB (R247E) to ARNT PasB was also observed in protein competition studies using the AlphaScreen assay where an IC50 value of 1140 n_M was measured for the HIF-2α (construct 949) (Table III).
Table III.
AlphaSreen and SPR Analysis of HIF-1a-COMPOUND 5 Conjugate
| PAS-B Protein | Construct | Domain boundary | Mutation | AlphaLISA IC50 n_M_ ± sem (N) | SPR K_d (μ_M) |
|---|---|---|---|---|---|
| HIF-1α | 900 | 238–349 | none | >10 | ND |
| HIF-1α | 947 | 238–349 | R245E | 43 ± 15 (6) | 1.2 ± 0.3 (7) |
| HIF-2α | 949 | 240–350 | R247E | 1140 ± 400 (8) | 7.4 ± 1.6 (3) |
| HIF-1α | 1106 | 238–349 | R245E, E266H, R311H, S330L | 58 ± 24 (4) | 0.5 ± 0.05 (3) |
| 1106_NMR* (WT) | 1106 | 238–349 | R245E, E266H, R311H, S330L | 75 ± 18 (3) | 1.1 ± 0.1 (2) |
| 1106_ COMPOUND 5 NMRa (conjugate) | 1106 | 238–349 | R245E, E266H, R311H, S330L | 1250 ± 44 (3) | 10 ± 1 (2) |
Table I.
Thermodynamic Data From ITC for HIF Proteins Binding to ARNT at 10 and 20°C
| HIF | Temperature (°C) | K_D (n_M) | Δ_G_ (kcal mol−1) | Δ_H_ (kcal mol−1) | T.Δ_S_ (kcal mol−1) |
|---|---|---|---|---|---|
| 1106 | 10 | 83 ± 20 | −9.2 | 1.1 ± 0.1 | 10.3 |
| 947 | 10 | 125 | −8.9 | 2.4 | 11.3 |
| 949 | 10 | 1400 ± 100 | −7.6 | 3.8 ± 1.1 | 11.4 |
| 1106 | 20 | ND | ND | ∼0.1 | ND |
| 947 | 20 | 190 | −9.0 | 1.5 | 10.5 |
| 949 | 20 | 1400 | −7.8 | 2.9 | 10.7 |
HIF-1α and HIF-2α PasB domains have 76% overall sequence identity (91% identity at the ARNT heterodimerization interface). We determined the X-ray crystal structure of the HIF-1α/ARNT complex revealing very high similarity of the HIF-1α/ARNT and HIF-2α/ARNT PasB heterodimer structures (Supporting Information Fig. S1). The C-alpha root-mean-square deviation (RMSD) for the core secondary structure is 0.61 Å, and for all residues (243–343 using HIF-2α numbering) is 0.72 Å. The protein–protein interface is also very similar between these two heterodimers, which share the common interaction partner ARNT. There are three amino acid differences at the interface (Fig. 1). The central and most buried residue is Val336 (Met338 in HIF-2α), located in the middle of strand β-I. This side chain contributes to the complementary surface between the HIF-α and ARNT. In HIF-2α the longer methionine side chain folds back against the HIF surface (three possible rotamers are actually observed), while nearby side chains on ARNT (directly across the PPI) have essentially identical orientations in both heterodimers. We have made all three HIF-2α to HIF-1α changes (M338V, V342L, and T327P) and analyzed binding of these mutants to ARNT using AlphaScreen and ITC. A single substitution of Met338 to Val in HIF-2α PasB domain results in ∼10-fold higher binding affinity to ARNT E362R PasB domain (Supporting Information Table S2 and Fig. S3) and can explain the lower affinity of the HIF-2α complex with ARNT compared to HIF-1α.
Figure 1.
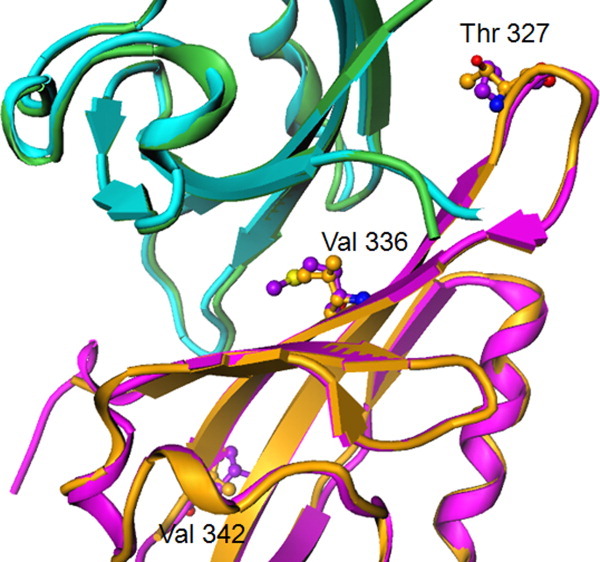
HIF/ARNT heterodimer, highlighting amino acids of HIF-1α (PDB ID 4H6J) that differ from HIF-2α (PDB ID 3F1N) at the PPI region. HIF-1α/ARNT (orange/green ribbons) and HIF-2α/ARNT (magenta/blue ribbons). HIF-1α T327, V336, and V342 are replaced, respectively, by HIF-2α residues P329, M338, and L344. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Additionally, the HIF-1α/ARNT heterodimer structure reveals the presence of a small internal cavity in HIF-1α PasB domain similar to HIF-2α, which is occupied by three well-defined water molecules. Interestingly, these three overlap with half of the six waters found in (non-fragment bound) HIF-2α structures (Fig. 2).17 Alternate orientations of His291, plus sequence differences of Ala275 versus Ile, and Leu317 versus Val, create different cavity shapes and water occupancies. In short, the shape of the cavity in HIF-1α is different, and somewhat smaller than in HIF-2α.
Figure 2.
