2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated glioma patients (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 3.
Published in final edited form as: Nat Med. 2012 Jan 26;18(4):624–629. doi: 10.1038/nm.2682
Abstract
Mutations in isocitrate dehydrogenase 1 and 2 (IDH1, 2) have been demonstrated in the majority of World Health Organization grade 2 and grade 3 gliomas in adults. These mutations are associated with the accumulation of 2-hydroxyglutarate (2HG) within the tumor. Here we report the noninvasive detection of 2HG by proton magnetic resonance spectroscopy (MRS). The pulse sequence was developed and optimized with numerical and phantom analyses for 2HG detection. The concentrations of 2HG were estimated using spectral fitting in the tumors of 30 patients. Detection of 2HG correlated with mutations in IDH1 or IDH2 and with increased levels of D-2HG by mass spectrometry of resected tumor. Noninvasive detection of 2HG may prove to be a valuable diagnostic and prognostic biomarker.
Introduction
Isocitrate dehydrogenase converts isocitrate to α-ketoglutarate (αKG) in the cytosol (IDH1) and mitochondria (IDH2). The recent identification of mutations in IDH1 and IDH2 among the majority of patients with WHO grade 2 and 3 gliomas1,2 has directed attention to the role of abnormal metabolism in the pathogenesis and progression of these primary brain tumors. The mutations are confined to the active site of the enzyme and result in gain of function that generates D-2-hydroxyglutarate (2HG)3 and induces DNA hypermethylation4,5. This metabolite, normally present in vanishingly small quantities, can be elevated by orders of magnitude in gliomas harboring IDH1 or IDH2 mutations. Intracellular concentrations on the order of several micromoles per gram of tumor have been reported3.
While the metabolic consequences and downstream molecular effects of these mutations are yet to be elucidated, their potential value as diagnostic and prognostic markers in gliomas has been established from their clear association with improved overall survival when outcomes are compared between IDH mutated and IDH wild-type tumors2,6. Immunohistochemistry (IHC), using a commercially available antibody to the R132H mutation of IDH1, identifies approximately 93% of the mutations but the remaining 7% of tumors harboring a different IDH1 or an IDH2 mutation require direct sequencing for detection7. Since 2HG is produced by all known IDH mutant enzymes, evaluation of 2HG is an alternative indirect method for determining IDH status. The finding that 2HG is present at very high levels in _IDH_-mutated gliomas has raised the possibility that this metabolite could be detected noninvasively by MRS. Since brain MRI is the primary modality for clinical evaluation of glioma patients, the ability to detect 2HG by MRS could provide important diagnostic and prognostic information.
Here we report the noninvasive detection of 2HG in glioma in vivo by MRS at 3tesla. The point-resolved spectroscopy (PRESS)8 and difference editing9 sequences were optimized with quantum-mechanical and phantom analyses for detection of 2HG in the human brain and were applied to tumor masses in 30 adults with all grades of gliomas. Analysis of MRS data was blinded to IDH status. For each case in which 2HG was detected by MRS, an IDH1 or 2 mutation in the tumor was confirmed. Failure to detect 2HG by MRS was associated with the detection of wild type IDH1 and 2 in each case. The sensitivity and specificity of the method described here and the ease with which it could be incorporated into standard MR imaging suggests that 2HG detection by MRS may be an important biomarker in the clinical management of these patients.
RESULTS
Optimization of MRS methods
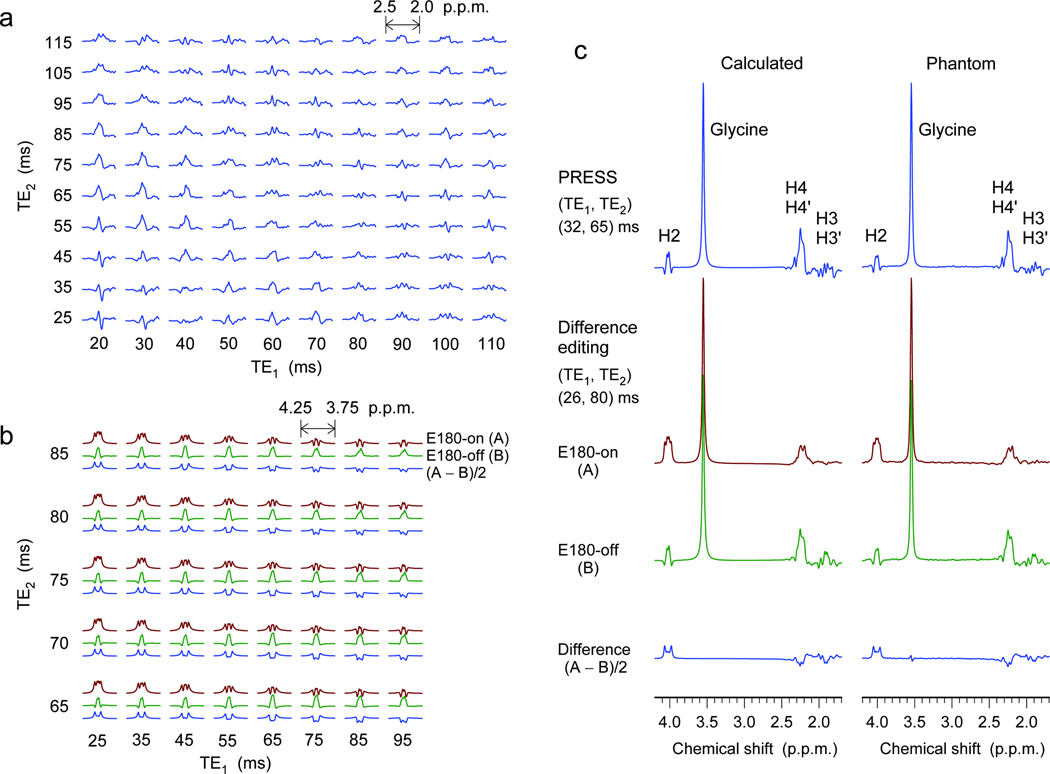
A 2HG molecule has five non-exchangeable scalar-coupled protons, resonating at 4.02, 2.27, 2.22, 1.98 and 1.83 p.p.m.10, giving rise to multiplets at approximately three locations at 3T; i.e., 4.02 p.p.m. (H2), approximately 2.25 p.p.m. (H4 and H4′), and approximately 1.9 p.p.m. (H3 and H3′) (Supplementary Fig. 1). The 2HG resonances are all scalar coupled and consequently the spectral pattern and signal strength vary with changing echo time of MRS sequence. A maximum 2HG signal may be expected at approximately 2.25 p.p.m. where the H4 and H4′ spins resonate proximately to each other. With its capability of full refocusing, a PRESS sequence was employed as a major tool for 2HG measurement in the present study. We conducted quantum mechanical simulations to search for optimal experimental parameters. The simulation