A pseudogene long noncoding RNA network regulates PTEN transcription and translation in human cells (original) (raw)
. Author manuscript; available in PMC: 2013 Oct 1.
Published in final edited form as: Nat Struct Mol Biol. 2013 Feb 24;20(4):440–446. doi: 10.1038/nsmb.2516
Abstract
PTEN is a tumor suppressor gene that has been shown to be under the regulatory control of a PTEN pseudogene expressed noncoding RNA, PTENpg1. Here, we characterize a previously unidentified PTENpg1 encoded antisense RNA (asRNA), which regulates PTEN transcription and PTEN mRNA stability. We find two PTENpg1 asRNA isoforms, alpha and beta. The alpha isoform functions in trans, localizes to the PTEN promoter, and epigenetically modulates PTEN transcription by the recruitment of DNMT3a and EZH2. In contrast, the beta isoform interacts with PTENpg1 through an RNA:RNA pairing interaction, which affects PTEN protein output via changes of PTENpg1 stability and microRNA sponge activity. Disruption of this asRNA-regulated network induces cell cycle arrest and sensitizes cells to doxorubicin, suggesting a biological function for the respective PTENpg1 expressed asRNAs.
Keywords: Pseudogene, PTEN, PTENp1, PTENpg1, antisense RNA, noncoding RNA, Epigenetics, Transcriptional regulation, DNMT3a, EZH2
Introduction
The tumor suppressor gene phosphatase and tensin homolog (PTEN) is a negative regulator of the PI3K-Akt pathway and is epigenetically silenced in several cancers1. The dosage of PTEN expression has been found to correlate with the severity of epithelial cancers2, indicating that a fine-tuned regulation of the PTEN gene is critical for maintaining cellular homeostasis. PTEN expression has been found to be post transcriptionally regulated by the action of a PTEN pseudogene (PTENpg1, also known as PTENp1, PTEN2 and PTENΨ1)3. The PTENpg1 is a long noncoding RNA (lncRNA), which was found to sequester numerous _PTEN_-targeting microRNAs (miRNAs) by acting as a miRNA sponge. The observed miRNA sponge effect from over-expressing the 3’UTR of the PTENpg1 lncRNA resulted in increased PTEN mRNA stability and increased amounts of PTEN protein, presumably due to miRNA sequestration away from the PTEN protein coding transcripts. In contrast, suppression of the PTENpg1 lncRNA released miRNAs targeting PTEN, which instead lead to destabilization of PTEN.
Expressed pseudogenes are conserved across millions of years of primate evolution4 and have traditionally been considered “non-functional DNA” that litters the human genome. Pseudogenes are almost as numerous as protein-coding genes5 and recent evidence show some of these to be transcribed into RNA3,6. An emerging paradigm suggests that some pseudogenes may be functional in directly regulating their protein-coding counterparts at either the transcriptional or post-transcriptional level3,7–9. In contrast to PTENpg1 mediated post-transcriptional regulation of PTEN, the pluripotency-associated transcription factor OCT4 was reported to be under the epigenetic regulation of an antisense RNA (asRNA) to the OCT4 pseudogene 57. Taken together, these previous observations encouraged us to investigate whether there was an asRNA also encoded from the PTENpg1 locus and to what extent such an asRNA was involved in epigenetically regulating the tumor suppressor gene PTEN.
Results
The PTEN pseudogene, PTENpg1, encodes asRNA
To investigate the genomic landscape and the presence of asRNA transcription at the PTENpg1 locus, we assessed Expressed Sequence Tags (EST) in the UCSC genome browser and also carried out an independent analysis of ENCODE Chromatin Immunoprecipitation (ChIP) sequencing data. The assessment of EST reads indicated asRNA transcription from the PTENpg1 locus (Supplementary Fig. 1a). In addition, our analysis of ENCODE ChIP sequencing data for the presence of the active transcriptional histone mark H3K4me3 indicated differential patterns among different cell lines (Supplementary Fig. 1b). Moreover, analysis of H3K4me3 and RNA Polymerase II (RNAPII) localization in human embryonic stem cells (H1-hESC) and K562 cells, showed overlap and binding at two different loci, indicating promoter activity and two different transcriptional start sites (TSS) at the PTENpg1 locus (Supplementary Fig. 1b).
Next, we set out to investigate whether the ChIP sequencing peaks for H3K4me3 and RNAPII corresponded to the TSS for the indicated PTENpg1 asRNA transcripts (Supplementary Fig. 1a–b). To this end we carried out 5’ RACE (Supplementary Table 1a) and primer walk (Supplementary Fig. 2a–b) analysis. These analyses indicated two different TSSs at the PTENpg1 locus that initiate asRNA transcription. In total, three dominant PTENpg1 asRNA isoforms were identified (unspliced, α, and β) (Fig. 1a, Supplementary Fig. 2a–e, and Supplementary Table 1a) as well as alternative splicing of the PTENpg1 asRNA exon 3 (Supplementary Fig. 2c and Supplementary Table 1b). Cellular fractionation showed that the spliced α and β isoforms were expressed at high levels in the cytoplasm, whereas the unspliced PTENpg1 asRNA α isoform was exclusively found in the nuclear fraction (Supplementary Fig. 2d–e). Depletion of polyadenylated (poly(A)) RNA from total cellular RNA confirmed the spliced α and β isoforms to be poly(A) positive in contrast to the PTENpg1 sense and unspliced PTENpg1 asRNA α isoforms, which were mainly poly(A) negative transcripts (Fig. 1b and Supplementary Fig. 2f).
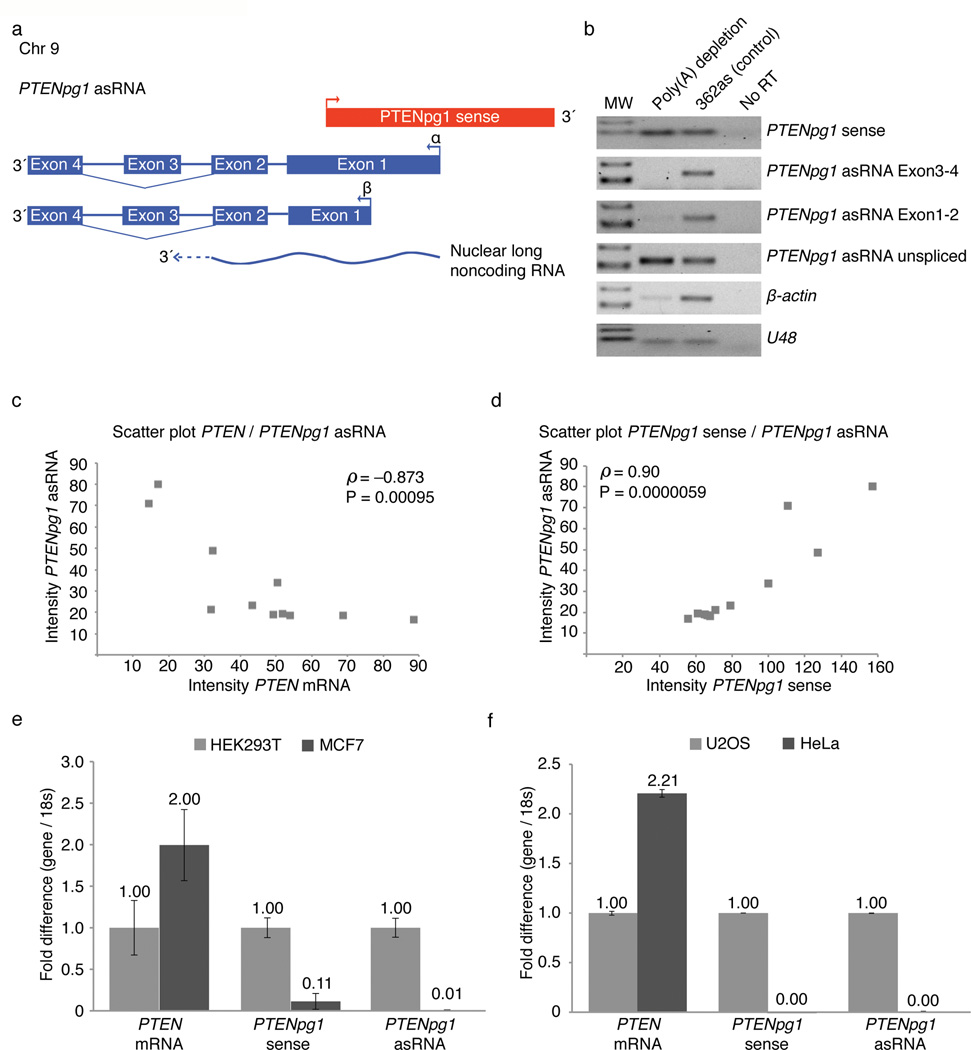
Figure 1. High expression of PTENpg1 asRNA correlates with low PTEN mRNA expression.
