Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides (original) (raw)
Abstract
The study of genetic disease mechanisms relies mostly on targeted mouse mutants that are derived from engineered embryonic stem (ES) cells. Nevertheless, the establishment of mutant ES cells is laborious and time-consuming, restricting the study of the increasing number of human disease mutations discovered by high-throughput genomic analysis. Here, we present an advanced approach for the production of mouse disease models by microinjection of transcription activator-like effector nucleases (TALENs) and synthetic oligodeoxynucleotides into one-cell embryos. Within 2 d of embryo injection, we created and corrected chocolate missense mutations in the small GTPase RAB38; a regulator of intracellular vesicle trafficking and phenotypic model of Hermansky-Pudlak syndrome. Because ES cell cultures and targeting vectors are not required, this technology enables instant germline modifications, making heterozygous mutants available within 18 wk. The key features of direct mutagenesis by TALENs and oligodeoxynucleotides, minimal effort and high speed, catalyze the generation of future in vivo models for the study of human disease mechanisms and interventions.
Keywords: gene targeting, mouse genetics, knockout, knockin
Gene targeting in embryonic stem (ES) cells is routinely applied to modify the mouse genome and establish the mouse as the most commonly used genetic disease model (1). Nevertheless, the production of targeted mutants is a laborious and time-consuming task that requires the construction of targeting vectors with selection markers, the isolation of mutant ES cells, and the generation of germ-line chimaeras. Moreover, advanced allele design strategies, like the creation of missense mutations, require an additional working step to eliminate selection marker genes from targeted loci (2). Therefore, the establishment of such mutants is presently a low-throughput and long-term procedure, restricting rapid advancements in disease modeling. In contrast, a burst of nucleotide replacements, small deletions and insertions in coding regions that underlie Mendelian or complex human diseases are discovered by high-throughput genomic analysis such as whole-exome sequencing (3–5). The faithful reproduction of such mutations in mouse models will be instrumental in understanding disease mechanisms and to develop therapeutic interventions. This demand is neither covered by available mutagenesis technologies nor by large-scale mouse mutagenesis programs that aim for complete gene inactivation in classical or conditional knockout mice (6). To cope with this challenge, we aimed to create mutations within the shortest time directly in the genome of one-cell mouse embryos, avoiding the time-consuming handling of ES cells, gene-targeting vectors, selection markers, and chimaeras. Our approach is based on transcription activator-like (TAL) effector nucleases (TALENs) to create targeted double-strand breaks that are either sealed by homologous recombination (HR) with mutant, synthetic oligodeoxynucleotides (ODNs) or closed by nonhomologous end-joining (NHEJ) repair that frequently elicits deletion and insertion mutations. We previously demonstrated that zinc-finger nucleases (ZFNs) can be used for direct gene editing in one-cell mouse embryos (7, 8), but limitations of the zinc-finger DNA recognition code precludes a wider application of this technology to any genomic coding sequence (9, 10). In contrast to ZFNs, the recognition code of the transcription activator-like (TAL) effector proteins enables the construction of new DNA-binding domains with unprecedented freedom (11, 12). The TAL DNA recognition code is based on four variants of 34-residue peptide elements (TAL repeats) that each mediate the binding to one of the four nucleotides of a DNA target sequence. As specified by a selected target sequence, TAL repeats can be combined in the appropriate order and function as sequence-specific TAL nucleases (TALENs) by fusion with the FokI nuclease domain. A typical, complete TALEN target site includes binding sites for two TAL-Fok fusion proteins of 14–18 bp that are each preceded by a T and separated by a spacer region of 14–16 bp (13, 14). Upon expression of TALENs in mammalian cell lines, mouse, rat, and pig embryos knockout alleles were obtained through NHEJ-mediated sequence deletion (15–17).
Here, we apply TALENs for the expedited creation of mouse disease models from one-cell embryos. We developed tools for the genome-wide analysis of TALEN target sites, the modular construction of TALEN coding vectors, and a nuclease reporter assay for quality control. TALEN mRNAs are microinjected together with synthetic, single-stranded ODNs into one-cell embryos that develop into heterozygous founder mutants. We applied this technology to the Rab38 gene, a small GTPase that regulates intracellular vesicle trafficking in melanocytes, retinal pigment epithelial cells, alveolar pneumocytes, and platelets (18, 19). Mutant chocolate mice (18) (_Rab38_cht) exhibit a missense, and ruby rats (20) exhibit a nonsense mutation within Rab38 and are considered to be phenotypic models of Hermansky–Pudlak syndrome (21, 23), a disease characterized by oculocutaneous albinism (OCA), progressive pulmonary fibrosis, and platelet storage disease. For the human Rab38 gene, more than 50 missense, nonsense, and deletion mutations were identified by large-scale genomic analysis (24). By using a _Rab38_-specific TALEN and a mutagenic ODN in one-cell embryos, we generated founders harboring targeted nucleotide replacements or various deletion/insertion mutations within a single microinjection experiment. Individual founders enabled the establishment of heterozygous progeny within 18 wk after microinjection. Using the same TALEN and a repair template with wild-type sequence, we were further able to correct the Rab38 G19V mutation in the genome of chocolate mutants. These results demonstrate that TALENs combined with ODNs in one-cell embryos provide a powerful tool for the expedited creation and reversion of mutations, able to boost the generation of future in vivo disease models.
Results
Genome-Wide Analysis for TALEN Target Sites.
