RNA pseudouridylation: new insights into an old modification (original) (raw)
. Author manuscript; available in PMC: 2014 Apr 1.
Published in final edited form as: Trends Biochem Sci. 2013 Feb 4;38(4):210–218. doi: 10.1016/j.tibs.2013.01.002
Abstract
Pseudouridine is the most abundant posttranscriptionally modified nucleotide in various stable RNAs of all organisms. Pseudouridine is derived from uridine via base-specific isomerization, resulting in an extra hydrogen bond donor that distinguishes it from other nucleotides. In eukaryotes, uridine-to-pseudouridine isomerization is catalyzed primarily by box H/ACA RNPs, ribonucleoproteins that act as pseudouridylases. When introduced into RNA, pseudouridine contributes significantly to RNA-mediated cellular processes. It was recently discovered that pseudouridylation can be induced by stress, suggesting a regulatory role for pseudouridine. It has also been reported that pseudouridine can be artificially introduced into mRNA by box H/ACA RNPs and that such introduction can mediate nonsense-to-sense codon conversion, thus demonstrating a new means of generating coding/protein diversity.
Pseudouridine is an abundant and distinctly modified nucleotide
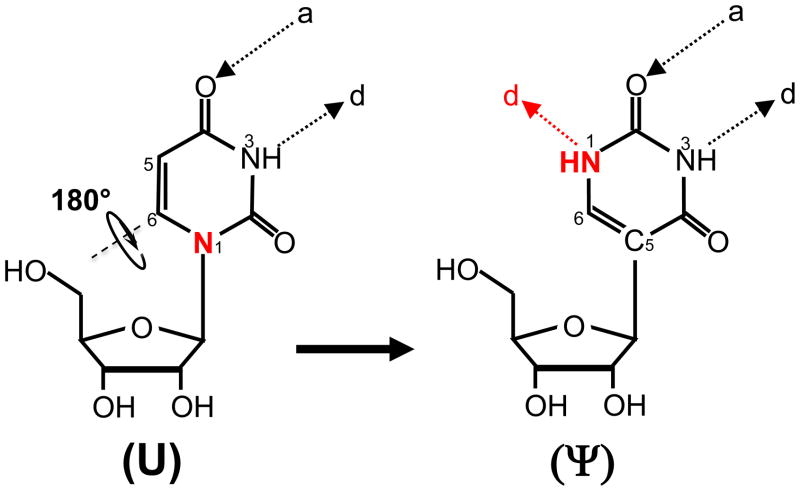
Among the ~100 different types of posttranscriptional modifications that have been identified in various RNAs of all organisms [1–3], pseudouridine (Ψ) was the first to be discovered and is by far the most abundant [4–6]. Ψ is a uridine isomer (5–ribosyluracil) formed via isomerization (Fig. 1). Initially, the nitrogen–carbon (N1–C1′) bond linking the uracil base (in uridine) to the sugar is broken. The liberated uracil base is then turned 180° along the N3–C6 axis, establishing a new carbon carbon–(C5–C1′) bond between the base and the sugar (Fig. 1). As a result, the modification creates an extra hydrogen bond donor at its non-Watson-Crick edge (Fig. 1), endowing Ψ with chemical properties distinct from those of uridine and all other known nucleotides.
Fig. 1.
Schematic representation of U-to-Ψ isomerization. The structures of uridine (U) and Ψ are shown. Ψ is derived from U through the isomerization reaction where the base is rotated 180° along the N3-C6 axis and the C5-C1′ bond forms. The nitrogen at position 1 in U and the extra hydrogen bond donor in Ψ are indicated (red). Hydrogen bond acceptor (a) and hydrogen bond donor (d) are also indicated.
It has been known for many decades that Ψ is present in a wide range of cellular RNAs, from tRNA [7–10] to rRNA [11–14] to a variety of small nuclear RNAs (snRNAs) [15–19]. Based on its abundant, widespread, and highly conserved nature, Ψ is believed to be functionally important. Still, for a long time the study of RNA pseudouridylation (and RNA modifications in general) was hampered by a lack of appropriate pseudouridylation assays and effective experimental systems. In the past ~15 years, however, several labs have made substantial progress towards developing convenient, yet sensitive, modification assays and experimental systems that have produced exciting results. It is now known that eukaryotic RNA pseudouridylation (rRNA and snRNA pseudouridylation in particular) is catalyzed chiefly by ribonucleoproteins (RNPs) of the box H/ACA class, the most complex pseudouridylases known to date [14, 20–25]; in some rare cases, pseudouridylation of eukaryotic snRNA is catalyzed by stand-alone protein pseudouridylases [26, 27]. When incorporated into RNA, Ψ can alter RNA structure [28, 29], increase base stacking [30], improve base-pairing [31], and rigidify the sugar-phosphate backbone [28, 32, 33]. Studies have also linked Ψ, either directly or indirectly, to human diseases. For instance, an increased level of oxidized Ψ has been associated with neurodegenerative diseases, such as Alzheimers and Parkinsons [34]. Mutations in a box H/ACA RNP have been linked to the X-linked form of the bone marrow failure syndrome dyskeratosis congenita [35, 36].
Owing to its unique structural and chemical properties and its proven biological relevance, Ψ has increasingly attracted researchers’ attention, which has resulted in several recent important discoveries. Here, we discuss the mechanisms and functions of box H/ACA RNA guided–pseudouridylation, focusing on recent advances in this fascinating posttranscriptional modification.
Mechanism of RNA-guided RNA pseudouridylation
Box H/ACA RNPs catalyze pseudouridylation of eukaryotic and archaeal rRNAs
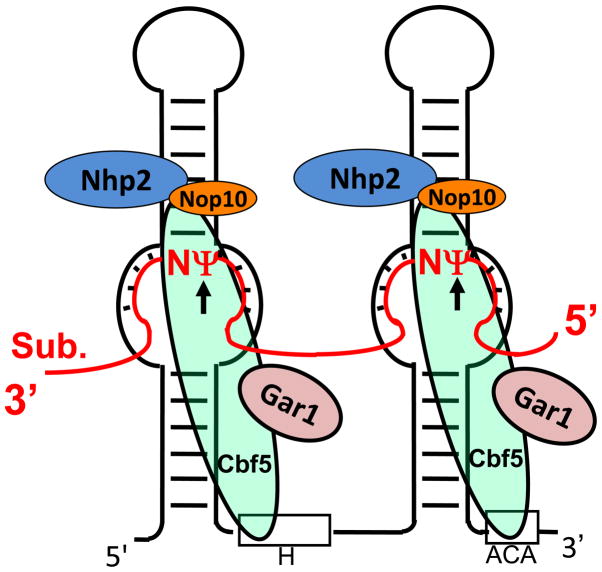
In 1996, box H/ACA RNAs were identified as one of the major families of small RNAs in the nucleolus [37]. One year later, the Kiss and Fournier groups further demonstrated that box H/ACA RNAs function as guide RNAs that target specific uridines in rRNA for pseudouridylation [20, 21]. Specifically, each box H/ACA RNA folds into a conserved “hairpin-hinge-hairpin-tail” structure, exposing a conserved H box in the hinge region and a conserved ACA box in the tail region (Fig. 2). Importantly, each hairpin contains a single-stranded internal loop (often referred to as the pseudouridylation pocket) that is complementary to a short specific sequence in substrate RNA. Upon base-pairing interactions between the complementary sequences, the target uridine in the substrate RNA is positioned precisely at the base of the upper stem of the hairpin and thus undergoes pseudouridylation (Fig. 2).
