Ocean currents shape the microbiome of Arctic marine sediments (original) (raw)
Abstract
Prokaryote communities were investigated on the seasonally stratified Alaska Beaufort Shelf (ABS). Water and sediment directly underlying water with origin in the Arctic, Pacific or Atlantic oceans were analyzed by pyrosequencing and length heterogeneity-PCR in conjunction with physicochemical and geographic distance data to determine what features structure ABS microbiomes. Distinct bacterial communities were evident in all water masses. Alphaproteobacteria explained similarity in Arctic surface water and Pacific derived water. Deltaproteobacteria were abundant in Atlantic origin water and drove similarity among samples. Most archaeal sequences in water were related to unclassified marine Euryarchaeota. Sediment communities influenced by Pacific and Atlantic water were distinct from each other and pelagic communities. Firmicutes and Chloroflexi were abundant in sediment, although their distribution varied in Atlantic and Pacific influenced sites. Thermoprotei dominated archaea in Pacific influenced sediments and Methanomicrobia dominated in methane-containing Atlantic influenced sediments. Length heterogeneity-PCR data from this study were analyzed with data from methane-containing sediments in other regions. Pacific influenced ABS sediments clustered with Pacific sites from New Zealand and Chilean coastal margins. Atlantic influenced ABS sediments formed another distinct cluster. Density and salinity were significant structuring features on pelagic communities. Porosity co-varied with benthic community structure across sites and methane did not. This study indicates that the origin of water overlying sediments shapes benthic communities locally and globally and that hydrography exerts greater influence on microbial community structure than the availability of methane.
Keywords: Arctic, biogeography, methane, microbiome, sediment, water
Introduction
Many studies have characterized the composition and ecological role of microorganisms in methane-containing marine sediments around the world (Inagaki et al., 2006; Heijs et al., 2007; Parkes et al., 2007; Pernthaler et al., 2008; Wegener et al., 2008; Harrison et al., 2009; Hamdan et al., 2012). These works assist in understanding the biological controls on oceanic methane and may contribute to establishing baselines to monitor change in marine sediments. This may be of particular importance in the Arctic Ocean due to its sensitivity to climate change and abundance of methane (Kvenvolden et al., 1993; McGuire et al., 2009).
Methane biogeochemistry in marine sediment involves ancient enzymatic pathways (for example, sulfate reduction, anaerobic methanotrophy), and geographically cosmopolitan microorganisms (Battistuzzi et al., 2004). Along with sulfate reducers, methanogens and methanotrophs, the Japan Sea candidates, Obsidian pool candidates and Chloroflexi, all of which have unclear biogeochemical roles, are routinely observed in high abundance in methane-containing sediments (Orphan et al., 2001; Webster et al., 2004; Inagaki et al., 2006; Liao et al., 2009; Hamdan et al., 2011). These observations suggest that communities in methane-containing sediments in the global ocean bear similar characteristics (Harrison et al., 2009; Knittel and Boetius, 2009). The phylogenetic and biogeochemical commonalities in these habitats imply that lineages have been conserved over time, possibly due to physical isolation and stable environmental conditions. An alternate explanation is recruitment of taxa from overlying waters. The latter agrees with the paradigm in microbial ecology that states ‘Everything is everywhere; but the environment selects' (Baas-Becking, 1934). Due to their high densities, small size and absence of migration barriers, the probability of global dispersal of aquatic prokaryotes is high (Finlay, 2002; Fenchel, 2003; Martiny et al., 2006; Caporaso et al., 2012). Therefore, study is needed to ask the question if the apparent cosmopolitan nature of microbiomes in methane-containing marine sediments is the result of dispersal of cells in the hydrosphere or co-evolution in isolation. With this question in mind, the dispersal of sediment microbiomes was investigated on the Alaska Beaufort Shelf (ABS).
The ABS was selected for this study because it receives input from the Pacific and Atlantic Oceans and has a resident water mass derived from river drainage and sea ice melting (McLaughlin et al., 2004). Pacific water enters the Arctic through the Bering Strait and flows east as a boundary layer along the ABS. Atlantic water enters the Arctic through the Fram Strait, and moves counter-clockwise along the ABS break. During summer, as ice melts and vertical mixing is reduced, water on the ABS is density stratified, resulting in a layer of Pacific origin water sandwiched between an Arctic surface layer, and a deep, cold and saline layer of Atlantic origin water (Carmack and Macdonald, 2002; McLaughlin et al., 2004; Shimada et al., 2005). The halocline (40–100 m) between surface and Pacific water and pycnocline (200 m) between Pacific and Atlantic water physically separates these masses (McLaughlin et al., 2004; Pickart, 2004). During stratified periods, free-living microorganisms are resistant to sinking through boundary layers (Jonas, 1997; Hamdan and Jonas, 2006) and will become entrained in a water mass (Agogue et al., 2011). Over an annual cycle, microorganisms from all layers may thermodynamically disperse and reach the sea floor. However, because planktonic prokaryotes are primarily free-living on the ABS, (Garneau et al., 2009) it is reasonable to propose that the stratified period provides an opportunity to observe hydrographic influences on sediment microbiomes. Another factor making the ABS a candidate for study is the abundant methane in shelf and slope sediments (Kvenvolden et al., 1993; McGuire et al., 2009), which permits examination of methane-containing sediments in contact with water of Atlantic or Pacific origin but in close geographic proximity. Because of these features, the influence of water dispersal, the physical environment and local geochemistry on microbiome structure can be parsed.
The goals of this work were to study how water column stratification impacts microbiome composition, and to determine which environmental factors correlate with benthic and pelagic microbiome structure. Data from this study and previous works on continental margins in the Pacific Ocean (Hamdan et al., 2011, 2012) were used in a comparative assessment of microbial communities to address the role of the hydrosphere in shaping benthic microbiomes and distributing phylotypes to methane-containing sediments.
Materials and methods
Sample collection
Sediments were collected with a piston corer (PC) described in Hamdan et al. (2012). Cores were sampled at 20–70 cm intervals. For methane analysis, 2-cm holes were drilled into PC liners and 3-ml plugs were transferred to gas-tight 20 ml vials. Temperature was measured simultaneously. Cores were split horizontally, and sediment for genetic and gravimetric porosity analyses was taken from one half and stored at −80 and −20 °C, respectively. Pore-water was Rhizon extracted (Rhizosphere Research Products, Wageningen, NL, USA) from the other half.
Water samples and hydrodynamic profiles of temperature, salinity, density and dissolved oxygen (DO) were obtained with a Seabird 911 CTD rosette fitted with 10 l Niskin bottles. For methane analysis, water was placed in 20 ml vials and a 4-ml headspace of nitrogen was used for analysis. A minimum of 2 l of water for phylogenetic analysis was filtered through Sterivex filters (Millipore, Billerica, MA, USA). Remaining water was removed and filters were sealed, and frozen at −80 °C.