indicated that the 2HG H4 resonances give rise to a maximum multiplet at total echo time of 90 – 100 ms, for which the first subecho time, TE1, is shorter than the second subecho time, TE2 (Fig. 1a). Given the large spectral distance of the H2 resonance from its weak coupling partners (H3 spins), we also measured the H2 resonance by means of difference editing. Selective 180° rotation of the H3 spins was switched on and off within a PRESS sequence in alternate scans to induce unequal H2 multiplets in subspectra. Subtraction between the spectra generated an edited 2HG H2 multiplet, canceling other resonances which were not affected by the editing 180° pulses. The computer simulation indicated that a large edited H2 signal can be obtained using a short-echo time set in which TE1 should be the shortest possible (Fig. 1b). The echo times of the PRESS and difference editing sequences were optimized as (TE1, TE2) = (32, 65) and (26, 80) ms, respectively. We tested these optimized MRS sequences in an aqueous solution with in-house synthesized 2HG. The spectral pattern and signal intensity of 2HG were consistent between calculation and experiment (Fig. 1c).
Figure 1.
(a) Quantum-mechanically calculated spectra of the 2HG H4 resonances, at 3T, are displayed vs. TE1 and TE2 of PRESS (subecho times of the first and second slice-selective 180° radio-frequency pulses, respectively). (b) Calculated difference-edited multiplets of the 2HG H2 resonance are plotted vs. subecho times TE1 and TE2 of scalar difference editing. Shown for each TE1-TE2 pair are, top to bottom, E180-on (brown) and E180-off (green) subspectra, and difference between the two subspectra (blue). Here E180 denotes editing 180° pulses tuned to 1.9 p.p.m. PRESS and edited spectra are all broadened to singlet line width of 4 Hz. Spectra in (a) and (b) are scaled equally for direct comparison. Signal reduction due to T2 relaxation effects was not included in the calculations. (c) Calculated and phantom spectra of 2HG for PRESS and difference editing. The echo times were 97 and 106 ms for PRESS and editing. The concentrations of 2HG and glycine in the phantom were both 10 mM (pH = 7.0). Spectra are scaled with respect to the glycine singlet at 3.55 p.p.m..
The optimized PRESS provided a 2HG multiplet at approximately 2.25 p.p.m. with maximum amplitude among echo times greater than 40 ms (Supplementary Fig. 2a,b). Moreover, the optimized echo time gave rise to narrowing of the multiplet and substantial reduction of 2HG signals at approximately 1.9 p.p.m.. Similar signal modulation occurred in the glutamate multiplets, allowing 2HG to be measured with high selectivity against the background signals of adjacent resonances (Supplementary Fig. 2c,d). The optimized echo time is relatively long, so signal loss due to transverse relaxation effects may be considerable in vivo. However, given that 2HG does not exhibit a well defined spectral pattern at short echo times (e.g., 30 ms), the 2HG signals can be better resolved at the optimized long echo time, benefiting from the suppressed complex baseline signals of macromolecules. The signal yield of 2HG in difference editing was low (38%) compared to PRESS (Fig. 1), but the editing provides an evident tool for proving 2HG elevation since the edited signal at 4.02 p.p.m. is uniquely generated via the coupling connections of 2HG. For in vivo, since the difference editing utilizes spectral difference induced by selective 180° rotations tuned at approximately 1.9 p.p.m., the 4.15-p.p.m. resonance of the glutamate moiety of N-acetyl-aspartyl-glutamate11 is coedited, but the resonance is relatively distant from the 2HG 4.02-p.p.m. resonance and thus does not interfere with 2HG editing (Supplementary Fig. 3). The lactate resonance at 4.1 p.p.m.12 is not coedited since the coupling partners at 1.31 p.p.m. are not influenced by the editing 180° pulse.
For spectral fitting, the present study employed model spectra which were calculated including the effects of the volume-localized radio-frequency pulses used for in vivo, allowing spectral fitting by signal patterns identical to experiment. Calculation of spectra at numerous echo times for MRS sequence optimization was efficiently accomplished using the product-operator-based transformation-matrix algorithm in the quantum-mechanical simulations13–15 (Supplementary Methods). The spectral pattern of 2HG is pH-dependent10 with significant shifts noted for pH < 6.0 (Supplementary Fig. 4). Computer simulations and MRS sequence optimization for 2HG detection were performed assuming pH approximately 7 in tumors16–18.