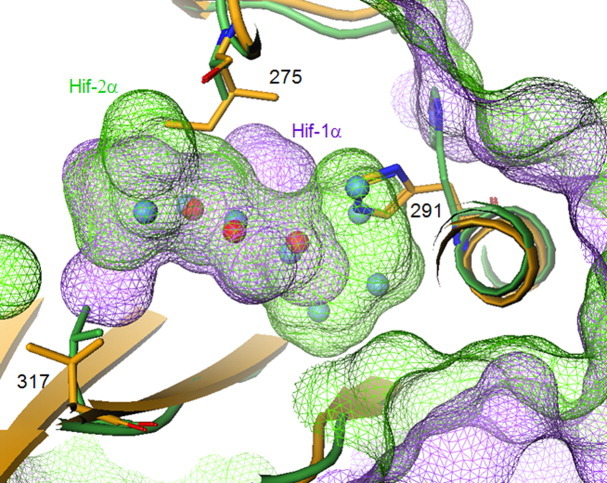
Trapped within the internal cavity, three waters are common between HIF-1α (yellow protein; magenta surface; red spheres) and HIF-2α (green protein; green surface; blue spheres), with similar hydrogen bonding networks. Alternate orientations of His291, plus sequence differences of Ala versus Ile at 275, and Leu versus Val at 317, create different cavity shapes and water occupancies. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Reactomic strategy for identification of HIF-1α cysteine residues critical for HIF-1α/ARNT PPI interaction
Examination of the HIF-1α/ARNT heterodimer structure revealed the presence of three Cys residues in areas potentially critical for the HIF-1α/ARNT PPI. Of the three cysteines in HIF-1α, Cys255 and Cys337 are buried and yet potentially accessible from the internal cavity, while Cys334 is located at the heterodimer interface and can directly affect the PPI [Fig. 3(A)]. Cys337 is clearly the most accessible within the internal cavity of HIF-1α PasB, its sulfhydryl actually forming part of the cavity wall [Fig. 3(B)]. However, the sulfur of Cys255 while seemingly inaccessible, actually lies within 8 Å of a water molecule located inside the cavity, and a concerted movement of a few side chains might permit the approach of a electrophile from a small reactive fragment present within the cavity [Fig. 3(B)]. These two cysteines, found in both HIF-1α and HIF-2α, reside on the beta sheet that forms a PPI with ARNT, and while their side chains point toward the protein interior (precluding direct contact with ARNT), modification of either could modulate beta sheet topology and/or dynamics sufficiently to change affinity for ARNT.
Figure 3.
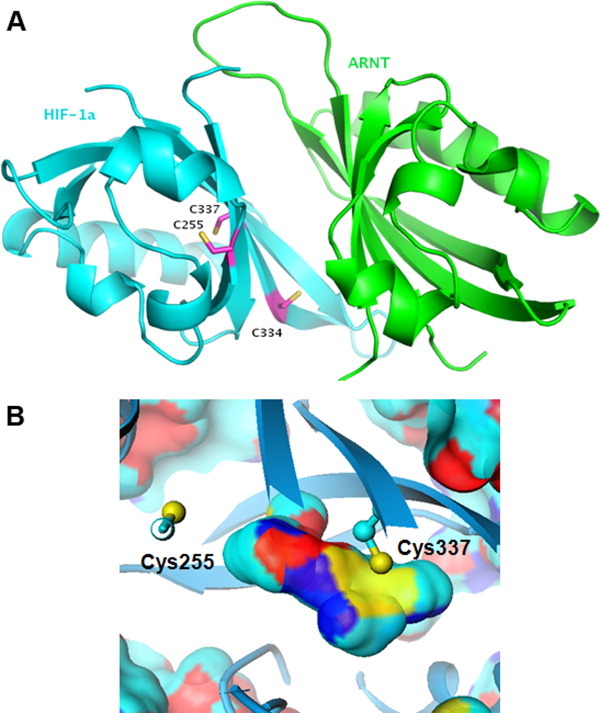
A: Location of Cys255, Cys334, and Cys337 on the HIF-1α/ARNT heterodimer (PDB ID 4H6J). B: Position of Cys255 and Cys337 relative to interior cavity of HIF-1α. The cavity is depicted as a surface and colored by protein atom types defining that surface (cyan = carbon; dark blue = nitrogen; red = oxygen; yellow = sulfur). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Building on recent renewed interest in identification of covalent inhibitors and taking into account properties of the HIF-1α internal cavity, we designed a “reactomics library” to explore covalent modification of the cysteine residues in the HIF-1α PasB domain as a starting point for identification of modulators of the HIF-1α/ARNT PPI. Compounds with at least one electrophilic group from the Pfizer compound collection were selected. These electrophiles were filtered by (1) passing quality control for purity >95 %, (2) MW <350 but >125, (3) clogP <3.5, (4) total polar surface area <140, and (5) number of total rotatable bond <9. Ward's hierarchical clustering method was applied to compounds that passed this criteria.25 Compounds from each cluster were picked randomly as representatives and combined for screening. In this case, the library was chosen to give as diverse a representation of chemical space as possible. Since these compounds have reactive functionalities, we were only interested in compounds that had been recently synthesized and purified. We also sought compounds with a MW above 125 to avoid overly reactive fragments.