Multitag pyrosequencing and phylogenetic analysis
Genomic DNA was extracted from sediment and filters using the FastDNA SPIN kit for soil (MP Biomeicals Inc., Santa Ana, CA, USA). Filter capsules were cracked and the filter was removed aseptically and placed in the Lysing tubes. DNA was visualized on a 1% agarose gel with ethidium bromide and diluted with diethylpyrocarbonate (DEPC) treated water. Approximately 10 ng of DNA was used as PCR template. Prior to multitag pyrosequencing (MTPS), samples were analyzed by length heterogeneity-PCR (LH-PCR) as a quality control for linear amplification and to normalize PCR yield before pooling. Amplification of V1 and V2 of the small subunit rRNA gene was performed using the primers 6-FAM-27F and 355R for bacteria and 1HK and 589R for archaea (Hamdan et al., 2011, 2012). Universal archaeal primers UA571F and UA1204R were evaluated; however, limited product was obtained. Positive (Escherichia coli and Sulfolobus solfataricus) and negative controls (DEPC water) accompanied reactions. The PCR mixture and run conditions are described in Hamdan et al. (2012). Product was visualized on an agarose gel with ethidium bromide and quantitated. Products were diluted, mixed with ILS-600 (Promega, Madison, WI, USA) and HiDi formamide, and analyzed on an ABI3130.
For MTPS, 60 forward bacterial primers (27F) tagged on the 5′ end with a 8-base barcode along with the emulsion PCR-A titanium adapter were used with a reverse fusion primer (355R) and emulsion PCR-B titanium adapter (Hamdan et al., 2011, 2012). Thirty forward archaeal barcoded primers (1HKF) and a reverse fusion primer (589R) with titanium adapters were also synthesized. Samples were amplified for 30 cycles with a 10-min final extension step (Hamdan et al., 2012) using barcoded primers. Amplified product was visualized and quantitated, diluted for normalization and pooled. Pooled product was quantitated by Quant-iT PicoGreen assay (Invitrogen, Grand Island, NY, USA) on a Beckman Coulter DTX 880 and diluted for use in 454 emulsion PCR. Sequencing was performed on the Roche (Branford, CT, USA) GS-FLX Jr. Data were analyzed by the GS run browser with signal processing for shotgun or paired end and processed in the QIIME pipeline (qiime.sourceforge.net). Sequences <200 bp long, with quality scores <25, with multiple Ns and chimeras (identified with ChimeraSlayer) were excluded. Operational taxonomic units (OTUs) were clustered with CD-HIT at 97% similarity. Sequences were aligned with PyNAST against the Greengenes core set and phylogeny was assigned with RDP. BLAST searches against GenBank were also performed to provide information on the numerous OTUs that were not classified by RDP. The abundance of OTUs was annotated with the number of reads and relative abundance was calculated using the total reads from each barcode.
Geochemical analysis
Methane was determined with a Shimadzu 14-A gas chromatograph and flame ionization detector (Hamdan et al., 2011). Equilibration concentrations for water were calculated as described in Wiesenburg and Guinasso (1979). Chloride concentration measured with a Dionex DX-120 ion chromatograph was used to calculate salinity in sediments. Phosphate, nitrate and nitrite were determined according to Murphy and Riley (1962) and Grasshoff et al. (1983).
Statistical analysis
UniFrac was used to quantify the relatedness of phylogenies (Lozupone et al., 2006). The weighted UniFrac metric was computed, and values were Bonferroni corrected. PCoA was conducted on UniFrac distances to display results. MDS (multidimensional scaling analysis) and hierarchal cluster analysis were conducted on Bray-Curtis similarity data for LH-PCR results from this study and previous works (Hamdan et al., 2011, 2012) to sample display assemblages. MTPS and LH-PCR similarity data were applied to the Biota-Environmental matching procedure (Bio-Env, Primer-E), to determine the abiotic factors that co-varied with community structure. A geographic distance matrix was also used in Bio-Env. Bio-Env provides a rank correlation coefficient (ρ, high=1, low=0) to attribute covariance to specific variables. The similarity percentage (SIMPER, Primer-E) analysis was used to determine the sequences that contributed most to similarity within, and dissimilarity between groups of samples identified by water mass and sample type. SourceTracker (Knights et al., 2011) was used to identify sources and sinks of OTUs in samples. The analysis was conducted at a rarefaction depth of 1000. Results are reported as proportion estimates.
Results
Study area
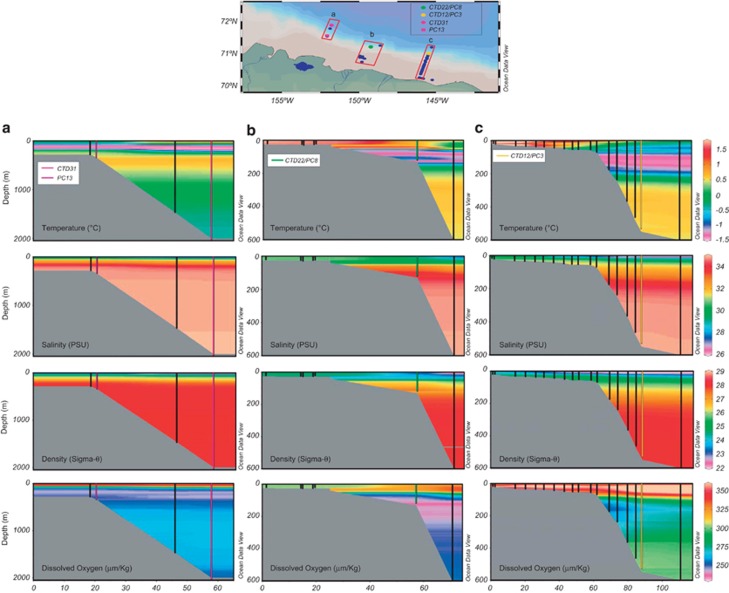
Samples were collected on board the USCGC Polar Sea along three transects located north and east of Barrow Alaska (Figure 1; Table 1). This area receives freshwater input primarily from the Coleville River (Figure 1), and secondarily from the Kuparuk and Meade Rivers. Seismic images identified areas with gas in the sediment, and the bottom simulating reflector, the seismic signature of gas below the hydrate stability zone. Sediments were collected from water depths ranging from 136 to 490 m (Table 1). Sediment samples discussed are from the upper 1 m of the sediment column from PC3, PC8 and PC13 so that hydrographic influences could be constrained. The age of sediments from PC13 and a core collected 1 km from PC8 was determined by radiocarbon analysis of bacterial FAMEs (fatty acid methyl esters) (Dougherty, 2012). FAMEs in the upper 1 m of the sediment column at these locations averaged 2518 and 3613 radiocarbon years, respectively, and the fraction modern was 0.63 and 0.73. Radiocarbon age increased and the fraction modern decreased significantly below 1 m depth at both sites. The observed fractions modern indicates recent organic matter deposition (Griffith et al., 2010). Radiocarbon ages are consistent with another study in the same region, which reported that sedimentation is controlled by current flow across the ABS (Darby et al., 2009).
Figure 1.
Hydrographic properties of the ABS. The upper map depicts the three regions sampled (a–c). Lower panels are contour plots of temperature, salinity, density and DO in areas (a–c) from the map. Water column locations used to construct contour plots but not used for discrete sample analysis are depicted as blue dots on the map and black vertical lines in a–c. Piston core (PC) and water column (CTD) discrete sampling locations are identified as colored dots on the map, and vertical lines in panels a–c. Contour plots and map were generated in Ocean Data View.