Fig. 2.
Schematic depiction of eukaryotic box H/ACA RNP-catalyzed pseudouridylation. Box H/ACA RNP, consisting of one guide RNA with a hairpin-hinge-hairpin-tail-structure (black line) and four core proteins, Cbf5, Nhp2, Nop10 and Gar1 (color-coded ovals), is shown. The substrate RNA (red line), which is paired with the guide sequences in the pseudouridylation pockets of box H/ACA RNA, is also shown. Ψ (red) is the target nucleotide converted from uridine, and N (red) represents any nucleotide. Boxes H and ACA of the guide RNA are indicated. Although box H/ACA RNA is usually a double-hairpin molecule in eukaryotic cells, it appears that the two hairpins function independently (each is an independent functional unit).
Box H/ACA RNAs exist in eukaryotic and archaeal cells as RNPs (ribonucleoproteins) [17]. Each box H/ACA RNP consists of one unique box H/ACA RNA and four common core proteins—Nhp2 (L7Ae in archaea), Gar1, Nop10, and Cbf5 (NAP57 or dyskerin in mammals) (Fig. 2). Whereas the RNA component serves as a guide that specifies the target uridine, Cbf5 is the pseudouridylase that catalyzes the chemical reaction of U-to-Ψ isomerization. The unique base-pairing between the guide sequence and the substrate RNA (Fig. 2) can therefore be used to predict the pseudouridylation site [14]. Interestingly, however, recent work by Bernick et al. suggests the presence of novel small RNAs in some archaeal species that show atypical Ψ-guiding features [38].
Box H/ACA RNPs also catalyze pseudouridylation of eukaryotic snRNAs
The discovery of the mechanism of box H/ACA RNA–guided rRNA pseudouridylation sparked a wave of interest in searching for additional box H/ACA RNAs. Both computer-based algorithms and experimental approaches were developed, resulting in the discovery of hundreds of new box H/ACA RNAs in several different organisms [14, 39–42]. One specific experimental approach, dubbed RNomics, proved very effective [41, 42]. Although RNomics involves a series of steps, including small-RNA purification, end-tailing, PCR amplification, cloning, and sequencing [41–43], it does not rely on predictions or known Ψ-containing sequences and is therefore unbiased. Thus, unsurprisingly, many of the large number of potential box H/ACA RNAs identified by this approach have no known rRNA targets [42]. Interestingly, some of the non-rRNA guides exhibit complementarity with spliceosomal snRNAs [42], suggesting that the box H/ACA RNAs may also guide pseudouridylation of snRNAs, which also contain many Ψs [15–19].
To experimentally verify that box H/ACA RNAs directs spliceosomal snRNA pseudouridylation, several labs tested the guide activity of newly identified snRNA-specific box H/ACA RNAs using several independent systems. For instance, using Xenopus oocytes, Zhao et al. demonstrated that a box H/ACA RNA containing two pseudouridylation pockets could direct pseudouridylation of U2 snRNA (one of the five major spliceosomal snRNAs) at two different sites (positions 34 and 44) [44]. The Kiss lab showed that U85, a special type of mammalian box H/ACA RNP, specifically guides U5 snRNA pseudouridylation at position 46 in mammalian cells [45]. Using yeast, Ma at al. also demonstrated that snR81 RNP, a box H/ACA RNP, uses its 5′ pseudouridylation pocket to catalyze Ψ42 formation in U2 [25] (The 3′ pseudouridylation pocket of snR81 is responsible for the formation of Ψ1051 in 25S rRNA [40]). Interestingly, it was also found that snRNA pseudouridylation occurs in Cajal bodies rather than in the nucleoli where rRNA modification occurs, and that the box H/ACA RNAs that are responsible for snRNA pseudouridylation are unique in that they all contain a Cajal body-localization signal (the CAB box) [46]. It was subsequently discovered that Wdr79 binds to the CAB box and targets the box H/ACA RNPs to Cajal bodies where snRNAs transiently reside [47, 48]. It is thus clear that cells use box H/ACA RNPs to modify both rRNAs and snRNAs.
Although box H/ACA RNPs are believed to be the only pseudouridylases that modify snRNAs in higher eukaryotes, S. cerevisiae also uses stand-alone protein pseudouridylases [26, 27]. For instance, there are three Ψ sites in S. cerevisiae U2 snRNA (Ψ35, Ψ42, Ψ44); Ψ42 formation is catalyzed by snR81 RNP (RNA-dependent mechanism) [25], whereas formation of Ψ44 and Ψ35 is catalyzed by the stand-alone pseudouridylases Pus1 [26] and Pus7 [27], respectively (RNA-independent mechanism). Interestingly, both Pus1 and Pus7 catalyze tRNA pseudouridylation as well [26, 49].
The molecular mechanisms of pseudouridylation by box H/ACA RNPs
Until recently, the detailed molecular mechanism(s) underlying box H/ACA RNPs–mediated catalysis of U-to-Ψ conversion remained elusive. To address the mechanism, biochemical, genetic and structural approaches have been used to good effect. Early work mutating, deleting and/or depleting individual components from cells indicated that each box H/ACA RNP component (RNA core proteins) is absolutely required for pseudouridylation and RNP stability [50–54]. Subsequently, a great deal of effort went into dissecting the box H/ACA RNA in detail. Initial work, which was carried out at the cellular level, indicated that the conserved boxes H (hinge region) and ACA (tail region) are important for nucleolar localization as well as for proper pseudouridylation [55–57]. Further work led to the development of several pseudouridylation reconstitution systems where functional box H/ACA RNP could be assembled and the function of box H/ACA RNA assayed [22–25]. These systems allowed identification of three important sequence and structural elements: the stability of the hairpin structure harboring the guide sequence (the pseudouridylation pocket), the stability of base-pairing between the guide sequence and target RNA, and the distance (obligatory 14–16 nt) between the target uridine and box H or ACA [22].