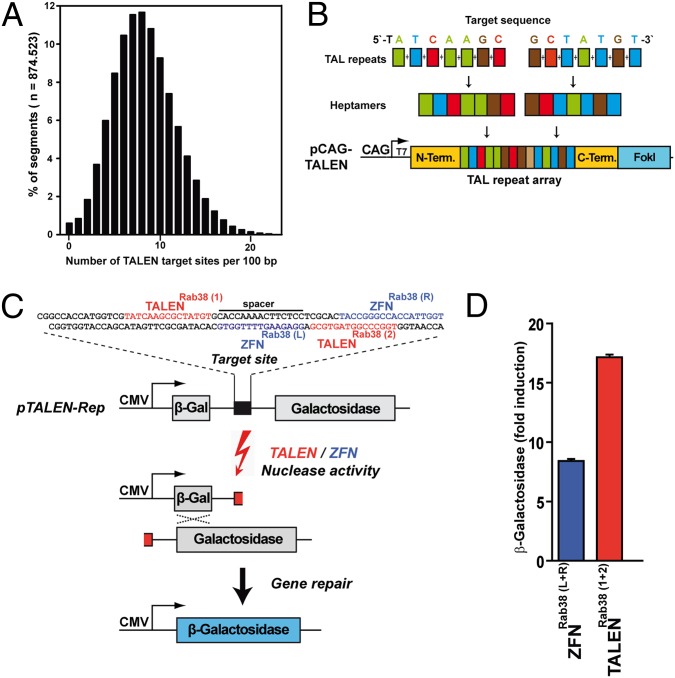
For the convenient identification of TALEN target sequences, we developed a software tool and a public Web page (www.talen-design.de; Fig. S1) that provide genome-wide analyses of all mouse coding regions and of user-specific sequences. The frequency and distribution of TALEN target sites in the mouse genome was analyzed using chromosome 18 as a representative example. Within the sequence of 87.5 Mb, our TALEN-Designer tool identifies 7.27 million target sites that occur at a mean frequency of one site per 12 bp. As analyzed in segments of 100 bp, 99.4% of segments exhibit one or more TALEN target sites, the majority of segments shows five to nine target sites, and only 0.6% of segments are free of a single site (Fig. 1_A_). For targeting of the mouse exome the TALEN-Designer offers 12.7 million predesigned TALEN target sites within 176.2-Mb coding sequence of 416.230 exons. Because TALEN target sites are found at an average spacing of 14 bp, the entire coding space of the mouse genome can be addressed with TALENs at a resolution that is amenable for targeted mutagenesis by synthetic ODNs. The distribution of target sites within the first exon of the mouse Rab38 gene is shown in Fig. S2.
Fig. 1.
Target-site analysis, TALEN construction, and nuclease activity. (A) Frequency of TALEN target sites in 100-bp segments of mouse chromosome 18, using bipartite 15-bp target sequences, separated by a spacer region of 15 bp. (B) Construction of TALEN expression vectors. For targeting of a 15-bp sequence [here: TALENRab38 (1)] a pair of ligations, each combining seven TAL repeat coding DNA segments, is assembled in the order specified by the variable 14-bp sequence following the invariable T at the first position. Full-length ligation products are inserted into the vector pCAG-TALEN, providing a CAG promoter (CAG), a T7 promoter (T7), invariable N- and C-terminal TAL sequences and a FokI nuclease domain. (C) Outline of the nuclease reporter assay. TALENRab38 (shown in red) and ZFNRab38 (shown in blue) target sequences are cloned in between the β-galactosidase gene segments of the pTALEN-Rep plasmid. Cotransfection of the reporter plasmid with TALENRab38 or ZFNRab38 vectors into HEK293 cells leads to nuclease dependent gene repair and β-galactosidase expression. (D) Reporter activity of TALENRab38 with the Rab38 reporter (c) compared with ZFNRab38. Values are expressed as nuclease stimulated increase of β-galactosidase activity compared with background levels without nuclease vectors. CMV, CMV promoter region.
TALEN Construction and Activity.
Using the TALEN-Designer, we constructed a TALEN pair against a target sequence within the first exon of Rab38 (TALENRab38; Fig. 1_C_) and TALENs against 20 other genes by selection of bipartite 15-bp target sequences, separated by spacer regions of 15 bp. For each target sequence, we constructed TALEN coding regions using a pair of ligation reactions that each combine seven TAL repeat coding DNA segments, in the order specified by the variable 14-bp sequence that follow the invariable T at the first position. Complete coding regions for TALEN proteins are obtained by the insertion of both full-length ligation products into the generic expression vector pCAG-TALEN (Fig. 1_B_). Because the activity of TALENs can vary by one order of magnitude (25), we controlled the quality of our nucleases, being able to use highly active TALENs for the application in one-cell embryos. For this purpose, the TALEN target regions were inserted into a nuclease reporter plasmid (pTALEN-Rep) in between a partly duplicated, nonfunctional β-galactosidase gene (Fig. 1_C_). Upon cotransfection of the reporter and TALEN expression vectors into HEK293 cells, nuclease-induced double-strand breaks (DSBs) stimulate the repair of the gene segments into a functional reporter gene, the activity of which is determined in cell lysates. As a positive control, we used a ZFN (ZFNRab38) (8) recognizing a target sequence next to TALENRab38 (Fig. 1_C_) that leads to an 8.4-fold increase of reporter activity in this assay (Fig. 1_D_). We found that all of our TALENs showed nuclease activity and that 70% of these TALENs (14/20) were comparable or superior to the ZFNRab38 positive control (Fig. S3). TALENRab38 recognizing a target sequence in the first exon of the Rab38 gene showed a 17.2× increase of reporter activity (Fig. 1_D_) and was used for the application in one-cell embryos to create targeted gene modifications.