(a) A schematic depicting the PTENpg1 sense and asRNA divergently transcribed locus. (b) Semi-qRTPCR of poly(A) depleted fractions was assessed for PTENpg1 sense and different isoforms of the PTENpg1 asRNA. (c–d) Semi-qRTPCR (Supplementary Fig. 3d) was quantified using ImageJ software. A Spearman rank correlation (n=11) and scatter plot was carried out on the quantified PCR products. (c) Scatter plot contrasting PTEN and PTENpg1 asRNA. (d) Scatter plot contrasting PTENpg1 sense and PTENpg1 asRNA. (e) QRTPCR assay on HEK293T cells (high PTENpg1 asRNA) contrasted the MCF7 cell line (low PTENpg1 asRNA) (n=2). (f) QRTPCR assay on U2OS cells (high PTENpg1 asRNA) contrasted the HeLa cell line (low PTENpg1 asRNA) (n=2).
Characterization of the PTENpg1 asRNA
The dominantly spliced α and β asRNA transcripts appeared to emanate from two different TSSs that curiously overlap with the PTENpg1 sense promoter (Fig. 1a, Supplementary Fig. 2a–b and Supplementary Table 1a). These regions also appeared functional as divergently transcribed promoters, as determined in luciferase expression assay (Supplementary Fig. 3a–c). In a screen among different human cell lines, highly expressed PTENpg1 asRNA significantly correlated with low expression of PTEN mRNA based on a Spearman rank correlation analysis (Fig. 1c and Supplementary Fig. 3d). Surprisingly, the opposite correlation was observed for PTENpg1 sense and asRNA, which appeared to be co-expressed (Fig. 1d and Supplementary Fig. 3d). The discordant expression between PTEN and PTENpg1 asRNA was further supported by qRTPCR analysis on a subset of cell lines (Fig. 1e–f) and also by western blot analysis of PTEN protein levels, with the only exception being the PTEN deleted PC3 prostate cancer cell line (Supplementary Fig. 3e). Furthermore, the absolute expression of PTENpg1 sense and asRNA transcripts was also measured by cloning the cDNA of each transcript. Defined amounts of these cDNA clones were used for standard curve analysis on qRTPCR and the PTENpg1 asRNA was in general expressed at higher copy numbers as compared to the sense counterpart (Supplementary Fig. 3f).
PTENpg1 asRNA α is a negative regulator of PTEN expression
The observed discordant expression across disparate cell lines was suggestive of a negative regulatory association between the PTENpg1 asRNA and PTEN mRNA expression. To investigate this notion further, HEK293T cells were transfected with siRNAs (pg1as α siRNA) or shRNAs (pg1as α shRNA) targeting the PTENpg1 asRNA α transcript (Supplementary Fig. 4a). Suppression of PTENpg1 asRNA α with these si- or shRNAs, resulted in a significant increase in PTEN mRNA expression in HEK293T cells (Fig. 2a and Supplementary Fig. 4b–c). The activation of PTEN did not appear to involve any changes in the levels of the previously reported miRNA sponge acting PTENpg1 (Fig. 2b). The discordant regulation between PTEN and PTENpg1 asRNA α was further confirmed in the U2OS cell line (Fig. 2c) and the HeLa cell line (Fig. 2d). Notably, HeLa cells lack detectable levels of PTENpg1 sense (Fig. 1f and Supplementary Fig. 3d), suggesting that PTENpg1 asRNA α functions independently of PTENpg1 sense in all investigated cell lines. The observed induction of PTEN after targeting of the PTENpg1 asRNA α was transcriptional in nature based on changes in the expression of PTEN pre-mRNA (Fig. 2a). The transcriptional activation of PTEN was further supported by Run-On analysis (Fig. 2e) and ChIP for RNAPII binding at the PTEN promoter (Fig. 2f). Moreover, the activation of PTEN did not appear to involve any off target effects mediated by the siRNAs (Supplementary Fig. 4d–e), or shRNAs (Supplementary Fig. f). The si- or shRNA-constructs targeting the PTENpg1 asRNA α did not affect PTEN expression in HCT116 or MCF7 cells, which do not express appreciable levels of the PTENpg1 asRNA (Supplementary Fig. 4d–f), suggesting the PTEN activation to be mediated by specific targeting of the PTENpg1 asRNA α. Taken together, these observations suggest that PTEN is susceptible to PTENpg1 asRNA mediated transcriptional regulation, possibly by similar mechanisms as previously observed for other genes where noncoding RNAs (ncRNA), in particular asRNAs10,11, have been found to play a role in the recruitment of repressive chromatin remodelers such as Enhancer of Zeste (EZH2)7,12, DNA methyltransferase 3A (DNMT3a)13 and the methyl transferase G9A7.
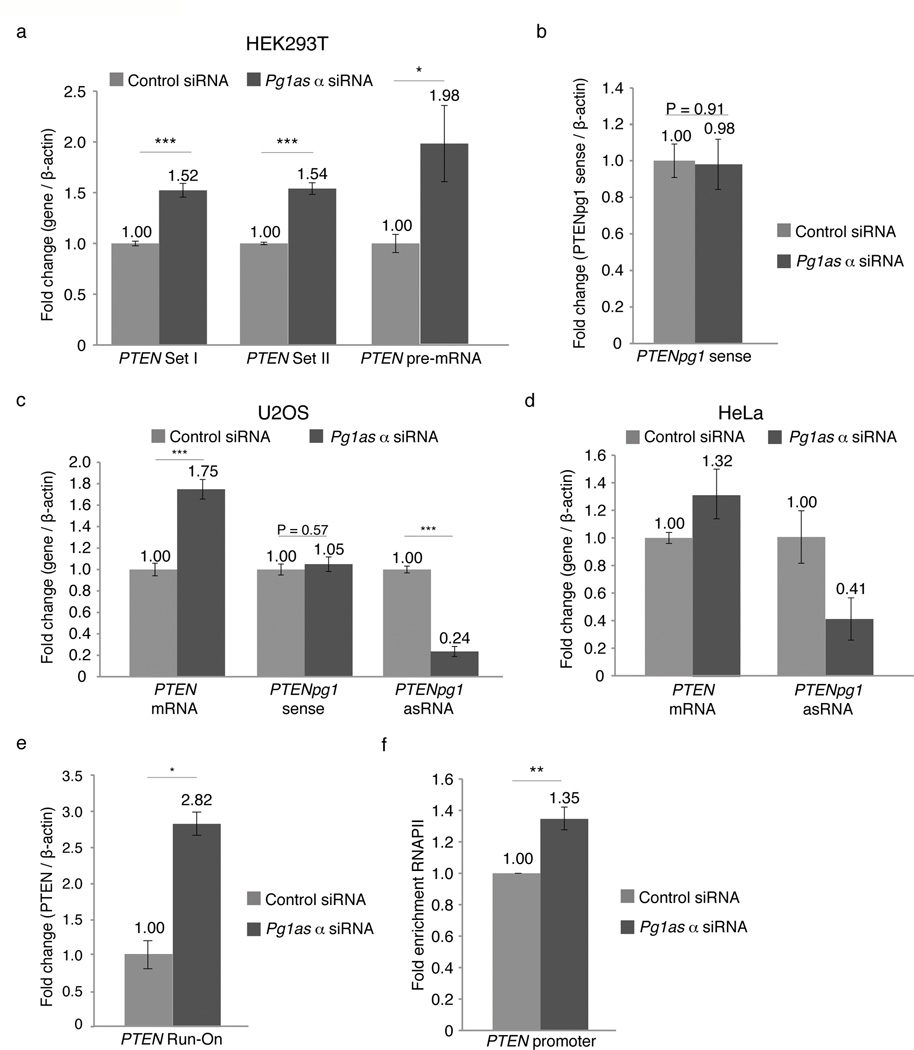
Figure 2. Functional characterization of PTENpg1 asRNA α.