Although biochemical and genetic approaches are effective when defining essential protein components and specific RNA structural elements required for function, a complete understanding of the mechanism of box H/ACA RNP–catalyzed pseudouridylation requires structural analyses. In this regard, NMR was used to study the interactions between guide sequences and their substrate RNAs. Interestingly, the NMR solution structure indicated an unusual base-pairing topology: the substrate RNA base-pairs with the guide sequence of box H/ACA RNA from one side only (one face of the box H/ACA) instead of threading through the pseudouridylation pocket (guide), thereby resulting in a U- or Ω-shaped substrate structure [58, 59]. Such a mode of interaction seems to be advantageous in allowing successive loading of substrate RNAs onto and release of modified substrate RNAs from the box H/ACA RNP [59].
In recent years, remarkable progress has also been made in crystallizing box H/ACA RNPs. A number of different archaeal box H/ACA RNP crystal structures are currently available, including several partial complexes [60–62], a complete complex [63], and even some substrate-bound complexes (functional and non-functional)[64–66]. These structures have provided a detailed picture of H/ACA RNP function. Three of the four protein components, Cbf5, Nop10, and L7Ae (equivalent to eukaryotic Nhp2), interact directly with the upper stem of the box H/ACA RNA hairpin. Cbf5 also interacts with the lower stem of the hairpin and the ACA (or H) box, positioning its catalytic domain close to the pseudouridylation pocket. Cbf5 and Nop10 also exhibit extensive interactions amongst each other. Gar1, by contrast, binds only to Cbf5, making no contact with the RNA or other proteins.
When the RNA substrate is recruited, in addition to the guide-substrate base-pairing interaction, multiple new interactions occur between the proteins (especially Cbf5) and the substrate RNA, further locking the substrate RNA in place [64, 65]. It appears that substrate binding also induces additional conformational changes. Specifically, the upper stem of the box H/ACA RNA, along with Nop10 and L7Ae (both of which bind the upper stem), is brought even closer to the Cbf5 catalytic domain, resulting in further anchoring of the guide RNA [65]. Concurrently, the Cbf5-bound lower stem undergoes a noticeable rotation, apparently creating more room for substrate RNA binding [64]. These structural changes presumably ensure that the substrate is securely docked at the pseudouridylation pocket and that the target nucleotide is positioned precisely at the Cbf5 catalytic site for modification. Indeed, crystal structures show that the target nucleotide is effectively modified in the active site of Cbf5 [64, 65]. Although Gar1 is not involved in anchoring the substrate or guide RNA, it probably plays an important role in regulating the release of the modified RNA product [64].
Ψs are important for RNA function
Given that Ψ displays chemical properties distinct from those of other naturally occurring nucleotides, it is expected that this modified nucleotide would affect the function of the RNA in which it resides. Studies carried out over the past 15 years demonstrate that Ψ is indeed functionally important.
Characteristics of Ψ
Soon after its discovery many decades ago, Ψ modified nucleotide. So far, Ψ has been found in tRNAs [7–10] and is abundant in rRNAs [11–14] and spliceosomal snRNAs [15–19]. For example, there are ~50 Ψs (representing ~1.2% of total nucleotides) in S. cerevisiae rRNAs and twice as many in mammalian rRNAs (~1.4% of total nucleotides) [12, 14, 40]. In vertebrate U2 snRNA, Ψ accounts for ~7% of total nucleotides [15, 18].
Besides being abundant, many pseudouridylation sites are conserved as well. In fact, many Ψs are identified at identical or near-identical sites in rRNAs or snRNAs from various species; this is especially true among vertebrate snRNAs [15, 18]. Although S. cerevisiae snRNAs contain a relatively small number of Ψs, each Ψ has a counterpart in higher eukaryotic (e.g. vertebrate) snRNAs [16, 18, 25].
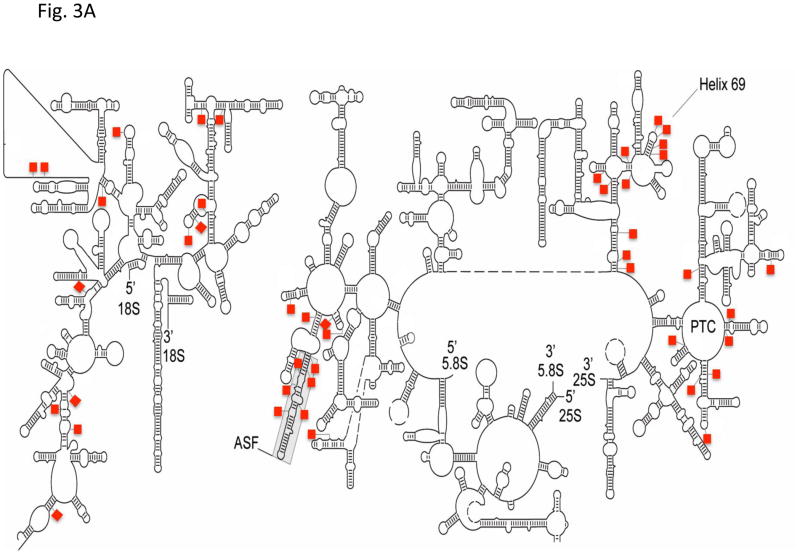
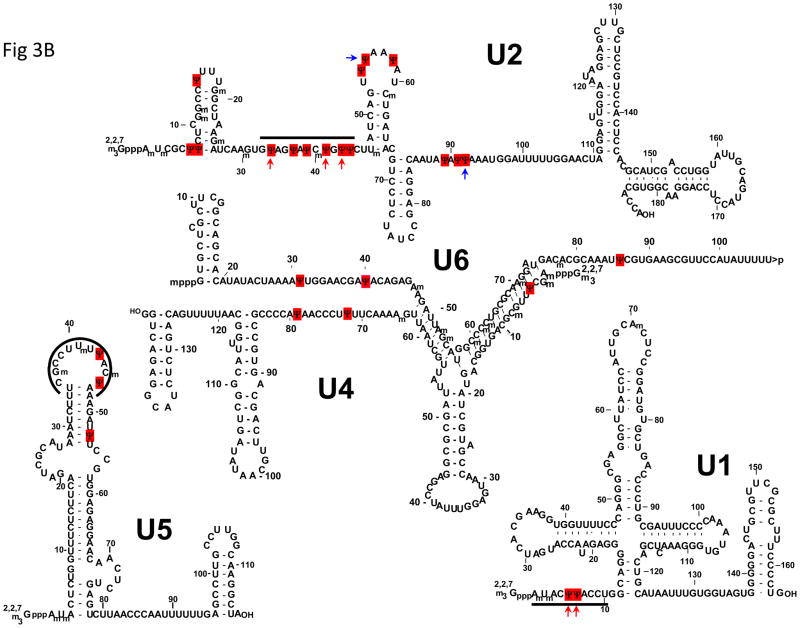
Careful inspection of the location of modified nucleotides in rRNAs and snRNAs further indicates that Ψs are almost always clustered in functionally important regions. For instance, Ψs are not distributed randomly in rRNAs at the secondary structural level [12, 67] (Fig. 3). At the three-dimensional level, Ψs remain, according to a ribosome structural model, highly concentrated in functionally important sites. Specifically, Ψs are clustered in the peptidyl transferase center (PTC), the decoding center, the A-site finger (ASF) region (directly above the A-site of the ribosome), and the sites where ribosomal subunits interact [68–72]. Likewise, Ψs are located mostly in the functionally important regions of spliceosomal snRNAs [15–18]. For instance Ψs are clustered in the 5′-end region of U1, the branch-site recognition sequence of U2, and the loop sequence of U5, all of which play important roles in spliceosome assembly and pre-mRNA splicing (Fig. 3).