Creation of a Rab38 Chocolate Mutation with TALENs and ODNs.
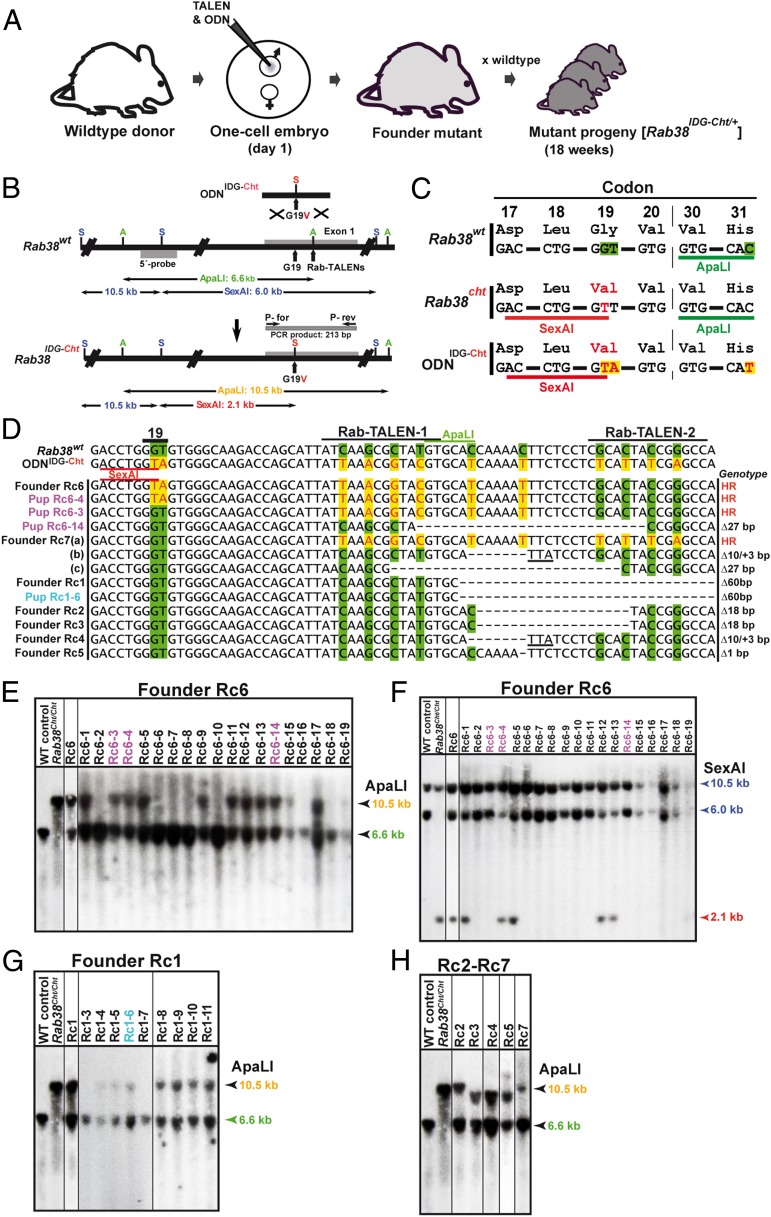
The workflow of TALEN-mediated, embryo-based gene targeting (Fig. 2_A_) starts with the microinjection of TALEN mRNA and a targeting ODN into one-cell embryos. Upon translation, the TALEN proteins are imported into the pronuclei and create DSBs in the paternal and maternal target loci. In the paternal genome, DSBs are sealed either by HR with the mutagenic ODN or become processed in both genomes by error-prone NHEJ repair. Mice derived from embryo injections are genotyped to identify founders harboring the desired mutation. The mating of founders produces heterozygous (1-mo-old) mutant progeny just 18 wk after embryo injection. By further breeding, first homozygote mutants can be obtained within 25 wk. As proof of this principle, we targeted the Rab38 gene to create a glycine-to-valine missense mutation at codon 19 (G19V), as found in the _Rab38_cht allele of chocolate mutants (18). As targeting molecule, we used a synthetic, single-stranded ODN (ODNIDG-Cht) of 144 nt that covers 47 bp of the lagging strand sequence upstream of codon 19 and 94 bp of downstream sequence (Fig. 2_B_). ODNIDG-Cht includes a G-to-T replacement at the second position of codon 19, creating a valine triplet and a SexAI restriction site, and a silent T-to-A exchange as an unique identifier of our targeted _Rab38_IDG-Cht allele (Fig. 2_C_). To map the proficiency of HR in relation to the distance between the DSB site and ODN coded replacements, the TALENRab38 spacer is located 40 bp downstream of codon 19 and 10 silent nucleotide replacements were included into ODNIDG-Cht (Fig. 2_D_). One of these replacements eliminates an ApaLI restriction site. ODNIDG-Cht was microinjected together with TALENRab38 mRNAs into one-cell embryos, and the resulting offspring were analyzed for gene-editing events by PCR amplification of a 213-bp region covering the first exon of Rab38 (Fig. 2_B_). Founder mice were identified by the digestion of PCR products with ApaLI. Digestion-resistant PCR products were recovered, cloned, and analyzed by sequencing. In addition, founder mutants were analyzed by Southern blot analysis of tail DNA using a hybridization probe located upstream of exon 1. Both NHEJ- and HR-mediated repair events led to the loss of the ApaLI site within exon 1, as indicated by a 10.5-kb ApaLI band in addition to the 6.6-kb wild-type fragment (Fig. 2_B_). HR events that include the G19V replacement can be specifically recognized by the presence of a diagnostic 2.1-kb SexAI band compared with the 6.0-kb wild-type fragment. In addition, a second, invariable 10.5-kb fragment is detected by the hybridization probe for both the Rab38 G19V and wild-type alleles (Fig. 2_B_). Among 117 pups derived from embryo injections of TALENRab38 and ODNIDG-Cht, we identified 7 mutant founders (Rc1 to Rc7) harboring 11 modified Rab38 alleles (Table S1). Founder Rc6 contained, as shown by sequencing of cloned PCR products, a recombined _Rab38_IDG-Cht allele that includes the G19V mutation and six additional nucleotide replacements from ODNIDG-Cht (Fig. 2_D_). A Southern blot analysis of Rc6 tail DNA showed the 2.1-kb SexAI fragment diagnostic for the _Rab38_IDG-Cht allele and the recombinant 10.5 kb ApaLI band, besides a wild-type locus (Fig. 2 E and F). Pups derived from matings of founder Rc6 were genotyped by Southern blot analysis of SexAI- or ApaLI-digested tail DNA and the sequencing of PCR products. Five pups showed the 2.1-kb SexAI band, demonstrating germ-line transmission of the _Rab38_IDG-Cht allele. (Fig. 2_F_). Pup Rc6-4 was further characterized by PCR and sequence analysis and