(a) QRTPCR analysis of PTEN transcripts in HEK293T cells 72h post-transfection with siRNAs targeting the PTENpg1 asRNA α isoform (n=3). (b) QRTPCR analysis of PTENpg1 sense in cultures treated as described above (n=3). (c) QRTPCR analysis of PTEN and PTENpg1 transcripts in U2OS cells treated as described above (n=3). (d) QRTPCR analysis of PTEN transcripts in HeLa cells treated as described above (n=2). (e) Nuclear Run-On analysis of PTEN 48h post-transfection with siRNAs targeting the PTENpg1 asRNA α isoform (n=3). (f) ChIP analysis of RNAPII enrichment at the PTEN promoter in cultures treated as described (a–d above) (n=3). The (*) indicates the significance p<0.05, (**) p<0.01 and (***) p<0.005 using a Student’s T-test. Error bars represent the standard errors of the mean.
PTENpg1 asRNA α localizes to the PTEN promoter
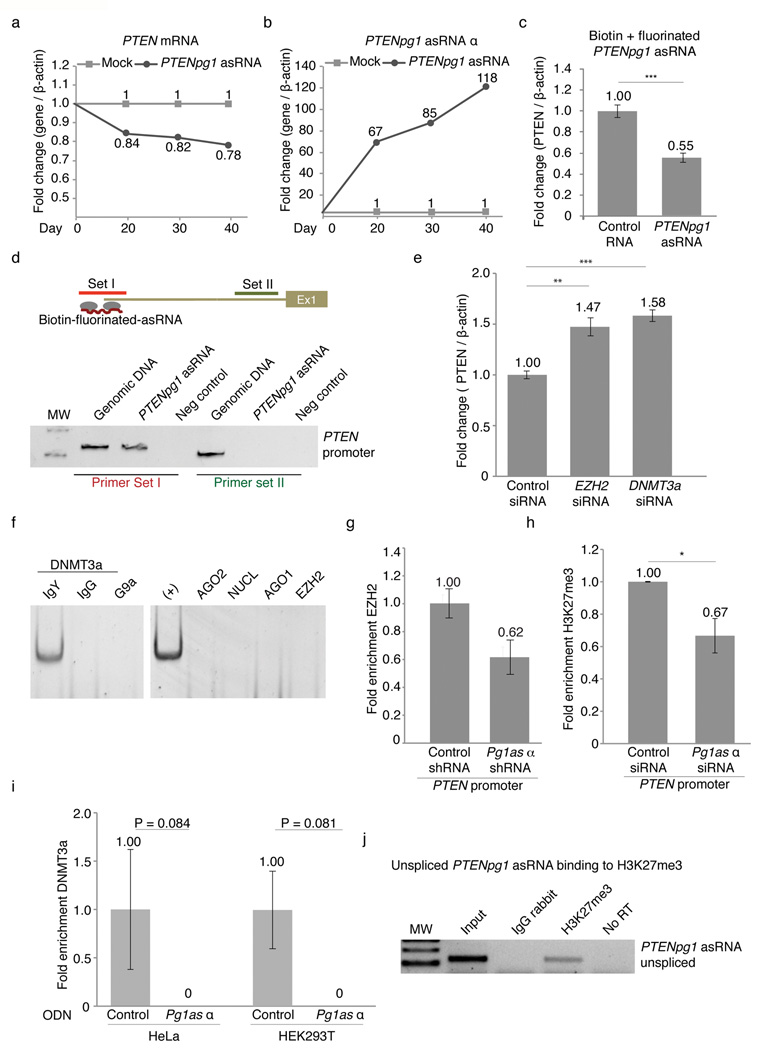
Previous observations implied a regulatory role for asRNAs in epigenetic-based regulation of gene transcription in human cells, but direct evidence supporting this notion has remained enigmatic7,10,11. To explore whether the induced expression of PTENpg1 asRNA α could suppress PTEN, we cloned the PTENpg1 asRNA α into GFP encoding lentiviral vectors with a U6 promoter driving its expression14. Stable Jurkat cell lines with the induced expression of PTENpg1 asRNA α were generated and followed over 40 days (Supplementary Fig. 5a). Expression of the PTENpg1 asRNA α was verified and a corresponding suppression of PTEN was observed, supporting the notion that the PTENpg1 asRNA α acts in trans as a negative regulator of PTEN (Fig. 3a–b). To further investigate the interaction between PTEN and PTENpg1 asRNA, PTENpg1 asRNA was in vitro transcribed using fluorinated and biotin linked nucleotides (Supplementary Fig. 5b). This modified in vitro transcript was successfully transfected into the target cells and had a half-life greater than 48 hours (Supplementary Fig. 5C). The fluorinated and biotin linked PTENpg1 asRNA was also found to suppress PTEN expression (Fig. 3c) and to localize to the PTEN promoter in the MCF7 cells (Fig. 3d). Overexpression of PTENpg1 asRNA did not induce any detectable levels of the IFNα-induced genes OAS1 and IL6, which potentially could be triggered by double-stranded RNA formation (Supplementary Fig. 5d). Altogether, these data show that PTENpg1 asRNA α localizes to the PTEN promoter and suppresses PTEN mRNA expression.
Figure 3. Mechanistic insights into PTENpg1 asRNA α regulation of PTEN.
(a–b) QRTPCR analysis of stable Jurkat cell lines. (a) PTEN mRNA expression and (b) PTENpg1 asRNA expression. (c) QRTPCR analysis of MCF7 cells transfected biotinylated-fluorinated PTENpg1 asRNA (n=3). (d) In vitro transcribed biotinylated-fluorinated PTENpg1 asRNA was transfected into MCF7 cells followed by a biotin-ChIP and semi-qRTPCR targeting different regions of the PTEN promoter. (e) QRTPCR analysis of siRNA depletion of DNMT3a and EZH2 in HEK293T cells (n=3). (f) RNA-IP followed by subsequent semi-qRTPCR for PTENpg1 asRNA α. Only the chicken (IgY) anti-DNMT3a antibody was found to IP with PTENpg1 asRNA α. The pulldown was contrasted to the non-specific protein G antibody.(g) QRTPCR analysis of ChIP-EZH2 samples after shRNA induced targeting of PTENpg1 asRNA α in HEK293T cells (n=2). (h) QRTPCR analysis of ChIP-H3K27me3 samples after siRNA induced targeting of PTENpg1 asRNA α in HEK293T cells (n=3). (i) QRTPCR analysis of ChIP-DNMT3a samples following single stranded ODN targeting of PTENpg1 asRNA α in HeLa and HEK293T cells (n=3). (j) Semi-qRTCPR of nuclear expressed and unspliced PTENpg1 asRNA α following a modified ChIP protocol for RNA bound to H3K27me3. The (*) indicates the significance p<0.05, (**) p<0.01 and (***) p<0.005 using a Student’s T-test. Error bars represent the standard errors of the mean.