Fig. 3.
Ψs are clustered in functionally important regions of rRNAs and snRNAs. The secondary structures of yeast 18S and 25S–5.8S rRNAs (A) and the primary sequences and secondary structures of vertebrate snRNAs (B) are depicted. The red squares represent Ψs. The peptidyl transferase center (PTC), the A-site finger (ASF), and Helix 69 of 25S rRNA are indicated. Some important regions of snRNAs, including the 5′ end of U1, the branch site-recognition region of U2, and the loop sequence of U5, are indicated by thick lines. The red arrows mark the Ψs (in U1 and U2) that are conserved across species (including yeast). The blue arrows indicate the Ψs (in U2) that can be induced by stress. The 5′-cap structures of snRNAs are also shown.
Taken together, the abundance, conservation, and strategic locations of Ψs strongly suggest that they contribute to rRNA and snRNA functions.
Functional roles of Ψs in rRNA and snRNA
Ψs were shown to have a functional role in rRNA processing and protein synthesis. The Fournier lab deleted, either individually or in combination, five box H/ACA RNAs responsible for Ψ formation in the peptidyl transferase center of the ribosome [68]. Although single deletions caused only a modest growth defect, combined deletion of all five box H/ACA RNAs had a profound impact on rRNA processing, protein synthesis, and cell growth, suggesting that these Ψs contribute to ribosome function in a synergistic manner [68]. Using similar approaches, the functional importance of the Ψs located in the decoding center of 18S rRNA [70], the Ψs in the ASF region of 25S rRNA [72], and the Ψs in Helix 69 of 25S rRNA that interacts with both A- and P-site tRNAs during translation was also shown [71][73]. Recently, Dinman and colleagues further reported that ribosomes containing un-pseudouridylated (or hypo-pseudouridylated) rRNAs show decreased affinity for tRNA compared to wild-type ribosomes, resulting in decreased translational fidelity. The effects of rRNA pseudouridylation defects on ribosome-tRNA binding and translational fidelity appear to be evolutionally conserved from yeast to human [74].
Ψs have also been shown to have important functional roles in snRNP biogenesis and function. Virtually all Ψs tested in a Xenopus oocyte reconstitution system were found to be functionally important for snRNP biogenesis and splicing, and Ψs in spliceosomal snRNAs also appear to function synergistically [75, 76], as was found for Ψs in rRNAs [68]. Consistently, modified nucleotides (including Ψs) in the 5′-end region of U2 snRNA were also found to contribute to splicing in a HeLa in vitro reconstitution system [77]. Further, Yang et al. found that Ψ35 in U2 snRNA, together with the U2 position 40 nucleotide, is required for pre-mRNA splicing in budding yeast [78]. This result is consistent with a solution structure showing that Ψ35 is favored over uridine for maintaining the bulge of the branch-point nucleotide adenosine, a structure that is important for splicing [29]. Further crystal structures of the RNA duplex formed between a pre-mRNA branch site and Ψ35-containing U2 indicate that the bulged branch-point nucleotide (or its 5′-adjacent nucleotide) adopts an extrahelical conformation, exposing its 2′-OH group for nucleophilic attack in the first step of splicing [79]. The importance of Ψ35 in splicing was further supported by the work of Valadkhan and Manley who showed that pseudouridylation at this position greatly enhances the production of X-RNA, a product generated by a splicing-related reaction in a cell- and protein-free system [80].
Pseudouridylation can be induced by stress
Until recently, RNA pseudouridylation, and RNA modifications in general, have been described only as constitutive processes; that is, soon after transcription an RNA becomes modified and remains modified thereafter. This presumed permanence of RNA modifications implies that they, unlike most DNA and protein modifications that are inducible and reversible, do not represent biochemically regulatable steps. This assumption also raises a long-standing question of why cells would not take advantage of post-transcriptional modifications to offer an inducible regulatory mechanism for RNA function. Despite huge efforts to address this question, no fruitful results were generated.
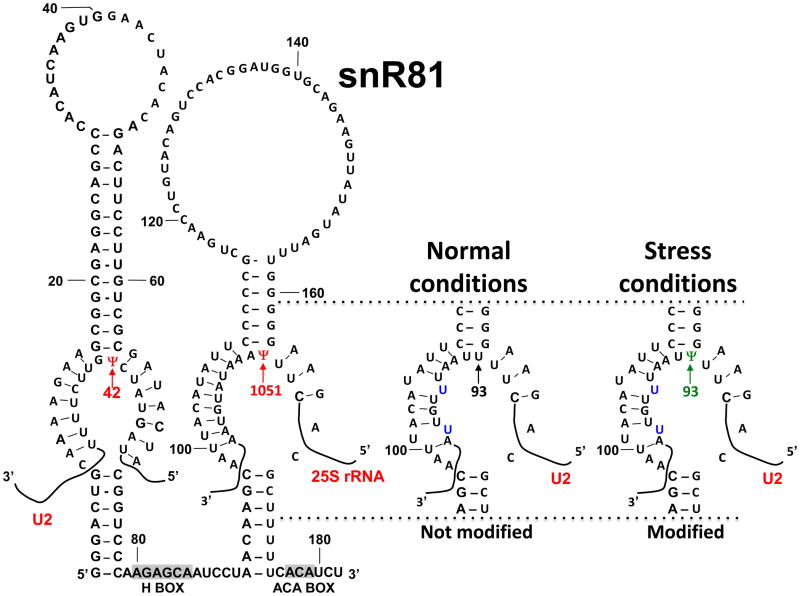
Recently, Wu et al. presented evidence for the first time that in S. cerevisiae U2 snRNA, in addition to the three apparently constitutive Ψs (Ψ35, Ψ42, Ψ44), there are at least two novel Ψs (Ψ56, Ψ93) that can be conditionally induced [81, 82]. In that study, yeast cells were exposed to two widely used stresses: heat-shock and nutrient deprivation [81]. Cells were collected before and after stress, and RNAs were isolated for pseudouridylation assays. Remarkably, two clear Ψ signals were detected at positions 56 and 93 in U2 snRNA isolated from confluent (or nutrient-deprived), but not control cells [81]. Additionally, when the cells were heat shocked Ψ56 (but not Ψ93) was detected [81]. These two positions had previously been identified as “unmodified” uridines in yeast U2 snRNA. Further detailed analyses indicated that while the stand-alone pseudouridylase Pus7 catalyzes Ψ56 formation, the snR81 RNP catalyzes Ψ93 formation (using the 3′-pocket of snR81) (Fig. 4). Pus7 and the 3′ pocket of snR81 had previously been shown to be responsible for constitutive Ψ formation of Ψ35 in U2 [27] and Ψ1051 in 25S rRNA, respectively [40]. Remarkably, the inducibility of U2 pseudouridylation at positions 56 and 93 can be attributed to their imperfect substrate sequences [81]. Specifically, the sequences surrounding positions 56 and 93 in U2 are similar but not identical to the sequences surrounding the constitutively pseudouridylated targets of Pus7 and snR81, respectively (Fig. 4), thus leading to less favorable interactions between the pseudouridylases and the substrate sequences. For instance, when guiding U2 pseudouridylation at position 93 during nutrient deprivation, the 3′-pocket of snR81 pairs imperfectly (two mismatches) with the substrate sequence. The imperfect base-pairing appears to be necessary for induced pseudouridylation [81].