confirmed the identity of its _Rab38_IDG-Cht allele to the founder Rc6 (Fig. 2_D_). Pup Rc6-4, carrying the _Rab38_IDG-Cht allele, was mated with C57BL/6 wild-type mice, and the derived heterozygous offspring were intercrossed to obtain homozygous _Rab38_IDG-Cht mice. Compared with their _Rab38_wt/wt littermates, homozygous _Rab38_IDG-Cht/IDG-Cht mice exhibited the expected dark brown coat color characteristic of chocolate mutants (18), confirming the functionality of the _Rab38_IDG-Cht (G19V) mutation (Fig. S4). Furthermore, seven pups derived from founder Rc6 showed a 10.5-kb ApaLI, but no 2.1-kb SexAI band (Fig. 2_F_), indicating the presence of additional, edited Rab38 alleles in the founder’s germ line. PCR and sequence analysis of pup Rc6-3 revealed the presence of a targeted Rab38 allele that includes all nucleotide replacements within the TALENRab38-binding region but excludes the replacements in codon 19. In contrast, pup Rc6-14 harbored a Rab38 allele exhibiting an in-frame deletion of 27 bp within the TALENRab38-binding region (Fig. 2_D_). In conclusion, the germ line of founder Rc6 constituted a mosaic of three mutant Rab38 loci, including two alleles that underwent HR with ODNIDG-Cht and one allele processed by NHEJ. In founder Rc7, the PCR and sequence analysis of tail DNA revealed a similar triple mutant genotype including a partially recombined _Rab38_IDG-Cht allele [Rc7(a); Fig. 2_D_], as found in pup Rc6-3, and two NHEJ-processed loci that exhibit a 27-bp in-frame deletion in allele Rc7(c) and the combined deletion and insertion of 10 and 3 bp, leading to a frameshift mutation within exon1 [Rc7(b); Fig. 2_D_]. Founders Rc1 to Rc5 harbored additional Rab38 alleles processed by NHEJ within the TALENRab38-binding region. Founder Rc1 exhibited an in-frame deletion of 60 bp that was germ line-transmitted and confirmed in pup Rc1-6 (Fig. 2_D_). Founders Rc2 and Rc3 harbored an identical in-frame deletion of six Rab38 codons, whereas founders Rc4 and Rc5 showed frame-shift mutations by the deletion/insertion of 10/3 bp [Rc4, identical to Rc7(b)] or the deletion of 1 bp (Rc5; Fig. 2_D_). The integrity of the modified Rab38 alleles in founders Rc1 to Rc7 and the offspring derived from Rc6 and Rc1 was analyzed by Southern blotting of SexAI- or ApaLI-digested tail DNA using the 5′-hybridization probe. In founders Rc6, Rc7, Rc1, and Rc2 and the Rc6- and Rc7-derived progeny, the presence of the 10.5-kb ApaLI and of the 2.1-kb SexAI bands indicate the genomic integrity of the modified Rab38 alleles (Fig. 2 E_–_H). In contrast, the NHEJ-processed Rab38 loci of founders Rc3, Rc4, and Rc5 exhibit a shortened, ∼9-kb ApaLI fragment, indicating the occurrence of additional sequence deletions. These results reveal four characteristic features of TALEN and ODN mediated mutagenesis in one-cell embryos: (i) HR is capable of transferring ODN encoded nucleotide replacements into a TALEN target region; (ii) nucleotide replacements occur preferentially in proximity to the DSB site but are found over a distance of up to 44 bp; (iii) a DSB site is either processed by HR into a targeted allele or by NHEJ repair into a variety of alleles containing undirected frame-shift or loss-of-codon mutations; and (iv) multiple alleles of both types may occur in a single founder and can be transmitted via the germ line. Taken together, these results provide proof of principle that TALEN and ODNs provide a versatile tool for the directed mutagenesis of the mouse germ line. The occurrence of seven loci processed by NHEJ is equal to a mutagenesis rate of 6%, whereas the presence of three homologous recombined alleles in two founders indicate an HR rate in the range of 2%.