PTENpg1 asRNA α induces chromatin remodeling
Next, we set out to investigate the molecular mechanisms whereby PTENpg1 asRNA α exerts its regulatory function on PTEN transcription. Based on previous work on transcriptional regulation mediated by ncRNAs, we investigated the involvement of a subset of previous identified proteins, such as Histone deacetylase 1 (HDAC1), Argonaute 1 (AGO1), Argonaute 2 (AGO2), DNMT3a, EZH2 and G9A7,12,13,15–17. SiRNA induced depletion showed that both EZH2 and DNMT3a appeared to be involved in the regulation of PTEN. The suppression of EZH2 and DNMT3a in cell lines with expression of PTENpg1 asRNA resulted in activation of PTEN (Fig. 3e and Supplementary Fig. 6a–c). In contrast, depletion of EZH2 and DNMT3a in the HCT116 cell line, which lack PTENpg1 asRNA expression (Supplementary Fig. 4d), had only a modest effect on PTEN levels (Supplementary Fig. 6b–d). In addition, simultaneous depletion of both EZH2 and PTENpg1 asRNA α did not prove additive in two different cell lines with regards to activating PTEN, further supporting the notion that these two factors function in concert (Supplementary Fig. 6e–f). These observations intrigued us to investigate whether EZH2 and DNMT3a may directly interact within an protein:RNA complex together with PTENpg1 asRNA α. RNA immunoprecipitation (IP) of DNMT3a showed DNMT3a and PTENpg1 asRNA α to co-IP and thus interact within the same protein:RNA complex (Fig. 3f and Supplementary Table 1c). Taken together, these data indicate EZH2 and DNMT3a are involved in the regulation of PTEN expression and importantly, that DNMT3a interacts directly with the with PTENpg1 asRNA α transcript.
Previous reports have suggested DNMT3a and EZH2 interact16 with one another and regulate levels of transcription by catalyzing the addition of methyl groups to the histone 3 lysine 27 (H3K27), which forms a negative repressive chromatin mark18,19. Therefore, we decided to investigate the presence of DNMT3a and EZH2 at the PTEN promoter and whether the PTENpg1 asRNA α actively recruits these factors. To explore this notion, we induced siRNA depletion of PTENpg1 asRNA α and investigated possible changes in the levels of H3K27me3, DNMT3a and EZH2 at the PTEN promoter. The suppression of the PTENpg1 asRNA α variant correlated with a loss of EZH2 and H3K27me3 at the PTEN promoter (Fig. 3g–h). In addition, we also set out to target the PTENpg1 asRNA α variant using single stranded phosphorothioate oligonucleotides (ODNs) (Supplementary Fig. 4a). In contrast to siRNAs, ODNs function by the RNase H pathway, which induces strand specific targeting of RNA followed by degradation of the RNA-ODN duplex20. Notably, the suppression of the PTENpg1 asRNA α variant using these ODNs demonstrated a substantial loss of DNMT3a at the PTEN promoter in two different cell lines (Fig. 3i).
Collectively, these data demonstrate that the PTENpg1 asRNA α binds and recruits the chromatin remodelers DNMT3a and EZH2 to the PTEN promoter and catalyzes the formation of H3K27me3. This intrigued us to investigate whether the endogenous PTENpg1 asRNA α localizes to the PTEN promoter, and more specifically to the H3K27me3 chromatin mark. We therefore carried out a modified ChIP protocol for H3K27me3 and analyzed whether the nuclear localized PTENpg1 asRNA associated directly with this epigenetic mark. Indeed, endogenous unspliced PTENpg1 asRNA was found to localize to this histone mark (Fig. 3j).
PTENpg1 asRNA β interacts with PTENpg1 sense
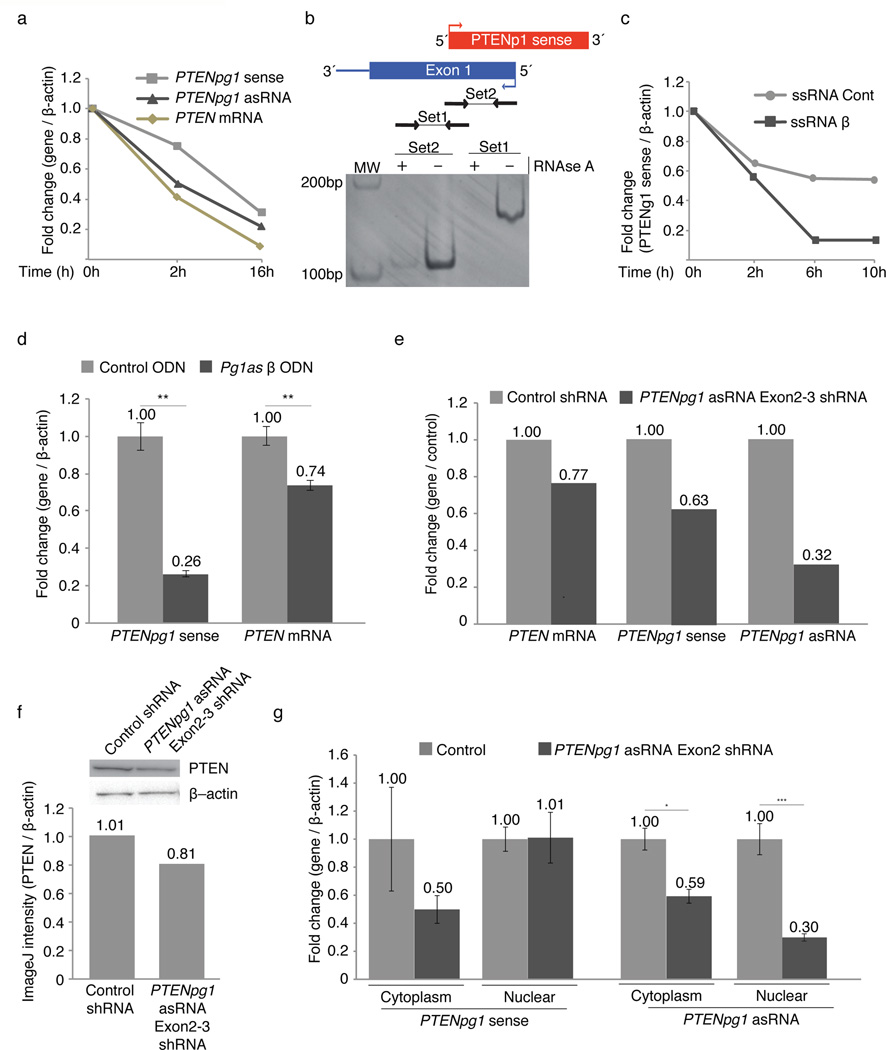
The observation that the PTENpg1 sense lacks a robust poly(A) tail (Fig. 1b and Supplementary Fig. 2f) was further validated by 3’ RACE. The cloned 3’ RACE end of PTENpg1 sense was sequenced, revealing the lack of a poly(A) tail (Supplementary Table 1d). Interestingly, poly(A) tailed transcripts have been implicated in RNA stability21 and transport of RNA from the nucleus to the cytoplasm22,23. Collectively, the lack of poly(A) tail suggested that the PTENpg1 sense might be an unstable transcript that is retained in the nucleus. However, contrary to this notion, blocking transcription by Actinomycin D treatment demonstrated similar stability of all three transcripts analyzed; PTENpg1 sense and the poly(A) positive transcripts PTEN and PTENpg1 asRNA (Fig. 4a). Therefore, we surmised that PTENpg1 sense RNA is stabilized via alternative means in the cell. One possible mechanism could involve sense:asRNA interactions, which have previously been reported to modulate RNA stability24. The notion that PTENpg1 sense and asRNA are co-expressed among different cell lines (Fig. 1d and Supplementary Fig. 3d) and that they also share a divergently transcribed promoter (Supplementary Fig. S3a–c), suggested that they might be co-regulated and possibly interacting with one another through a double-stranded RNA intermediate. In vitro RNAse A analysis reaffirmed this notion and indicated that the PTENpg1 sense and asRNA transcripts interact with one another in an RNA:RNA dependent manner (Fig. 4b).
Figure 4. Mechanistic insights into PTENpg1 asRNA β regulation of PTENpg1.
(a) QRTPCR analysis of the stability of PTENpg1 sense, PTENpg1 asRNA and PTEN after blocking transcription in HEK293T cells using Actinomysin D. The data were normalized to T=0h. (b) Semi-qRTPCR of RNase A analysis for the detection of RNA:RNA pairing. RNAse A treatment followed by RT and PCR with primers spanning the PTENpg1 sense and asRNA divergently transcribed locus (shown schematically). (c) QRTPCR analysis of the stability of PTENpg1 sense in HEK293T cells after interfering with the PTENpg1 sense:asRNA β interaction using U6 encoded ssRNAs. Transcription was blocked using Actinomysin D and the data were normalized to T=0h. (d) QRTPCR analysis of PTEN and PTENpg1 sense after interfering with the PTENpg1 sense:asRNA β interaction using single stranded ODNs (n=3). (e) QRTPCR analysis after shRNA targeting of the PTENpg1 asRNA transcripts. (f) Western blot showing the expression of PTEN after PTENpg1 asRNA depletion. The intensities were quantified using the ImageJ software. (g) QRTPCR analysis on fractionated HEK293T cells after depletion of PTENpg1 asRNA. The (*) indicates the significance p<0.05, (**) p<0.01 and (***) p<0.005 using a Student’s T-test. Error bars represent the standard errors of the mean.