Table 1. Description of samples collected from the Alaska Beaufort Shelf during September 2009.
| Station ID | Date | Time (UTC) (hours) | Sample ID | Water mass | Water column depth (m) | Sediment depth (mbsf) | Max water depth (m) | Latitude (N) | Longitude (W) | Temp (°C) | Salinity (PSU) | Density (Sigma-Θ) | Methane (μℳ) | PO 4 (μℳ) | NO 2 (μℳ) | NO 3 (μℳ) | DO (μℳol _kg_−1) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Water column | |||||||||||||||||
| CTD12 | 20-Sep-09 | 1959 | CTD12.05a | Surface | 10 | — | 550 | 70.99 | 145.47 | −0.49 | 27.2 | 21.8 | 0.01 | 0.6 | 0.0 | 0.0 | 407.0 |
| CTD12.06 | Pacific | 150 | — | 550 | 70.99 | 145.47 | −1.40 | 33.0 | 26.6 | 0.01 | 1.8 | 0.3 | 12.0 | 287.5 | |||
| CTD12.07 | Atlantic | 316 | — | 550 | 70.99 | 145.47 | 0.56 | 34.7 | 27.8 | 0.02 | 1.0 | 0.0 | 13.6 | 291.9 | |||
| CTD12.08 | Atlantic | 526 | — | 550 | 70.99 | 145.47 | 0.53 | 34.8 | 27.9 | 0.00 | 0.9 | 0.0 | 13.2 | 301.3 | |||
| CTD22 | 23-Sep-09 | 0341 | CTD22.09 | Surface | 3 | — | 130 | 71.20 | 149.22 | 0.57 | 29.2 | 23.4 | 0.01 | 0.3 | 0.0 | 0.0 | 323.4 |
| CTD22.10 | Pacific | 60 | — | 130 | 71.20 | 149.22 | −1.13 | 32.8 | 26.3 | 0.02 | ND | ND | ND | 247.0 | |||
| CTD22.11 | Pacific | 115 | — | 130 | 71.20 | 149.22 | −0.57 | 34.2 | 27.5 | 0.05 | ND | ND | ND | 240.2 | |||
| CTD31 | 24-Sep-09 | 1622 | CTD31.12 | Surface | 5 | — | 2017 | 71.87 | 151.76 | 0.72 | 26.4 | 21.1 | 0.01 | ND | ND | ND | 327.3 |
| CTD31.13 | Atlantic | 1,000 | — | 2017 | 71.87 | 151.76 | −0.01 | 34.8 | 28.1 | 0.01 | ND | ND | ND | 264.3 | |||
| CTD31.14 | Atlantic | 2,017 | — | 2017 | 71.87 | 151.76 | −0.50 | 34.9 | 28.0 | 0.00 | ND | ND | ND | 253.1 | |||
| Sediment | |||||||||||||||||
| PC3 | 20-Sep-09 | 2319 | 3.59 | Atlantic influenced | — | 0.2 | 490 | 70.97 | 145.49 | −0.70 | 32.2b | 25.9 | 0.29 | ND | ND | ND | ND |
| 3.58 | Atlantic influenced | — | 0.5 | 490 | 70.97 | 145.49 | −0.40 | 32.5b | 26.1 | 0.44 | ND | ND | ND | ND | |||
| 3.57 | Atlantic influenced | — | 1.1 | 490 | 70.97 | 145.49 | −1.20 | 31.6b | 25.4 | 0.41 | ND | ND | ND | ND | |||
| PC8 | 22-Sep-09 | 0320 | 8.110 | Pacific influenced | — | 0.1 | 136 | 71.20 | 149.22 | 1.20 | 30.9b | 24.7 | 0.64 | ND | ND | ND | ND |
| 8.109 | Pacific influenced | — | 0.3 | 136 | 71.20 | 149.22 | 1.00 | 31.8b | 25.5 | 1.14 | ND | ND | ND | ND | |||
| 8.108 | Pacific influenced | — | 0.5 | 136 | 71.20 | 149.22 | 0.80 | 31.8b | 25.5 | 2.23 | ND | ND | ND | ND | |||
| 8.107 | Pacific influenced | — | 0.7 | 136 | 71.20 | 149.22 | 0.70 | 31.7b | 25.4 | 3.73 | ND | ND | ND | ND | |||
| PC13 | 24-Sep-09 | 0628 | 13.212 | Atlantic influenced | — | 0.2 | 342 | 71.55 | 152.06 | 0.90 | 32.5b | 26.0 | 2.94 | ND | ND | ND | ND |
| 13.211 | Atlantic influenced | — | 0.5 | 342 | 71.55 | 152.06 | 1.10 | 32.5b | 26.0 | 11.09 | ND | ND | ND | ND | |||
| 13.210 | Atlantic influenced | — | 0.7 | 342 | 71.55 | 152.06 | 0.50 | 32.3b | 25.9 | 27.15 | ND | ND | ND | ND | |||
| 13.209 | Atlantic influenced | — | 0.9 | 342 | 71.55 | 152.06 | 0.20 | 32.0b | 25.7 | 127.53 | ND | ND | ND | ND |
With the exception of PC13, sediment samples were obtained in proximity to water samples (Figure 1a). Although samples were not obtained from the water column directly above PC13, CTD data demonstrate that hydrodynamic properties in the upper 600 m of the water column at CTD31 were equivalent to that observed above PC13.
Determination of hydrographic water masses
Water masses were determined from depth profiles of temperature, salinity, density and DO according to McLaughlin et al. (2004) (Table 1; Figure 1). Arctic surface water (<40 m) at all sites had water temperatures <0.7 °C, salinity ranging from 26 to 29 PSU, and density <24 kg m−3 (Figure 1). A sharp increase in salinity was observed below ∼20 m at all stations concomitant with an increase in temperature and DO. The temperature maximum was between 20 and 50 m. Between 40 and 60 m strong gradients in temperature, DO and density were evident marking the upper boundaries of the Pacific layer. Between 50–115 m at CTD22, and 100–180 m at CTD12 and CTD31, the temperature minimum was observed in association with a secondary halocline and pycnocline denoting the depths of the Pacific water mass. Below these depths, at CTD12 and CTD31, salinity gradually increased to 34.8 PSU. A tertiary thermocline was observed between ∼200 and 300 m at CTD12 and CTD31 along with the DO minimum, marking the transition between Atlantic and Pacific water. Below 400 m at CTD12 and CTD31 temperature declined and salinity, DO and density moderately increased towards the sea floor. With these data, samples were placed in three categories indicative of their oceanic origin (Table 1) and sediments were coded according to their immediately overlying water mass.