Fig. 2.
Creation of a Rab38 chocolate mutation with TALENs and ODNs. (A) Outline of TALEN-mediated gene targeting in one-cell embryos. Upon TALENRab38 and ODN microinjections (day 1), founders and progeny (4-wk-old) heterozygous for _Rab38_IDG-Cht alleles are obtained within 18 wk. (B) Insertion of a G19V mutation into the first exon of Rab38. The position of the targeting oligonucleotide (ODNIDG-Cht); the structure of the Rab38 wild-type (wt) locus and the targeted _Rab38_IDG-Cht allele; the location of the TALEN-binding sites, the Rab38 5′-hybridization probe, PCR primers P-for and P-rev, SexAI (S), and ApaLI (A) restriction sites; and the size of genomic fragments are shown. (C) Comparison of codons of _Rab38_wt, _Rab38_cht, and ODNIDG-Cht. Nucleotides and amino acids that deviate from wild type are shown in red color. (D) Sequence comparison within the first exon of _Rab38_wt and ODNIDG-Cht and PCR products amplified with primers P-for and P-rev from tail DNA of founder Rc6, which includes the G19V replacement in codon 19, and its pups Rc6-4, Rc6-3, and Rc6-14; of founder Rc7, founder Rc1, and its pup Rc1-6; and of founders Rc2 to Rc5. The position of codon 19 and the TALENRab38-binding sites are indicated, nucleotides deviating from wild type (green background) are shown in red letters on yellow background, deleted nucleotides are dashed, and sequence insertions are underlined. The genotype describes the mutant allele as a product of HR- or NHEJ-associated deletion (Δ). (E) Southern blot analysis of ApaLI-digested tail DNA of founder Rc6 and of 19 Rc6 derived pups using the Rab38 5′-hybridization probe. Pups with Rab38 alleles that were sequence-analyzed (D) are highlighted in color. (F) Southern blot analysis of SexAI-digested tail DNA of founder Rc6 and of 19 Rc6-derived pups. The presence of a _Rab38_IDG-Cht allele is indicated by a 2.1-kb SexAI fragment. (G) Southern blot analysis of ApaLI-digested tail DNA from founder Rc1 and its offspring Rc1-3 to Rc1-11. (H) Southern blot analysis of ApaLI-digested tail DNA from founders Rc2, Rc3, Rc4, Rc5, and Rc7. Founders Rc3, Rc4, and Rc5 exhibit smaller bands, as expected from the sequence analysis of exon 1 (D), indicating additional deletion events. WT control, C57BL/6 DNA; _Rab38_Cht/Cht, DNA from a homozygous (ApaLI resistant) _Rab38_IDG-Cht mouse generated with ZFNs (8).
Correction of the chocolate Mutation in _Rab38_cht Mice.
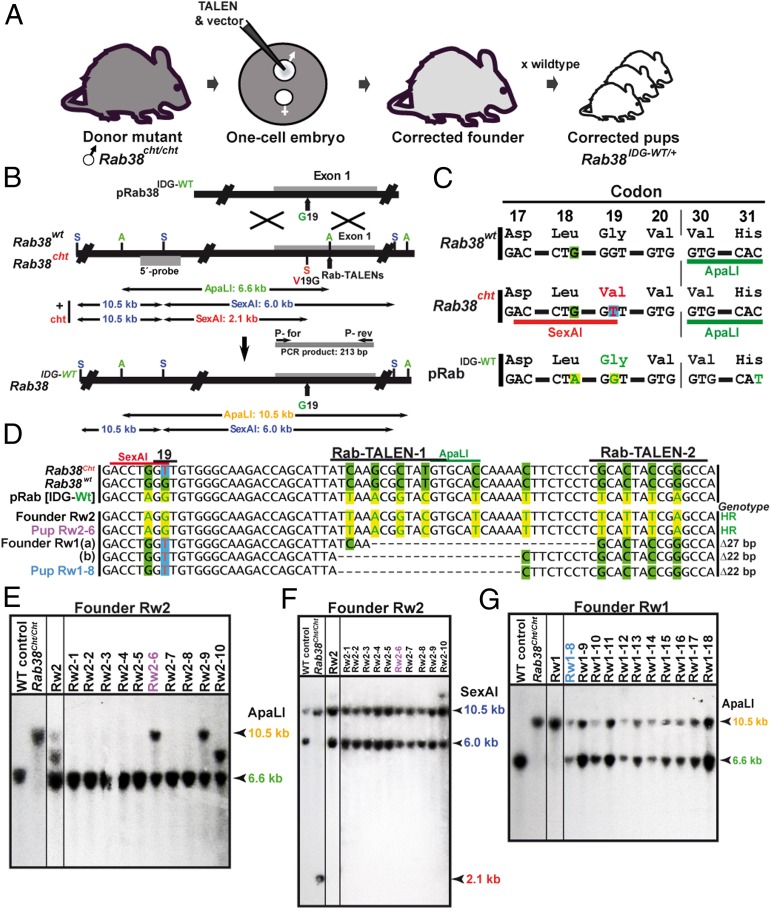
In a second experiment, we used TALENRab38 to correct the _Rab38_cht allele in the genome of chocolate mutants (18) using a repair template with a wild-type sequence. By this means, the phenotypic contribution of a single missense mutation can be evaluated within a complex genetic background. TALENRab38 mRNAs and the correction targeting vector pRab38IDG-WT were microinjected into the male pronucleus of one-cell embryos fertilized by _Rab38_cht/cht males (Fig. 3_A_). The resulting offspring were screened for founders harboring a corrected paternal Rab38IDG-WT allele and further mated to establish a colony of reverted mutants. The pRab38IDG-WT-targeting vector reverses the valine codon 19 of _Rab38_cht into a wild-type glycine codon (V19G) and eliminates a SexAI restriction site (Fig. 3_C_). In addition, we introduced a neutral nucleotide replacement into codon 18, serving as a unique identifier, and included further silent exchanges within the TALENRab38-binding region that also remove an ApaLI site (Fig. 3 B and C). A fully corrected Rab38IDG-WT allele is characterized by the absence of the SexAI and ApaLI restriction site within exon 1. Because editing of Rab38 by HR or NHEJ will both lead to the loss of the ApaLI site, the offspring derived from microinjections were first genotyped by ApaLI digestion of PCR products covering exon 1. Digestion-resistant PCR products were cloned and analyzed by sequencing. Among 50 offspring derived from embryo injections of TALENRab38 and pRab38IDG-WT, we identified two mutant founders harboring three modified Rab38 alleles (Table S1). Founder Rw2 contained, as shown by