A sequence in the Drosophila H3-H4 promoter triggers 1 Histone Locus Body assembly and biosynthesis of replication-coupled histone mRNAs (original) (raw)
. Author manuscript; available in PMC: 2014 Mar 25.
Abstract
Compartmentalization of RNA biosynthetic factors into nuclear bodies (NBs) is a ubiquitous feature of eukaryotic cells. How NBs initially assemble and ultimately affect gene expression remains unresolved. The histone locus body (HLB) contains factors necessary for replication-coupled histone mRNA transcription and processing and associates with histone gene clusters. Using a transgenic assay for ectopic Drosophila HLB assembly, we show that a sequence located between, and transcription from, the divergently transcribed H3-H4 genes nucleates HLB formation and activates other histone genes in the histone gene cluster. In the absence of transcription from the H3-H4 promoter, “proto-HLBs”, containing only a subset of HLB components, form and the adjacent histone H2a-H2b genes are not expressed. Proto-HLBs also transiently form in mutant embryos with the histone locus deleted. We conclude that HLB assembly occurs through a stepwise process involving stochastic interactions of individual components that localize to a specific sequence in the H3-H4 promoter.
Keywords: nuclear bodies, Drosophila, histone gene expression, mRNA processing
Introduction
In the last decade, compartmentalization of nuclear processes has emerged as an important organizing principle of the genome. Nuclei contain a host of distinct compartments or “nuclear bodies” such as nucleoli, speckles, paraspeckles, Cajal bodies, PML bodies, and histone locus bodies (HLBs) where factors involved in processes such as transcription, RNA processing and maturation, and DNA replication and repair are concentrated (Carmo-Fonseca and Rino, 2011; Handwerger and Gall, 2006; Matera et al., 2009; Misteli, 2007). Despite a role for NBs in a wide range of biological processes, a complete understanding of the relationship between NB formation and the associated biochemical reactions (e.g. transcription and pre-mRNA splicing/processing) is lacking.
NBs are thought to enhance the efficiency of reactions by concentrating reaction components (Matera et al., 2009; Misteli, 2007). While there is some evidence for this idea (Chen et al., 2010; Strzelecka et al., 2010; Wagner et al., 2007), the importance of the contribution that NBs make to their associated processes is not always clear. The Cajal body, a NB involved in snRNP biogenesis, provides a good example. Mutation of the gene encoding Coilin, a critical assembly component of Cajal bodies, is lethal in zebrafish due to failure to form sufficient snRNPs (Strzelecka et al., 2010), but not in flies (Liu et al., 2009). Moreover, while coilin mutant mice are not fully viable or fertile (Tucker et al., 2001; Walker et al., 2009), coilin mutant flies, which lack detectable Cajal bodies, are fertile and correctly perform several snRNA modifications that are specified by scaRNAs normally localized to Cajal bodies (Deryusheva and Gall, 2009).
Determining how NBs form is critical for understanding how NBs affect their associated biochemical processes. Current evidence suggests that NBs form by a process of “self-organization” in which individual factors encounter other NB components through random molecular collisions, and remain in proximity due to binding affinities (Handwerger and Gall, 2006; Misteli, 2001, 2005; Nizami et al., 2010). The high affinity between factors associated with specific processes results in the formation of microscopically visible structures. Two contrasting models of self-organization have been proposed (Matera et al., 2009; Misteli, 2007). Tethering experiments that artificially localize individual nuclear body components support a stochastic self-organization model, where nuclear body components can assemble in any order (Kaiser et al., 2008; Shevtsov and Dundr, 2011). In contrast, genetic evidence supports a hierarchical model, which posits that NBs assemble in a particular order with assembly of some components predicated on prior assembly of other components (Rajendra et al., 2011; White et al., 2011). Recently, a hybrid model has emerged from studies of the HLB and paraspeckles suggesting that the stochastic and hierarchical models of nuclear body formation are not mutually exclusive (Dundr, 2011; Mao et al., 2011; White et al., 2011).
The HLB is an excellent model for investigating both the mechanism and function of NBs. HLBs assemble at replication-coupled histone genes in animal cells and contain factors associated with the transcription and processing of histone mRNA (Bongiorno-Borbone et al., 2010; Bongiorno-Borbone et al., 2008; Frey and Matera, 1995; Ghule et al., 2008; Liu et al., 2006; Nizami et al., 2010; White et al., 2011; White et al., 2007). HLBs contain factors such as U7 snRNP and FLASH that are necessary for an endonucleolytic cleavage of histone pre-mRNA resulting in a unique 3′ stem-loop structure that mediates all aspects of histone mRNA regulation, rather than a poly(A) tail (Marzluff et al., 2008). In fact, HLBs were originally defined through studies of the localization of the U7 snRNP specific proteins, Lsm10 and Lsm11, as well as U7 snRNA (Liu et al., 2006). HLBs also contain the protein NPAT, a substrate of Cyclin E/Cdk2 that is concentrated at the two clusters of human histone genes (Ma et al., 2000; Zhao et al., 2000). NPAT is essential for entry into S-phase and for expression of histone mRNA, although the precise molecular basis of NPAT action is not understood (Ma et al., 2000; Miele et al., 2005; Wei et al., 2003; Ye et al., 2003; Zhao et al., 2000). There is no evidencethat NPAT directly binds DNA, and more likely it acts as a cofactor for histone gene transcription and possibly coordinates the multiple steps in histone mRNA biosynthesis.
In metazoans, the accumulation of replication-dependent histone mRNAs is confined to S-phase when histone proteins are required for chromatin assembly (Marzluff and Duronio, 2002; Marzluff et al., 2008). The tight regulation of histone accumulation during the cell cycle is essential for genetic stability (Gunjan and Verreault, 2003; Marzluff, 2010; Meeks-Wagner and Hartwell, 1986), suggesting that the S phase role of the HLB in histone biosynthesis is likely to impact a wide range of genomic functions. The HLB is also present during G1 and G2 phase, when histone mRNAs are not actively synthesized (White et al., 2011; White et al., 2007), indicating that HLB assembly and/or maintenance is not strictly dependent on active transcription and/or mRNA processing. Histone gene expression is activated in cycle 11 of Drosophila embryogenesis, the same time as the HLB forms. A subset of HLB components, FLASH and Mxc, the Drosophila ortholog of NPAT, accumulates at the histone locus before the onset of histone gene expression in the early Drosophila embryo, and we term this complex a “proto-HLB” (White et al., 2011).
Because HLBs assemble only at histone genes, we hypothesized that a sequence element(s) within or associated with the histone locus would drive HLB assembly. Here we present the surprising result that despite the fact that all five replication-dependent histone genes are coordinately transcribed and processed, only a single element in the Drosophila histone gene locus is capable of nucleating the HLB. We demonstrate that formation of the Drosophila HLB depends on a sequence in the 300 nt histone H3-H4 bidirectional promoter, and that this sequence is essential for expression of other histone genes in the cluster. A proto-HLB assembles on the minimal sequence in the absence of transcription, but transcription driven by this sequence is necessary for formation of a complete HLB. In addition, we show that proto-HLBs form transiently even in the absence of histone genes, indicating that some HLB components have self-organization properties. Together these results support a model whereby transcription-dependent ordered assembly and stochastic self-organization of components both contribute to HLB assembly during development.