Geochemistry and porosity
Sediment methane concentration was highest in PC13 where concentrations exceeded 100 μℳ (Table 1). In PC3, methane was <1 μℳ and up to 4 μℳ was observed in PC8. At all sites, water column methane was significantly lower than sediment concentrations. Water column methane was highest in CTD22 (0.05 μℳ). Nitrate, nitrite and phosphate data were available for CTD12 and CTD22. Phosphate and nitrite (<0.6 and 0.0 μℳ) were depleted in surface water at both sites (Table 1), and elevated (1.8 and 0.3 μℳ) at 150 m in CTD12. Nitrate was also depleted at the surface (0.0 μℳ), although it exceeded 12 μℳ deeper in CTD12. Sediment porosity ranged from 0.5% to 0.8% (Supplementary Table S1) and was lowest in samples from PC8.
Microbial diversity
On average, 4000 sequences were obtained per sample (Table 2). The number of OTUs and diversity was generally higher for bacteria than archaea. Sequence coverage varied substantially between bacteria and archaea, especially in sediment samples. The Chao 1 statistic was used to estimate the depth of coverage provided by MTPS with the understanding that the statistic scales with sequencing effort. According to the Chao 1 estimate, bacteria were under sequenced by 21–55%. Archaeal OTUs were near or exceeded the number predicted by the Chao1 estimate. The number of archaeal sequences per sample varied considerably (37–6526) and in general, more sequences were obtained from water than sediment. Bacterial sequences were more evenly distributed (∼1472 per sample) across sample types. Archaeal sequence abundance was low in surface water samples (<10 m) (Table 2), and template concentration in CTD12.05 was too low for MTPS. Diversity estimates in general were not significantly different between water and sediment. Sample 8.110 had the highest diversity (Table 2) and surface water samples had the lowest diversity.
Table 2. Descriptive statistics and diversity estimates for bacterial and archaeal sequences.
| Summary statistics | Count |
|---|---|
| Raw input sequences | 223 119 |
| Length outside bounds of 200 and 1000 | 20 212 |
| Ambiguous bases | 4296 |
| Missing quality score | 0 |
| Qual score below minimum of 25 | 6545 |
| Max homopolymer run exceeds limit of 6 | 2167 |
| Sequences written to selected samples | 87 008 |
| Raw length min/max/avg | 40/957/371 |
| Wrote length min/max/avg | 200/479/344 |
| Sample statistics |
| Sample ID | Water mass | Sequence count | OTUs/sample | Chao1 | Simpsons | Shannon | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacteria | Archaea | Bacteria | Archaea | Bacteria | Archaea | Bacteria | Archaea | Bacteria | Archaea | ||
| Water | |||||||||||
| CTD12.06 | Pacific | 3107 | 6526 | 477 | 226 | 758 | 181 | 0.04 | 0.12 | 4.58 | 3.23 |
| CTD12.07 | Atlantic | 1995 | 2424 | 364 | 167 | 666 | 192 | 0.02 | 0.05 | 4.68 | 3.72 |
| CTD12.08 | Atlantic | 2408 | 5625 | 488 | 258 | 778 | 189 | 0.02 | 0.05 | 4.99 | 3.82 |
| CTD22.09 | Surface | 2316 | 935 | 255 | 35 | 386 | 54 | 0.07 | 0.49 | 3.99 | 1.26 |
| CTD22.10 | Pacific | 2172 | 3641 | 408 | 176 | 697 | 169 | 0.05 | 0.06 | 4.48 | 3.56 |
| CTD22.11 | Pacific | 2212 | 8059 | 475 | 304 | 859 | 214 | 0.03 | 0.07 | 4.83 | 3.74 |
| CTD31.12 | Surface | 4578 | 63 | 331 | 20 | 419 | 17 | 0.07 | 0.14 | 4.04 | 2.43 |
| CTD31.13 | Atlantic | 3074 | 4059 | 551 | 210 | 836 | 201 | 0.02 | 0.05 | 5.04 | 3.79 |
| CTD31.14 | Atlantic | 2135 | 4109 | 573 | 157 | 1039 | 131 | 0.01 | 0.06 | 5.41 | 3.48 |
| Sediment | |||||||||||
| 3.59 | Atlantic | 2160 | 139 | 857 | 32 | 1471 | 41 | 0.00 | 0.10 | 6.19 | 2.77 |
| 3.58 | Atlantic | 3102 | 37 | 1006 | 17 | 1452 | 11 | 0.01 | 0.08 | 6.20 | 2.55 |
| 3.57 | Atlantic | 2011 | 515 | 726 | 52 | 1154 | 57 | 0.01 | 0.08 | 5.90 | 3.18 |
| 8.110 | Pacific | 2749 | 1092 | 1097 | 113 | 1600 | 47 | 0.01 | 0.02 | 6.30 | 4.13 |
| 8.109 | Pacific | 1271 | 178 | 354 | 28 | 595 | 32 | 0.02 | 0.08 | 4.96 | 2.83 |
| 8.108 | Pacific | 2648 | 941 | 459 | 35 | 734 | 38 | 0.03 | 0.11 | 4.65 | 2.65 |
| 8.107 | Pacific | 2899 | 128 | 601 | 12 | 822 | 13 | 0.02 | 0.21 | 5.26 | 1.85 |
| 13.212 | Atlantic | 1019 | 872 | 426 | 115 | 926 | 179 | 0.02 | 0.04 | 5.18 | 3.89 |
| 13.211 | Atlantic | 1033 | 370 | 475 | 70 | 1037 | 91 | 0.01 | 0.04 | 5.60 | 3.62 |
| 13.210 | Atlantic | 1339 | 1323 | 463 | 133 | 879 | 61 | 0.03 | 0.11 | 4.97 | 3.25 |
| 13.209 | Atlantic | 1472 | 272 | 458 | 39 | 914 | 51 | 0.02 | 0.08 | 5.20 | 2.92 |
| Average | 2285 | 2065 | 542 | 110 | 901 | 98 | 0.02 | 0.10 | 5.12 | 3.13 | |
| Total | 45 700 | 41 308 | 6204 | 901 |
Phylogenetic composition
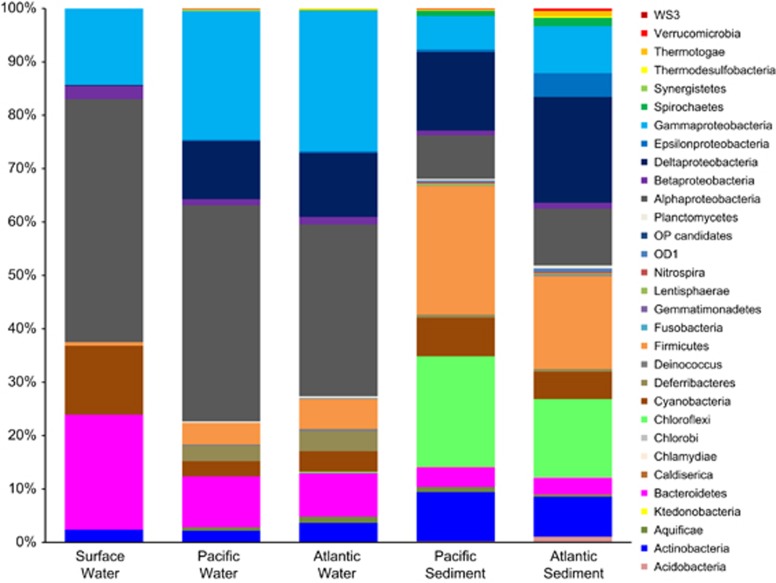
The most abundant pelagic bacterial group was the Alphaproteobacteria (Figure 2), with the majority associated with the SAR11 clade. These were most abundant in surface water and least abundant in Atlantic water (Figure 2; Supplementary Figure S1). Alphaproteobacteria were <10% of sediment OTUs. Gammaproteobacteria and Bacteroidetes were well represented in the water column (Figure 2). Bacteroidetes abundance decreased with depth and Gammaproteobacteria increased with depth (Figure 2; Supplementary Figure S1). Gammaproteobacteria were related to 11 orders and the Bacteroidetes were associated with four orders.