sequencing of PCR products, a fully recombined _Rab38_IDG-WT allele that includes the V19G reversion and all neutral replacements within the TALEN-binding region (Fig. 3_D_). A Southern blot analysis of Rw2 tail DNA showed a weak, but diagnostic, 10.5-kb ApaLI band besides a 6.6-kb fragment derived from the maternal wild-type locus. The presence of an additional, unexpected 8.5-kb band indicates further processing of the paternal Rab38cht allele in a fraction of cells (Fig. 3_E_). In founder Rw2, gene correction occurred by HR with the paternal _Rab38_cht allele, as shown by the absence of the diagnostic 2.1-kb SexAI fragment (Fig. 3_F_). To achieve germ-line transmission of the corrected allele, 10 pups derived from matings of founder Rw2 were genotyped by Southern blot analysis of ApaLI- or SexAI-digested tail DNA. Two pups (Rw2-6, -9) showed the 10.5-kb ApaLI band, demonstrating germ-line transmission of the _Rab38_IDG-WT allele (Fig. 3_E_). Pup Rw2-6 was further characterized by PCR and sequence analysis and confirmed the identity of its _Rab38_IDG-WT allele to the founder Rw2 (Fig. 3_D_). None of the pups derived from founder Rw2 showed a 2.1-kb SexAI band characteristic for the _Rab38_cht allele (Fig. 3_F_). In contrast, founder Rw1 harbored, as shown by PCR and sequence analysis, two NHEJ-processed Rab38 alleles characterized by the in-frame deletion of 27 bp [Rw1(a)] or the loss of 22 bp [Rw1(b)] (Fig. 3_D_) within the TALENRab38-binding region. Both of these alleles were derived from the paternal _Rab38_cht allele, as shown by the presence of the G19V mutation. Nevertheless, the Southern blot analysis of tail DNA from founder Rw2 revealed the absence of a wild-type 6.6-kb ApaLI band (Fig. 3_G_), indicating that also the maternal Rab38+ allele was processed by NHEJ, but this allele was not detected by our PCR analysis. Mating of founder Rw1 showed the germ-line transmission of NHEJ-processed Rab38 alleles to all of 11 pups, as indicated by the presence of 10.5-kb ApaLI band (Fig. 3_G_). Pup Rw1-8 was further characterized by PCR and sequence analysis and confirmed the presence of an Rw1(b) allele (Fig. 3_D_). Taken together, we demonstrate the reversion of a missense mutation in the paternal allele of an early embryo and the establishment of gene corrected progeny. A single targeted allele was identified, indicating a HR rate of ∼2%, similar to the introduction of replacement mutations.
Fig. 3.
Correction of the chocolate mutation in _Rab38_cht mice. (A) Outline of TALEN-mediated gene correction. (B) Correction of the G19V mutation in the first exon of the _Rab38_cht allele. The targeting vector (pRab38IDG-WT), the _Rab38_wt and _Rab38_cht loci, the corrected _Rab38_IDG-WT allele, the TALEN-binding sites, the Rab38 5′-hybridization probe, PCR primers, SexAI (S) and ApaLI (A) sites, and the size of genomic fragments are shown. (C) Comparison of codons of _Rab38_wt, _Rab38_cht, and pRab38IDG-WT. Sequences deviating from wild type in _Rab38_cht are shown in red, and residues reverted in pRab38IDG-WT are shown in green. (D) Sequence comparison within the first exon of _Rab38_cht, _Rab38_wt, and pRab38IDG-WT and of PCR products from tail DNA of founder Rw2, its pup Rw2-6, and of founder Rw1 and its pup Rw1-8. Nucleotides deviating from wild type (green background) are shown in green letters on yellow background, the G19V mutation is shown in red on blue background, and deleted nucleotides are shown as dash. (E) Southern blot analysis of ApaLI-digested DNA of founder Rw2 and of 10 pups using the 5′-hybridization probe. Rw2 is mosaic for a reverted _Rab38_IDG-WT allele, transmitted to pups Rw2-6 and Rw2-9, indicated by a 10.5-kb band and a rearranged allele at 8.5 kb. (F) Southern blot analysis of SexAI-digested DNA of founder Rw2 and its pups that do not show the 2.1-kb band predicted from a paternal _Rab38_cht allele. (G) Southern blot analysis of ApaLI-digested DNA from founder Rw1 and its pups. WT control, C57BL/6 DNA; _Rab38_cht/cht, DNA from a homozygous (ApaLI resistant) _Rab38_IDG-Cht mouse generated with ZFNs (8).