2HG detected in MRS spectra from patients with gliomas
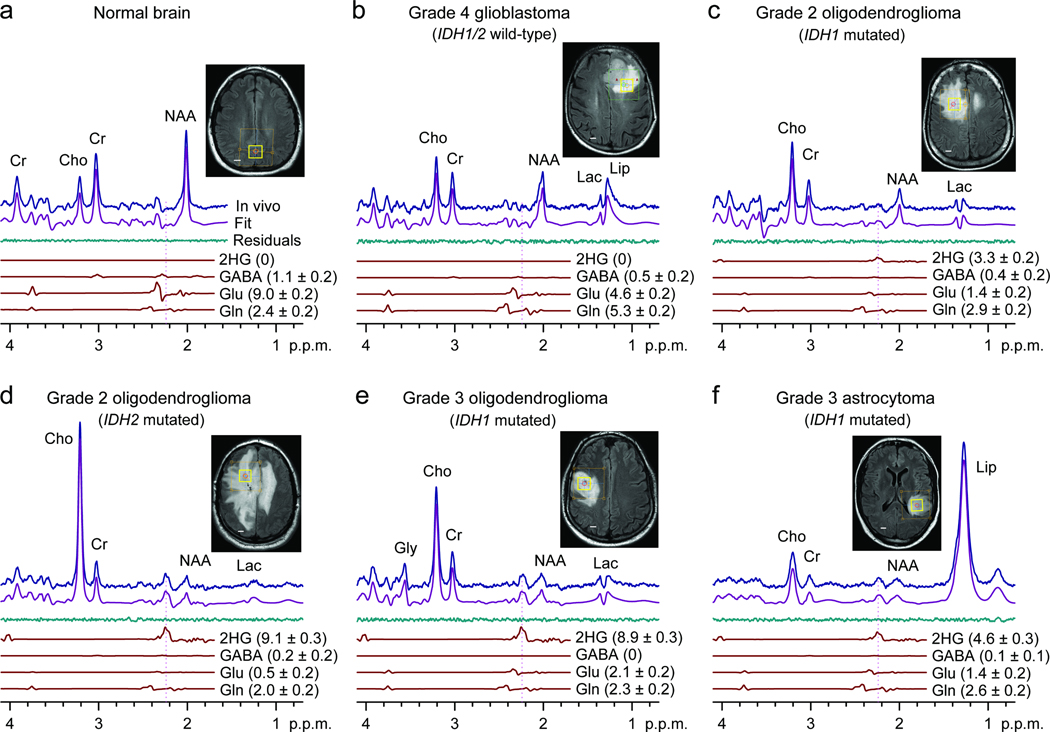
We included 29 glioma patients in the 2HG MRS analysis (Table 1). The optimized PRESS was applied to the tumor mass. A representative normal brain spectrum (Fig. 2a) demonstrated the expected pattern of choline, creatine, and N-acetylaspartate (NAA) without evidence of 2HG. In contrast, the classic pattern of elevated choline with decreased creatine and NAA was observed in all glioma grades (Fig. 2b-f). A signal attributed to 2HG was discernible at 2.25 p.p.m. in the WHO Grade 2 and 3 tumors (Fig. 2c-f), but not in the glioblastoma (Fig. 2b). An IDH1 or 2 mutation was identified in each of these cases. We analyzed the single-voxel localized PRESS data with linear combination of model (LCModel) software 19, using spectra of 20 metabolites as basis sets, calculated incorporating the volume-localized pulses. We estimated the concentration of 2HG using the brain water signal from the voxel as reference and adjusted the relaxation effects on the observed metabolite signals using published relaxation times of brain metabolites for 3Tesla (T) 20–22. With a 2 min scan on 2×2×2 cm3 brain tissues, 2HG was measurable for concentrations higher than 1.5 mM, with Cramér-Rao lower bound (CRLB) less than 18% (Table 1). With the use of precisely calculated model spectra for spectral fitting, the LCModel fits reproduced the in vivo spectra closely, resulting in residuals at the noise levels which did not exhibit considerable chemical-shift dependences.
Table 1.
Correlation between 2HG detection by MRS PRESS and IDH1 and 2 mutational status. The MRS measures labeled ‘Not detected’ were 2HG estimates ≤ 0.08 mM with CRLB ≥ 85%. The MRS estimates of 2HG concentrations were significantly different between IDH mutated and wild-type (unpaired t-test; p = 6×10−8).
| Histological Diagnosis | 2HG by MRSmM (CRLB) | IDH1, 2 Mutationsby DNA Sequencing | IDH1 (R132H) ByImmunohistochemistry |
|---|---|---|---|
| Oligodendroglioma (WHO Grade 2) | 2.7 (13%) | IDH2 (R172K) | Negative |
| Oligodendroglioma (WHO Grade 2) | 3.3 (11%) | IDH1 (R132H) | Positive |
| Oligodendroglioma (WHO Grade 2) | 2.6 (14%) | IDH1 (R132C) | Negative |
| Oligodendroglioma (WHO Grade 2) | 1.7 (17%) | IDH1 (R132H) | Positive |
| Oligodendroglioma (WHO Grade 2) | 3.3 (7%) | IDH1 (R132C) | Negative |
| Astrocytoma (WHO Grade 2) | 4.2 (10%) | IDH1 (R132H) | Positive |
| Astrocytoma (WHO Grade 3) | 2.1 (16%) | IDH1 (R132H) | Positive |
| Oligoastrocytoma (WHO Grade 3) | 3.9 (6%) | IDH1 (R132H) | Positive |
| Oligodendroglioma (WHO Grade 3) | 8.9 (3%) | IDH1 (R132H) | Positive |
| Oligoastrocytoma (WHO Grade 3) | 3.4 (8%) | IDH2 (R172W) | Negative |
| Astrocytoma (WHO Grade 3) | 2.7 (11%) | IDH2 (R172G) | Negative |
| Astrocytoma (WHO Grade 3) | 5.3 (6%) | IDH1 (R132H) | Positive |
| Astrocytoma (WHO Grade 3) | 2.5 (16%) | IDH1 (R132C) | Negative |
| Astrocytoma (WHO Grade 3) | 2.2 (15%) | IDH1 (R132C) | Negative |
| Astrocytoma (WHO Grade 3) | Not Detected | None | Negative |
| Sec Glioblastoma (WHO Grade 4) | 2.1 (15%) | IDH1 (R132H) | Positive |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
| Glioblastoma (WHO Grade 4) | Not Detected | None | Negative |
Figure 2.