To explore this notion further, we first developed U6 expressed single stranded RNAs (ssRNAs) targeting the PTENpg1 sense and asRNA β overlap (Supplementary Fig. 4a). Interestingly, interfering with this interaction resulted in a decreased half-life of PTENpg1 sense (Fig. 4c). Next, single stranded ODNs, which specifically target the PTENpg1 sense and asRNA β overlap, and shRNAs targeting the PTENpg1 asRNA exon 2 and 3 were designed (Supplementary Fig. 4a). Both of these approaches resulted in a concordant reduction of PTENpg1 sense, and interestingly also suppressed PTEN mRNA (Fig. 4d–e) and protein levels (Fig. 4f). This observed suppression of PTEN is in line with previous reports, where PTENpg1 sense has been described to bind and sequester miRNAs3. More precisely, the RNA:RNA interaction is lost upon depletion of PTENpg1 asRNA β, whereby PTENpg1 sense is destabilized and its expression decreased. Consequently, miRNAs, which are normally bound and sequestered by PTENpg1 sense, may be released and free to bind the PTEN mRNA instead. Taken together, these data suggest that the RNA:RNA interaction between PTENpg1 sense and asRNA β are required for PTENpg1 sense to function as a miRNA sponge. Interestingly, this also shows that the PTENpg1 asRNA α and β isoforms have different functions; the α isoform regulates PTEN transcription while the β isoform appears to be involved as a miRNA sponge, ultimately affecting post-transcriptional regulation of PTEN.
We further set out to investigate whether this RNA:RNA interaction occurred in the nucleus or in the cytoplasm. To this end PTENpg1 asRNA β was knocked down using shRNA targeting followed by fractionation of cytoplasm and nuclei. The PTENpg1 asRNA β shRNA targeting resulted in a reduction in PTENpg1 sense levels exclusively in the cytoplasm, while the levels were unaffected in the nucleus (Fig. 4g). In summary, these data suggest that the PTENpg1 sense:asRNA β interaction is required for maintaining stable levels of PTENpg1 sense in the cytoplasm, and ultimately PTENpg1 sense sponge activity.
PTENpg1 asRNA α regulates the cell cycle and apoptosis
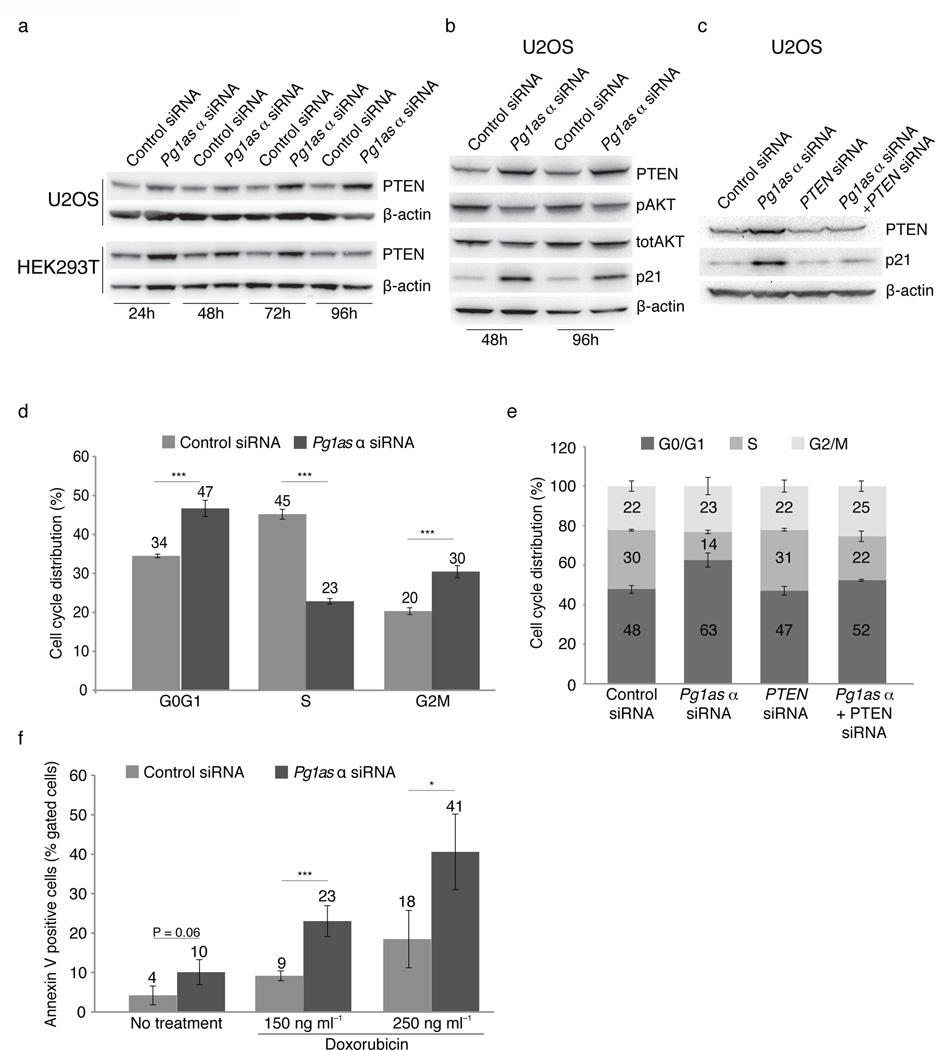
The mechanistic studies presented here suggest a regulatory interaction between PTEN, PTENpg1 sense and PTENpg1 asRNA. Since perturbations of PTENpg1 asRNA α levels markedly affected PTEN mRNA expression, we decided to investigate the effects of disrupting this lncRNA network on PTEN protein expression. Western blot analysis following siRNA depletion of PTENpg1 asRNA α resulted in a clear induction of PTEN protein levels at several time points in various cell lines (Fig. 5a). Furthermore, the elevated PTEN protein levels lead to a concomitant downregulation of the PTEN downstream target pAKT (Fig. 5b). Since PTEN is a negative regulator of the cell cycle, we studied the effect on the CDK inhibitor p21 following depletion of PTENpg1 asRNA α. Depletion of PTENpg1 asRNA α resulted in a marked induction of p21, both 48 and 96h post transfection (Fig. 5b)25. Notably, p21 was not induced upon PTENpg1 asRNA α depletion in cell lines lacking expression of PTENpg1 asRNA α, arguing against off target effects of the siRNA (Supplementary Fig. 6g). More specifically, PTEN dependent induction of p21 was verified by simultaneous siRNA depletion of PTEN and PTENpg1 asRNA α, which did not result in an induction of p21 protein levels (Fig. 5c). This negative effect on the cell cycle was further supported by FACS analysis, (Fig. 5d), demonstrating a significant G0/G1-arrest in the PTENpg1 asRNA α suppressed U2OS cells. In contrast, simultaneous depletion of PTEN and PTENpg1 asRNA α partially rescued the G0/G1 cell cycle arrest (Fig. 5e).
Figure 5. Functional impact of PTENpg1 asRNA α.
(a) Western blot showing PTEN expression after knockdown of PTENpg1 asRNA α in two different cell lines at different time points. The data were standardized to β-actin.(b) Western blot at two different time points after knockdown of PTENpg1 asRNA α. (c) Western blot showing the effect on PTEN and p21 after simultaneous knockdown of the PTENpg1 asRNA α and PTEN. (d) Cell cycle distribution in U2OS cells following knockdown of PTENpg1 asRNA α (n=3). (e) Cell cycle distribution after simultaneous knockdown of PTENpg1 asRNA α and PTEN in U2OS cells (n=3) (f) Cell death analysis by AnnexinV / PI staining after 72h knockdown of PTENpg1 asRNA α and 48h treatment with doxorubicin (n=3). The (*) indicates the significance p<0.05, (**) p<0.01 and (***) p<0.005 using a Student’s T-test. Error bars represent the standard errors of the mean.