Discussion
Gene targeting in one-cell embryos by TALENs and ODNs provides an advanced technology for the rapid and direct modification of the mouse genome. We show that the coinjection of TALEN mRNAs and a targeting ODN leads to the coincident generation of targeted alleles through HR and a variety of loss-of-function alleles by error-prone NHEJ repair. Among the 167 pups derived from embryo microinjection of TALENs for editing of Rab38, we obtained nine founders, eight of which harbored NHEJ-mediated loss-of-function alleles (1/21 pups; 4.8%) and three contained targeted alleles (1/56 pups; 1.8%). Because three of these founders represented a mosaic of multiple, up to three HR- or NHEJ-processed alleles, a total number of 14 edited Rab38 alleles were generated (Table S1). Within our settings, the injection of 120 embryos within a single day results in ∼30 live pups. Thus, 2 d of microinjection, resulting in 60 pups, are presently required to generate founders containing one targeted allele and three loss-of-function mutations. These rates are comparable to our previous results from embryo microinjection of ZFNs (7, 8). Both TALEN- and ZFN-induced DSBs are preferentially repaired by NHEJ, exceeding the rate of HR more than twofold under the present settings. The current low rate of HR events may be improved in the future by the inhibition of central components of the NHEJ pathway, as demonstrated by the inactivation of DNA ligase IV in Drosophila embryos (26). All of the four founders we tested for germ-line contribution could transmit one or more mutant alleles to 20–64% of their offspring, indicating that embryonic TALEN expression does not interfere with fertility and that a fraction of founders were not fully heterozygous but mosaic for the individual modified alleles. As we noticed earlier from embryo injections of ZFNs (7, 8), founders can either represent fully heterozygous mutants, harboring a single modified allele in all body cells or represent mosaic mutants that bear one or more mutations only in a fraction of cells. Fully heterozygous founders are obtained if HR occurs in the pronucleus before replication of the target gene, whereas after replication, the processing at only one chromatid results in a mosaic founder that contains the modified allele in half of its cells. ODNs were successfully used together with ZFNs to achieve targeted gene modifications in mammalian cell lines (27). Our result with ODNIDG-Cht demonstrates that ODNs, together with TALEN, induced DSBs present a versatile tool to create targeted replacements in the mouse genome. To evaluate the extent of HR-mediated sequence conversion we used an ODN of 144 residues that includes 12 nucleotide exchanges over a distance of 62 nucleotides. None of the three recombined alleles contained all mutations, but replacements occurred over a distance of 44, 40, or 22 bp and transferred up to 10 exchanges into the genome, preferentially in proximity to the DSB site. Therefore, we recommend positioning TALENs such that their spacer region is located close by the targeted nucleotides with an upper working distance of 40 bp. Because ODNs were also shown to mediate targeted sequence insertions and deletions in vitro, it will be interesting to further explore these applications in one-cell embryos. Our TALEN-Designer tool identifies TALEN target sites in genomic and coding sequences of the mouse at an average distance of 13–14 bp. Therefore, the entire sequence space of the mouse genome is accessible for targeted mutagenesis by TALENs together with synthetic ODNs. Compared with an established ZFN, 70% of our TALENs showed an equal or higher nuclease activity such that most target sites can be covered by the construction of a single vector pair. Whether all nucleases that meet this activity standard will lead to equally efficient recombination in vivo requires further experimentation to explore the potential influence of the chromosomal context on target accessibility. Using a TALEN against the Fus gene, we observed a targeting rate comparable to TALENRab38 in mouse embryos, and a variety of target sites were shown to be competent for TALEN-induced NHEJ repair in mouse, rat, and pig embryos (15, 17, 28). These findings suggest that a substantial number of chromosomal target sites are accessible to nuclease-induced targeting in embryos, but the potential fraction of resistant sites, compromising the generation of mutants, remains to be defined. In recent years, an increasing number of missense and nonsense mutations are discovered, by high-throughput genomic analysis, to cause or contribute to human Mendelian or complex diseases (3–5). Genetic mouse models, which reproduce such disease mutations, will be important to decipher disease mechanisms and for development of therapeutic interventions. This demand can be well matched by the TALEN and ODN technology that requires only 1–2 wk of preparatory work for TALEN construction and ODN synthesis and 2 d of embryo injections and delivers the first heterozygous mutants within 18 wk. In addition, the use of one-cell embryos enables the creation and reversion of genetic modifications in the genetic background of any wild-type or mutant mouse strain. Vice versa, it is possible, as exemplified by our Rab38IDG-WT allele, to reverse a mutation for evaluation of its phenotypic contribution within a complex disease background [e.g., in nonobese diabetic (NOD) mice]. Taken together, we show that TALENs, combined with ODNs, provide an advanced tool for targeted mutagenesis directly in the mouse germ line. Minimal effort and shortest time to the delivery of mutant mice are key features of this technology that will catalyze the generation of future in vivo models to study human diseases.
Materials and Methods
Nuclease Expression Vectors.
The _Rab38_-specific ZFN expression vectors pCAG-Rab-ZFN-L and -R were described previously (8). For the expression of TALENs in mammalian cells, we designed the generic expression vector pCAG-TALEN, which contains a CAG hybrid promoter region and a transcriptional unit comprising a sequence coding for the N-terminal amino acids 1–176 of TAL nucleases, located upstream of a pair of BsmBI restriction sites. Further details on TALEN construction, sequences, activity, TALEN reporters, and activity testing are given in SI Materials and Methods.