Figure 2.
Bacterial community composition in sample groups from the ABS identified by water mass (Atlantic, Pacific, surface) and sample type (sediment, water).
The Chloroflexi were 20% and 14% of OTUs in Pacific and Atlantic influenced sediments, respectively (Figure 2). The Firmicutes were more abundant in Pacific than Atlantic influenced sediments and averaged 5% in Pacific and Atlantic waters. Deltaproteobacteria were concentrated in Atlantic influenced sediments (∼20%), and were at least 10% of sequences from all sample groups except surface water. Epsilonproteobacteria were most abundant in Atlantic influenced sediments, and OTUs affiliated with the Sulfurovum and Sulfurimonas accounted for up to 13% of sequences in PC13 (Supplementary Figure S1).
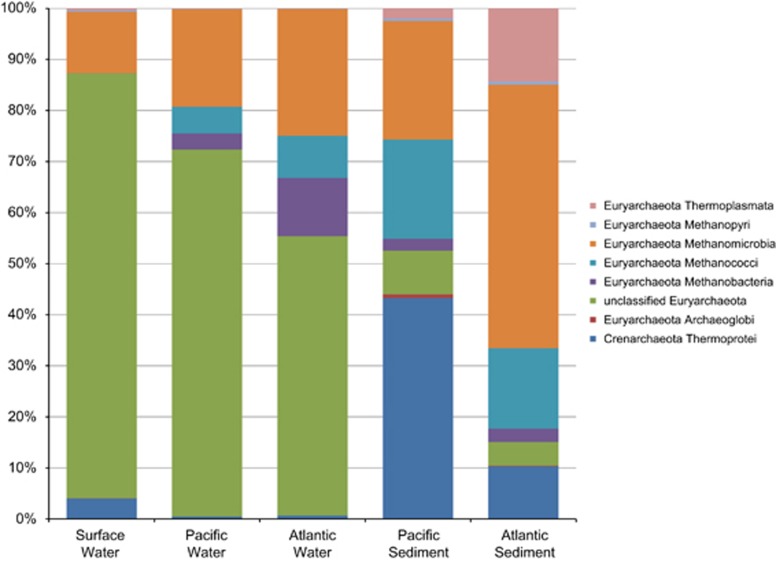
The Euryarchaeota dominated archaea in terms of OTU abundance (Figure 3) and diversity. Over half of water sequences fell into a category that could not be annotated beyond Euryarchaeota with either RDP or GenBank. Over 80% of surface waters sequences and over 400 OTUs fell in this category. Sequences related to the methanogenic Methanomicrobia were most abundant in Atlantic influenced sediments. In Pacific influenced sediments, Crenarchaeota related to the Thermoprotei were abundant, accounting for 43% of sequences (Figure 3). Crenarchaeota were ∼10% of Atlantic influenced sediment OTUs, and minimally abundant in pelagic samples (Supplementary Figure S2).
Figure 3.
Archaeal community composition in sample groups from the ABS identified by water mass (Atlantic, Pacific, surface) and sample type (sediment, water).
Community structure
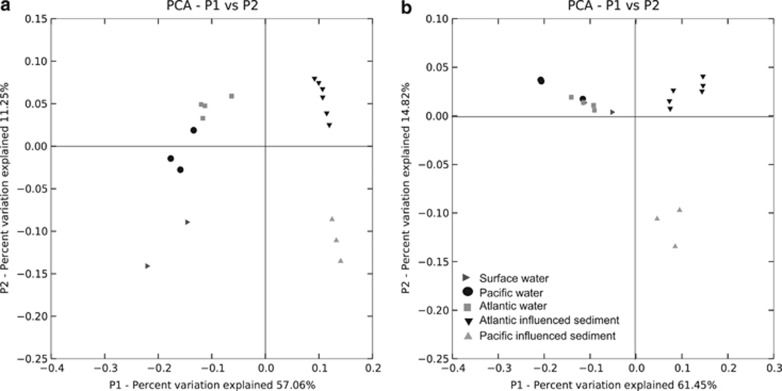
The weighted UniFrac metric was computed for bacteria and archaea separately. Bacterial samples distributed into five groups (Figure 4a). Surface samples from CTD12 and CTD31 were co-located. Pacific and Atlantic water samples grouped near each other, but formed separate groups. PC3 and PC13, from areas with immediately overlying Atlantic water clustered together. PC8 collected within the Pacific water mass formed a distinct cluster. There was no correlation between sediment and pelagic communities. A similar clustering pattern for archaea in sediment was observed (Figure 4b). The majority of pelagic archaeal samples were in a single group, however, Pacific samples CTD12.06 and CTD22.11 collected from similar depths (150 and 115 m, respectively; Table 1) grouped apart.
Figure 4.
Weighted UniFrac distances for sediment and water column Multitag Pyrosequenced data for bacteria (a) and archaea (b) from the ABS.
SIMPER analysis revealed that differences in bacterial and archaeal communities were explained by one or a few OTUs (Supplementary Tables S2 and S2). Dissimilarity was generally lower between groups of the same sample type (water or sediment) (Supplementary Table S3), and highest between sediment and surface water. Bacteria in Atlantic and Pacific sediment had the lowest dissimilarity relative to other comparisons (Supplementary Table S3) because both groups were influenced by two shared OTUs, a Chloroflexi and a Firmicutes (Supplementary Table S2). The former accounted for 18% of OTUs in Pacific sediments and controlled dissimilarity between this and other groups. The Firmicutes OTU was also important to similarity in Atlantic water (6–12% of sequences), and was influential in all dissimilarity comparisons with Atlantic water or sediment (Supplementary Table S3). However, similarity in Atlantic water was driven by an unclassified Deltaproteobacteria (Supplementary Table S2), which was the main discriminator between Atlantic sediment and water (Supplementary Table S3). A SAR11 affiliated OTU contributed most to bacterial similarity in Pacific and surface water samples. While the SAR11 OTU may explain the lower dissimilarity between Pacific and surface water (Supplementary Table S2), it drove dissimilarity in all other surface water pairings.
Archaeal within group similarity was lower than observed for bacteria (Supplementary Table S2). In all groups, unclassified marine isolates were most influential on SIMPER comparisons. The most abundant archaeal OTUs generally did not provide the greatest contribution to similarity, and unlike the bacteria, the most influential OTUs generally were not shared across groups. This was most evident in sediment groups, and explains their high dissimilarity to each other (Supplementary Table S3). An unclassified Euryarchaeote accounted for >10% of within group similarity in all pelagic groups (Supplementary Table S3). The abundance of this OTU in Pacific and Atlantic water explains their low dissimilarity to each other (Supplementary Table S3). Conversely, it drove dissimilarity between all pelagic and sediment pairings.