Mass spectrometry identification of HIF-1α PasB binders
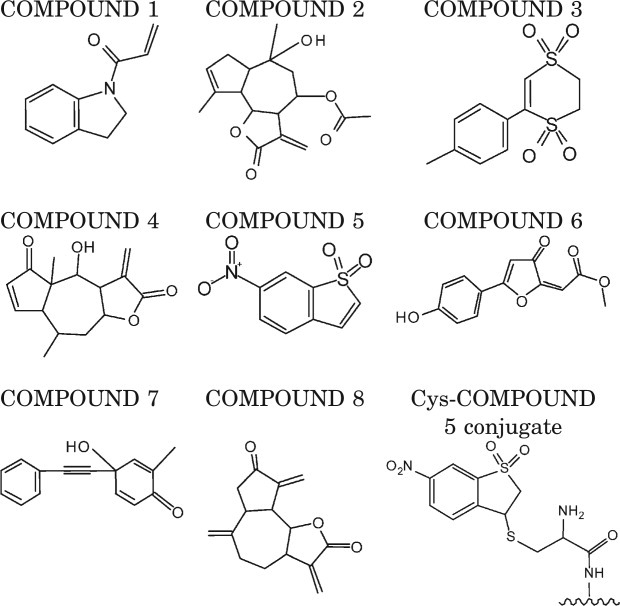
Mass spectrometry (MS) was employed to identify compound binding sites within the HIF-1α PasB domain. The HIF-1α PasB domain stability mutant (1106) was utilized for MS analysis due to its superior solution behavior.24 ITC analysis indicated that the 1106 stability mutant has similar ARNT solution binding constant as the 947 interface mutant (Table I, Supporting Information Fig. S3). One hundred seventy-seven compounds were screened at 200 μ_M_ concentration as described in Material and Methods for the ability to covalently modify Cys255 in the 1106 stability mutant. The seven compounds summarized in Table II met our requirement of >50% incorporation of compound on total HIF-1α protein signal, while COMPOUND 1 was identified during pilot screen of 20 compounds and had 25% incorporation at Cys255 (Table II). After the compounds were identified, MS/MS mapping studies were performed to locate the site of modification. MS spectra revealed adducts of COMPOUND 2, COMPOUND 3, and COMPOUND 4 on the intact protein but not on the Cys255, Cys334, or Cys337 containing peptides from the protein digest and therefore were eliminated from further analysis (Table II). The remaining four compounds reacted primarily with Cys255. However, additional adducts on His246, His266, Lys297, and Cys334 residues were also observed for COMPOUNDS 6–8 at 10–20% incorporation and complicate the potential biophysical characterization of these protein–compound conjugates. An example of MS/MS spectra of peptide liberated tryptic digest of COMPOUND 5 conjugated at Cys255 of 1106 stability mutant is presented in Supporting Information Figure S4. Full sequence coverage confirmed Cys255 conjugation, whereas the unmodified peptide form of Cys255 was not observed. Consequently, because COMPOUND 5 reacted completely and only with Cys255, its conjugate with HIF-1α PasB domain was chosen for further biophysical characterization.
Table II.
Compounds from the “Reactomics” Library Screen Incorporated into >50% of HIF-1α PasB Domain and Schematic Representation of Cys-COMPOUND 5 Conjugate
Irreversible binding of COMPOUND 5 to HIF-1α modulates the HIF-1α/ARNT PasB domain interaction
To characterize the protein–compound conjugate we used the 1106 stability mutant conjugated to COMPOUND 5 at Cys255. A 1:1 protein compound complex was isolated. This was characterized using liquid chromatography-mass spectrometry (LC-MS), analytical size exclusion chromatorgraphy (SEC), and differential scanning calorimetry (DSC) which indicated the presence of a well folded monodisperse protein in solution in the concentration range up to 80 μ_M_. We next examined the effect of COMPOUND 5 modified HIF-1α PasB domains on PPI properties using AlphaScreen, and surface plasmon resonance (SPR) assays.
Purified forms of 1106 with covalent bound COMPOUND 5 show a greater than 16-fold shift in AlphaScreen IC50 (1250 ± 440 n_M_, n = 3) compared to unconjugated 1106 prepared in parallel (75 ± 18 n_M_, n = 3). The significant shift in IC50 was consistent from two independent preparations of COMPOUND 5 irreversibly bound to 1106 HIF-1α [Fig. 4(A), Table III]. Similarly, SPR studies with the 1106-COMPOUND 5 conjugate binding to ARNT showed an order of magnitude higher K_D (10 ± 1 μ_M, n = 2) compared to the un-modified form of the protein (1.1 ± 0.1 μ_M_, n = 2) [Fig. 4(B), Table III] consistent with modulation of binding affinity by covalent binding of COMPOUND 5 to HIF-1α.
Figure 4.
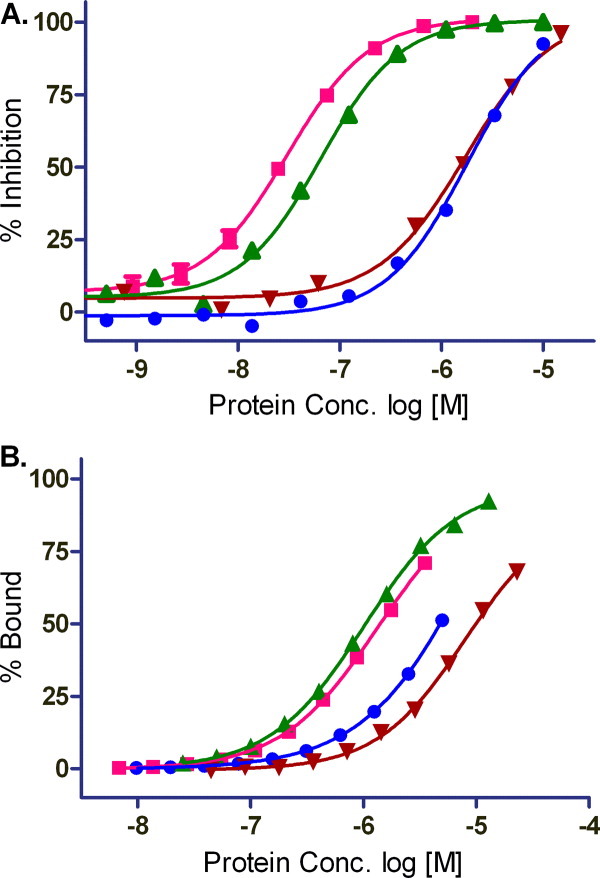
Binding studies with HIF-1α PasB constructs with COMPOUND 5. A: AlphaScreen protein competition study. Competitive proteins lacking biotinyl or Flag were incubated with biotinyl HIF-1α R245E and Flag-ARNT E362R prior to addition of streptavidin and anti-Flag AlphaScreen beads. B: SPR isotherms for ARNT binding HIF constructs at 10°C. Proteins: 947, HIF-1α R245E (pink square); HIF-1α 1106, stability mutant (green triangle); 1106-conjugate, COMPOUND 5 irreversibly bound 1106 (brown triangle); 949, HIF-2α (blue circle). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Covalent binding of COMPOUND 5 to HIF-1α modulates local structure of HIF-1α
Structural characterization of the COMPOUND 5 HIF-1α conjugate is hindered by the dependence for homo- and heterodimerization of HIF-1α and ARNT to obtain crystals suitable for X-ray analysis. Our unpublished 1106 crystal structure forms a homodimer utilizing the same HIF residues involved in HIF-1α heterodimerization with ARNT while homodimer crystals could be obtained only for constructs containing a combination of stability and interface mutations. The 10-fold reduction in affinity induced by conjugation of COMPOUND 5 to the HIF-1a precludes isolation of a stable conjugated HIF-1α/ARNT complex and crystallization of COMPOUND 5 conjugated complex. To overcome this challenge and gain insights to the mechanism of heterodimerization inhibition by COMPOUND 5, we conducted comparative NMR analysis of unconjugated and conjugated 1106 HIF-1a PasB construct. NMR backbone assignment of the HIF-1α PasB 1106 stability mutant were previously reported.24 Figure 5 shows the overlay of HSQC spectra of 35 μ_M_ apo 1106 stability mutant (red) and the same concentration of COMPOUND 5 conjugated 1106 protein (blue) recorded under identical conditions. Large chemical shift changes were detected on several amino acids, such as Glu245, His246, Ile324, Tyr325, Asn326, Gln333, Cys334, and I335, as indicated by the arrows. The majority of the other residues maintained their resonance frequencies. There was also a minor perturbation at Cys255, the conjugation site of the compound. The chemical shift change Δδobs of each amino acid was measured according to Eq. (1) and shown in Figure 6. Large chemical shift changes were observed within three defined regions, 244–246, 324–326, and 333–338, while the other amino acids show only small perturbations. These data indicate that the irreversible binding of COMPOUND 5 to HIF-1α PasB induced local but not global conformational change.