RESULTS
An HLB can assemble at a single, active histone gene repeat
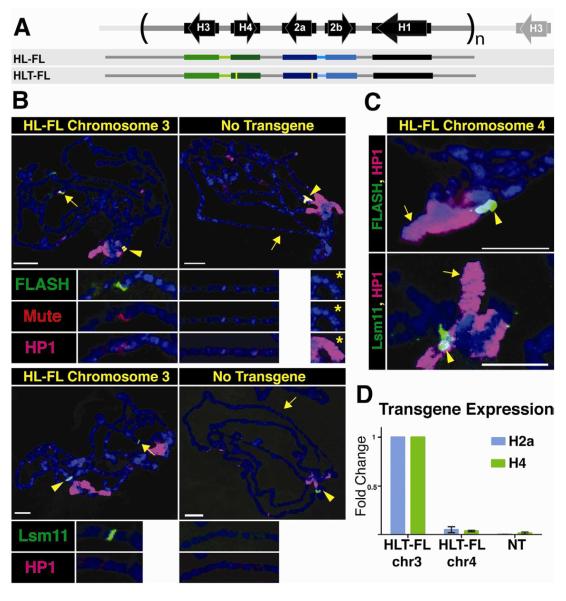
The Drosophila replication-coupled histone genes are present in a single locus on chromosome 2L as a tandem 5 kB repeat present in about 100 copies. Each repeat unit contains one copy of each of the five histone genes (Fig. 1A). The H2a-H2b and H3-H4 gene pairs are divergently transcribed, while the H1 gene is located about 1.5 kB 3′ of the H3 gene, and ends about 300 nts before the 3′ end of the H2b gene (Fig. 1A). To determine whether sequences within a single repeat unit were sufficient to direct the formation of an HLB, we used a construct containing 1.2 copies of the repeat unit such that all contiguous sequences 500 nts long were represented in the construct (_H_istone _L_ocus-_F_ull _L_ength, or HL-FL; Fig. 1A) and generated transgenes at specific loci in the Drosophila genome by ΦC31-mediated integration (Bateman et al., 2006; Bischof et al., 2007). To test whether the chromatin environment around the ectopic histone genes can influence expression, HL-FL was inserted into two specific sites: a euchromatic site on chromosome 3 (86Fb) and a heterochromatic site on chromosome 4 (102D).
Figure. 1. An HLB forms at an ectopic locus containing one histone gene repeat unit.
A. Diagram of the histone repeat (chromosome 2). The 5.1 kB histone repeat unit is indicated by parentheses. A fragment containing 1.2 repeat units (HL-FL) was cloned and inserted into the Drosophila genome at either site 86Fb on chromosome 3, or at site 102D on chromosome 4. The yellow bars in the HLT-FL construct represent N-terminal FLAG tags in H2a and H4.
B. Chromosome squashes from salivary glands of third instar larvae containing the HL-FL at 86Fb (left; n=15) or no transgene (right; n=7) stained with Mute (red), FLASH (green) and HP1 (pink, top panel) or Lsm11 (green) and HP1 (pink, bottom panel). The insets show a higher magnification of the 86Fb chromosome region except the panel with * which shows chromosome 4 (102D). The arrowhead indicates the endogenous HLB and the arrow indicates chromosomal position 86Fb. Bars = 10 μm.
C. Chromosome 4 from salivary glands of 3rd instar larvae containing the HL-FL transgene at position 102D (arrow; n=8) stained with HP1 (pink) and either FLASH (green, top) or Lsm11 (green, bottom). The endogenous histone locus near the chromocenter is indicated by the arrowhead.
D. RT-PCR analysis of H2a and H4 expression from HLT-FL located at 86Fb (chr3) and 102D (chr4) compared to no transgene (NT). Histone gene expression was normalized relative to the expression of actin mRNA. Error bars represent SEM.
We visualized HLBs using antibodies to four components of the HLB: Multi sex combs (Mxc), the Drosophila orthologue of NPAT that we recently identified (White et al., 2011); FLASH, a histone pre-mRNA processing factor (Burch et al., 2011a; Yang et al., 2009); Mute, an essential protein of unknown function homologous to YY1 associated protein (Bulchand et al., 2010); and Lsm11, a component of U7 snRNP (Azzouz and Schumperli, 2003; Pillai et al., 2003). For some experiments we utilized a Drosophila line expressing V5 tagged Lsm11, which rescues an Lsm11 null mutant, and visualized U7 snRNP in the HLB using an anti-V5 antibody (Godfrey et al., 2009). This panel of reagents includes factors that are first detected in the HLB before (Mxc and FLASH) and after (Mute and Lsm11) the onset of zygotic histone transcription (White et al., 2011; White et al., 2007).
We analyzed chromosome spreads from 3rd instar larval salivary gland cells. In these polyploid cells the genome reaches more than 1000C and individual chromatids line up in register, resulting in polytene chromosomes that provide high resolution for cytological experiments (Fig. 1B). Using antibodies to Lsm11, Mute, and FLASH, we observed HLB assembly at the ectopic HL-FL locus at 86Fb on chromosome 3, as well as at the endogenous histone locus at 39D-E on chromosome 2 (Fig. 1B). In contrast, when the repeat was located at 102D on chromosome 4, HLB assembly was not observed (Fig. 1C), although its genomic presence was confirmed by PCR (not shown). We conclude that one copy of the histone repeat is sufficient to assemble an HLB at a euchromatic but not a heterochromatic site.
To assess whether ectopic genes were expressed, identical transgenic lines were generated containing 5′ FLAG tags on the H2a and H4 genes (_H_istone _L_ocus _T_agged-_F_ull _L_ength; HLT-FL, Fig. 1A). We used a FLAG specific qRT-PCR primer to determine the expression of the ectopic H2a and H4 mRNAs relative to actin mRNA, and normalized these results to the HLT-FL insertion at 86Fb on chromosome 3. We detected expression of both genes from the ectopic repeat located at 86Fb but not from the transgene inserted at 102D on chromosome 4 (Fig. 1D). Thus, sequences present in the histone repeat are sufficient to direct HLB assembly and histone gene expression, and no sequences flanking the histone locus are necessary. However, other factors such as local chromatin structure influence HLB assembly and histone gene expression.
Histone gene expression correlates with HLB assembly
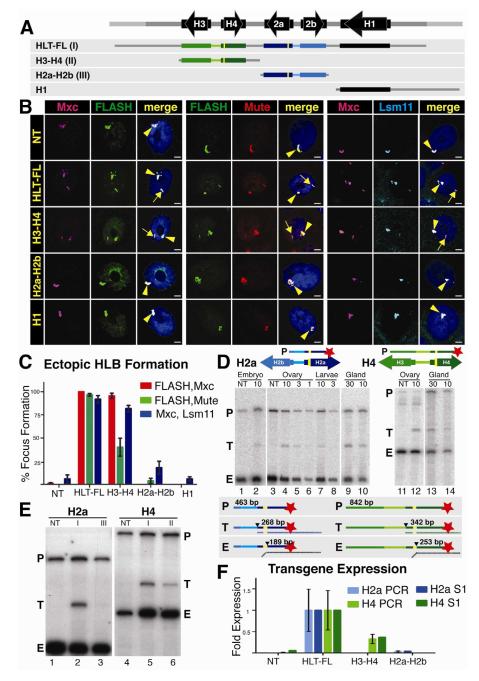
The formation of HLBs at ectopic sites that expressed histone genes provided us with an opportunity to define sequences within the histone gene repeat that direct HLB assembly and histone gene expression, and to determine how these two processes are functionally related. We made transgenic flies with constructs inserted at 86Fb that contain only the H3-H4 gene pair, the H2a-H2b gene pair, or the histone H1 gene plus the long intergenic region between it and the 3′ end of the H3 gene (Fig. 2A). We assessed HLB formation by quantifying the presence of ectopic HLBs in intact salivary gland nuclei (Wagner et al., 2007) using multiple pairs of HLB markers (Fig. 2B). Ectopic HLBs were defined by co-localization of two or three HLB components in a focus (arrows, Fig. 2B), in addition to the endogenous HLB (arrowhead, Fig. 2B). For each experiment, ectopic HLBs were quantified using 1μm sections of a 150μm2 area through the posterior portion of the salivary gland, and the data are presented as percent mean ectopic focus formation of 7-10 individuals (>100 cells for each construct) (Fig. 2C).