Apart from causing a negative cell cycle effect, PTEN levels are also important in regulating apoptosis sensitivity26. Suppression of PTENpg1 asRNA α caused a modest effect on the basal levels of cell death but clearly sensitized cells to the DNA-damaging agent doxorubicin (Fig. 5f). Together, these data suggest that the regulatory impact of PTENpg1 asRNA α on PTEN expression is physiologically relevant and may function in the sensitization of tumor cells to chemotherapeutic treatments, such as doxorubicin.
Discussion
The data presented here argue for a more complex role for lncRNAs in human cellular gene regulation than had previously been appreciated. We find here that lncRNAs can form higher ordered biologically relevant functional structures through RNA:protein as well as RNA:RNA interactions. Earlier studies have reported that PTENpg1 sense lncRNA functions as a miRNA sponge, an interaction that has typically been thought to occur in the cytoplasm3,27. Mechanistically, it was suggested that PTENpg1 sense lncRNA binds to the PTEN targeted miRNAs whereby the miRNA targeting of PTEN is diminished, ultimately resulting in increased PTEN mRNA and protein expression. These previous studies however overlooked the presence of PTENpg1 associated asRNA transcripts.
We report here that the PTENpg1 locus expresses mechanistically functional asRNAs and that these transcripts appear to function as major regulators of PTEN expression, both at the transcriptional and translational levels.
Within this body of work, we investigate three PTEN regulatory lncRNAs; PTENpg1 sense, PTENpg1 asRNA α and PTENpg1 asRNA β and observe that these transcripts are expressed from two divergently transcribed promoters (Supplementary Fig. 3a–c). We find that a complex regulatory network of lncRNAs is operative whereby PTENpg1 asRNA α physically localizes to and directs epigenetic remodeling proteins to the PTEN promoter to control PTEN transcription. In contrast, the PTENpg1 asRNA β transcript forms an RNA:RNA interaction with PTENpg1 sense, altering cellular compartmental localization of the PTENpg1 sense transcripts, miRNA sequestration, and ultimately PTEN protein levels (Fig. 6). While more complex than previously appreciated it still remains unknown as to how the transcriptional regulation of the PTENpg1 locus is controlled and if any regulatory switch or interplay is functional between PTENpg1 asRNA α and PTENpg1 asRNA β. In addition, even though we find the PTENpg1 asRNA α to directly localize at the PTEN promoter and to co-IP with DNMT3a, the details of these interactions remain to be explored.
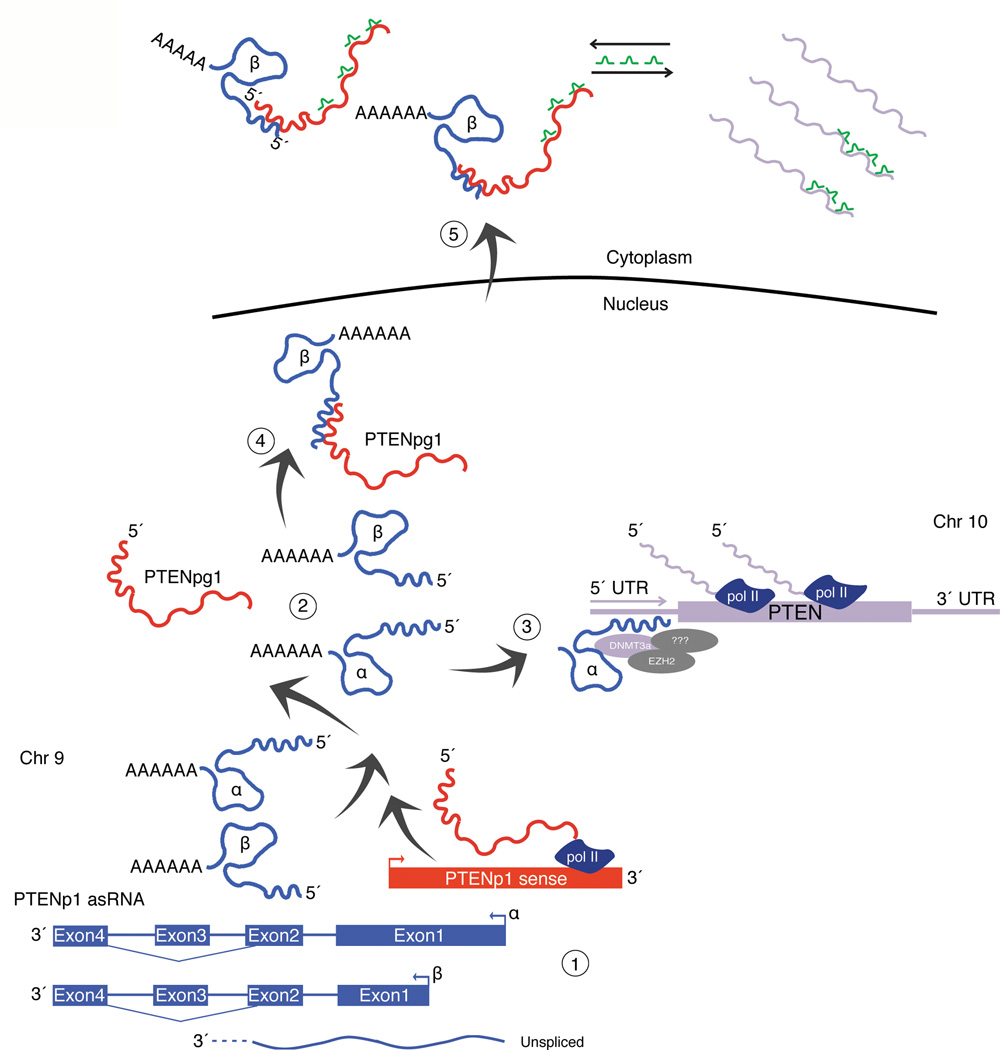
Figure 6. Model of PTENpg1 asRNA based regulation of PTEN.
(1) The PTENpg1 locus is transcribed in both sense (red) and antisense orientation (blue). (1–2) Three different isoforms of the PTENpg1 asRNA are transcribed; PTENpg1 asRNA α, β and the unspliced nuclear localized PTENpg1 asRNA α. (3) PTEN transcription is suppressed by the PTENpg1 asRNA α and unspliced isoforms, which act in trans by recruiting the chromatin repressor proteins EZH2 and DNMT3a to the PTEN promoter. (4) The PTENpg1 sense lacks a poly(A) tail and the stability and export of PTENpg1 sense to the cytoplasm is facilitated by the interaction with the spliced PTENpg1 asRNA β transcripts. (5) Such interactions lead to an increase of PTENpg1 sense sponge activity and thus increased PTEN mRNA stability and translation. This mechanism (1–5) results in both transcriptional and post-transcriptional regulation of PTEN by its pseudogene. The model indicates a level of synchronization between the pseudogene and its protein-coding counterpart.
These observations presented here suggest a model whereby a pseudogene, PTENpg1, contains bimodal functionality that directly and simultaneously affects PTEN transcription and translation (Fig. 6). This model, built on RNA:RNA and RNA:protein interactions, might highlight a mechanism whereby organisms lacking an RNA dependent RNA polymerase (RdRP), such as primates, can utilize a different paradigm such as pseudogene expressed lncRNAs to regulate gene expression. A functional RdRP would be expected to result in amplification of these RNA:RNA complexes and ultimately RNAi based mechanisms of regulation. However, the lack of a viable RdRP offers increased functional complexities in both the nucleus and cytoplasm, as the lncRNA complexes would not be processed by RdRP but rather functional in a different context, such as is demonstrated here whereby lncRNA complexes simultaneously govern both gene transcription and translation. Such observations add a significant layer of complexity to our current understanding of gene regulation and highlight a biological role for pseudogene-expressed lncRNAs in human cells. Such a system, as is determined here (Fig. 6), provides for a useful cellular adaptation strategy that allows for fine control and balance of the output of PTEN proteins, ultimately resulting in control of cell growth.