Figure 5.
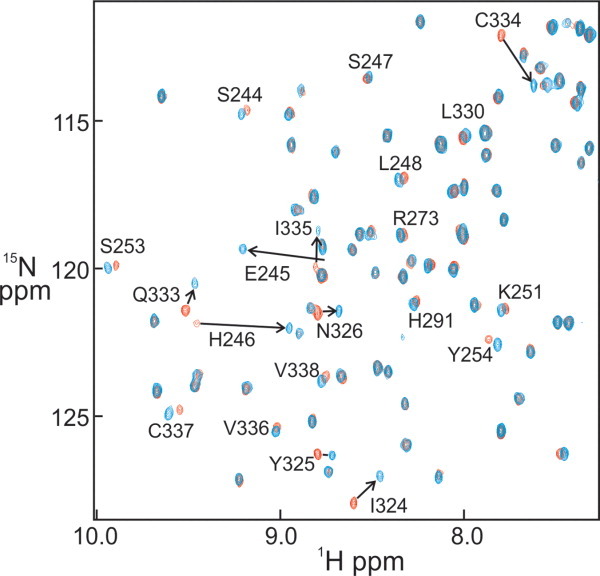
Overlay of 15N–1H HSQC spectra of apo HIF-1α (red) and conjugated HIF-1α (blue). Selected backbone assignments for resonances that show large chemical shift changes due to the conjugation are labeled and the changes indicated by arrows. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 6.
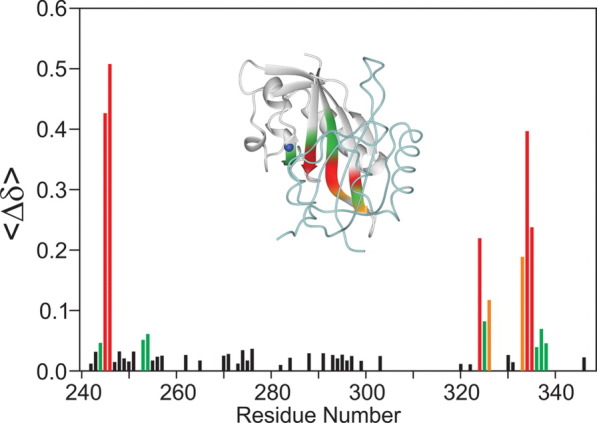
Plot of the HSQC chemical shift changes of HIF-1α PasB 1106 stability mutant between the monomer and COMPOUND 5 conjugate as a function of residue number. The inset shows the crystal structure of HIF-1α (gray ribbon)/ARNT (light blue backbone rod) heterodimer colored based on chemical shift changes from COMPOUND 5 conjugation. Blue atom: the backbone nitrogen of the conjugated Cys255 residue. HSQC chemical shift changes: Red, Δδav > 0.2 ppm; Orange, 0.2 > Δδav > 0.1 ppm; Green, 0.1 > Δδav > 0.04 ppm.
Discussion
HIF proteins play a critical role in cancer progression and are attractive targets for the development of anticancer agents.26 A number of drugs have been shown to inhibit HIF via indirect molecular mechanisms and present an attractive case to test the incorporation of HIF inhibitors into current cancer therapies. With the exception of acriflavine which is thought to target heterodimerization, most of the agents target HIF transcription, protein synthesis, protein stabilization, DNA binding, and transactivation (for review see Ref. 27). Heterodimerization of the HIF/ARNT PasB has previously been characterized for the HIF-2α/ARNT interaction17 and in this work for HIF-1α/ARNT. While it is clear that there are additional contributions to overall binding affinity of HIF to ARNT from the PasA domains as well as from the final transcription complex assembly on hypoxia response element (HRE), the PasB domain portion of the heterodimer could represent a true molecular target to modulate the hypoxia-mediated phenotype in a therapeutic setting.
The structural analysis of the HIF-1α/ARNT heterodimer presented here confirms that two potential strategies exist to modulate the PasB domain heterodimerization. Firstly, identification of molecules which bind to the HIF-1α or ARNT beta sheet surface involved in the PPI. Secondly, identification of allosteric inhibitors that bind internal cavities of the HIF-1α and HIF-2α PasB domains in a similar approach to the one adopted by Gardner and coworkers.17, 18 Due to the similar features of the HIF-1α and HIF-2α PPI interfaces with ARNT, the present study required an engineered salt bridge as employed by others to isolate a stable HIF/ARNT PasB domain complex. PPI of the salt bridge engineered constructs reveal 10-fold higher affinity of the HIF-1α binding to ARNT compared to HIF-2α. By comparing the structures of HIF-1α and HIF-2α PPI interfaces with ARNT we have also identified a single amino acid substitution M338V in the HIF-2α PasB domain that increases HIF-2α PasB domain binding affinity to ARNT to the level similar to HIF-1α PasB domain. The Δ_S_ driven interactions for the HIF–ARNT complexes are consistent with the burial of significant non-polar surface at the complex interface. The relatively unfavorable Δ_H_ for the HIF-2α/ARNT interaction is consistent with a less optimal steric fit of a methionine, revealed in the X-ray structure, by the extra bulk of Met338 (Table I). Further studies will be required to delineate the biological significance of the different affinities of the HIFs/ARNT PasB heterodimers.