Fig 2. The H3-H4 genes assemble an ectopic HLB.
A. Diagram of the four constructs inserted into chromosomal location 86Fb. The yellow bars represent N-terminal FLAG tags in H2a and H4.
B. HLB assembly for each construct (indicated at left) was assessed by confocal microscopy of intact salivary gland nuclei stained with Mxc and FLASH (left), Mute and FLASH (center) or Mxc and Lsm 11 (right). The endogenous HLB contained Mxc (pink), FLASH (green), Mute (red) and Lsm11 (blue) in all samples (arrowhead). Note the assembly of an ectopic HLB with each marker for nuclei containing the HLT-FL or the H3-H4 transgenes (arrow). Scale bar indicates 10 μm.
C. Quantification of ectopic HLB formation. Error bars depict SEM.
D. Expression of histone mRNA from the HLT-FL transgene was assessed throughout development by 5′ S1 nuclease protection assay using a 32P end-labeled (red star) probe (P) complementary to either the H2a or H4 endogenous and ectopic transcripts. Numbers above the gel indicate the amount PTE of RNA (μg, except glands which were total number of glands) in each reaction. The S1 nuclease assay is diagrammed below the gel. Numbers indicate the length in nt of the probe (P), ectopic (T) and endogenous (E) protected H2a or H4 transcripts. The black triangle indicates nuclease cleavage of the probe at the point where the RNA (vertical dashed line below probe) is not complementary.
E. Expression of histone mRNA was assessed in salivary glands by 5′ S1 nuclease protection assay. Roman numerals indicate the transgene inserted in each sample (depicted in A). Note that ectopic histone expression (T) was detected from constructs carrying HLT-FL and H3-H4.
F. Relative histone mRNA expression was measured for H2a (light blue columns) and H4 (light green columns) by qRT-PCR and quantification of the S1 protection assay (dark columns). Both assays are presented as fold expression compared to HLT-FL, which was set at 1.0. Error bars depict SEM.
HLT-FL supported HLB formation in nearly 100% of nuclei with all marker pairs (Fig. 2B,C). The H3-H4 construct formed ectopic HLBs in 40% to 95% of the cells depending on the marker pair examined (Fig. 2B,C). In contrast, less than 20% of cells containing either the H2a-H2b gene pair or the H1 gene formed an ectopic HLB, similar to non-transgenic controls (Fig. 2C). All four markers were present in the HLBs that formed on the H3-H4 gene pair, which was similar in size to the _H2a_-H2b or the H1 transgenes. These data indicate that a specific sequence(s) within the H3-H4 gene pair directs HLB assembly.
To test whether HLB assembly was important for histone gene expression, we developed an S1 nuclease protection assay using a 5′ end-labeled probe (P) containing the FLAG-tagged H2a or H4 genes (Fig. 2D). These probes detect both the endogenous (E) H2a and H4 histone mRNAs and the longer mRNAs produced by the transgenes (T) (Fig. 2D). This assay is quantitative and allowed us to determine the relative level of ectopic versus endogenous histone mRNA accumulation by comparing signal intensities between the S1 nuclease protected fragments in each sample. To validate the assay, we analyzed varying amounts of total RNA isolated from dissected salivary glands, 3-6 hr old embryos (diploid cells), and ovaries and whole 3rd instar larvae (mixed diploid and polyploid cells). The ectopic H2a and H4 genes in HLT-FL were expressed at ~7% the level of the endogenous genes in salivary glands, compared to 2.5% in ovaries and 1% in embryos and whole larvae (Fig. 2D). While the basis for these differences is not known, they may be due to under-replication of the endogenous histone genes relative to the rest of the salivary gland genome (Hammond and Laird, 1985). The relatively high expression of the ectopic histone mRNA as measured by S1 nuclease protection assay made salivary gland RNA the best source to carry out subsequent experiments.
We determined the expression of the FLAG tagged H2a and H4 genes in the H3-H4 and H2a-H2b transgenic lines relative to the expression of the corresponding gene in the HLT-FL full repeat unit. The H4 gene in the H3-H4 line was expressed at 35% of the level of the H4 gene in HLT-FL (Fig. 2E lanes 4-6). In contrast, the H2a gene in the H2a-H2b gene pair was expressed at <5% of the level in the full-length transgene (Fig. 2E lanes 1-3). These S1 nuclease assays were consistent with data obtained by qRT-PCR (Fig. 2F). These data correlate well with HLB assembly, which occurred with the H3-H4 gene pair but not H2a-H2b (Fig. 2B,C), suggesting that HLB assembly contributes to histone gene expression.
The H3-H4 promoter is necessary and sufficient for HLB formation
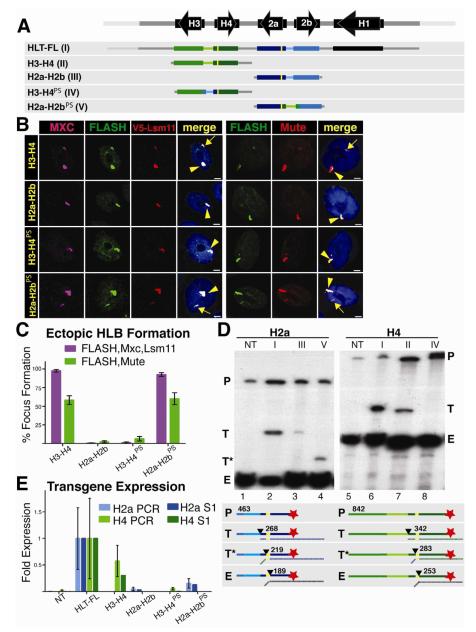
Both the H3-H4 and H2a-H2b constructs contain a ~300 nt bidirectional promoter, the two coding regions, and the mRNA 3′ end processing signals. To determine the sequences responsible for HLB formation, we swapped the intergenic promoter region (i.e. from start codon to start codon) of the H2a-H2b genes with the corresponding region of the H3-H4 genes (Fig. 3A) and generated transgenic insertions at 86Fb. We kept the FLAG tag on the N-terminus of the H2a and H4 genes to allow us to assess expression of the ectopic genes. Strikingly, the H2a-H2b gene pair containing the H3-H4 promoter (H2a-H2bPS) now formed an HLB, while the H3-H4 gene with the H2a-H2b promoter (H3-H4PS) did not (Fig. 3B). Moreover, the H2a-H2bPS transgene formed an HLB with the same efficiency as the H3-H4 transgene (Fig. 3C). Thus the H3-H4 intergenic region containing the bidirectional promoter is the critical element for HLB formation.