Fig. 4.
Pseudouridylation can be induced by stress. As indicated, snR81, a yeast box H/ACA RNA, normally uses its 5′ pseudouridylation pocket to direct the formation of Ψ42 (red arrow) in U2, and its 3′ pseudouridylation pocket to guide the formation of Ψ1051 (red arrow) in 25S rRNA. Under normal conditions, the 3′ pseudouridylation pocket cannot direct the conversion of U93 (black arrow) into Ψ93 in U2 snRNA. However, under stress conditions, the 3′ pseudouridylation pocket becomes capable of directing the formation of Ψ93 (green arrow), despite the fact that there are two U-U mismatches (indicated) between the 3′ pseudouridylation pocket and the U2 sequence flanking position 93. The sequence of snR81 and partial sequences of U2 and 25S rRNA are shown. Boxes H and ACA within snR81 are indicated.
How can Pus7 and snR81 RNP, which under normal conditions do not recognize positions 56 and 93, respectively, relax their specificities to include these inducible sites upon stress? One possibility, for which there currently is no evidence, is that each enzyme becomes covalently modified (or a co-factor that is capable of facilitating substrate binding is expressed and recruited) when the cellular environment changes, thereby altering the substrate specificity. In this regard, SUMO-modified box H/ACA RNP proteins have been identified [83].
Induction of Ψ56 and Ψ93 also negatively impacts U2 function [81]. This observation is consistent with the well-established notion that, to survive harsh conditions such as starvation or heat shock, cells downregulate gene expression in general. It is conceivable that induction of Ψ56 and Ψ93 in U2, an important spliceosomal component, may contribute to this general downregulation by negatively impacting pre-mRNA splicing. In addition, it is also possible that induced pseudouridylation of snRNAs under stress conditions may significantly alter the landscape of alternatively spliced mRNAs in higher eukaryotes.
mRNA pseudouridylation directed by box H/ACA RNA
Box H/ACA RNA can guide mRNA pseudouridylation
The fact that box H/ACA RNPs are capable of introducing pseudouridylation into two different types of RNAs (rRNA in nucleoli and snRNA in Cajal bodies) raises an interesting question: can this RNA-guided pseudouridylation mechanism be used to modify other RNA types, in particular mRNA? Despite a great deal of effort, the low abundance of mRNAs has made it extremely difficult to analyze their modification.
The abundance problem was overcome through co-expression of a reporter mRNA and an artificial box H/ACA RNA (derived from snR81) that targeted the stop codon (UAA, UAG, or UGA) within the reporter mRNA [84]. Because the reporter was highly expressed, it became possible to measure site-specific U-to-Ψ conversion in the mRNA. The reporter mRNA was pseudouridylated at the expected site only upon co-expression of the box H/ACA RNA containing the correct guide sequence. Although the pseudouridylation efficiency was relatively low (~7–10%), the result leaves no doubt that mRNA can be pseudouridylated at specific sites by box H/ACA RNPs [84]. In an independent Xenopus oocyte study, Chen and Yu injected artificial box H/ACA RNAs targeting a pre-mRNA and found that the pre-mRNA was pseudouridylated at the target sites [85].
Given that the artificial box H/ACA RNA was constructed based on naturally occurring box H/ACA RNAs and that only the short guide sequences were changed [84], it is conceivable that naturally occurring box H/ACA RNAs will naturally pseudouridylate their mRNA substrates if the guide sequences match their respective substrate sequences. In this regard, given that there are a large number of known box H/ACA RNAs, it is quite possible that some of the guide sequences will find their matches (or potential naturally occurring pseudouridylation sites) in mRNAs. Along this line, taking into account that there are a large number of novel box H/ACA RNAs that are recently identified in cells [38, 86], and that induced pseudouridylation requires imperfect pairing between the guide sequences and their substrate sequences [81], naturally occurring mRNA pseudouridylation is likely to be widespread.
Pseudouridylation of mRNA alters codon specificity
Encouraged by the fact that box H/ACA RNAs can guide mRNA pseudouridylation and that the chemical properties of Ψ differ distinctly from those of known nucleotides, Karijolich and Yu tested whether U-to-Ψ conversion in an mRNA affects its coding potential [84]. Given that uridine appears in all three stop/nonsense codons (UAA, UAG, UGA) and that each uridine contacts the release factor during translation termination [87], it seemed probable that the uridine in stop codons is crucial for translation termination. Thus, Karijolich and Yu focused on the pseudouridylation of stop codons in mRNA, speculating that pseudouridylation would alter their translation termination function.
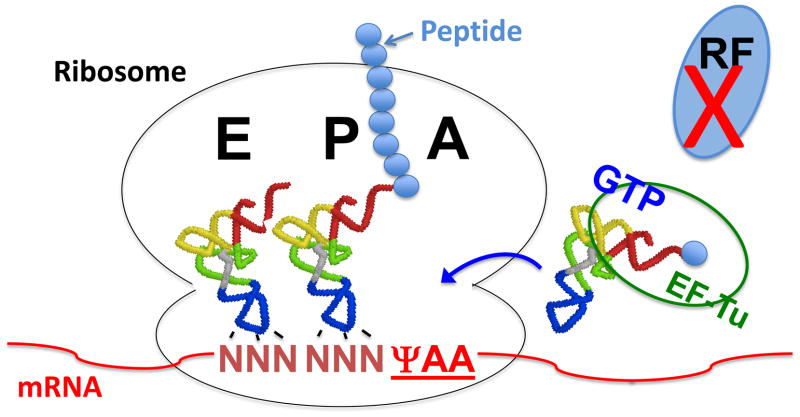
To examine whether pseudouridylation could alter translation termination, Karijolich and Yu modified the well-established yeast CUP1 reporter system [88] by changing the second codon (TTC) to a stop codon (TAA). Consequently, this reporter gene could not produce the functional translation product, Cup1, and hence cells were copper sensitive. An artificial box H/ACA RNA was then co-expressed to target the pre-mature termination codon (PTC) of the reporter CUP1-PTC mRNA for pseudouridylation. Remarkably, the expression of the PTC-specific box H/ACA RNA rescued the growth phenotype in copper-containing medium, a measure of production of read-through Cup1p [84]. Although the rescue was not robust owing primarily to the low level of pseudouridylation (~7–10%), the nonsense suppression by targeted stop codon pseudouridylation is clear. Consistently, this result could be reproduced in a mammalian in vitro system [84]. These results thus indicate that during translation pseudouridylated stop codons are no longer recognized by release factors (RFs). Instead, they are recognized by specific aminoacylated tRNAs (Fig. 5). Using mass spectrometry sequencing, Karijolich and Yu indeed identified the amino acids (and presumably their cognate tRNAs) encoded by the pseudouridylated nonsense codons. Specifically, ΨAA and ΨAG each code for both serine and threonine, whereas ΨGA codes for tyrosine and phenylalanine [84]. Using the currently available ribosome crystal structures as well as other relevant techniques and analyses, the Pan group has recently offered an elegant rationalization for selective decoding of these pseudouridylated stop codons [89].