In vivo single-voxel localized PRESS spectra from normal brain (a) and tumors (b-f), at 3T, are shown together with spectral fits (LCModel) and the components of 2HG, GABA, glutamate, and glutamine, and voxel positioning (2×2×2 cm3). Spectra are scaled with respect to the water signal from the voxel. Vertical lines are drawn at 2.25 p.p.m. to indicate the H4 multiplet of 2HG. Shown in brackets is the estimated metabolite concentration (mM) ± standard deviation. Abbreviations: Cho, choline; Cr, creatine; NAA, N-acetylaspartate; Glu, glutamate; Gln, glutamine; GABA, γ-aminobutyric acid; Gly, glycine; Lac, lactate; Lip, lipids. Scale bars, 1 cm.
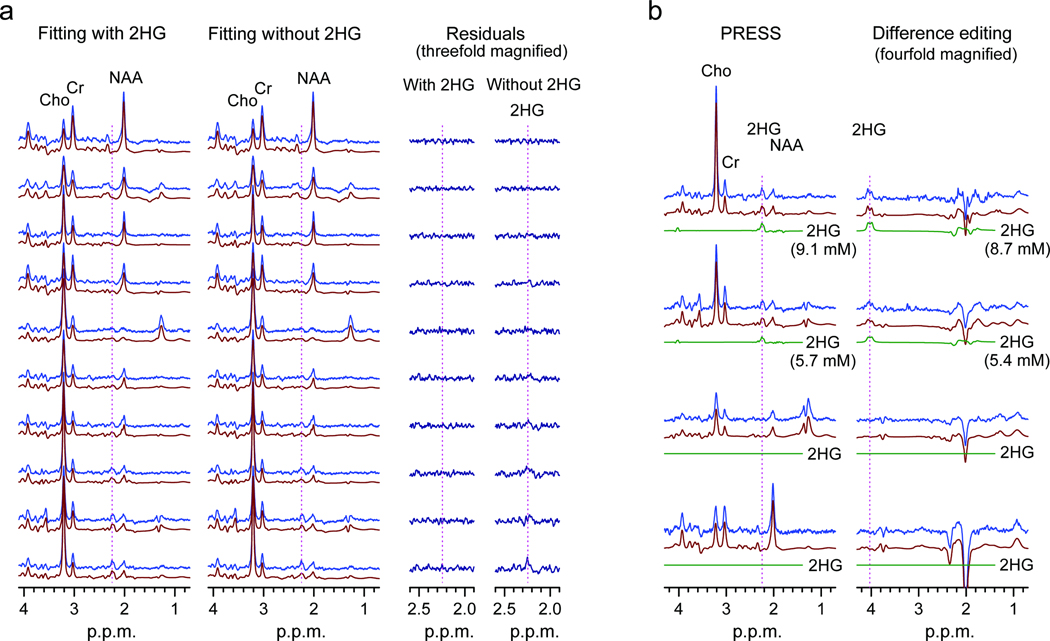
Gamma-Aminobutyric acid (GABA), glutamate and glutamine have resonances between 2.1 – 2.4 p.p.m.. Thus, the signals are partially overlapped with the 2HG 2.25-p.p.m. signal in PRESS spectra and can interfere with 2HG estimation depending on their signal strengths. Spectral fitting with the calculated spectra enabled resolution of the metabolites with CRLB < 20% for concentrations greater than approximately 2 mM (Fig. 2). We validated the PRESS detection of 2HG using two methods. First, we compared spectral fitting outputs from a basis set with or without a 2HG signal. For spectra without measurable 2HG signals (Fig. 3a, top traces), the residuals were essentially identical between the two fitting methods. However, spectra with a noticeable signal at 2.25 p.p.m., when fitted using a basis set without 2HG, resulted in large residuals at 2.25 p.p.m. (Fig. 3a, lower traces). For spectra with intermediate 2HG concentrations, the residuals were progressively larger with increasing 2HG estimates. This result demonstrates that the signal at 2.25 p.p.m. is primarily attributed to 2HG without substantial interference from the neighboring resonances. Second, we used difference editing to confirm the PRESS measures of 2HG in 7 subjects. When a signal at 2.25 p.p.m. was discernible in PRESS spectra, an edited H2 signal at 4.02 p.p.m. was detected (Fig. 3b, top traces). When 2HG was not measurable in PRESS spectra, no edited peak was observed at 4.02 p.p.m. (Fig. 3b, lower traces). This co-detection of the PRESS 2.25 p.p.m. peak and the edited 4.02 p.p.m. signal supports that the signals are both attributed to 2HG. The similarity between the 2HG concentrations estimated by PRESS and editing provides evidence that the PRESS measurement of 2HG is valid since the edited 2HG signal at 4.02 p.p.m. was generated without substantial interference via the scalar coupling connection between 4.02 and approximately 1.9 p.p.m. resonances, which is a unique feature of 2HG among known brain metabolites12.
Figure 3.
(a) LCModel fitting results (fits and residuals) of PRESS spectra obtained with basis set with or without 2HG. Data are displayed in the order of increasing 2HG estimates, top to bottom. (b) PRESS and difference-edited spectra from four subjects are shown in pairs, together with LCModel fits and 2HG signal components. Vertical lines are drawn at 2.25 and 4.02 p.p.m. in the PRESS and edited spectra, respectively.
Validation of MRS measures of 2HG by tissue analysis of IDH gene status
We analyzed each tumor for IDH gene status by immunohistochemistry (IHC) for the IDH1 R132H mutation and gene sequencing of IDH1 and 2 (Table 1). Of the 30 subjects studied, 15 had measurable 2HG by MRS and in each case, an IDH1 (12 of 15 subjects) or an IDH2 mutation (3 of 15 subjects) was confirmed. The remaining 15 subjects did not have detectable 2HG by MRS (≤ 0.08 mM, CRLB > 85%) and analysis of IDH1 and 2 revealed no mutations. The MRS estimates of 2HG concentrations were significantly different between IDH mutated and wild-type (unpaired t-test; p = 6×10−8).