Figure 3. The H3-H4 Promoter Assembles an Ectopic HLB.
A. Diagram of the five constructs inserted into chromosomal location 86Fb. The yellow bars represent N-terminal FLAG tags in H2a and H4. The promoter swap (PS) includes the bidirectional promoter and 5′UTR from each gene pair.
B and C. HLB assembly for the indicated constructs was assessed with the indicated markers and quantified as in Figure 2. V5 antibody was used to detect Lsm11 in a strain where V5-Lsm11 replaces the endogenous protein. Note that the H3-H4 promoter (H3-H4, H2a-H2bPS) assembles an HLB, regardless of the associated transcript.
D and E. Histone gene expression was assessed and quantified as in Figure 2. Numbers in the diagram indicate the length in nt of the probe and possible protected fragments. Roman numerals refer to the depicted transgene, and NT is the no transgene control. Note robust histone mRNA expression from HLT-FL, H3-H4 (T) and H2a-H2bPS (T*)
In addition to transferring the capability for HLB assembly, the promoter swap also resulted in expression of the H2a transgene as determined by S1 nuclease protection and qRT-PCR (Fig. 3D,E) The H2a gene in H2a-H2bPS was expressed at levels similar to the H4 gene in the H3-H4 transgene (about 15% of the intact repeat; Fig. 3D, E), while the H4 gene in H3-H4PS was no longer expressed. Note that the promoter swap results in a smaller protected fragment with the H2a probe (Fig. 3D, lane 4) because the chimeric H2a gene now contains the H4 5′UTR, causing the S1 nuclease to cleave at the end of the FLAG tag rather than at the end of the H2a 5′ UTR (Fig. 3D, diagram). Also note that with the overloading of the endogenous histone mRNA, a low-level of expression (~3%) of the H2a gene in the original H2a-H2B gene pair is detected by the S1 nuclease assay (Fig. 3D, lane 3). Correspondingly, expression of the H4 gene in H3-H4PS was very low, and essentially undetectable with the S1 protection or the qPCR assay (Fig. 3D, lane 8; 3E). These results demonstrate that there is a sequence in the H3-H4 promoter that directs HLB assembly and high level histone gene expression.
mRNA Processing Signals are Dispensable for HLB Assembly
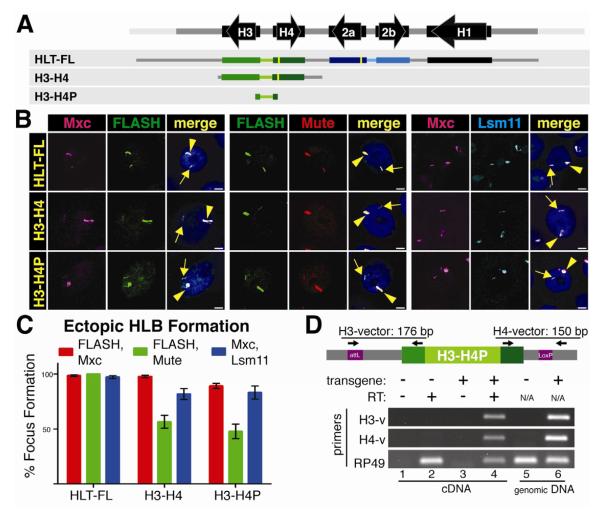
Our results thus far reveal an HLB assembly element in the H3-H4 intergenic region and a strong correlation between HLB formation and histone gene expression, but we cannot conclude a causal relationship between these two activities. For example, do the unique 3′ processing elements of a histone pre-mRNA contribute to HLB assembly? To determine whether mRNA produced from an intact histone gene influences HLB formation, we introduced just the 300 nt histone H3-H4 intergenic region (from start codon to start codon) into 86Fb (Fig. 4A). This fragment, H3-H4P, efficiently recruited Mxc, FLASH, and Mute, as well as U7 snRNP (Lsm11) (Fig. 4B,C), and was transcriptionally active. Hybrid transcripts containing either the H3 or H4 5′UTR and respective flanking vector sequences were detected by RT-PCR (Fig. 4D). H3-H4P generates transcripts containing only the 57 and 59 nt 5′ UTRs of the histone H3 and H4 mRNAs followed by flanking vector sequence, but contains no histone ORF, 3′ UTR or pre-mRNA processing signals. Hence, the observation that both FLASH and U7 snRNP were recruited by the H3-H4P construct rules out the possibility that the histone processing factors are recruited to the HLB by interacting with cis elements in the nascent transcript. They must be recruited directly to the HLB. However, as with the intact histone genes, there is still strong correlation between HLB assembly and transcription.
Figure 4. The H3 and H4 coding region and 3′ processing signals are not required for HLB assembly.
A. Diagram of the three constructs inserted into chromosomal location 86Fb. The yellow bars represent N-terminal FLAG tags in H2a and H4.
B and C. HLB assembly for the indicated constructs was assessed with the indicated markers and quantified as in Figure 2. Note that all three constructs assemble an HLB
D. Transcription from the H3-H4P transgene was assessed by RT-PCR using a primer in either the H3 or H4 5′UTR and corresponding flanking vector sequence as diagramed above the gel. Transcripts were detected in the H3-H4P strain (lane 4) and not the NT control. RP49 transcripts were detected in all cDNA preparations. Genomic DNA was analyzed in lanes 5 and 6, confirming the presence of the transgene.
Transcription Stimulates HLB Maturation
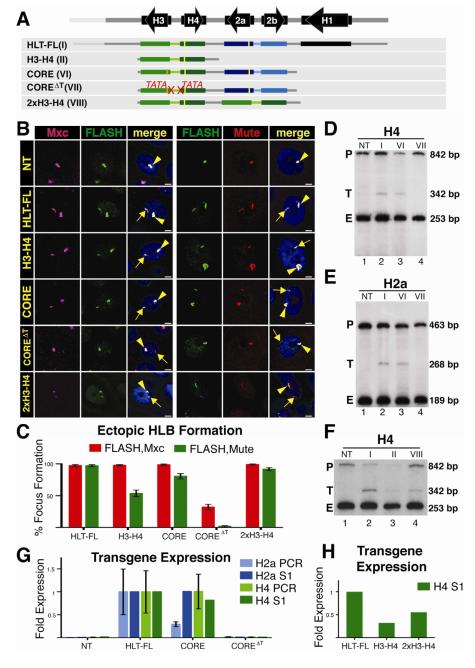
In the developing embryo, the transition from proto-HLB to a mature HLB occurs at the onset of zygotic transcription (White et al., 2011). To test whether transcription plays a direct role, we In the developing embryo, the transition from the H3-H4 promoter on HLB formation. We also asked if transcription from this promoter was necessary for expression of other histone genes in the cluster. To address these questions we inserted a transgene (CORE) at 86Fb containing just the 4 core histone genes (i.e. H3-H4 and H2a-H2b gene pairs) and a nearly identical transgene (COREΔT) different only in that both TATA boxes in the H3-H4 promoter were mutated (Fig. 5A). FLAG tags on the H4 and H2a genes and an HA-tag on the H3 gene allowed us to measure expression from these transgenes. The CORE construct assembled an ectopic HLB as efficiently as the full repeat unit (HLT-FL) and more efficiently than the H3-H4 gene pair, and both the H4 and H2a genes were expressed at levels close to that of the full repeat (Fig. 5B, C). As expected there was no expression of the H4 gene or the H3 gene from the COREΔT construct, as analyzed by S1 nuclease mapping (Fig. 5D, lane 4) or RT-PCR (not shown), respectively. Interestingly, Mxc and FLASH were recruited to the COREΔT construct, although the intensity of the signals and the frequency of HLB assembly were substantially lower than with the CORE construct (Fig. 5B,C; Fig. S1). In addition, Mute was not recruited to this construct (Fig. 5B,C). Thus, some HLB components, including Flash a pre-mRNA processing factor, assemble into a proto-HLB containing Mxc and FLASH in the absence of H3-H4 transcription.