Fig. 5.
Schematic depiction of Ψ-mediated nonsense suppression. When the invariant uridine of a stop codon is converted into Ψ, the modified stop codon ΨAA in the ribosomal A-site is no longer recognized by release factors (RF). Instead, ΨAA is recognized by a specific aminoacylated tRNA (depicted as EF-Tu/GTP/aa-tRNA ternary complex), allowing the incorporation of a specific amino acid into the elongating peptide and resulting in nonsense suppression. The ribosome and the ribosomal A-, P- and E-sites are indicated. tRNAs, mRNA and elongating peptide are also depicted. The small solid blue circles represent amino acids.
The fact that pseudouridylation of nonsense codons results in nonsense suppression prompts speculation about the possible effect of pseudouridylation on coding specificity of U-containing sense codons. Such speculation is also consistent with early reports indicating that pseudouridylated anticodons in tRNA read alternate codons that would otherwise not have been recognized if the anticodons were not modified [90]. Given the large number of U-containing sense codons (34 of the 61 sense codons contain one or more uridines), targeted mRNA pseudouridylation portends an expansion of the genetic code (Fig. 6). Interestingly, recent work by the Kariko group indicated that the presence of Ψ in mRNA increased translation efficiency [91–93]. Although neither the detailed mechanism of this effect nor the effect of Ψ-mediated codon specificity change (e.g., nonsense suppression) has been clearly defined, Ψappears to profoundly impact mRNA function during translation.
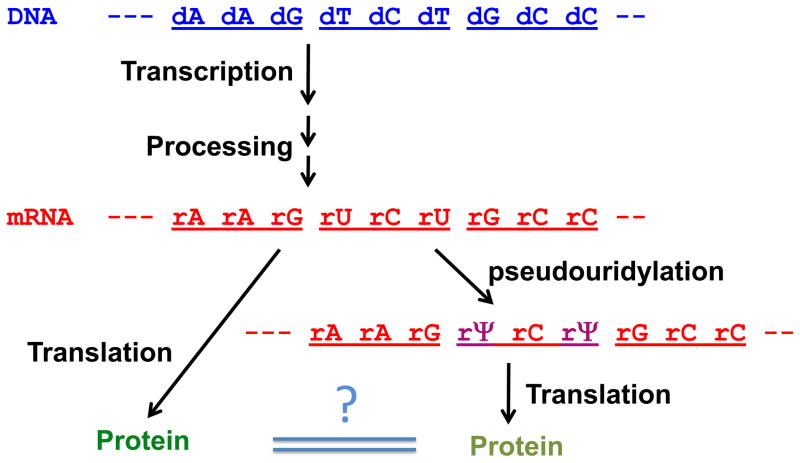
Fig. 6.
mRNA pseudouridylation as a novel means to produce protein diversity. The pathway of gene expression from DNA (blue) to mature mRNA (red) to protein is depicted. Each codon consists of three nucleotides and is indicated by an underline. When mRNA is pseudouridylated, the modified mRNA (purple) may encode a protein different from that encoded by the un-modified mRNA (red). The question mark indicates the possible differences between the two proteins.
Concluding remarks
It has been more than six decades since the birth of the field of RNA modification and more than 15 years since the discovery of the box H/ACA RNA family. Over this long period, RNA pseudouridylation has been extensively studied, and we now know that a large number of box H/ACA RNPs function as sophisticated and complex enzymes that catalyze U-to-Ψ conversion in RNAs and that Ψ contributes significantly to RNA functions. Clearly we have come a long way in our understanding of the mechanisms underlying RNA-guided RNA pseudouridylation and its function.
Although we have accumulated a wealth of knowledge on the mechanisms and functions of Ψ, recent efforts to address inducible pseudouridylation and mRNA pseudouridylation have generated exciting results and have thus formed the basis for two new areas of study: (1) regulated pseudouridylation, and (2) Ψ-mediated alternative coding and protein diversity. A number of new questions arise. For instance, how exactly is pseudouridylation induced under stress? Is induced pseudouridylation reversible? How are pseudouridylated codons recognized in the ribosome decoding center? How many new viable codons can be generated by targeted mRNA pseudouridylation? Do cells use this strategy to modulate protein diversity (see Fig. 6)? There is no doubt that addressing these questions will require a great deal of work and a combination of experimental techniques. However, the results generated from these experimental efforts will significantly advance our knowledge of the breadth of Ψ function.