It is also interesting to speculate whether the PTENpg1 asRNA α could act as an ncRNA oncogene due to its negative regulatory effect on PTEN. In future studies, it would be of great interest to profile human cancer tumors with diminished PTEN expression, and investigate whether increased expression of PTENpg1 asRNA could promote epigenetic silencing of PTEN and tumor development. If so, knowledge of this PTENpg1 and other pseudogene ncRNA regulatory pathways may prove useful in therapeutically relevant approaches to controlling gene expression as well as informative with regards to tumor biology.
Online methods
Cell cultures
HEK293, HEK293T and U2OS were cultured in 5% CO2 at 37 °C in Dulbecco modified MEM medium, supplemented with 10% heat-inactivated fetal bovine serum, 2 mM glutamine, 50 µg/ml of streptomycin and 50 µg/ml of penicillin.
Bioinformatics
ChIP sequencing data were aligned to PTEN and PTENpg1 using Bowtie 0.12.81 with the parameter options -v0 and -m1. The alignments were visualized using IGV 2.12. Data for H3K4me3 (K562 and H1-hESC cells): http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeBroadHistone/ Data for RNAPII (K562 and H1-hESC cells): http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeHaibTfbs/ Data for H3K4me3 (HCT116 and HEK293 cells): http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwHistone/
ShRNAs, ssRNAs and siRNAs
ShRNA and ssRNA expressing constructs were generated and cloned into the U6M2 construct using the Bgl II and Kpn I restriction sites (Supplementary Table 2) as previously described3. ShRNA and ssRNA constructs were transfected using Lipofectamine 2000 (Life Technologies). SiRNAs were ordered from the respective manufactures (Supplementary Table 2) and transfected using Lipofectamine 2000 (Life Technologies) or HiPerFect (Qiagen) at 10-20 nM unless stated otherwise.
RNA extraction and cDNA
RNA was extracted using Qiagen RNeasy mini kit (Qiagen) and DNase treated (Ambion Turbo DNA-free, Life Technologies). RNA (~500ng) was used for the generation of cDNAs using MuMLV (Life Technologies) and a mixture of oligo(dT)18 with nanomers.
PCR and qRTPCR
PCR was performed using the KAPA2G FAST mix (Kapa Biosystems) with the corresponding oligos (Supplementary Table 2). QRTPCR quantification was carried out using the KAPA 2G SyberGreen (Kapa Biosystems) on the Eppendorf RealPlex or Applied Biosystem 7900HT platform with the following cycling conditions: 95°C for 3min, 95°C for 3sec, 60°C for 30sec.
Nuclear Run-On
HEK293T cells were transfected with 50nM of siRNA using HiPerFect (Qiagen). Pellets were collected 48h later and lysed (10mM Tris-HCl, pH 7.4, 10 mM NaCl, 3mM MgCl2, 0.5% NP-40). The nuclei were resuspended in Glycerol storage buffer (50mM Tris-HCl, pH 8.3, 0.1 mM EDTA, 40% glycerol, 5 mM MgCl2) and flash frozen in liquid nitrogen. Glycerol storage buffer (50 µL) was defrosted, mixed with 60 µL transcription buffer (20mM Tris-HCl, pH8, 5mM MgCl2, 300mM KCl, 4mM DTT, 2mM ATP, 2mM CTP, 2mM GTP, 1mM biotin-16-AA-5’-UTP, 100U RNase out), incubated at 30°C for 45min followed by DNase treatment and nuclear lysis (50mM Tris-HCL pH 7.4, 5% SDS, 0.125M EDTA) with proteinase K treatment. The RNA was extracted and biotinylated RNA captured using Dynabeads MyOne Streptavidin beads (Life Technologies). The beads were resuspended in H2O and cDNA prepared directly on beads.
Protein analysis
Samples were lysed in 50 mM Tris–HCl, pH 7.4, 1% NP40, 150 mM NaCl, 1mM EDTA, 1% glycerol, 100 _µ_M vanadate, protease inhibitor cocktail and PhosSTOP (Roche Diagnostics GmbH). Lysates were subjected to SDS-PAGE and transferred to PVDF membranes. The proteins were detected by Western blot analysis using an enhanced chemiluminescence system (Western Lightning–ECL, PerkinElmer). Antibodies used: PTEN (Cell signaling, Cat# 9552, 1:1000), AKT (Cell signaling, Cat# 9272, 1:1000), phospho-AKT (Cell signaling, Cat# 4060S, 1:1000), p21 (BD Biosciences Sparks, Cat# 610234, 1:1000), β-actin (Sigma-Aldrich, Cat# A5441, 1:5000).
Fractionation
Fractionation of HEK293 cells was performed using the PARIS kit according to the manufacturer’s recommendations (Life Technologies).
Promoter activity
HEK293T cells were co-transfected with the Renilla/Firefly bidirectional promoter construct4 and GFP using lipofectamine 2000 (Life Technologies). The expression of GFP and Luminescence was measured 24h post transfection using the Dual-Glo Luciferase Assay System (Promega) and detected by the The GloMax®-Multi+ Detection System, (Promega). The expression of Luminescence was normalized to GFP.
ChIP; RNAPII and H3K27me3
The ChIP was carried out using the ChIP Assay Kit (Upstate/Millipore). Cells were crosslinked in 1% formaldehyde, quenched and lysed according to the manufacturer’s recommendations. The samples were sonicated with a Bioruptor Sonicator (Diagenode) and incubated over night at 4°C with the appropriate antibody. Salmon Sperm DNA/Protein A Agarose (Upstate/Millipore) was used to pulldown the antibody. DNA was eluted in Elution buffer (1% SDS, 100mM NaHCO3), followed by reverse crosslinking, RNaseA and protease K treatment. The DNA was eluted using Qiagen PCR purification kit. The following antibodies were used (4 _µ_g/sample); Polymerase II (Santa Cruz biotechnologies, Cat# sc899x) and H3K27me3 (Upstate/Millipore, Cat# 17-622).
PTENpg1 asRNA binding to H3K27me3
ChIP protocol, as described above, was carried out without LiCl wash and RNaseA treatment. The beads were resuspended in 95µL proteinase K buffer (100 mM NaCl, 10mM Tris-HCl, 1mM EDTA, 0.5% SDS) with 5 µL proteinase K (20mg/ml), incubated for 60min at 50°C and heated to 95°C for 10min. The RNA was eluted using the miRNeasy mini kit (Qiagen) and DNase treated and cDNA was prepared as previously.
ChIP and IP-RT-sequencing- DNMT3a
ChIP analysis of ODN treated cells were carried out in an automated fashion using the EpiMotion (Eppendorf) and previously published protocols5,6 adapted to this system. The resultant ChIPs were carried out with one modification, no RNAse A was added. The resultant elutes were DNAse treated and cDNA prepared as described above. PCR was carried out on the IP-RTs using PTENpg1 sense or antisense specific primer sets 3 and 2, respectively. Only the sequencing results for primer set 2 (Supplementary Table 2), PTENpg1 asRNA, are shown as all primer set 3 sampling proved negative. The antibodies used were; DNMT3a (ProSci, Cat# xw-7148), AGO-2 (Cell signaling, Cat# C34C6), Nucleolin (Abcam, Cat# ab50279), AGO-1 (Upstate/Millipore, Cat# 07-599), G9a (Upstate/Millipore, Cat# 07-55) and EZH2 (Upstate/Millipore, Cat# 07-689).
RNAse A
RNA from HEK293 cells was exposed to RNAse A (0.1µg) for 30 min at 37°C. The samples were heat inactivated (95°C for 10min) and cDNA prepared as described above. The resultant cDNAs were PCR amplified with PTENpg1 sense and PTENpg1 asRNA primers (Sets 1–2, Supplementary Table 2).
5´ RACE
RNA from several human cell lines was pooled and cDNA prepared according to 5´ RACE system kit (Life Technologies) using primer PTENpg1 asRNA R1 (Supplementary Table 2). PCR was carried out using forward primer AAP with the reverse primer PTENpg1 asRNA R5. Nested PCR was followed using forward primer AUAP with reverse primer PTENpg1 asRNA R3 (Supplementary Table 2). PCR products were gel purified and sequenced.