Further validation was done by measuring D- and L-2HG concentrations in tumor samples by liquid chromatography/tandem mass spectrometry for 13 of the 30 subjects for whom sufficient frozen material from the initial tumor resection was available (Supplementary Table 1). Samples from brain adjacent to tumor were available for analysis from 3 patients and thus this tissue can serve as relative normal controls (Supplementary Fig. 5). L-2HG and D-2HG were clearly differentiated in the spectra. Five WT IDH1/2 GBM tumors had similar concentrations of L- and D-2HG. In all tumor samples, L-2HG was less than 1.0 nmol mg−1 protein. In marked contrast, D-2HG levels in IDH1 and 2 mutated tumors were 20-2000 fold higher than WT IDH GBMs (Supplementary Fig. 6). Together these data demonstrate that the presence of a 2HG peak in MRS is 100% correlated with the presence of a mutation in IDH1 or 2 and elevated concentrations of D-2HG in the tumor. Moreover, the absence of a 2HG peak when MRS is performed within a tumor mass is 100% correlated with wild type IDH1 and 2 and lack of accumulation of D-2HG in the tumor tissue. Thus, the ability to detect 2HG by MRS within a tumor mass is both highly sensitive and specific.
Development of multi-voxel imaging of 2HG
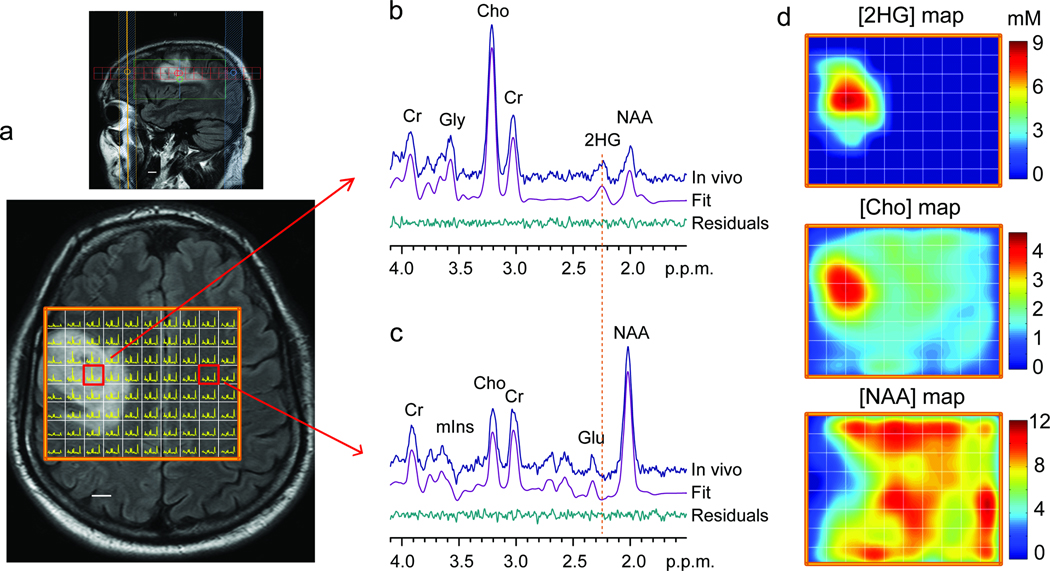
We extended the optimized PRESS echo time method to multi-voxel imaging of 2HG. The Grade 3 oligodendroglioma patient, whose single-voxel MRS data is shown in Fig. 2e, was scanned with 1×1 cm2 resolution on a 1.5 cm thick slice that included the tumor mass (Fig. 4a). The patterns of the single-voxel acquired spectra were reproduced in spectra obtained with the multi-voxel MRS method. The 2HG signal at 2.25 p.p.m. was clearly discernible in spectra from the tumor regions (Fig. 4b). Spectra from contralateral normal brain showed no 2HG signals at 2.25 p.p.m. (Fig. 4c). A map of 2HG concentrations (Fig. 4d) showed that 2HG was concentrated at the center of the increased T2w-FLAIR region. The 2HG concentrations were estimated using the normal brain NAA concentration of 12 mM12 as reference, giving 2HG concentration approximately 9 mM at the center of the tumor mass, in agreement with the 2HG estimates by single-voxel MRS. The spatial distribution pattern of choline was similar to that of 2HG within the region of increased T2w-FLAIR but, as expected, is found throughout the brain whereas 2HG had rapid drop off in normal brain. The NAA concentrations were low within the tumor mass, and the choline-to-NAA ratio showed high contrast between tumors and normal brain With its capability of 2HG detection in small volumes, the metabolic measures by the multi-voxel MRS method may contain reduced partial volume effects compare to the single-voxel MRS. Since 2HG is unique to tumor cells, the specificity of detection is an important advance in clinical MRS for IDH-mutated gliomas.
Figure 4.
(a) Multi-voxel imaging spectra from a patient with a WHO Grade 3 oligodendroglioma patient are displayed on top of the T2w-FLAIR image. The grid size is 1×1 cm, with slice thickness 1.5 cm. The spectra are displayed between 4.1 – 1.8 p.p.m. (left to right). (b,c) Two representative spectra (one from the tumor and another from the contralateral normal brain) are shown together with LCModel fits and residuals. (d) The estimated concentrations of 2HG, choline and NAA in individual voxels were color coded for comparison. The NAA level in gray matter in normal brain was assumed to be 12 mM. Scale bars, 1 cm.