The strategy of identification of allosteric binders has shown early promise through identification of a fragment capable of binding to a HIF-2α PasB internal cavity and modulating the PPI with ARNT.17, 18 The structural analysis of HIF-1α presented in this work confirms the presence of a similar internal cavity in the HIF-1α PasB domain. However, a key difference between HIF-1α and HIF-2α cavities is the smaller internal volume in HIF-1α which could present opportunities for selectivity versus HIF-2α as well as challenges in drugability of HIF-1α. During examination of our HIF-1α/ARNT crystal structure, we noticed the location of Cys255 and Cys337 in the internal cavity. We hypothesized that these amino acids could serve as attractive target sites for a covalent-based screening strategy and provide a starting point for development of inhibitors of HIF-1α/ARNT interaction. Covalent inhibitors have many desirable features, including increased biochemical efficiency of target disruption, less sensitivity toward pharmacokinetic parameters, and increased duration of action that outlasts the pharmacokinetics of the compound. There are numerous examples of covalent inhibitors in clinical and preclinical development (for review see Ref. 28). Recent advances in identification of covalent kinase inhibitors based on acrylamide warheads29–33 led us to design a prototypical reactomics library to explore the role of Cys residues in HIF-1α/ARNT PPI with the potential that the small fragments attached to the warhead could serve as a starting point for further development of selective covalent inhibitors or for design of non-covalent allosteric binders. Mass spec analysis of an iodoacetamide modified 1106 stability mutant indicated that both Cys255 and Cys337 sulfhydryl groups in the internal cavity could be accessed using acetamide chemistry, while probing with β-mercaptoethanol revealed slow reaction kinetics (data not shown). Coupled with MS/MS mapping of amino acid modification sites, MS provided a robust way to detect fragment covalent binding to the HIF-1α PasB domain. A high degree of selectivity was achieved as all fragments from the current library reacted with Cys255. Additionally, we were not able to identify any fragments that reacted with the solvent exposed Cys334, which could be explained by the lack of a pocket surrounding this residue sufficient to provide significant residence time for a fragment to react with Cys334.34 These results suggest that the local environment around specific Cys residues, and the chemical nature of the reactive moiety is likely responsible for specificity to Cys255 which was not predicted from prior structural analysis.
We further explored uniqueness of identified hits using 2D similarity comparison to shed light on preferred reactivity of those compounds to Cys255 HIF-1α PasB domain. We compared COMPOUNDS 1–8 to the entire library of 178 compounds using extended connectivity fingerprints (ECFP6)35 and setting various levels of Tanimoto similarity, a commonly used technique for identifying compounds similar to a reference compound from a collection of compounds. We did not identify compounds related to the hits summarized in Table II at 80% Tanimoto similarity which would have been expected to show similar biological reactivity.
We isolated a stable conjugate of the 1106 stability mutant with COMPOUND 5 and conducted detailed binding studies. AlphaScreen and SPR analysis indicated that the modification of Cys255 on HIF-1α PasB with COMPOUND 5 results in ∼10-fold reduction of HIF-1α binding to ARNT PasB domain. Based on the results of two independent binding assays together with folding/stability analysis of purified 1106-COMPOUND 5 conjugate, we conclude that the observed reduction in PPI affinity is a specific effect of the compound binding to HIF-1α PasB domain. Comparison of the HSQC spectra the HIF-1α PasB domain in the presence and absence of COMPOUND 5 suggests that the observed reduction of PPI affinity is a result of a localized conformation change in the HIF-1α PasB domain. When the chemical shift perturbations were mapped on the HIF-1α crystallographic structure in the HIF-1α/ARNT complex (Fig. 6), the regions of greatest chemical shift change induced by COMPOUND 5 were adjacent to another on the four parallel β-strands on the edge of the PPI interface. Thus, based on the NMR analysis of HIF-1α PasB conformation and structural analysis of PasB domain interaction, we conclude that the modification of Cys255 by conjugation with COMPOUND 5 causes a defined and localized conformational change at the adjacent beta-strand. This strand contains the Glu245 residue, which is critical for the interaction between the PasB domains of HIF-1α and ARNT. Further studies are required to elucidate the exact mechanism of this conformational change. Cysteine mutations were introduced into the HIF-1α PasB 1106 stability mutant to assess the dependence on these residues for productive PPI with ARNT E362R and to address the contribution of the fragment warhead of COMPOUND 5 in the overall reduction of binding affinity. Replacement of Cys255 with either Ser or Ala in stabilized 1106 resulted in a protein with extremely low solubility and stability which limited our ability to do further protein binding studies with ARNT. Poor solution behavior of C255A mutant suggests that homodimerization of HIF-1α PasB is likely affected by this mutation since we have previously reported that homodimerization is required for production of stable HIF-1α PasB domain.24 Additionally, the overlap in HSQC chemical shift changes on residues 242–246 and 334–337 of HIF-1α PasB domain induced by both Cys255 modification and homodimerization of the 1106 stability mutant allows us to suggest that modification of Cys255 has an allosteric effect on the formation of stable beta-sheet interactions in both the homo- and heterodimer interfaces. These observations further validate the mechanism of COMPOUND 5 mediated inhibition of HIF-1α/ARNT heterodimerization.
Quantitative reactive cysteine profiling was recently described on a proteome wide scale using an iodoacetamide-based probe where it was suggested that hyper-reactive cysteines likely perform important catalytic and/or functional roles in their parent proteins.36 Moreover, there are recent reports where cysteine alkylation of specific residues in target proteins has significant therapeutic effect in addition to well-known examples of irreversible kinase inhibitors. For example, the modification of Cys179 in IKKβ by CDDO (2-cyano-3,12-dioxooleana-1,9,-dien-28-oic acid) was shown to inhibit IKKβ kinase activity, which in turn fails to activate NF-κB.37, 38 Additionally, a recent report showed that specific alkylation of Cys463 in Stat3 by virtual ligand screening derived compounds selectively blocked Stat3–DNA interaction.39 The reactomics approach described here takes advantage of differential cysteine reactivity on the recombinant target protein within the constraints of our current library and allows systematic interrogation of cysteine-based hotspots in proteins thereby providing a strategy to identify novel points of alkylation with therapeutic potential. The successful identification of Cys255 as an allosteric hotspot capable of modulating HIF-1α/ARNT PasB domain PPI validates feasibility of this approach in this challenging target space.