Microinjection of One-Cell Embryos and Southern Blot Analysis.
The injection of TALEN mRNA and targeting molecules and Southern blotting was performed as described previously for ZFNs (7, 8). See SI Materials and Methods for further details.
PCR, Digestion, and Sequence Analysis.
To further analyze the Rab38 alleles, we amplified exon 1 from mice using the PCR primer pair P-forward (P-for) (5′-GGCCTCCAGGATGCAGACACC-3′) and P-reverse (P-rev) (5′-CCAGCAATGTCCCAGAGCTGC-3′). Amplification was performed using Herculase II polymerase (Agilent Technologies) in 25-µL reactions with 30 cycles of 95 °C for 20 s, 60 °C for 15 s, and 72 °C for 15 s. Afterward, the PCR products were directly digested with 10 units of ApaLI and analyzed on agarose gels. The undigested fragments, which correspond to recombined alleles, were extracted with the Qiaquick Gel Extraction Kit (Qiagen), cloned into pSC-B (Stratagene), and sequenced.
Supplementary Material
Supporting Information
Acknowledgments
We thank R. Kneuttinger, P. Kunath, A. Krause, A. Tasdemir, and S. Weidemann for technical assistance; B. Lentes for technical server and network support; M. Seabra for _Rab38_cht mice; and Lillian Garret for critically reading the manuscript. This work was supported by the European Union within the EUCOMM project (LSHG-CT-2005-018931) (to W.W.), the German Ministry of Education and Research within the projects DIGTOP (01GS0858) (to W.W. and R.K.), and the German Mouse Clinic (01GS0850) (to W.W. and M.H.A.) of the NGFN-Plus program, and by the European Union Seventh Framework Programme (FP7/2007-2013) (to O.O.) under Grant 251864.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Capecchi MR. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6(6):507–512. doi: 10.1038/nrg1619. [DOI] [PubMed] [Google Scholar]
- 2.Wefers B, Wurst W, Kühn R. Design and generation of gene-targeting vectors. Curr Protoc Mouse Biol. 2011;1(1):199–211. doi: 10.1002/9780470942390.mo100179. [DOI] [PubMed] [Google Scholar]
- 3.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12(11):745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 4.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13(8):565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 5.Bras J, Guerreiro R, Hardy J. Use of next-generation sequencing and other whole-genome strategies to dissect neurological disease. Nat Rev Neurosci. 2012;13(7):453–464. doi: 10.1038/nrn3271. [DOI] [PubMed] [Google Scholar]
- 6.Collins FS, Rossant J, Wurst W. International Mouse Knockout Consortium A mouse for all reasons. Cell. 2007;128(1):9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 7.Meyer M, de Angelis MH, Wurst W, Kühn R. Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci USA. 2010;107(34):15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer M, Ortiz O, Hrabé de Angelis M, Wurst W, Kühn R. Modeling disease mutations by gene targeting in one-cell mouse embryos. Proc Natl Acad Sci USA. 2012;109(24):9354–9359. doi: 10.1073/pnas.1121203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23(8):967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 10.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 11.Bogdanove AJ, Voytas DF. TAL effectors: Customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 12.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326(5959):1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 13.Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 14.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung YH, et al. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31(1):23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 16.Reyon D, et al. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30(5):460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tesson L, et al. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 18.Loftus SK, et al. Mutation of melanosome protein RAB38 in chocolate mice. Proc Natl Acad Sci USA. 2002;99(7):4471–4476. doi: 10.1073/pnas.072087599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wasmeier C, et al. Rab38 and Rab32 control post-Golgi trafficking of melanogenic enzymes. J Cell Biol. 2006;175(2):271–281. doi: 10.1083/jcb.200606050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oiso N, Riddle SR, Serikawa T, Kuramoto T, Spritz RA. The rat Ruby ( R) locus is Rab38: Identical mutations in Fawn-hooded and Tester-Moriyama rats derived from an ancestral Long Evans rat sub-strain. Mamm Genome. 2004;15(4):307–314. doi: 10.1007/s00335-004-2337-9. [DOI] [PubMed] [Google Scholar]
- 21.Di Pietro SM, Dell’Angelica EC. The cell biology of Hermansky-Pudlak syndrome: Recent advances. Traffic. 2005;6(7):525–533. doi: 10.1111/j.1600-0854.2005.00299.x. [DOI] [PubMed] [Google Scholar]
- 22.Lopes VS, Wasmeier C, Seabra MC, Futter CE. Melanosome maturation defect in Rab38-deficient retinal pigment epithelium results in instability of immature melanosomes during transient melanogenesis. Mol Biol Cell. 2007;18(10):3914–3927. doi: 10.1091/mbc.E07-03-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osanai K, et al. Altered lung surfactant system in a Rab38-deficient rat model of Hermansky-Pudlak syndrome. Am J Physiol Lung Cell Mol Physiol. 2010;298(2):L243–L251. doi: 10.1152/ajplung.00242.2009. [DOI] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39(12):e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bozas A, Beumer KJ, Trautman JK, Carroll D. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics. 2009;182(3):641–651. doi: 10.1534/genetics.109.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen F, et al. High-frequency genome editing using ssDNA oligonucleotides with zinc-finger nucleases. Nat Methods. 2011;8(9):753–755. doi: 10.1038/nmeth.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson DF, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information