Figure 5. Transcription is required for HLB assembly.
A. Diagram of the constructs inserted into chromosomal location 86Fb. HLB assembly was assessed and quantified as in Figure 2. The orange bar indicates the HA tag added to the H3 T gene. The 2867 bp CORE construct contains both the H2a-H2b and H3-H4 gene pairs. The COREΔT construct contains mutations in both the H3 and H4 TATA (ΔT) boxes. 2xH3-H4 contains a duplication of the H3-H4 gene pair in which only one of the H4 transgenes contains a FLAG tag.
B and C. HLB assembly for the indicated constructs was assessed with the indicated markers and quantified as in Figure 2. Note that while the CORE construct assembles an HLB, mutating the H3 and H4 TATA boxes reduces Mxc/FLASH assembly and results in undetectable Mute accumulation. Also note that increasing the number of transcription units increases Mute recruitment (compare H3-H4 and 2xH3-H4).
D-H. Histone gene expression was assessed and quantified as in Figure 2. Roman numerals refer to the depicted transgene, and NT indicates a no transgene control. Note the expected lack of transgenic H4 transcription (Fig.5 D) and absence of ectopic H2a (Fig.5 E) from the COREΔT construct. Also note that ectopic H4 mRNA levels increased upon addition of another H3-H4 gene pair (F).
See also Supplemental Figure 1.
Surprisingly, while H2a was expressed equally well from the HLT-FL and CORE constructs (Fig. 5E, lanes 2 and 3), there was little expression of the histone H2a gene from the COREΔT construct (Fig. 5E, lane 4). H2a expression from COREΔT was at least 10-fold less than that from the histone CORE construct (Fig. 5F), and similar to both non-transgenic controls (Fig. 5F) and the low level found from the single H2a-H2b gene pair (Fig. 2E,F). Thus, we conclude that transcription from the histone H3-H4 promoter is essential for activation of transcription of the histone H2a gene and likely the H2b gene as well, and for the stable recruitment of Mute to the HLB. Furthermore, these data suggest that HLB assembly nucleated at the H3-H4 intergenic region, together with expression from these promoters, is required for complete assembly of the HLB and full expression of all core histone genes in the repeat. The formation of a proto-HLB rather than a complete HLB, evidenced by failure of Mute to accumulate on the ectopic TATA mutant transgene, suggests that Mute recruitment, and hence HLB maturation, depends on transcription initiation from the H3-H4 promoter, and is not solely directed by a sequence element in the histone locus. We also observed a reduction in, but not the absence of, Mute localization to the H3-H4 construct (Fig. 2C, 5C). We hypothesize that the amount of Mute recruitment to the HLB may be related to the amount of transcription of the histone gene cluster, and that expression from two genes recruits insufficient amounts of Mute for us to detect it in all HLBs. Alternatively, Mute recruitment might require sequences from both H3-H4 and H2a-H2b genes. To distinguish between these possibilities, we replaced the H2a-H2b gene pair in the CORE construct with a second copy of H3-H4 (Fig. 5A). In order to directly compare the level of H4 gene expression from this transgene to our other transgenes, only one of the two H4 genes contained a FLAG sequence. Mute recruitment to the H3-H4/H3-H4 construct was higher than to the H3-H4 construct and comparable to that of the H3-H4/H2a-H2b CORE transgene (Fig. 5B,C). H4 expression from the H3-H4/H3-H4 construct also increased 1.6 fold compared to the H3-H4 construct (Fig. 5G,H). We conclude that Mute recruitment to the HLB positively correlates with the number of active histone gene promoters, and suggest that transcription from the H3-H4 promoter at the histone locus is an essential step in the development of a mature and stable HLB.
Stable assembly of the HLB during development requires the histone gene cluster
Our transgenic experiments in salivary glands have determined which sequences from the histone locus are sufficient to nucleate ectopic HLB formation. To determine whether formation of the HLB requires histone genes, we took advantage of a Drosophila mutation, Df(2L)Ds6, in which the entire histone gene cluster is deleted (Moore, 1983). Heterozygote Df(2L)Ds6/+ females are viable and fertile, and deposit sufficient maternal histone protein into the egg (shown for WT in Fig.6G) such that when mated to Df(2L)Ds6/+ males, the resulting homozygous mutant embryos lacking histone genes develop normally through S phase of cycle 14 (~3 hours of development), after which zygotic histone expression is required for normal S phase and cell cycle progression (Gunesdogan et al., 2010). If the histone locus is essential for all HLB components to assembly into a nuclear body, then we should not observe nuclear focus formation with any HLB marker in Df(2L)Ds6 homozygous mutant embryos. However, if any HLB components have self-organizational properties, we may detect nuclear foci with particular HLB markers even in the absence of histone genes.
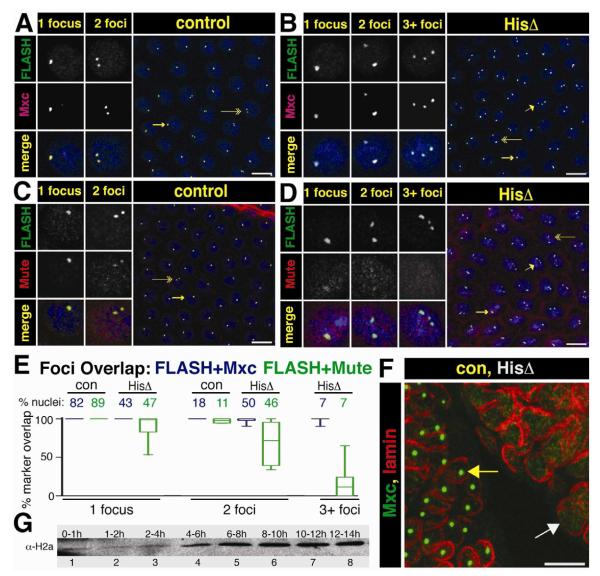
Figure 6. HLB assembly in the absence of the Histone Locus.
A.-D. Syncytial Df(2L)Ds6 and control sibling embryos from a 1.5 to 2.5 hr collection were stained with αFLASH and αMxc (panels A,B) or αFLASH and αMute (panels C,D. Arrows indicate the nuclei chosen for split channel magnification to highlight cells with 1 focus (long arrow), 2 foci (double arrow) and 3+ foci (short arrow). Scale bar = 10 μm.