Acknowledgments
We thank the members of the Yu lab for valuable discussions. Our work was supported by grants GM104077 and AG39559 (to Yi-Tao Yu) from the National Institute of Health, by the University of Rochester CTSA award UL1TR000042 (to Yi-Tao Yu) from the National Center for Advancing Translational Sciences of the National Institute of Health, and by grant 81071865 from the National Natural Science Foundation of China (to Junhui Ge).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FA, Fabris D, Agris PF. The RNA Modification Database, RNAMDB: 2011 update. Nucleic acids research. 2011;39(Database issue):D195–201. doi: 10.1093/nar/gkq1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic acids research. 2009;37(Database issue):D118–21. doi: 10.1093/nar/gkn710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, Texas: Landes Bioscience; 2009. [Google Scholar]
- 4.Cohn WE, Volkin W. Nucleoside-5′-Phosphates from Ribonucleic Acid. Nature (London) 1951;167:483–484. [Google Scholar]
- 5.Davis FF, Allen FW. Ribonucleic acids from yeast which contain a fifth nucleotide. J Biol Chem. 1957;227(2):907–15. [PubMed] [Google Scholar]
- 6.Lane BG. Historical Perspectives on RNA Nucleoside Modifications. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. ASM Press; Washington, D. C: 1998. pp. 1–20. [Google Scholar]
- 7.Bjork GR. Biosynthesis and function of modified nucleotides. In: Soll D, RajBhandary U, editors. tRNA: Structure, biosynthesis, and function. ASM Press; Washington, DC: 1995. pp. 165–205. [Google Scholar]
- 8.Grosjean H, Sprinzl M, Steinberg S. Posttranscriptionally modified nucleosides in transfer RNA: their locations and frequencies. Biochimie. 1995;77(1–2):139–41. doi: 10.1016/0300-9084(96)88117-x. [DOI] [PubMed] [Google Scholar]
- 9.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26(1):148–53. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17(2):162–80. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- 11.Branlant C, Krol A, Machatt MA, Pouyet J, Ebel JP, Edwards K, Kossel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic acids research. 1981;9(17):4303–24. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ofengand J, Fournier M. The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In: Grosjean H, Benne R, editors. Modification and Editing of RNA. ASM Press; Washington, DC: 1998. pp. 229–253. [Google Scholar]
- 13.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1990;39:241–303. doi: 10.1016/s0079-6603(08)60629-7. [DOI] [PubMed] [Google Scholar]
- 14.Schattner P, Barberan-Soler S, Lowe TM. A computational screen for mammalian pseudouridylation guide H/ACA RNAs. RNA. 2006;12(1):15–25. doi: 10.1261/rna.2210406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy R, Busch H. Small nuclear RNAs: RNA sequences, structure, and modifications. In: Birnsteil ML, editor. Structure and function of major and minor small nuclear ribonucleoprotein particles. Sringer-Verlag Press; Heidelberg: 1988. pp. 1–37. [Google Scholar]
- 16.Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In: Grosjean H, editor. Modification and Editing of RNA. ASM Press; Washington, DC: 1998. pp. 201–228. [Google Scholar]
- 17.Yu YT, Terns RM, Terns MP. Mechanisms and functions of RNA-guided RNA modification. In: Grosjean H, editor. Topics in Current Genetics. Springer-Verlag; New York: 2005. pp. 223–262. [Google Scholar]
- 18.Yu AT, Ge J, Yu YT. Pseudouridines in spliceosomal snRNAs. Protein & cell. 2011;2(9):712–25. doi: 10.1007/s13238-011-1087-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu G, Yu AT, Kantartzis A, Yu YT. Functions and mechanisms of spliceosomal small nuclear RNA pseudouridylation. Wiley interdisciplinary reviews. RNA. 2011;2(4):571–81. doi: 10.1002/wrna.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89(4):565–73. doi: 10.1016/s0092-8674(00)80238-x. [DOI] [PubMed] [Google Scholar]
- 21.Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell. 1997;89(5):799–809. doi: 10.1016/s0092-8674(00)80263-9. [DOI] [PubMed] [Google Scholar]
- 22.Xiao M, Yang C, Schattner P, Yu YT. Functionality and substrate specificity of human box H/ACA guide RNAs. RNA. 2009;15(1):176–86. doi: 10.1261/rna.1361509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baker DL, Youssef OA, Chastkofsky MI, Dy DA, Terns RM, Terns MP. RNA-guided RNA modification: functional organization of the archaeal H/ACA RNP. Genes & development. 2005;19(10):1238–48. doi: 10.1101/gad.1309605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic acids research. 2005;33(10):3133–44. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. Embo J. 2005;24(13):2403–2413. doi: 10.1038/sj.emboj.7600718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol. 1999;19(3):2142–54. doi: 10.1128/mcb.19.3.2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. Embo J. 2003;22(8):1889–97. doi: 10.1093/emboj/cdg191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry. 1994;33(24):7560–7. doi: 10.1021/bi00190a008. [DOI] [PubMed] [Google Scholar]
- 29.Newby MI, Greenbaum NL. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol. 2002;9(12):958–65. doi: 10.1038/nsb873. [DOI] [PubMed] [Google Scholar]
- 30.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res. 1995;23(24):5020–6. doi: 10.1093/nar/23.24.5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newby MI, Greenbaum NL. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. Rna. 2001;7(6):833–45. doi: 10.1017/s1355838201002308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newby MI, Greenbaum NL. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(20):12697–702. doi: 10.1073/pnas.202477199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49(5):341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 34.Lee SH, Kim I, Chung BC. Increased urinary level of oxidized nucleosides in patients with mild-to-moderate Alzheimer’s disease. Clinical biochemistry. 2007;40(13–14):936–8. doi: 10.1016/j.clinbiochem.2006.11.021. [DOI] [PubMed] [Google Scholar]
- 35.Grozdanov PN, Fernandez-Fuentes N, Fiser A, Meier UT. Pathogenic NAP57 mutations decrease ribonucleoprotein assembly in dyskeratosis congenita. Human molecular genetics. 2009;18(23):4546–51. doi: 10.1093/hmg/ddp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature genetics. 1998;19(1):32–8. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 37.Balakin AG, Smith L, Fournier MJ. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86(5):823–34. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 38.Bernick DL, Dennis PP, Hochsmann M, Lowe TM. Discovery of Pyrobaculum small RNA families with atypical pseudouridine guide RNA features. RNA. 2012;18(3):402–11. doi: 10.1261/rna.031385.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic acids research. 2005;33(Web Server issue):W686–9. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schattner P, Decatur WA, Davis CA, Ares M, Jr, Fournier MJ, Lowe TM. Genome-wide searching for pseudouridylation guide snoRNAs: analysis of the Saccharomyces cerevisiae genome. Nucleic acids research. 2004;32(14):4281–96. doi: 10.1093/nar/gkh768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Curr Opin Chem Biol. 2002;6(6):835–43. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 42.Huttenhofer A, Kiefmann M, Meier-Ewert S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. Embo J. 2001;20(11):2943–53. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu YT, Tarn WY, Yario TA, Steitz JA. More Sm snRNAs from vertebrate cells. Experimental cell research. 1996;229(2):276–81. doi: 10.1006/excr.1996.0372. [DOI] [PubMed] [Google Scholar]
- 44.Zhao X, Li ZH, Terns RM, Terns MP, Yu YT. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. Rna. 2002;8(12):1515–25. [PMC free article] [PubMed] [Google Scholar]