Community structuring features
Bio-Env analysis indicates that salinity (_ρ_>0.66) and density (_ρ_>0.62) were the principal structuring elements on bacterial and archaeal pelagic microbiomes. However, temperature and methane had minimal influence on pelagic community structure (ρ<0.01). A suite of sediment geochemical variables was used to determine the features that structured sediment communities (Supplementary Table S1). Bio-Env indicates that porosity, wt% carbon and % nitrogen, all provided _ρ_-values >0.40 for bacteria and archaea.
The impact of vertical and horizontal geographic distance on microbiome structure was addressed with Bio-Env. The distance in km from CTD31 was calculated for all sites. Geographic distance, the maximum water depths for sediment samples and the depth of collection for water samples were considered in the analysis. The test indicated that distance was not an important structuring element on pelagic microbiomes (_ρ_=0.007); however, water depth was (_ρ_=0.60, bacteria; _ρ_=0.50, archaea). Distance moderately co-varied with sediment archaea (_ρ_=0.30) and to a lesser extent, sediment bacteria (_ρ_=0.18). The maximum water depth co-varied consistently with sediment bacteria and archaea (_ρ_>0.37).
SourceTracker was used to address the influence of water masses on sediment microbiomes. CTD samples were source terms and sediment samples were sink terms, and the proportion of OTUs from Atlantic, Pacific and Surface water, and unknown sources were calculated. The source of OTUs to ABS sediments is largely unknown (Supplementary Figure S3). However, the influence of Atlantic water was evident on all Atlantic influenced sediments. Up to 9% of bacterial OTUs at PC3 and PC13 were attributed to Atlantic water. However, the Pacific influence was not evident in PC8 bacterial samples. In only one archaeal sample from PC8 a proportion of OTUs were related to Pacific water. The proportion of OTUs shared between water masses was also estimated (Supplementary Figure S4). Bacterial OTUs in surface and bottom water samples were dominated by their respective source. This was also the case for Archaeal OTUs with the exception of surface sample CTD31.12 in which 35% of OTUs were attributed to the underlying Pacific water mass. Most samples from intermediate depths had a mix of sources from adjacent water masses.
Discussion
Community composition
Few previous works have explored microbial diversity in Arctic pelagic communities (Bano et al., 2004; Galand et al., 2009, 2010; Kirchman et al., 2010) and none has examined sediment communities. Others have observed Proteobacteria including the SAR11 and Bacteroidetes to be prominent in Arctic surface waters (Galand et al., 2010; Kirchman et al., 2010) and abundant Deltaproteobacteria in deeper waters (Galand et al., 2010). The dominance of unclassified Euryarchaeota in Beaufort Shelf waters and minimal abundance of Crenarchaeota has also been observed (Galand et al., 2006, 2009). Despite the distinct water masses on the ABS, the pelagic archaeal community was relatively homogeneous. Recent works highlight inconsistent patterns in pelagic archaea around the world (Martin-Cuadrado et al., 2008) and the general lack of knowledge regarding archaeal population dynamics in the ocean. Although archaeal composition in stratified waters on the ABS requires further study, due to the consistency between this and previous studies of prokaryotes in the Arctic, these results are a robust characterization of ABS communities, which permits examination of the features that shape them.
Hydrographic influences on ABS communities
Prokaryotic community structure in open water is dictated by physical constraints on dispersal and environmental variability (Martiny et al., 2006; Follows et al., 2007). A previous study observed that water masses are a factor explaining the distribution of pelagic microbiomes in the Arctic ocean (Galand et al., 2010). Our data agree with that previous work where bacteria are concerned; however, no consistent separation by water mass was observed for archaea. The SourceTracker analysis suggests that there is a greater degree of mixing of archaeal OTUs in different water masses than for bacteria (Supplementary Figure S4). However, this may be a function of the relative homogeneity of abundant archaeal OTUs in pelagic samples.
Salinity is a significant structuring element on microbial composition in coastal environments (Crump et al., 2004; Auguet et al., 2010) and density gradients limit the physical dispersion of plankton (Galand et al., 2009). Salinity and density are related in open water, and while both impact physical dispersal, salinity also exerts physiological control on the expression of phylotypes. Bio-Env indicates that salinity and density most strongly co-varied with pelagic microbiome structure. As in previous works (Auguet et al., 2010), temperature, a physiological constraint on enzyme activity, was not an important structuring element on pelagic communities. Despite the presence of methane in the water column (Table 1) and methanogen-related OTUs (Figure 3), methane had no observable influence on community structure. Overall, these results suggest that physical isolation of water masses not physiological constraints is responsible for pelagic microbiome structure on the ABS and agree with the findings of Galand et al. (2010).
The sampling of nearby (<100 km) sediments influenced by different water masses provided a unique opportunity to determine the influence of hydrography on benthic microbiomes. SourceTracker provided some evidence for water deposition of OTUs in sediments, particularly at PC3 and PC13, and Bio-Env indicated the covariance between microbiome structure and water depth. A separate cluster of samples was evident for PC8 (Figure 4). Because PC8 was in physical contact with Pacific origin water, it may be assumed that hydrography was primarily responsible for this cluster. However, the influence of the in situ environment must be considered. SourceTracker reported that a lower proportion of OTUs in PC8 were derived from the water column relative to PC3 and PC13 (Supplementary Figure S3). The lower porosity in PC8 (Supplementary Table S1) compared with PC3 and PC13 and the strong covariance between sediment communities and porosity as indicated by Bio-Env suggests that porosity may physically influence OTUs deposition in sediment. However, because of the covariance between nitrogen and carbon with sediment community structure, a combined influence of permissible physical and geochemical conditions for OTU deposition and recruitment is most likely.
Microbiome structure in methane-containing marine sediments
Because this study indicates that hydrography influences nearby sediment microbiomes on the ABS, an attempt was made to explore this in locations at greater geographic distance. Microbiomes in methane-containing sediment from the mid-Chilean margin (Hamdan et al., 2012) and the Hikurangi margin (Hamdan et al., 2011) were studied previously. LH-PCR data from these previous works and the ABS were used in MDS and Bio-Env analysis to evaluate which features (that is, physical constraints, dispersal, in situ environment) are responsible for community composition across a larger geographic area. LH-PCR data were used because previous works employed older MTPS platforms and thus, data were not comparable. Samples from <1 m sediment depth from the Chilean and Hikurangi margins and ABS were considered.