DISCUSSION
We have detected 2HG noninvasively by optimized MRS methods in patients with gliomas and have shown concordance with mutations in IDH1 and 2 as well as accumulation of 2HG in tumor tissue. Currently, this is the only direct metabolic consequence of a genetic mutation in a cancer cell that can be identified through noninvasive imaging. The signal overlaps of 2HG with GABA, glutamate and glutamine, which occurs in short echo time standard data acquisitions, were overcome with multiplet narrowing by MRS sequence optimization and spectral fitting using precisely calculated basis spectra of metabolites. The methods presented here estimated metabolite concentrations using the brain water signal as reference in tumors, assuming an equal contribution of gray and white matter. The metabolite estimation may be valid only when the water concentration is similar among regions of the brain and between normal brain and tumor. The water concentration in tumors could be increased due to the effects of high cellularity or brain edema which would result in an underestimate of metabolite concentrations in the present study. Given a maximum possible water concentration of 55.6 M (bulk water), the true metabolite concentrations could be higher by up to 30% than our estimates, which were obtained using a water concentration of 42.3 M, calculated from the published values for the water concentrations in gray and white matter23. Although uncertainties in metabolite estimates can be theoretically minimized by employing an external reference signal such as from a phantom24, referencing with respect to brain water signals may be a realistic means of estimating metabolite concentrations in tumors25.
Two studies have previously reported in vivo detection of 2HG in the brains of patients with 2-hydroxyglutaric aciduria26,27. Large singlet-like signals at 2.5 – 2.6 p.p.m. were assigned to 2HG although this chemical shift assignment is not consistent with in vitro high resolution MR spectra of 2HG at neutral pH10. A 2HG signal at approximately 2.5 p.p.m., which is actually a multiplet, can occur only at very low pH (≈ 2.5) as reported previously10 and confirmed in this work (Supplementary Fig. 4). The pH measured noninvasively in a wide range of tumors ranges between 6.8 and 7.2. Even using microelectrode studies the lowest pH was approximately 6.0. Intracellular pH of approximately 7.0 has been reported in cancer cells16–18. The chemical shifts and coupling constants, used for MRS data analysis in the present study, were measured at pH 710. Since the proton NMR spectrum of 2HG is approximately constant between pH 6.5 – 7.5 (Supplementary Fig. 4), the efficiency of detecting 2HG in gliomas should not be sensitive to tumor pH.
A PRESS sequence used for 2HG measurement in the present study is commonly available in clinical MR systems. The field strength, 3T, is becoming more commonly used in the academic and clinical MR community and the data acquisition method could be implemented on standard hardware already in place in many MRI centers. Without the need for more specialized instrumentation or the production of expensive exogenous probes, the detection of 2HG by MRS is a method that could be quickly adopted for clinical use. Since the presence of an IDH1 or 2 mutation makes the diagnosis of glioma when evaluating a brain mass, the ability to detect 2HG by MRS will be a valuable diagnostic tool. Although not obviating the need for a surgical procedure to determine tumor grade, the presence of 2HG on MRS would differentiate tumor from a non-neoplastic process such as demyelinating disease. Moreover, the association of IDH1 and 2 mutations with improved survival among gliomas makes the detection of 2HG an important prognostic marker as well. The additional clinical value of this biomarker may be in its dynamic measurement over the time course of treatment and follow up. If 2HG concentrations reflect changes in tumor cellularity, then proliferation would lead to increased 2HG and tumor cells killed by radiation or chemotherapy would lead to decreased 2HG. Stable disease would be expected to have stable 2HG. While these correlations are still speculative, the potentially important uses include the follow up of WHO grade 2 glioma patients who typically have a long period of minimal progression followed rapidly by aggressive growth and transformation to high grade. Similarly, the availability of an imaging biomarker to follow for the detection of recurrent disease would be a major advance in the clinical management of glioma patients. Additional studies of dynamic properties of 2HG measurement will address these important potential uses.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/. Supplementary information is available on the Nature Medicine website.
Patient inclusion
We selected subjects from 2 University of Texas Southwestern Medical Center (UTSW) Institutional Review Board-approved brain tumor clinical protocols that have MR imaging and include MRS as part of the study procedures. We obtained informed consent for each subject. Scans from 53 subjects were screened and were included for analysis of 2HG if there was 1) a visible tumor mass by standard MR sequences (gadolinium enhancement or T2w-FLAIR signal abnormality), 2) MRS had been performed in at least 1 voxel in the tumor that was of acceptable spectral quality (singlet line width less than 6 Hz), and 3) there was adequate tissue available for IDH gene sequencing. Scans from 30 patients met these criteria and Twenty-nine patients had been imaged prior to initial surgery or after a limited surgical procedure (biopsy or subtotal resection) and had not been treated with chemotherapy or radiation. A patient with secondary GBM was imaged at the time of recurrence, 3 years after treatment with radiation. A search was done in each case for the availability of frozen tissue. In 13 of 30 cases, a frozen tumor sample was identified with 3 cases having both tumor and a sample of adjacent, non-tumor bearing, brain available for analysis.
MRS data acquisition
Experiments were carried out on a 3T whole-body scanner (Philips Medical Systems, Best, The Netherlands). A body coil was used for radio-frequency transmission and an 8-channel head coil for reception. Data were acquired according to our published methods28. PRESS8 and scalar difference editing9 were used for measuring 2HG in brain tumors. For editing, two 20-ms Gaussian 180° pulses, tuned to 1.9 p.p.m., were switched on and off in alternate scans to generate an edited H2 signal at 4.02 p.p.m. in difference spectra. The echo times of PRESS and editing were 97 and 106 ms, respectively. The quantum-mechanical simulations were carried out by means of the product-operator-based transformation matrix algorithm (Supplementary Methods). For in vivo MR scans, following the survey imaging, T2w-FLAIR images were acquired to identify tumor regions. For single-voxel localized data acquisition, a 2×2×2 cm3 voxel was positioned within the tumor mass. PRESS acquisition parameters included sweep width = 2500 Hz, 2048 sampling points, repetition time = 2 s, and 64 averages (scan time 2.1 min). Editing data were acquired with 384 averages (scan time 13 min). An unsuppressed water signal was acquired with echo time = 14 ms and repetition time = 20 s for use as reference in metabolite quantification. Spectroscopic imaging data were acquired, using the optimized PRESS echo time, from a 1.5-cm thick slice with resolution of 1×1 cm2. Undersampling of k-space data by 20% was employed, the scan time being approximately10 min (2 averages; repetition time = 1.3 s). Data were zero filled for the un-acquired k-space points and filtered with a cosine function prior to Fourier transformation.
