Aire-dependent thymic development of tumor-associated regulatory T cells (original) (raw)
. Author manuscript; available in PMC: 2013 Sep 8.
Published in final edited form as: Science. 2013 Mar 8;339(6124):1219–1224. doi: 10.1126/science.1233913
Abstract
Despite considerable interest in the modulation of tumor-associated Foxp3+ regulatory T cells (Tregs) for therapeutic benefit, little is known about the developmental origins of these cells and the nature of the antigens that they recognize. Here, we identified an endogenous population of antigen-specific Tregs (termed “MJ23” Tregs) found recurrently enriched in the tumors of mice with oncogene-driven prostate cancer. MJ23 Tregs were not reactive to a tumor-specific antigen, but instead recognized a prostate-associated antigen that was present in tumor-free mice. MJ23 Tregs underwent Aire-dependent thymic development in both male and female mice. Thus Aire-mediated expression of peripheral tissue antigens drives the thymic development of a subset of organ-specific Tregs, which are likely co-opted by tumors developing within the associated organ.
Treg cells are critical for the prevention of autoimmunity, the maintenance of immune homeostasis, and the suppression of anti-tumor immune responses (1, 2). For many human cancers, the density of Tregs within tumor lesions is predictive of poor clinical outcome (3), suggesting that Tregs play a functional role in cancer progression. In this study, we set out to establish a tractable animal model in which a single specificity of naturally occurring tumor-associated Tregs could be studied in the context of a genetically driven mouse model of autochthonous cancer. In order to identify an endogenous tumor-associated Treg response, we analyzed the immune response in TRAMP mice, which develop prostatic adenocarcinoma due to the transgenic expression of the model oncogene SV40 T antigen in the prostate (4, 5). Unlike the prostates of tumor-free mice, which contain very few Treg cells (which are identified as CD4+Foxp3+), a substantial population of Tregs can be detected in the prostate tumors of TRAMP mice (fig. S1). We employed an experimental system involving T cell antigen receptor alpha chain (TCRα) repertoire analysis of T cell populations from TRAMP mice expressing the Foxp3_gfp_ reporter (6) and a fixed (transgenic) TCRβ chain. The fixed TCRβ used in this study was a TCRβ chain that was found to be recurrently expressed by CD4+Foxp3+ Tregs isolated from the prostates of TRAMP mice (fig. S2), and will be referred to hereafter as “TCRβtg”. TCR complementarity determining region 3 (CDR3) length distribution analysis of cDNA from purified CD4+Foxp3+ and CD4+Foxp3neg T cells from the prostate tumors of TRAMP+/− Foxp3gfp TCRβtg males revealed a substantial overrepresentation of CD4+Foxp3+ T cells expressing a Vα2 (TRAV14) TCRα chain with a CDR3 of nine amino acids in length (as defined by IMGT, http://www.imgt.org) (Fig. 1A, denoted in red). Deep sequencing of these samples revealed that the identical TCRα chain, of CDR3 sequence LYYNQGKLI, was recurrently expressed by Foxp3+ Tregs (Fig. 1B), indicating that Tregs of a single specificity are recurrently enriched within TRAMP prostate tumors. Strikingly, in many prostate samples, the Vα2-LYYNQGKLI TCR chain was encoded by a single nucleotide sequence (fig. S3), suggesting that in many cases, tumor-infiltrating Tregs of this specificity may originate from a single clone.
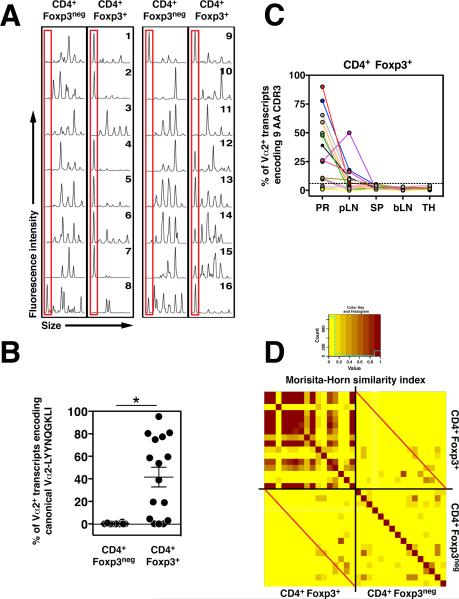
Fig. 1. CD4+Foxp3+ Tregs expressing a canonical TCR are recurrently enriched in TRAMP prostate tumors.