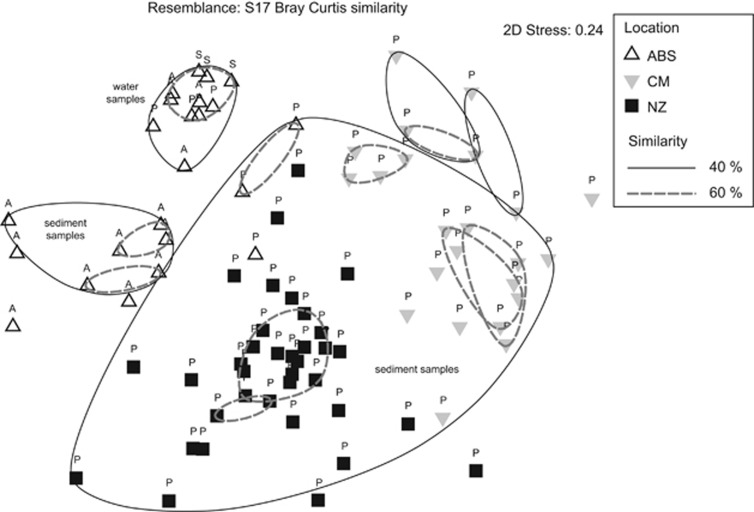
The MDS analysis revealed that samples from each individual area generally coordinated (Figure 5). Bio-Env suggests that small geographic distance between samples from individual areas co-varied well with community composition (_ρ_=0.47). As observed for ABS sediments, porosity and water depth were also important physical structuring features (_ρ_=0.36 and _ρ_=0.31, respectively). Carbon and nitrogen content could not be evaluated across study areas, but carbon co-varied with bacteria from the Hikurangi margin (Hamdan et al., 2011). Although samples from the Chilean margin, Hikurangi margin and PC13 contained methane, it did not co-vary with community composition (_ρ_=0.03).
Figure 5.
MDS of length heterogeneity-PCR data for samples collected during this study on the ABS and from the Chilean margin (CM) and the Hikurangi margin of New Zealand (NZ) during previous studies. Contours were generated by a hierarchal cluster analysis on Bray-Curtis similarity data. Samples are coded according to the water mass they were collected in (A—Atlantic, P—Pacific and S—Arctic surface).
All ABS water samples clustered apart from sediment samples (Figure 5). Atlantic influenced sediments formed a distinct cluster and three of four Pacific ABS sediment samples coordinated within the cluster containing most Pacific samples from the Chilean and Hikurangi margins despite the fact that ABS sites were >12 000 km from the other Pacific sites. Sample 8.110 from the surface of PC8 (Table 2) grouped with water samples from the ABS. This is intriguing as SourceTracker suggested that this sample had greater interaction with OTUs from the overlying water column than others from PC8.
This comparative analysis provides evidence that historical events and the contemporary environment both contribute to marine sediment microbiomes. Specifically, Bio-Env indicates that near proximity is the principal structuring feature on sediment microbiomes. However, SourceTracker results for the ABS suggest that marine sediments are a sink for OTUs in the water transiting them. The grouping of Pacific ABS samples with other Pacific samples (Figure 5) indicates that phylogenetic integrity of pelagic microbiomes is retained during dispersal across long distance. This agrees with studies that demonstrate high bacterial dispersal rates within water masses (Schauer et al., 2010). Bio-Env results for depth and porosity also indicate that local physical conditions influence microbiomes in methane-containing sediments to a greater extent than local geochemistry, possibly by regulating the rate of pelagic OTU deposition.
In keeping with ‘Everything is everywhere; but the environment selects' (Baas-Becking, 1934), this study indicates that OTUs are transported in oceanic masses over long distances, and local sediment features control recruitment from the water column. Because of the limited influence of local geochemistry, in particular methane concentration, this work indicates that co-evolution in isolation does not explain similarity in microbiome composition in methane-containing sediments. Instead, it suggests that dispersal and the physicochemical environment shape benthic microbiomes on a continual basis. This work is a first attempt at describing hydrographic influences on microbiomes in methane-containing sediments. In order to firmly establish the theories posited here, a comprehensive study and samples from a range of geographic distances are needed. This work demonstrates the importance of establishing ecological baselines for communities in marine sediments so that environmental change and any resulting effects that modify the structure of communities involved in methane regulation in the ocean can be monitored and assessed.
Acknowledgments
LJH was supported by the Naval Research Laboratory (NRL) Chemistry Division Young Investigator Program. Ship time was funded by the NRL platform support program. TT was supported by the Cluster of Excellence ‘The Future Ocean' funded by the German Research Foundation. We thank the captain and crew of USCGC Polar Sea and C Verlinden, R Downer and L Bryant for field assistance. We thank R Plummer, D Gustafson, the MITAS shipboard scientific party and the nutrient laboratory at Royal NIOZ for laboratory assistance. We thank M Dougherty, K Grabowski, T Lorenson and W Wood for helpful discussions.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
References
- Agogue H, Lamy D, Neal PR, Sogin ML, Herndl GJ. Water mass-specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol Ecol. 2011;20:258–274. doi: 10.1111/j.1365-294X.2010.04932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auguet JC, Barberan A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- Baas-Becking LGM.1934Geobiologie of inleiding tot de milieukunde (Geobiology as Introduction for Environmental Research) Vol. 18/19W. P. van Stockum & Zoon: The Hague, Netherlands; p263. [Google Scholar]
- Bano N, Ruffin S, Ransom B, Hollibaugh JT. Phylogenetic Composition of Arctic Ocean Archaeal Assemblages and Comparison with Antarctic Assemblages. Appl Environ Microbiol. 2004;70:781–789. doi: 10.1128/AEM.70.2.781-789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistuzzi F, Feijao A, Hedges SB. A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land. BMC Evolutionary Biol. 2004;4:44. doi: 10.1186/1471-2148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. The Western English Channel contains a persistent microbial seed bank. ISME J. 2012;6:1089–1093. doi: 10.1038/ismej.2011.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmack EC, Macdonald RW. Oceanography of the Canadian shelf of the Beaufort Sea: a setting for marine life. Arctic. 2002;55:29–45. [Google Scholar]
- Crump BC, Hopkinson CS, Sogin ML, Hobbie JE. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol. 2004;70:1494–1505. doi: 10.1128/AEM.70.3.1494-1505.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby DA, Ortiz J, Polyak L, Lund S, Jakobsson M, Woodgate RA. The role of currents and sea ice in both slowly deposited central Arctic and rapidly deposited Chukchi-Alaskan margin sediments. Glob Planet Change. 2009;68:56–70. [Google Scholar]
- Dougherty MR. Compound Specific Carbon Isotope Analysis for Biomarkers Associated with Methanotrophy in the Arctic. Doctoral Dissertation. Department of Chemistry and Biochemistry. University of Maryland: College Park; 2012. [Google Scholar]
- Fenchel T. Biogeography for bacteria. Science. 2003;301:925–926. doi: 10.1126/science.1089242. [DOI] [PubMed] [Google Scholar]
- Finlay BJ. Global dispersal of free-living microbial eukaryote species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. [DOI] [PubMed] [Google Scholar]