In summary, we have presented a novel approach for identification of reactive fragments that modulate the HIF-1α/ARNT PPI via covalent modification of Cys255 in the HIF-1α PasB domain. Further studies are required to understand whether COMPOUND 5 or its analogues will achieve potency and selectivity to modulate HIF-1 driven transcription in cells.
Materials and Methods
Expression and purification of human HIF-1α and ARNT PasB wild-type and mutant proteins
Expression and purification of HIF and ARNT proteins was performed as described24 (detailed description is provided in Supporting Information).
Purification, crystallization, and structure determination of HIF-1α/ARNT PasB Heterodimer
Purification of HIF-1α/ARNT complex started with binding of 6xHis-GST-HIF-1α R245E to GST beads (GE Healthcare) for 1 h at 4°C in 25 m_M_ Hepes pH 7.4, 20 m_M_ NaCl, 1 m_M_ tris(2-carboxyethyl)phosphine (TCEF), 2% glycerol (binding buffer). The beads were then extensively washed with binding buffer and incubated with 1.5× molar excess of ARNT E362R 3 h at 4°C. Following the ARNT binding to HIF-1α, the beads were washed with binding buffer and the HIF-1α/ARNT complex was released by overnight incubation at 4°C with at least 10 units of glutatione S-transferase tagged tobacco etch virus (GST-TEV) protease/mg of complex. Fractions containing heterodimer complex were verified by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis and loaded on a Superdex 75 26/60 column for a final purification step. The HIF-1α/ARNT complex was then concentrated to 6.1 mg/mL and crystallized by the vapor-diffusion method in 20% polyethylene glycol (PEG) 8 kDa, 0.2_M_ MgCl2, 0.1 M Tris pH 8.5 at 4°C. Crystals of the HIF-1α/ARNT PasB domain heterodimer belong to space group C2 with unit cell dimension 68.6, 44.9, 78.1 Å and β = 96.5°, with one heterodimer per asymmetric unit. Diffraction data to 1.52 Å were collected using a Pilatus 6M detector on beamline 17-ID at the Advanced Photon Source (Argonne National Laboratory) from cryogenically frozen crystals, and processed with autoProc.40 Data statistics include _R_merge of 0.05 (high resolution shell 1.60–1.52 Å at 0.43); I/σ(I) of 21 (3.5); completeness of 97% (80); and redundancy of 6.4 (5.0). The structure was determined using the coordinates of our previously solved HIF-1α PasB domain (data not shown), which crystallizes as a homodimer, along with the published structure of ARNT PasB domain.41 The HIF-1α/ARNT complex and 155 water molecules were refined using CNS42 to R/_R_free values of 21.5/24.8, with rmsd bond and angle deviations of 0.004 Å and 0.80°, respectively.
Reactive fragment screening and preparation of compound protein conjugate
A library of 177 thiol reactive compounds was selected and prepared at 30 m_M_ concentration each in dimethyl sulfoxide (DMSO). A 20-fold molar excess of each compound was added to 50 μL of 10 μ_M_ 1106 stability mutant in 50 m_M_ Tris pH 8.0, 50 m_M_ NaCl for 24 h reaction time at ambient temperature in the dark. COMPOUND 1, a compound identified in preliminary studies as a potent Cys modifier, was used as a positive control. Reactivity of compounds was assessed by a novel matrix-assisted laser desorption/ionization time-of-flight MS method (MALDI-TOF MS, Bruker Ultraflextreme™). Detailed protocol used for identification of reactive compounds is described in Supporting Information.
HIF-1α-PasB-COMPOUND 5 conjugate was prepared by desalting the reaction mixture using a Zeba column (Thompson Scientific, IL) then adding to an appropriate buffer depending on a downstream assay. The final samples were reanalyzed by electrospray ionization time of flight mass spectrometry (ESI-TOF-MS) to confirm the presence of conjugated product with appropriate stoichiometry then the modification site was reconfirmed by LC-MS/MS analysis. The monodispersity of protein compound-conjugate was also confirmed by analytical SEC analysis.
AlphaScreen for PPI
AlphaScreen bead reagents, 384-well Optiplate and Envision™ plate reader were purchased from PerkinElmer. Biotin-HIF-1α R245E and Flag-ARNT E362R protein suspended in 50 m_M_ HEPES pH 7.4, 10 m_M_ NaCl, 1 m_M_ dithiothreitol (DTT), 0.1% bovine serum albumin (BSA), 0.01% Tween20 assay buffer were transferred to the Optiplate test well. AlphaScreen streptavidin-coated donor beads and anti-Flag antibody coated acceptor beads at a final concentration of 20 μg/mL each were added to the protein solution for 1 h incubation at 22°C in the dark before measure of the AlphaScreen signal using an Envision™ plate reader. Assay background was defined as the signal in the presence of 200 n_M_ free biotin that blocks capture of biotin conjugated HIF-1α protein. Protein competition studies used reagents incapable of associating with the AlphaScreen donor or acceptor beads. Competitive proteins serially diluted in assay buffer were transferred to the test well prior to addition of biotinyl-HIF-1α R245E and Flag-ARNT E362R at a final assay concentration of 5 n_M_ each followed by addition of AlphaScreen beads for 1h incubation. The percentage of competitor protein inhibition was calculated from the total assay signal in the absence of competitor protein and subtraction of assay background signal in the presence of free biotin. Dose response data was fit to a 4th order polynomial equation (GraphPad Prism) to obtain IC50 values. Competitor proteins were tested in two or more independent experiments.