Poly(G) tailing
RNA was 3´ poly(G) tailed using yeast poly(A) polymerase (Affymetrix) and guanosine 5´-triphosphate (GE Healthcare). Poly(G) tailed RNA was used for first strand cDNA synthesis, followed by nested PCR (Supplementary Table 2). The PCR product was cloned into StrataClone PCR cloning Kit (Agilent Technologies) and sequenced.
Poly(A) depletion
Dynabeads MyOne Streptavidin beads (Life Technologies) were pre-loaded with 5’-biotinylated oligo(Dt)18 or control (biotin-362as)7. DNase treated RNA from HEK293T cells was depleted of poly(A) transcripts and used to generate cDNA.
Generation of promoter constructs
The PTENpg1 promoter sequences were PCR amplified using primers containing Kpn1 restriction sites (Supplementary Table 2). The resultant PCR products were cloned into the pLucRLuc Bidirectional vector4.
Generation of U6 expressed PTENpg1 asRNA α lentiviral constructs
The U6 promoter was amplified from the U6M2 cloning plasmid3 and ligated into the Not1 restriction site of the pHIV7-IMPDH2 vector8. PTENpg1 asRNA α was PCR amplified and subsequent cloned into the Nhe1 and Pac1 restriction sites in the pHIV7-IMPDH2-U6 plasmid (Supplementary Table 2).
Lentiviral production, transduction and selection
Lentiviral particles were produced and titrated as previously described8. Jurkat cells were infected at a multiplicity of infection = 1 by spinoculation for 30min. Transduced cells were placed under selection with 1–2 _µ_M mycophenolic acid (Sigma-Aldrich) for 20 days.
Generation of biotin linked PTENpg1 asRNAs
PTENpg1 asRNA α was PCR amplified using primers with Nhe1 or Kpn1 restriction sites (Supplementary Table 2) and cloned into pcDNA 3.1. T7 transcripts were generated by linearization of pcDNA 3.1 with Kpn1 and T7 transcribed with the Durascribe T7 kit (Epicenter, Madison WI, USA) using fluorinated CTPs and UTPs (TriLink). Biotin UTPs were incorporated at a ratio of 1:10. The resultant transcripts were DNase treated and dephosporylation using Antartic Phosphatase (New England Biolabs). Transcripts were precipitated using phenol-chloroform and EtOH precipitation. The T7 transcripts were transfected into target cells at 25-50nM using Lipofectamine 2000 (Life Technologies). For in vivo pulldown, MCF7 cells were transfected, 30h later collected and avidin IPs performed as described5,6.
PI-annexin V staining
Treated cells were harvested, washed in PBS, pelleted, and resuspended in 100_µ_L Annexin V incubation buffer (10 mM HEPES/NaOH, pH 7.4, 140 mM NaCl, 5 mM CaCl2) containing 1% annexin V FLOUS (Annexin V FLUOS, Roche Molecular Biochemicals) and 500 _µ_g/_µ_l PI staining. The samples were incubated for 15min followed by the addition of 400 µL of Annexin V incubation buffer and subsequent analysis on a cytometry machine using the Cell Quest software.
Cell cycle distribution
Cells were washed in PBS and fixed in 4% PFA at room temperature over night. PFA was removed and cells were resuspended in 95% EtOH. The samples were then rehydrated in distilled water, stained with DAPI and analyzed by flow cytometry on a FACS Calibur (Becton Dickinson) using the cell Quest software.
ODN targeting of PTENpg1 asRNA transcripts
The ODNs were produced (IDT) and transfected at 100nM using RNAiMax (Life Technologies).
Supplementary Material
1
Acknowledgements
The project was supported by the National Institute of Allergy and Infectious Disease (NIAID) R56 AI096861-01 and P01 AI099783-01 to KVM and the National Cancer Institute (NCI) R01 CA151574, National Institutes of Health (NIH) R01 CA153124 to Peter K. Vogt. The Swedish Childhood Cancer Foundation, The Swedish Cancer Society, Radiumhemmets Forskningsfonder, the Karolinska Institutet PhD support programme, and Vetenskapsrådet to DG. The Erik and Edith Fernstrom foundation for medical research to P.J.
Footnotes
Author contributions
P.J., and K.V.M., designed, performed and supervised the experiments. A.A., and L.V., performed experiments. M.C., provided helpful discussions. W.-O.L., supervised ChIP-sequencing analysis. D.G., supervised the experiments. P.J., D.G., and K.V.M., wrote the paper.
References for main text
- 1.Xing M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid : official journal of the American Thyroid Association. 2010;20:697–706. doi: 10.1089/thy.2010.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alimonti A, et al. Subtle variations in Pten dose determine cancer susceptibility. Nature genetics. 2010;42:454–458. doi: 10.1038/ng.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khachane AN, Harrison PM. Assessing the genomic evidence for conserved transcribed pseudogenes under selection. BMC genomics. 2009;10:435. doi: 10.1186/1471-2164-10-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pei B, et al. The GENCODE pseudogene resource. Genome biology. 2012;13:R51. doi: 10.1186/gb-2012-13-9-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suo G, et al. Oct4 pseudogenes are transcribed in cancers. Biochemical and biophysical research communications. 2005;337:1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 7.Hawkins PG, Morris KV. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010;1:165–175. doi: 10.4161/trns.1.3.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H, Shabbir A, Molnar M, Lee T. Stem cell regulatory function mediated by expression of a novel mouse Oct4 pseudogene. Biochemical and biophysical research communications. 2007;355:111–116. doi: 10.1016/j.bbrc.2007.01.106. [DOI] [PubMed] [Google Scholar]
- 9.Piehler AP, et al. The human ABC transporter pseudogene family: Evidence for transcription and gene-pseudogene interference. BMC genomics. 2008;9:165. doi: 10.1186/1471-2164-9-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS genetics. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Molecular cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberg MS, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Human gene therapy. 2012;23:473–483. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. The Journal of biological chemistry. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 16.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 17.Malecova B, Morris KV. Transcriptional gene silencing through epigenetic changes mediated by non-coding RNAs. Current opinion in molecular therapeutics. 2010;12:214–222. [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 20.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Molecular and cellular biology. 1989;9:659–670. doi: 10.1128/mcb.9.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuke H, Ohno M. Role of poly (A) tail as an identity element for mRNA nuclear export. Nucleic acids research. 2008;36:1037–1049. doi: 10.1093/nar/gkm1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brodsky AS, Silver PA. Pre-mRNA processing factors are required for nuclear export. RNA. 2000;6:1737–1749. doi: 10.1017/s1355838200001059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahmoudi S, et al. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Molecular cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 25.Chang F, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2003;17:590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 26.Weng L, Brown J, Eng C. PTEN induces apoptosis and cell cycle arrest through phosphoinositol-3-kinase/Akt-dependent and -independent pathways. Human molecular genetics. 2001;10:237–242. doi: 10.1093/hmg/10.3.237. [DOI] [PubMed] [Google Scholar]
- 27.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nature methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for online methods
- 1.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarzguioui M, Rossi JJ, Kim D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS letters. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- 4.Polson A, Durrett E, Reisman D. A bidirectional promoter reporter vector for the analysis of the p53/WDR79 dual regulatory element. Plasmid. 2011;66:169–179. doi: 10.1016/j.plasmid.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg MS, et al. The antisense strand of small interfering RNAs directs histone methylation and transcriptional gene silencing in human cells. RNA. 2006;12:256–262. doi: 10.1261/rna.2235106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner AM, De La Cruz J, Morris KV. Mobilization-competent Lentiviral Vector-mediated Sustained Transcriptional Modulation of HIV-1 Expression. Molecular therapy : the journal of the American Society of Gene Therapy. 2009;17:360–368. doi: 10.1038/mt.2008.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner AM, Ackley AM, Matrone MA, Morris KV. Characterization of an HIV-targeted transcriptional gene-silencing RNA in primary cells. Human gene therapy. 2012;23:473–483. doi: 10.1089/hum.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1