E. Percent overlap was calculated by measuring complete overlap of FLASH foci and either Mxc (blue) or Mute (green) in each nucleus. The percent of total nuclei represented in each class (control or histone deletion for each antibody pair) is given across the top X axis (FLASH+Mxc, con N=943, HisΔ N=758, FLASH+Mute, con N=1144, HisΔ N=1397). The box represents the 25th to 75th percentiles, the line in the box represents the median value, and the bars extend to the 10th and 90th percentiles.
F. MXC foci (green) are not present in germ band extended mutant embryos (cell cycle 15 and 16). Mxc HLBs are present in the GFP positive sibling control nuclei, as outlined by lamin (red, left embryo, yellow arrow), whereas no foci are present in the GFP negative histone deletion embryo (right, white arrow).
G. Western blot analysis of histone H2a levels from an equal number of staged wild type embryos.
See also Supplemental Figure 2 and Supplemental Table 1.
To address these questions, we stained populations of syncytial blastoderm staged embryos collected from Df(2L)Ds6/+ heterozygous parents with antibodies against FLASH and Mxc or with antibodies against FLASH and Mute. These embryos were also stained with MPM-2 monoclonal antibodies, which detects a phosphoepitope on Mxc present in cells with active Cyclin E/Cdk2 (Fig. 6A-E, Fig. S2A,B) (White et al., 2011; White et al., 2007). Interestingly, all embryos at cycles 11 (when mature HLB assembly occurs and histone transcription normally begins) and 12 contained nuclei with foci of co-localizing Mxc and FLASH that were all MPM-2 positive, even though 25% of these embryos lack histone genes (Fig. 6A,B; Fig. S2A,B). However, the pattern of HLB marker staining differed between embryos collected from Df(2L)Ds6/+ parents versus wild type parents. In wild type embryos, all nuclei contained either 1 or 2 FLASH/Mxc foci or FLASH/Mute foci, which represent paired (1 focus) and unpaired (2 foci) homologous histone loci, respectively (Fung et al., 1998; White et al., 2007). In Df(2L)Ds6/+ collections, ~75% of the embryos contained nuclei with either 1 or 2 Mxc + FLASH foci (Fig. 6A) and the remaining ~25% of embryos contained a small fraction (7%) of nuclei with 3 or more Mxc + FLASH foci, along with nuclei containing 1 or 2 foci (Fig. 6B,E). Because we never see nuclei with three or more Mxc + FLASH foci in wild type embryos, we conclude from these data that this phenotypic class represents the homozygous histone deletion genotype (Table S1; p < 0.05 via Chi-squared analysis).
In the embryos lacking histone genes, the FLASH foci were smaller than in controls (Fig. S2C). None of these foci stained intensely with Mute, in contrast to the cells in embryos containing histone genes at cycle 11, although weak Mute staining was detected in cells with one or two foci (and hence a greater amount of accumulated FLASH) (Fig. 6C-E). These results suggest that the Mxc/FLASH foci in histone deletion embryos are proto-HLBs. The percentage of nuclei with either one or two Mxc/FLASH foci in successive nuclear cycles was statistically similar between wild type and histone deletion embryos (Fig. S2D; p<0.05). These percentages reflect the degree of homologous chromosome pairing during early embryogenesis (Fung et al., 1998). This result suggests that Mxc/FLASH foci may be assembling on chromatin in the absence of histone genes. In addition, MPM-2 staining (and thus Cyclin E/Cdk2 phosphorylation of Mxc) occurs on these foci independent from histone transcription. Finally, by germ band extended stages (i.e. cycles 15-16) there were no nuclear foci present in the histone deletion embryos as assessed by staining for Mxc (Fig. 6F) or FLASH (not shown). These data indicate that Mxc and FLASH can self organize into NBs (proto-HLBs) in the absence of histone genes. Without the scaffold of the H3-H4 promoter sequence, these proto-HLBs are not stable, mature HLBs, and do not persist through embryogenesis.
DISCUSSION
In most organisms the genes encoding the five replication-coupled histone proteins are physically linked, suggesting that there has been selective pressure to maintain this linkage, which is not the case for most sets of genes (e.g. globins, ribosomal proteins) that are coordinately expressed. One reason for tight linkage of the histone genes might be to promote the localization of factors required for histone mRNA biosynthesis, particularly those needed to form the unique histone mRNA 3′ end. A second reason might be to help control the coordinated, cell cycle-regulated expression of all the histone genes via a cis-acting element(s) at the locus. Our studies in Drosophila provide evidence for a ~300 nt sequence containing only the H3-H4 bi-directional promoter that mediates the concentration of transcription and processing factors in the HLB and promotes expression of all the replication-coupled histone genes.
How do HLBs form during development?
Several lines of evidence suggest that Mxc and FLASH self-organize into a proto-HLB and participate in the earliest steps of HLB assembly. First, an Mxc/FLASH-containing body (that does not contain Mute) forms in cycle 10 of wild-type Drosophila embryos, one cycle prior to initiation of histone gene transcription (White et al., 2011). Similarly, in embryos that lack histone genes we observe nuclear foci containing Mxc and FLASH, but not robust or consistent staining of Mute, in cycles 11 and 12 when wild-type embryos have formed a complete HLB and initiate histone gene expression. In the absence of histone DNA, the initial proto-HLBs are unstable. Furthermore, FLASH and Mxc, but not Mute, were recruited to the ectopic transgene (COREΔT) in which H3 and H4 transcription was ablated. Finally, during mitosis Mxc and FLASH remain associated with the mitotic chromosomes, while Mute and U7 snRNP do not (White et al., 2011), and likely serve to nucleate formation of a mature HLB after each cell division. We suggest that the proto-HLB represents an initial intermediate in HLB assembly that associates tightly with the H3-H4 promoter.
Although many NBs are not associated with a genomic locus, it has been proposed that a nucleotide sequence (often RNA) “seeds” formation of the structure (Dundr, 2011). Here we show that a ~300 nt sequence containing the bi-directional H3-H4 promoters is sufficient to recruit multiple HLB components, while the H2a-H2b and H1 promoters, or other regions of the histone repeat, are not. Interestingly, the H3-H4 promoter fragment alone formed an HLB, indicating that the generation of full-length transcripts is dispensable for HLB assembly. However, preventing H3-H4 transcription suspends HLB development at the proto-HLB step. The inactivated H3-H4 promoter stabilizes this intermediate in HLB assembly. We conclude that while the 300nt sequence between the H3 and H4 genes provides a scaffold for HLB assembly, activity from the H3-H4 promoter is necessary to form the mature HLB.
The precise role of transcription from the H3-H4 promoter in HLB formation is not clear from our studies. Recruitment of the additional factors (Mute and U7 snRNP) to the HLB could be mediated through the assembly of the core transcription machinery at the H3-H4 promoter, a change in the phosphorylation status of RNA Polymerase II, and/or active transcription opening up the adjacent chromatin. Since many components of the RNA Pol II machinery assemble on promoters and remain “poised” even at times when transcripts are not being actively generated (Nechaev et al., 2010; Zeitlinger et al., 2007), one possibility is that components of the transcription machinery bind the H2a-H2b promoter and require a signal from the H3-H4 promoter for initial activation. We also cannot distinguish whether activation of the histone H2a-H2b genes either simply requires transcription from the H3-H4 promoter or formation of the mature HLB. We speculate that the initial transcription from the H3-H4 promoter stimulates mature HLB formation encompassing the entire histone repeat by facilitating the recruitment of additional Mxc and FLASH to the chromatin.