Surface plasmon resonance binding studies
Surface plasmon resonance-based binding studies were carried out using a Biacore 3000 instrument (GE Healthcare) at 10°C in 150 m_M_ NaCl, 25 m_M_ HEPES pH 7.5, 10% glycerol, 0.005% P20, 4 m_M_ TCEP. HIF-1α was immobilized on a CM5 chip by standard amine coupling chemistry at 25°C. Concentration of modified and unmodified HIF-1α was determined spectrophotometrically by Coomasie analysis using an _A_280 of 17420 _M_−1 cm−1. Fifty injections of 50 μL per minute were made. Protein injections were referenced to a blank surface and by a buffer blank. Data analysis was performed using the Scrubber2 software (BioLogic Software, Pty., Australia). The observed equilibrium dissociation constant _K_D was determined using Origin 5.0 (OriginLab, MA) using the following equation: R = _K_A[compound]_R_max/(_K_A[compound]+1) where R is the equilibrium response at a specific compound concentration, _R_max is the response at saturating compound concentration, and _K_A is 1/_K_D.
Isothermal titration calorimetry
ITC experiments were carried out on a VP ITC instrument (GE Healthcare) at 10 and 20°C. Samples were extensively dialyzed into a buffer containing 150 m_M_ NaCl, 25 m_M_ HEPES, pH 7.5, 10% glycerol, 2 m_M_ TCEP. Concentrations were determined spectrophotometrically using an _A_280 of 15,470 M_−1 cm−1 for ARNT (E362R), 18,910 M_−1 cm−1for HIF-1α 947 (R245E) and 1106 (R245E/E266H/R311H/S330L) mutant proteins, and 17,420 M_−1 cm−1 for HIF-2α. In a typical experiment, nineteen 15 μL injections of 200 μ_M ARNT was made into a 20 μ_M HIF solution. Data were analyzed using the ORIGIN software provided with the instrument. Heats of dilution determined for ligand titrated into buffer were of similar magnitude to the Δ_H of binding, in some cases, and accounted for the apparent endothermic to exothermic trend in the raw data. After correction for dilution heats, isotherms were fit to a simple 1:1 binding model as described by Wiseman.43 Errors were calculated based on data from experiments carried out in duplicates.
NMR structural characterization
The apo HIF-1α and conjugated HIF-1α (conj-HIF-1α) proteins were dialyzed in the identical buffer (25 m_M_ Tris pH 7.4, 150 m_M_ NaCl, 0.2 m_M_ TCEP) at 4°C before use. 1H–15N HSQC experiments were recorded at 30°C on a Bruker Avance 700 MHz spectrometer equipped with a TCI cryo-probe. Mnova 7.0 (Mestrelab Research) was used in data analysis based on the sequential assignments.24 The total chemical shift change Δδobs of the 1H–15N cross peak was calculated according to the formula,
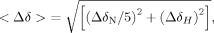 |
(1) |
|---|
where ΔδN and ΔδH are the 15N and 1H chemical shift changes. Chemical shift changes were mapped onto the co-crystal structure of HIF-1α/ARNT. For details on protein expression and purification, AlphaScreen assay development and Mass Spectrometry based identification of reactive compounds see Additional Supporting Information (HIFsup.doc).
Acknowledgments
The authors would like to acknowledge Michelle Arkin and Jim Wells for discussion and evaluation of sulfhydryl tethering strategy for HIF-1α PasB domain. The authors would like to acknowledge Lakshmi Narasimhan, Ben Bolanos, Oleg Brodsky for discussions and critical review of the manuscript. They are also thankful to Martin Edwards, Al Stewart, and Stephan Grant for their support and attention to this work.
Supplementary material
Additional Supporting Information may be found in the online version of this article.
References
- 1.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 2.Stolze IP, Mole DR, Ratcliffe PJ. Regulation of HIF: prolyl hydroxylases. Novartis Found Symp. 2006;272:15–25. 25–36. [PubMed] [Google Scholar]
- 3.Haase VH. The VHL tumor suppressor: master regulator of HIF. Curr Pharm Des. 2009;15:3895–3903. doi: 10.2174/138161209789649394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 5.Palayoor ST, Mitchell JB, Cerna D, Degraff W, John-Aryankalayil M, Coleman CN. PX-478, an inhibitor of hypoxia-inducible factor-1alpha, enhances radiosensitivity of prostate carcinoma cells. Int J Cancer. 2008;123:2430–2437. doi: 10.1002/ijc.23807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harada H, Inoue M, Itasaka S, Hirota K, Morinibu A, Shinomiya K, Zeng L, Ou G, Zhu Y, Yoshimura M, McKenna WG, Muschel RJ, Hiraoka M. Cancer cells that survive radiation therapy acquire HIF-1 activity and translocate towards tumour blood vessels. Nat Commun. 2012;3:783. doi: 10.1038/ncomms1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan C, de Noronha RG, Roecker AJ, Pyrzynska B, Khwaja F, Zhang Z, Zhang H, Teng Q, Nicholson AC, Giannakakou P, Zhou W, Olson JJ, Pereira MM, Nicolaou KC, Van Meir EG. Identification of a novel small-molecule inhibitor of the hypoxia-inducible factor 1 pathway. Cancer Res. 2005;65:605–612. [PubMed] [Google Scholar]
- 8.Choi HJ, Song BJ, Gong YD, Gwak WJ, Soh Y. Rapid degradation of hypoxia-inducible factor-1alpha by KRH102053, a new activator of prolyl hydroxylase 2. Br J Pharmacol. 2008;154:114–125. doi: 10.1038/bjp.2008.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mooring SR, Jin H, Devi NS, Jabbar AA, Kaluz S, Liu Y, Van Meir EG, Wang B. Design and synthesis of novel small-molecule inhibitors of the hypoxia inducible factor pathway. J Med Chem. 2011;54:8471–8489. doi: 10.1021/jm201018g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberger LM, Horak ID, Filpula D, Sapra P, Westergaard M, Frydenlund HF, Albaek C, Schroder H, Orum H. A RNA antagonist of hypoxia-inducible factor-1alpha, EZN-2968, inhibits tumor cell growth. Mol Cancer Ther. 2008;7:3598–3608. doi: 10.1158/1535-7163.MCT-08-0510. [DOI] [PubMed] [Google Scholar]