Analysis of Vα2 (TRAV14)+ TCRα chains in tumor-bearing TRAMP+/− Foxp3_gfp_ TCRβtg mice. CD4+Foxp3neg and CD4+Foxp3+ T cells were FACS-purified from different anatomical sites of ~27-week-old male mice, and cDNA was subjected to molecular analysis. (A) CDR3 length distribution analysis of Vα2+ TCRα chains of T cell subsets isolated from prostate tumors. The mouse number is indicated. The red boxes denote TCRα transcripts encoding a CDR3 of 9 amino acids (AA) in length. (B) Vα2+ TCRα transcripts from the indicated populations were PCR amplified and subjected to deep sequencing (see Methods (24)). The percentage of all Vα2+ TCRα transcripts encoding the canonical Vα2-LYYNQGKLI chain is plotted for each sample. The mean ± SEM is indicated. The asterisk indicates p < 0.05 (t-test). (C) Plots of the percentage of Vα2+ transcripts encoding a CDR3 of 9 AAs (based on peak area) for CD4+Foxp3+ T cells isolated from different anatomical sites of the mice depicted in panel (A). PR, prostate; pLN, periaortic lymph nodes; SP, spleen; bLN, brachial lymph nodes; TH, thymus. Samples from each mouse are color-coded. The plot does not depict data from mice 1 and 2, and includes data from two additional mice (numbers 17 and 18). Samples with values above the indicated threshold (dashed line) are considered “overrepresented”. This threshold is defined as the mean percent peak area plus three standard deviations for Vα2+ TCRα transcripts encoding a 9 AA CDR3 from the spleen of female TCRβtg mice. (D) Heat map of the Morisita-Horn (MH) similarity index for prostatic T cell subsets from mice 1–16. Samples are oriented in ascending numerical order, from top to bottom and left to right (not shown). A motif table of predicted amino acid sequences is presented in Table S1. CD4+Foxp3neg and CD4+Foxp3+ subsets from the same prostate tumor intersect at the red diagonal lines. Data are pooled from N = 3 independent FACS-sorting experiments.
A survey of different anatomical sites of TRAMP+/− Foxp3_gfp_ TCRβtg mice using CDR3 length distribution analysis revealed that the overrepresentation of Tregs expressing a Vα2+ TCRα chain of nine amino acids in length was observed in the prostate tumor and prostate-draining periaortic lymph nodes, but was not detected over background in non-draining brachial lymph nodes (Fig. 1C). Thus, Tregs of this specificity are not expanded systemically in tumor-bearing mice, but are instead selectively enriched in the prostate tumor environment.
In order to gain insight into the antigen specificities of polyclonal tumor-infiltrating CD4+ T cells, we determined the extent of overlap of the Vα2+ TCR repertoire for T cell subsets isolated from the prostate tumors of TRAMP+/− Foxp3gfp TCRβtg mice. Repertoire overlap was assessed using the Morisita-Horn (MH) similarity index (7–11), for which a value of 1 indicates identity, and a value of 0 denotes complete dissimilarity (Fig. 1D). The analysis revealed that the TCR repertoire of CD4+Foxp3+ and CD4+Foxp3neg populations isolated from a particular prostate tumor were largely distinct and non-overlapping (MH = 0.07 ± 0.10 SD, Fig. 1D, samples intersecting at red lines), implying that the antigens recognized by tumor-infiltrating Tregs are different from those recognized by conventional CD4+ T cells. Second, the TCR repertoire of CD4+Foxp3+ cells isolated from the prostates of different mice exhibited a high degree of similarity from mouse to mouse (MH = 0.38 ± 0.39 SD, Fig. 1D, upper left quadrant). While a substantial proportion of this similarity was due to the recurrent enrichment of the Vα2-LYYNQGKLI TCR, additional TCRα chains were identified that were recurrently expressed by prostatic Foxp3+ Tregs (fig. S4). This finding suggests that TRAMP prostate tumors do not recruit polyclonal Tregs of arbitrary specificity, but instead are associated with the reproducible enrichment of Tregs of distinct specificities.
To facilitate the study of T cells expressing the canonical Vα2-LYYNQGKLI TCRα chain paired with the fixed TCRβ chain (hereafter referred to as “MJ23” T cells), we generated transgenic (tg) mice expressing the MJ23 TCRαβ heterodimer. In female MJ23tg _Rag1_−/− mice, CD4+ T cells in the periphery were phenotypically naïve (Fig. 2A and fig. S5). Moreover, Foxp3+ MJ23tg cells were not detected above background in the thymus and periphery of these mice (Fig. 2, A and B, and fig. S5). In contrast, analysis of tumor-free male MJ23tg _Rag1_−/− mice revealed spontaneous accumulation of activated CD44hi CD4+ MJ23tg T cells in the prostate and prostate-draining periaortic lymph nodes (Fig. 2, A and B, and fig. S5), indicative of MJ23 reactivity to an autoantigen at these sites. In the prostate, T cell accumulation was observed in both the stroma and the epithelium, and was associated with disruption of the basement membrane of prostatic glands (Fig. 2C). The presence of activated CD4+ T cells was accompanied by the concomitant appearance of a percentage of CD4+Foxp3+ T cells (Fig. 2, A and B, and fig. S5). Foxp3+ cells exhibited in vitro suppressive activity (fig. S6) and phenotypic characteristics of Tregs (fig. S7), including expression of neuropilin-1 (supplementary text and figs. S7 and S8). Taken together, our data demonstrate that MJ23 Tregs, identified based on their enrichment in mouse prostate tumors, are not reactive to a unique tumor-specific antigen, but instead recognize a self antigen associated with the organ of cancer origin.