How might the 300nt between H3 and H4 activate transcription of the H2a-H2b genes? This sequence has some properties of an enhancer in that it activates genes from a distance. Since this sequence contains the core promoter for the H3-H4 genes it seems unlikely that it acts like a classical enhancer, by looping the chromatin between the H3-H4 and H2a-H2b promoters. In addition, the function of the putative enhancer would not be affected by mutation of the TATA box. It seems more likely that transcription from the H3-H4 promoter, which leads to recruitment of the additional HLB factors, results in activation of the H2a-H2b genes. This could result either from the HLB factors altering the chromatin structure throughout the histone locus or by them directly recruiting coactivators to the histone genes.
Model for HLB Assembly
In our model of HLB formation (Fig. 7), the initial event is a stochastic association of Mxc and FLASH that is triggered by an unknown mechanism at embryonic cycle 10 and that can occur independently of the histone genes. This Mxc/FLASH proto-HLB complex associates with the histone H3-H4 promoter prior to transcription of the histone locus, and provides a platform for the subsequent recruitment of the remaining HLB components. Recruitment of additional components, such as Mute, requires assembly of the core transcription complex or actual transcription from the promoter. Neither FLASH nor Mxc has obvious sequence-specific DNA binding domains, and current evidence suggest that in mammals the Mxc orthologue NPAT functions as a co-activator binding to some component of a complex present on the promoter, rather than binding directly to the DNA itself (Miele et al., 2005; Wei et al., 2003; Ye et al., 2003). There may be another, as yet undefined, component of the HLB that directly binds to the H3-H4 promoter as well as to Mxc/FLASH to form the initial complex on the histone gene repeat, or else a unique feature of the chromatin in this region recruits Mxc/FLASH. Close inspection of the 300 nt sequence in 12 Drosophila species did not reveal any highly conserved elements in the H3/H4 promoter other than the TATAA boxes (Fig. S3).
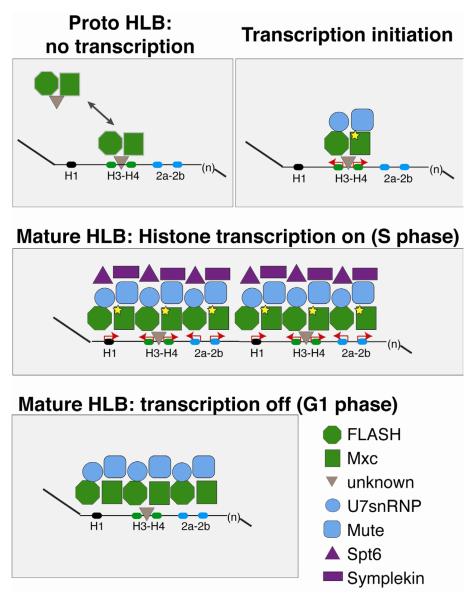
Figure 7. Model for HLB assembly and maintenance.
FLASH and Mxc form a “proto-HLB” in the absence of transcription. The proto-HLB is detected in syncytial stage embryos (cycle 10), histone deletion mutant embryos, and loci where histone transcription is abolished through mutation of the H3-H4 TATA boxes. While FLASH and Mxc are the earliest known components that begin to organize the HLB, it is possible that an additional factor(s) facilitates the interaction between Mxc/FLASH and the histone locus (grey triangle). Next, Cyclin E-mediated phosphorylation of Mxc (star) coincides with the onset of zygotic transcription during embryonic cycle 11. Transcription initiation from the H3-H4 promoter recruits Mute and U7 snRNP (Lsm11) to the HLB, and FLASH and Mxc continue to accumulate at the locus. S phase expression of all replication dependent histone genes transiently recruits to the locus other factors required for mRNA biogenesis, such as Spt6 and Symplekin. Finally, once established, the HLB remains associated with the locus in the absence of histone gene expression.
See Also Supplemental Figure 3.
HLBs compared with other nuclear bodies
The HLB differs from most NBs (e.g. Cajal bodies and PML bodies) because it is constitutively associated with a specific locus and the biosynthesis of a specific class of mRNAs. The nucleolus, which also assembles at a specific, repetitive gene locus, is one nuclear body with many similarities to the HLB. Indeed, a similar approach to ours that utilized ectopic rDNA genes in the Drosophila salivary gland has been used to address issues of nucleolar formation (Karpen et al., 1988). Transcription complexes including PolI, SL1 and Ubf form on arrays of rRNA promoters containing no rDNA coding regions, resulting in assembly of a body that has some features of the nucleolus (Prieto and McStay, 2008). Maintenance of morphologically complete nucleoli at rDNA genes requires both transcription and processing of rRNA (Hernandez-Verdun, 2006). Pre-nucleolar bodies form in experimentally induced micro-nuclei containing no rDNA (Hernandez-Verdun et al., 1991). Thus, there are likely to be similarities between the assembly process of the nucleolus and the HLB (Denissov et al., 2011). However, the incredible complexity of the nucleolus makes it difficult to comprehensively investigate the relationship between nucleolar structure and its multiple associated functions, some of which are not specifically involved in rRNA production (reviewed in (Boisvert et al., 2007)).
What are the functions of the HLB?
The HLB components that we have studied here (Mxc, FLASH, Mute and U7 snRNP) are all concentrated exclusively in the HLB and each is essential for proper Drosophila development (Bulchand et al., 2010; Godfrey et al., 2006; Godfrey et al., 2009; Saget et al., 1998) (D.C.T, W.F.M. and R.J.D, unpublished). However, we cannot conclude from this observation that the HLB itself is essential. FLASH and U7 snRNP have clearly defined biochemical functions in histone pre-mRNA processing (Burch et al., 2011b; Godfrey et al., 2006; Godfrey et al., 2009; Yang et al., 2009), while NPAT is essential for histone gene expression in mammals and Drosophila (White et al., 2011; Ye et al., 2003). The biochemical function of Mute is not known (Bulchand et al., 2010). HLBs may enhance the efficiency or rate of biochemical reactions associated with histone mRNA biosynthesis by increasing the local concentration of low-abundance factors at the histone locus. For instance, we previously found U7 snRNP is expressed but not localized to HLBs in Drosophila H2aV mutants, resulting in misprocessing of histone mRNA (Wagner et al., 2007). Concentration of histone biosynthetic factors in the HLB may also provide a mechanism for the coordination of gene expression. It is remarkable that the TATA mutation in the COREΔT construct not only blocked H3-H4 transcription, but also suppressed transcription from the neighboring H2a gene containing an intact promoter, suggesting a role for the HLB in coordinating expression of all the histone genes. Precisely defining the HLB nucleation sequence will allow us to dissect the molecular details of control of coordinate expression of the multiple genes in the histone locus, as well as the specific interactions that direct HLB assembly.
MATERIALS AND METHODS
Drosophila Strains
Histone locus sequences (Supplemental Experimental Procedures Table 1) were inserted into pattB (gift from K. Basler) and integrated into either 86Fb (BDSC 23648) or 102D (BDSC 24488) by ϕC31 mediated recombination (BestGene, Inc).