MRS data analysis
Data were analyzed as described previously28. Following a 1-Hz apodization, spectra were fitted with LCModel software19, using calculated spectra of 20 metabolites as basis functions. The basis set included spectra of 2HG, NAA, GABA, glutamate, glycine, creatine, myo-inositol, glutamine, lactate, alanine, acetate, aspartate, ethanolamine, glutathione, phosphorylethanolamine, _scyllo_-inositol, taurine, N-acetylaspartylglutamate, glucose, and choline. The metabolite concentrations were estimated with respect to the short echo time water signal. Assuming an equal composition of gray and white matter in tumors, we used a water concentration value of 42.3 M, calculated from the literature values23 for the water concentrations in gray and white matter. Relaxation effects on metabolite signals were corrected using published metabolite T2 and T1; T2 = 150, 230 and 280 for Cr, Cho and NAA, and 180 ms for other metabolites, respectively, and T1 = 1.2 for 2HG, glutamate, glutamine and myo-inositol, and 1.5 for other metabolites20–22.
IHC for detection of R132H mutation in IDH1
Paraffin sections were cut at 4 µm thickness and prepared according to standard clinical methods. The primary antibody for IDH1 R132H (Dianova Inc.) was diluted 1:20.
IDH1/2 DNA Sequencing
DNA was prepared from frozen tissue by standard methods or from formalin-fixed paraffin embedded (FFPE) samples according to a published method29. Primers for IDH1 and 2 sequencing were used according to published methods1,3.
Extraction and measurement of 2HG enantiomers by liquid chromatography-tandem mass spectrometry
D-2HG and L-2HG were extracted from tumor tissue and normal brain and liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses performed as previously described30,31.
Statistical analysis
Cramér-Rao lower bounds of metabolite estimates, which represent the lower bounds of the precision, were obtained with the built-in algorithm of the LCModel software19.
Additional Methods
Detailed methods for 2HG synthesis and transformation-matrix-incorporated density-matrix simulations are described in the Supplementary Methods.
Supplementary Material
1
Acknowledgements
This work was supported by NIH grant RC1NS0760675 and by the Cancer Prevention Research Institute of Texas (CPRIT) grant RP101243-P04. We are grateful to Christie Sheppard, BSc, for expert management of the patient database and coordinating research scans and tissue samples, Chan Foong, MS, for expert assistance with pathological analysis of tumor, and to Richard L. Boriack, senior research associate, (Pathology, Children's Medical Center, Dallas, Texas, USA) for the expert assistance with 2HG measurements by mass spectrometry.
Footnotes
Contributions
C.C. developed the MRS methodology for 2HG detection, designed and performed the MR experiments and data analysis, supervised the MRS study, prepared figures, and wrote the manuscript. E.A.M. led all aspects of the patient study, contributed to data analysis, preparation of figures and manuscript. S.K.G. carried out MR data acquisition and contributed to data analysis. D.R. performed mass spectrometry analysis on resected tumors and contributed to manuscript preparation. Z.K. synthesized 2HG and prepared a figure. R.J.D. and C.M. contributed to the conceptual approach, review of the data and manuscript preparation and contributed to manuscript preparation. K.J.H. and J.M.R contributed to tumor sample collection and validation, neuropathological evaluation and diagnosis, evaluation of immunohistochemical stains, and manuscript preparation. X.L.Y. and T.M. performed the tissue evaluation of IDH mutations. I.M-V and J.J.P contributed to conceptual analysis. B.E.M. recruited patients and contributed to clinical data analysis and manuscript preparation. C.M. recruited patients and contributed to manuscript preparation. R.M.B. contributed to the conceptual approach, led the tumor analysis workup, contributed to data analysis and manuscript preparation.
Competing financial interests
The authors declare no competing financial interests.
Contributor Information
Changho Choi, Email: changho.choi@utsouthwestern.edu, Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX; Department of Radiology, University of Texas Southwestern Medical Center, Dallas, TX.
Sandeep K. Ganji, Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX Department of Radiology, University of Texas Southwestern Medical Center, Dallas, TX.
Ralph J. DeBerardinis, Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX McDermott Center for Human Growth and Development, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Kimmo J. Hatanpaa, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Dinesh Rakheja, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX; Children’s Medical Center, Dallas, TX.
Zoltan Kovacs, Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX.
Xiao-Li Yang, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX; Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; McDermott Center for Human Growth and Development, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Tomoyuki Mashimo, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX; Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Jack M. Raisanen, Department of Pathology, University of Texas Southwestern Medical Center, Dallas, TX Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Isaac Marin-Valencia, Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX.
Juan M. Pascual, Department of Pediatrics, University of Texas Southwestern Medical Center, Dallas, TX Department of Neurology, University of Texas Southwestern Medical Center, Dallas, TX; Department of Physiology, University of Texas Southwestern Medical Center, Dallas, TX.
Christopher J. Madden, Department of Neurological Surgery, University of Texas Southwestern Medical Center, Dallas, TX Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Bruce E. Mickey, Department of Neurological Surgery, University of Texas Southwestern Medical Center, Dallas, TX Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Craig M. Malloy, Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX Department of Radiology, University of Texas Southwestern Medical Center, Dallas, TX; Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX; VA North Texas Health Care System, Dallas, Texas, USA.
Robert Bachoo, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX; Department of Neurology, University of Texas Southwestern Medical Center, Dallas, TX; Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
Elizabeth A. Maher, Email: elizabeth.maher@utsouthwestern.edu, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX; Department of Neurology, University of Texas Southwestern Medical Center, Dallas, TX; Annette Strauss Center for Neuro-Oncology, University of Texas Southwestern Medical Center, Dallas, TX; Harold C. Simmons Comprehensive Cancer Center, University of Texas Southwestern Medical Center, Dallas, TX.