- 45.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. Embo J. 2001;20(3):541–51. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. The EMBO journal. 2003;22(16):4283–93. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science. 2009;323(5914):644–8. doi: 10.1126/science.1165357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Molecular cell. 2009;34(1):47–57. doi: 10.1016/j.molcel.2009.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA. 2003;9(11):1371–82. doi: 10.1261/rna.5520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bousquet-Antonelli C, Henry Y, G’Elugne P, Caizergues-Ferrer JM, Kiss T. A small nucleolar RNP protein is required for pseudouridylation of eukaryotic ribosomal RNAs. The EMBO journal. 1997;16(15):4770–6. doi: 10.1093/emboj/16.15.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Henras A, Dez C, Noaillac-Depeyre J, Henry Y, Caizergues-Ferrer M. Accumulation of H/ACA snoRNPs depends on the integrity of the conserved central domain of the RNA-binding protein Nhp2p. Nucleic acids research. 2001;29(13):2733–46. doi: 10.1093/nar/29.13.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gelugne JP, Caizergues-Ferrer M. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. The EMBO journal. 1998;17(23):7078–90. doi: 10.1093/emboj/17.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J. Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Molecular and cellular biology. 1999;19(11):7461–72. doi: 10.1128/mcb.19.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lafontaine DL, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes & development. 1998;12(4):527–37. doi: 10.1101/gad.12.4.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narayanan A, Lukowiak A, Jady BE, Dragon F, Kiss T, Terns RM, Terns MP. Nucleolar localization signals of box H/ACA small nucleolar RNAs. The EMBO journal. 1999;18(18):5120–30. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lange TS, Ezrokhi M, Amaldi F, Gerbi SA. Box H and box ACA are nucleolar localization elements of U17 small nucleolar RNA. Molecular biology of the cell. 1999;10(11):3877–90. doi: 10.1091/mbc.10.11.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bortolin ML, Ganot P, Kiss T. Elements essential for accumulation and function of small nucleolar RNAs directing site-specific pseudouridylation of ribosomal RNAs. The EMBO journal. 1999;18(2):457–69. doi: 10.1093/emboj/18.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jin H, Loria JP, Moore PB. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Molecular cell. 2007;26(2):205–15. doi: 10.1016/j.molcel.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 59.Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(16):6655–60. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamma T, Reichow SL, Varani G, Ferre-D’Amare AR. The Cbf5-Nop10 complex is a molecular bracket that organizes box H/ACA RNPs. Nature structural & molecular biology. 2005;12(12):1101–7. doi: 10.1038/nsmb1036. [DOI] [PubMed] [Google Scholar]
- 61.Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H. Crystal structure of a Cbf5-Nop10-Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Molecular cell. 2006;21(2):249–60. doi: 10.1016/j.molcel.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 62.Manival X, Charron C, Fourmann JB, Godard F, Charpentier B, Branlant C. Crystal structure determination and site-directed mutagenesis of the Pyrococcus abyssi aCBF5-aNOP10 complex reveal crucial roles of the C-terminal domains of both proteins in H/ACA sRNP activity. Nucleic acids research. 2006;34(3):826–39. doi: 10.1093/nar/gkj482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443(7109):302–7. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 64.Duan J, Li L, Lu J, Wang W, Ye K. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Molecular cell. 2009;34(4):427–39. doi: 10.1016/j.molcel.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 65.Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nature structural & molecular biology. 2009;16(7):740–6. doi: 10.1038/nsmb.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liang B, Xue S, Terns RM, Terns MP, Li H. Substrate RNA positioning in the archaeal H/ACA ribonucleoprotein complex. Nature structural & molecular biology. 2007;14(12):1189–95. doi: 10.1038/nsmb1336. [DOI] [PubMed] [Google Scholar]
- 67.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends in biochemical sciences. 2002;27(7):344–51. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 68.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Molecular cell. 2003;11(2):425–35. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 69.Baudin-Baillieu A, Fabret C, Liang XH, Piekna-Przybylska D, Fournier MJ, Rousset JP. Nucleotide modifications in three functionally important regions of the Saccharomyces cerevisiae ribosome affect translation accuracy. Nucleic acids research. 2009;37(22):7665–77. doi: 10.1093/nar/gkp816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA. 2009;15(9):1716–28. doi: 10.1261/rna.1724409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Molecular cell. 2007;28(6):965–77. doi: 10.1016/j.molcel.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 72.Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ. Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. The Journal of biological chemistry. 2008;283(38):26026–36. doi: 10.1074/jbc.M803049200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9(7):771–9. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, Ruggero D, Dinman JD. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Molecular cell. 2011;44(4):660–6. doi: 10.1016/j.molcel.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhao X, Yu YT. Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. Rna. 2004;10(4):681–90. doi: 10.1261/rna.5159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. Embo J. 1998;17(19):5783–95. doi: 10.1093/emboj/17.19.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Donmez G, Hartmuth K, Luhrmann R. Modified nucleotides in the 5′ end of the human U2 snRNA are required for early spliceosome (E complex) formation in vitro. The 2004 RNA meeting abstract; 2004. p. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem. 2005;280(8):6655–62. doi: 10.1074/jbc.M413288200. [DOI] [PubMed] [Google Scholar]
- 79.Lin Y, Kielkopf CL. X-ray structures of U2 snRNA-branchpoint duplexes containing conserved pseudouridines. Biochemistry. 2008;47(20):5503–14. doi: 10.1021/bi7022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Valadkhan S, Manley JL. Characterization of the catalytic activity of U2 and U6 snRNAs. RNA. 2003;9(7):892–904. doi: 10.1261/rna.5440303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. The EMBO journal. 2011;30(1):79–89. doi: 10.1038/emboj.2010.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Meier UT. Pseudouridylation goes regulatory. The EMBO journal. 2011;30(1):3–4. doi: 10.1038/emboj.2010.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Westman BJ, Lamond AI. A role for SUMOylation in snoRNP biogenesis revealed by quantitative proteomics. Nucleus. 2011;2(1):30–7. doi: 10.4161/nucl.2.1.14437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature. 2011;474(7351):395–8. doi: 10.1038/nature10165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen C, Zhao X, Kierzek R, Yu YT. A flexible RNA backbone within the polypyrimidine tract is required for U2AF65 binding and pre-mRNA splicing in vivo. Molecular and cellular biology. 2010;30(17):4108–19. doi: 10.1128/MCB.00531-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jady BE, Ketele A, Kiss T. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes & development. 2012;26(17):1897–910. doi: 10.1101/gad.197467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korostelev A, Asahara H, Lancaster L, Laurberg M, Hirschi A, Zhu J, Trakhanov S, Scott WG, Noller HF. Crystal structure of a translation termination complex formed with release factor RF2. Proc Natl Acad Sci U S A. 2008;105(50):19684–9. doi: 10.1073/pnas.0810953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lesser CF, Guthrie C. Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics. 1993;133(4):851–63. doi: 10.1093/genetics/133.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA. 2012;18(3):355–67. doi: 10.1261/rna.031351.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomita K, Ueda T, Watanabe K. The presence of pseudouridine in the anticodon alters the genetic code: a possible mechanism for assignment of the AAA lysine codon as asparagine in echinoderm mitochondria. Nucleic acids research. 1999;27(7):1683–9. doi: 10.1093/nar/27.7.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kariko K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Molecular therapy: the journal of the American Society of Gene Therapy. 2012;20(5):948–53. doi: 10.1038/mt.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic acids research. 2010;38(17):5884–92. doi: 10.1093/nar/gkq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kariko K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Molecular therapy: the journal of the American Society of Gene Therapy. 2008;16(11):1833–40. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]