- 11.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 12.Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, Poellinger L, Pahlman S. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Zhang L, Erbel PJ, Gardner KH, Ding K, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J Biol Chem. 2005;280:36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 15.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix-loop-helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci USA. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham BC, Wells JA. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 17.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci USA. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jhoti H. Fragment-based drug discovery using rational design. Ernst Schering Found Symp Proc. 2007;3:169–185. doi: 10.1007/2789_2007_064. [DOI] [PubMed] [Google Scholar]
- 20.Fattori D, Squarcia A, Bartoli S. Fragment-based approach to drug lead discovery: overview and advances in various techniques. Drugs R D. 2008;9:217–227. doi: 10.2165/00126839-200809040-00002. [DOI] [PubMed] [Google Scholar]
- 21.Foloppe N. The benefits of constructing leads from fragment hits. Future Med Chem. 2011;3:1111–1115. doi: 10.4155/fmc.11.46. [DOI] [PubMed] [Google Scholar]
- 22.Erlanson DA, Wells JA, Braisted AC. Tethering: fragment-based drug discovery. Annu Rev Biophys Biomol Struct. 2004;33:199–223. doi: 10.1146/annurev.biophys.33.110502.140409. [DOI] [PubMed] [Google Scholar]
- 23.Singh J, Petter RC, Baillie TA, Whitty A. The resurgence of covalent drugs. Nat Rev Drug Discov. 2011;10:307–317. doi: 10.1038/nrd3410. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Martinez-Yamout M, Cardoso R, Yan J, Love RA, Grodsky N, Brooun A, Dyson HJ. Homodimerization of the PAS-B domains of hypoxia-inducible factors. J Phys Chem B. 2012;116:6960–6965. doi: 10.1021/jp300525n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward JHJ. Hierarchical grouping to optimize an objective function. J Am Stat Assoc. 1963;48:236–244. [Google Scholar]
- 26.Jones DT, Harris AL. Small-molecule inhibitors of the HIF pathway and synthetic lethal interactions. Expert Opin Ther Targets. 2012;16:463–480. doi: 10.1517/14728222.2012.674516. [DOI] [PubMed] [Google Scholar]
- 27.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33:207–214. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson DS, Weerapana E, Cravatt BF. Strategies for discovering and derisking covalent, irreversible enzyme inhibitors. Future Med Chem. 2010;2:949–964. doi: 10.4155/fmc.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honigberg LA, Smith AM, Sirisawad M, Verner E, Loury D, Chang B, Li S, Pan Z, Thamm DH, Miller RA, Buggy JJ. The Bruton tyrosine kinase inhibitor PCI-32765 blocks B-cell activation and is efficacious in models of autoimmune disease and B-cell malignancy. Proc Natl Acad Sci USA. 2010;107:13075–13080. doi: 10.1073/pnas.1004594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou W, Hur W, McDermott U, Dutt A, Xian W, Ficarro SB, Zhang J, Sharma SV, Brugge J, Meyerson M, Settleman J, Gray NS. A structure-guided approach to creating covalent FGFR inhibitors. Chem Biol. 2010;17:285–295. doi: 10.1016/j.chembiol.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry DW, Bridges AJ, Denny WA, Doherty A, Greis KD, Hicks JL, Hook KE, Keller PR, Leopold WR, Loo JA, McNamara DJ, Nelson JM, Sherwood V, Smaill JB, Trumpp-Kallmeyer S, Dobrusin EM. Specific, irreversible inactivation of the epidermal growth factor receptor and erbB2, by a new class of tyrosine kinase inhibitor. Proc Natl Acad Sci USA. 1998;95:12022–12027. doi: 10.1073/pnas.95.20.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou W, Ercan D, Chen L, Yun CH, Li D, Capelletti M, Cortot AB, Chirieac L, Iacob RE, Padera R, Engen JR, Wong KK, Eck MJ, Gray NS, Janne PA. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature. 2009;462:1070–1074. doi: 10.1038/nature08622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramstrom O, Lehn JM. Drug discovery by dynamic combinatorial libraries. Nat Rev Drug Discov. 2002;1:26–36. doi: 10.1038/nrd704. [DOI] [PubMed] [Google Scholar]
- 35.Muchmore SW, Debe DA, Metz JT, Brown SP, Martin YC, Hajduk PJ. Application of belief theory to similarity data fusion for use in analog searching and lead hopping. J Chem Inf Model. 2008;48:941–948. doi: 10.1021/ci7004498. [DOI] [PubMed] [Google Scholar]
- 36.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmad R, Raina D, Meyer C, Kharbanda S, Kufe D. Triterpenoid CDDO-Me blocks the NF-kappaB pathway by direct inhibition of IKKbeta on Cys-179. J Biol Chem. 2006;281:35764–35769. doi: 10.1074/jbc.M607160200. [DOI] [PubMed] [Google Scholar]
- 38.Shishodia S, Sethi G, Konopleva M, Andreeff M, Aggarwal BB. A synthetic triterpenoid, CDDO-Me, inhibits IkappaBalpha kinase and enhances apoptosis induced by TNF and chemotherapeutic agents through down-regulation of expression of nuclear factor kappaB-regulated gene products in human leukemic cells. Clin Cancer Res. 2006;12:1828–1838. doi: 10.1158/1078-0432.CCR-05-2044. [DOI] [PubMed] [Google Scholar]
- 39.Buettner R, Corzano R, Rashid R, Lin J, Senthil M, Hedvat M, Schroeder A, Mao A, Herrmann A, Yim J, Li H, Yuan YC, Yakushijin K, Yakushijin F, Vaidehi N, Moore R, Gugiu G, Lee TD, Yip R, Chen Y, Jove R, Horne D, Williams JC. Alkylation of cysteine 468 in Stat3 defines a novel site for therapeutic development. ACS Chem Biol. 2011;6:432–443. doi: 10.1021/cb100253e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vonrhein C, Flensburg C, Keller P, Sharff A, Smart O, Paciorek W, Womack T, Bricogne G. Data processing and analysis with the autoPROC toolbox. Acta Cryst D. 2011;67:293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Card PB, Erbel PJ, Gardner KH. Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization. J Mol Biol. 2005;353:664–677. doi: 10.1016/j.jmb.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 42.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Cryst D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 43.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.