References
- 1.Balss J, et al. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 2.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christensen BC, et al. DNA methylation, isocitrate dehydrogenase mutation, and survival in glioma. J Natl Cancer Inst. 2011;103:143–153. doi: 10.1093/jnci/djq497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Von Deimling A, Korshunov A, Hartmann C. The next generation of glioma biomarkers: MGMT Methylation, BRAF Fusions and IDH1 Mutations. Brain Pathology. 2011;21:74–87. doi: 10.1111/j.1750-3639.2010.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bottomley PA. US Patent. United States: 1984. Selective volume method for performing localized NMR spectroscopy. Vol. 4 228. [Google Scholar]
- 9.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Bal D, Gryff-Keller A. 1H and 13C NMR study of 2-hydroxyglutaric acid and lactone. Magnetic Resonance Chemistry. 2002;40:533–536. [Google Scholar]
- 11.Krawczyk H, Gradowska W. Characterisation of the 1H and 13C NMR spectra of N-acetylaspartylglutamate and its detection in urine from patients with Canavan disease. J Pharm Biomed Anal. 2003;31:455–463. doi: 10.1016/s0731-7085(02)00691-x. [DOI] [PubMed] [Google Scholar]
- 12.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000;13:129–153. doi: 10.1002/1099-1492(200005)13:3<129::aid-nbm619>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 13.Ernest RR, Bodenhausen G, Wokaun A. Principles of nuclear magnetic resonance in one and two dimensions. Oxford: Clarendon Press; 1987. [Google Scholar]
- 14.Thompson RB, Allen PS. Sources of variability in the response of coupled spins to the PRESS sequence and their potential impact on metabolite quantification. Magn Reson Med. 1999;41:1162–1169. doi: 10.1002/(sici)1522-2594(199906)41:6<1162::aid-mrm12>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Choi C, et al. Improvement of resolution for brain coupled metabolites by optimized (1)H MRS at 7T. NMR Biomed. 2010;23:1044–1052. doi: 10.1002/nbm.1529. [DOI] [PubMed] [Google Scholar]
- 16.Gillies RJ, Raghunand N, Garcia-Martin ML, Gatenby RA. pH imaging. A review of pH measurement methods and applications in cancers. IEEE Eng Med Biol Mag. 2004;23:57–64. doi: 10.1109/memb.2004.1360409. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths JR. Are cancer cells acidic? Br J Cancer. 1991;64:425–427. doi: 10.1038/bjc.1991.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean LA, Roscoe J, Jorgensen NK, Gorin FA, Cala PM. Malignant gliomas display altered pH regulation by NHE1 compared with nontransformed astrocytes. Am J Physiol Cell Physiol. 2000;278:C676–C688. doi: 10.1152/ajpcell.2000.278.4.C676. [DOI] [PubMed] [Google Scholar]
- 19.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30:672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 20.Mlynarik V, Gruber S, Moser E. Proton T (1) and T (2) relaxation times of human brain metabolites at 3 Tesla. NMR Biomed. 2001;14:325–331. doi: 10.1002/nbm.713. [DOI] [PubMed] [Google Scholar]
- 21.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging. 2004;19:537–545. doi: 10.1002/jmri.20053. [DOI] [PubMed] [Google Scholar]
- 22.Ganji SK, et al. T2 measurement of J-coupled metabolites in the human brain at 3T. NMR Biomed. 2011 doi: 10.1002/nbm.1767. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norton WT, Poduslo SE, Suzuki K. Subacute sclerosing leukoencephalitis. II. Chemical studies including abnormal myelin and an abnormal ganglioside pattern. J Neuropathol Exp Neurol. 1966;25:582–597. doi: 10.1097/00005072-196610000-00006. [DOI] [PubMed] [Google Scholar]
- 24.Keevil SF, et al. Absolute metabolite quantification by in vivo NMR spectroscopy: II. A multicentre trial of protocols for in vivo localised proton studies of human brain. Magn Reson Imaging. 1998;16:1093–1106. doi: 10.1016/s0730-725x(98)00118-0. [DOI] [PubMed] [Google Scholar]
- 25.Tong Z, Yamaki T, Harada K, Houkin K. In vivo quantification of the metabolites in normal brain and brain tumors by proton MR spectroscopy using water as an internal standard. Magn Reson Imaging. 2004;22:1017–1024. doi: 10.1016/j.mri.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Sener RN. L-2 hydroxyglutaric aciduria: proton magnetic resonance spectroscopy and diffusion magnetic resonance imaging findings. J Comput Assist Tomogr. 2003;27:38–43. doi: 10.1097/00004728-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 27.Goffette SM, et al. L-2-Hydroxyglutaric aciduria: clinical, genetic, and brain MRI characteristics in two adult sisters. Eur J Neurol. 2006;13:499–504. doi: 10.1111/j.1468-1331.2006.01282.x. [DOI] [PubMed] [Google Scholar]
- 28.Choi C, et al. Measurement of glycine in the human brain in vivo by (1) H-MRS at 3 T: application in brain tumors. Magn Reson Med. 2011 doi: 10.1002/mrm.22857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher EA, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66:11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 30.Rakheja D, Mitui M, Boriack RL, DeBerardinis RJ. Isocitrate dehydrogenase 1/2 mutational analyses and 2-hydroxyglutarate measurements in Wilms tumors. Pediatr Blood Cancer. 2011;56:379–383. doi: 10.1002/pbc.22697. [DOI] [PubMed] [Google Scholar]
- 31.Rakheja D, et al. Papillary thyroid carcinoma shows elevated levels of 2-hydroxyglutarate. Tumour Biol. 2011;32:325–333. doi: 10.1007/s13277-010-0125-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1