Histone Expression Analysis
Total RNA was extracted from tissues with Trizol (Invitrogen). cDNA, synthesized with RevertAid reverse transcriptase (Fermentas), was used for SYBR green (Fermentas) mediated qPCR quantification with an Applied Biosystems 7900HT PCR machine and detection of hybrid H3-vector and H4-vector transcripts. (Primers listed in Supplemental Experimental Procedures Table 2). qPCR results are presented as an average of at least 3 biological replicates; error bars represent SEM. Total salivary gland RNA or control yeast tRNA was hybridized to either a FLAG-H2a or FLAG-H4 radiolabeled probe (details in Supplemental Experimental Procedures) and subjected to S1 nuclease digestion and analysis as previously described (Lanzotti et al., 2002).
Immunofluorescence
Polytene squashes were prepared as described (Paro, 2000). Staining conditions and antibodies used are summarized in Supplemental Experimental Procedures Tables 3 and 4. Images were obtained on a Zeiss 510 confocal microscope. Ectopic HLBs were quantified by determining the percentage of ectopic foci in nuclei of 15-20 1μM sections of a 150 μM2 area in the posterior of 7-10 salivary glands. Graphs represent the mean and SEM for each indicated transgene.
Analysis of Histone Deletion Embryos
Post-blastoderm histone deletion embryos were collected from Df(2R)Ds6/CyO,twi-GFP parents and identified by lack of GFP expression. Syncytial stage histone deletion embryos were genotyped by the number of FLASH foci per nucleus. Briefly, nuclei of 20 WT embryos ranging from cycles 11-14 were counted to (1) ensure that 3+ foci are never present and (2) determine that WT embryos have at least 10% of nuclei containing 2 foci. Images from 59 embryos of the Df(2L)Ds6/CyO, twi-GFP collection were sorted into three classes based on foci present in the nuclei of each image. Images with only 1 or 2 foci in each nucleus were identified as control siblings. This group was further categorized by identifying embryos containing 90% or greater nuclei with a single foci as heterozygous for the histone deletion, the remaining being WT. Histone deletion embryos were identified by the presence of nuclei containing 3+ foci. Significance was assessed by chi-squared analysis. HLB formation was assessed for deletion and sibling control embryos by examining pairs of HLB markers. For each genotype, FLASH foci were identified and then scored for overlap with Mxc or Mute. A nucleus was only considered positive if all FLASH foci co localized with the other marker. Nuclei contained 1, 2 or 3+ foci and the graph presents the percent of total overlap out of the total number of nuclei counted for each of these classes within a genotype for either FLASH/Mxc or FLASH/Mute. The results are presented as a box (25th -75th quartiles) and whiskers (10-90th percentile) plot. (GraphPad Software, La Jolla California, USA)
Supplementary Material
01
ACKNOWLEDGEMENTS
We thank Dan McKay and Greg Matera for comments on the manuscript, Jess Nesmith for help analyzing histone deletion embryos, Dr. Joe Gall for Lsm11 antibody, Prem Fort for the histone western blot, and Best Gene for fly transgenesis. This work was supported by NIH grants GM58921 to W.F.M. and GM57859 to R.J.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzouz TN, Schumperli D. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA. 2003;9:1532–1541. doi: 10.1261/rna.5143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Barcaroli D, Knight RA, Di Ilio C, Melino G, De Laurenzi V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene. 2010;29:802–810. doi: 10.1038/onc.2009.388. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Bulchand S, Menon SD, George SE, Chia W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J Cell Sci. 2010;123:2697–2707. doi: 10.1242/jcs.063172. [DOI] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff W, Dominski Z. The interaction betwen FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 2011a doi: 10.1261/rna.2566811. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 2011b;17:1132–1147. doi: 10.1261/rna.2566811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Rino J. RNA seeds nuclear bodies. Nat Cell Biol. 2011;13:110–112. doi: 10.1038/ncb0211-110. [DOI] [PubMed] [Google Scholar]
- Chen M, Galvao RM, Li M, Burger B, Bugea J, Bolado J, Chory J. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell. 2010;141:1230–1240. doi: 10.1016/j.cell.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Lessard F, Mayer C, Stefanovsky V, van Driel M, Grummt I, Moss T, Stunnenberg HG. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011;12:231–237. doi: 10.1038/embor.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell. 2009;20:5250–5259. doi: 10.1091/mbc.E09-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol. 2011;193:605–606. doi: 10.1083/jcb.201104087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung JC, Marshall WF, Dernburg A, Agard DA, Sedat JW. Homologous chromosome pairing in Drosophila melanogaster proceeds through multiple independent initiations. J Cell Biol. 1998;141:5–20. doi: 10.1083/jcb.141.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. Rna. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, White AE, Tatomer DC, Marzluff WF, Duronio RJ. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre-mRNA processing and play an essential role in development. RNA. 2009;15:1661–1672. doi: 10.1261/rna.1518009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunesdogan U, Jackle H, Herzig A. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 2010;11:772–776. doi: 10.1038/embor.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism 658 that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- Hammond MP, Laird CD. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma. 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D. The nucleolus: a model for the organization of nuclear functions. Histochem Cell Biol. 2006;126:135–148. doi: 10.1007/s00418-006-0212-3. [DOI] [PubMed] [Google Scholar]
- Hernandez-Verdun D, Robert-Nicoud M, Geraud G, Masson C. Behaviour of nucleolar proteins in nuclei lacking ribosomal genes. A study by confocal laser scanning microscopy. J Cell Sci. 1991;98(Pt 1):99–105. doi: 10.1242/jcs.98.1.99. [DOI] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Terminating histone synthesis to preserve centromere integrity. Dev Cell. 2010;18:335–336. doi: 10.1016/j.devcel.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ. Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol. 2002;14:692–699. doi: 10.1016/s0955-0674(02)00387-3. [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol. 2005;25:6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Concepts in nuclear architecture. Bioessays. 2005;27:477–487. doi: 10.1002/bies.20226. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Moore G, Sinclair D, Grigliatti T. Histone gene multiplicity and position effect variegation in Drosophila melanogaster. Genetics. 1983;105:327–344. doi: 10.1093/genetics/105.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paro R. Mapping protein distribution on polytene chromosomes by immunostaining. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophila Protocols. Cold Spring Harbor; Cold Spring Harbor Press; NY: New York: 2000. pp. 131–140. [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto JL, McStay B. Pseudo-NORs: a novel model for studying nucleoli. Biochim Biophys Acta. 2008;1783:2116–2123. doi: 10.1016/j.bbamcr.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Rajendra TK, Praveen K, Matera AG. Genetic Analysis of Nuclear Bodies: From Nondeterministic Chaos to Deterministic Order. Cold Spring Harb Symp Quant Biol. 2011 doi: 10.1101/sqb.2010.75.043. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saget O, Forquignon F, Santamaria P, Randsholt NB. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics. 1998;149:1823–1838. doi: 10.1093/genetics/149.4.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell. 2007;28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Jin J, Harper JW. The cyclin E/Cdk2 substrate and Cajal body component p220(NPAT) activates histone transcription through a novel LisH-like domain. Mol Cell Biol. 2003;23:3669–3680. doi: 10.1128/MCB.23.10.3669-3680.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Burch BD, Yang XC, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol. 2011;193:677–694. doi: 10.1083/jcb.201012077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell. 2009;36:267–278. doi: 10.1016/j.molcel.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol. 2003;23:8586–8600. doi: 10.1128/MCB.23.23.8586-8600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01