- Follows MJ, Dutkiewicz S, Grant S, Chisholm SW. Emergent biogeography of microbial communities in a model ocean. Science. 2007;315:1843–1846. doi: 10.1126/science.1138544. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Galand PE, Lovejoy C, Vincent WF. Remarkably diverse and contrasting archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat Microb Ecol. 2006;44:115–126. [Google Scholar]
- Galand PE, Potvin M, Casamayor EO, Lovejoy C. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 2010;4:564–576. doi: 10.1038/ismej.2009.134. [DOI] [PubMed] [Google Scholar]
- Garneau ME, Vincent WF, Terrado R, Lovejoy C. Importance of particle-associated bacterial heterotrophy in a coastal Arctic ecosystem. J Marine Syst. 2009;75:185–197. [Google Scholar]
- Grasshoff K, Ehrhardt M, Kremling K, Almgren T.1983Methods of Seawater Analysis2nd rev. and extended ednVerlag Chemie: Weinheim; xxviii, p419. [Google Scholar]
- Griffith DR, Martin WR, Eglinton TI. The radiocarbon age of organic carbon in marine surface sediments. Geochim Cosmochim Acta. 2010;74:6788–6800. [Google Scholar]
- Hamdan LJ, Gillevet PM, Pohlman JW, Sikaroodi M, Greinert J, Coffin RB. Diversity and biogeochemical structuring of bacterial communities across the Porangahau ridge accretionary prism, New Zealand. FEMS Microbiol Ecol. 2011;77:518–532. doi: 10.1111/j.1574-6941.2011.01133.x. [DOI] [PubMed] [Google Scholar]
- Hamdan LJ, Jonas RB. Seasonal and interannual dynamics of free-living bacterioplankton and microbially labile organic carbon along the salinity gradient of the Potomac River. Estuaries Coasts. 2006;29:40–53. [Google Scholar]
- Hamdan LJ, Sikaroodi M, Gillevet PM. Bacterial community composition and diversity in methane charged sediments revealed by multitag pyrosequencing. Geomicrobiol J. 2012;29:340–351. [Google Scholar]
- Harrison BK, Zhang H, Berelson W, Orphan VJ. Variations in archaeal and bacterial diversity associated with the sulfate-methane transition zone in continental margin sediments (Santa Barbara Basin, California) Appl Environ Microbiol. 2009;75:1487–1499. doi: 10.1128/AEM.01812-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijs S, Haese R, van der Wielen P, Forney L, van Elsas J. Use of 16S rRNA gene based clone libraries to assess microbial communities potentially involved in anaerobic methane oxidation in a mediterranean cold seep. Microb Ecol. 2007;53:384–398. doi: 10.1007/s00248-006-9172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F, Nunoura T, Nakagawa S, Teske A, Lever M, Lauer A, et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments, on the Pacific Ocean Margin. Proc Natl Acad Sci USA. 2006;103:2815–2820. doi: 10.1073/pnas.0511033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas RB. Bacteria, dissolved organics and oxygen consumption in salinity stratified Chesapeake Bay, an anoxia paradigm. Am Zool. 1997;37:9. [Google Scholar]
- Kirchman DL, Cottrell MT, Lovejoy C. The structure of bacterial communities in the western Arctic Ocean as revealed by pyrosequencing of 16S rRNA genes. Environ Microbiol. 2010;12:1132–1143. doi: 10.1111/j.1462-2920.2010.02154.x. [DOI] [PubMed] [Google Scholar]
- Knights D, Kuczynski J, Charlson ES, Zaneveld J, Mozer MC, Collman RG, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–U107. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel K, Boetius A. Anaerobic oxidation of methane: progress with an unknown process. Annu Rev Microbiol. 2009;63:311–334. doi: 10.1146/annurev.micro.61.080706.093130. [DOI] [PubMed] [Google Scholar]
- Kvenvolden KA, Lilley MD, Lorenson TD, Barnes PW, Mclaughlin E. The Beaufort Sea Continental-Shelf as a Seasonal Source of Atmospheric Methane. Geophys Res Lett. 1993;20:2459–2462. [Google Scholar]
- Liao L, Xu X-w, Wang C-s, Zhang D-s, Wu M. Bacterial and archaeal communities in the surface sediment from the northern slope of the South China Sea. J Zhejiang University Sci B. 2009;10:890–901. doi: 10.1631/jzus.B0920181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. UniFrac—An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado AB, Rodriguez-Valera F, Moreira D, Alba JC, Ivars-Martinez E, Henn MR, et al. Hindsight in the relative abundance, metabolic potential and genome dynamics of uncultivated marine archaea from comparative metagenomic analyses of bathypelagic plankton of different oceanic regions. ISME J. 2008;2:865–886. doi: 10.1038/ismej.2008.40. [DOI] [PubMed] [Google Scholar]
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, Green JL, et al. Microbial biogeography: Putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- McGuire AD, Anderson LG, Christensen TR, Dallimore S, Guo LD, Hayes DJ, et al. Sensitivity of the carbon cycle in the Arctic to climate change. Ecol Monogr. 2009;79:523–555. [Google Scholar]
- McLaughlin FA, Carmack EC, Macdonald RW, Melling H, Swift JH, Wheeler PA, et al. The joint roles of Pacific and Atlantic-origin waters in the Canada Basin, 1997-1998. Deep Sea Res Part I. 2004;51:107–128. [Google Scholar]
- Murphy J, Riley JP. A modified single solution method for determination of phosphate in natural waters. Anal Chim Acta. 1962;26:31. [Google Scholar]
- Orphan VJ, Hinrichs KU, Ussler Iii W, Paull CK, Taylor LT, Sylva SP, et al. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl Environ Microbiol. 2001;67:1922–1934. doi: 10.1128/AEM.67.4.1922-1934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes RJ, Cragg BA, Banning N, Brock F, Webster G, Fry JC, et al. Biogeochemistry and biodiversity of methane cycling in subsurface marine sediments (Skagerrak, Denmark) Environ Microbiol. 2007;9:1146–1161. doi: 10.1111/j.1462-2920.2006.01237.x. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart RS. Shelfbreak circulation in the Alaskan Beaufort Sea: Mean structure and variability. J Geophys Res. 2004;109:C04024. [Google Scholar]
- Schauer R, Bienhold C, Ramette A, Harder J. Bacterial diversity and biogeography in deep-sea surface sediments of the South Atlantic Ocean. ISME J. 2010;4:159–170. doi: 10.1038/ismej.2009.106. [DOI] [PubMed] [Google Scholar]
- Shimada K, Itoh M, Nishino S, McLaughlin F, Carmack E, Proshutinsky A. Halocline structure in the Canada basin of the arctic ocean. Geophys Res Lett. 2005;32:L03605. [Google Scholar]
- Webster G, Parkes RJ, Fry JC, Weightman AJ. Widespread occurrence of a novel division of bacteria identified by 16S rRNA gene sequences originally found in deep marine sediments. Appl Environ Microbiol. 2004;70:5708–5713. doi: 10.1128/AEM.70.9.5708-5713.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener G, Shovitri M, Knittel K, Niemann H, Hovland M, Boetius A. Biogeochemical processes and microbial diversity of the Gullfaks and Tommeliten methane seeps (Northern North Sea) Biogeosciences. 2008;5:1127–1144. [Google Scholar]
- Wiesenburg DA, Guinasso NL. Equilibrium solubilities of methane, carbon-monoxide, and hydrogen in water and sea-water. J Chem Eng Data. 1979;24:356–360. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3