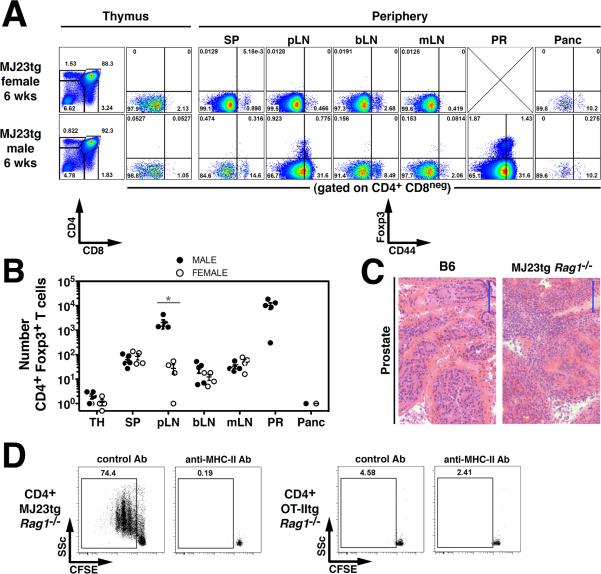
Fig. 2. MJ23 T cells recognize a prostate-associated self antigen.
(A–C) Spontaneous T cell autoreactivity in the prostates of tumor-free male MJ23tg _Rag1_−/− mice. (A) Representative flow cytometric analyses of T cells isolated from the indicated organs of 6-week-old male or female mice. SP, spleen; pLN, periaortic lymph nodes; bLN, brachial lymph nodes; mLN, mesenteric lymph nodes; PR, prostate; Panc, pancreas. (B) Summary plot of the absolute number of CD4+Foxp3+ T cells from the indicated organs of 6-week-old male or female mice. (C) Hematoxylin and eosin staining of dorsolateral prostatic lobes from 18-week-old MJ23tg _Rag1_−/− and B6 mice. Scale bar = 100 μm. (D) CFSE-labeled CD4+Foxp3neg MJ23tg T cells or OT-IItg cells were cultured with FACS-purified CD45+CD11c+F4/80neg cells isolated from TRAMP prostate tumors in the presence of recombinant mouse IL-2. In addition, MHC-II antibody or isotype control antibody was added to the culture. Dilution of CFSE was assessed by flow cytometry on day 4. The mean ± SEM is indicated. Asterisks indicate p < 0.05, ns = not significant. For (A–B), data are pooled from N = 2 independent experiments. For (B), t-tests were used to compare data from male and female mice at each site. Data in (D) are representative of N = 7 independent experiments.
In other experiments, CD11c+ dendritic cells isolated from TRAMP prostate tumors induced robust proliferation of MJ23tg T cells ex vivo (Fig. 2D), but did not stimulate OT-II transgenic T cells expressing an irrelevant class II-restricted TCR (12). These data provide direct evidence that prostate tumors contain the antigen recognized by MJ23 T cells.
In order to study the development of MJ23tg T cells at physiological clonal frequencies, we generated chimeric animals in which bone marrow cells from MJ23tg _Rag1_−/− donors were engrafted into host mice at low frequencies (<1%), and the development and distribution of MJ23tg T cells was assessed (Fig. 3 and fig. S9). Analysis of B6 male and tumor-bearing TRAMP male hosts revealed that Foxp3+ MJ23tg Tregs developed in the thymus (Fig. 3, A and B). The efficiency of development varied from mouse to mouse, and was inversely correlated with the frequency of MJ23tg precursors (Fig. 3B), a finding that is consistent with previously published studies (13, 14). In other experiments, OT-II _Rag1_−/− precursors did not develop efficiently into Foxp3+ cells in the thymus, demonstrating the specificity of MJ23tg Treg development (fig. S10). In the periphery of male chimeras, Foxp3+ MJ23tg cells were distributed throughout the secondary lymphoid organs, but were preferentially enriched in the prostate-draining periaortic lymph nodes (Fig. 3C), a finding consistent with evidence of reactivity to a prostate-associated antigen (Fig. 2). Very few donor-derived MJ23tg Tregs were observed in the prostate (Fig. 3, A and C), likely reflecting competition with endogenous MJ23 Tregs (supplementary text and fig. S11). Upon intravenous transfer of naïve CD4+Foxp3neg MJ23tg T cells into TRAMP males, the induction of Foxp3 expression by MJ23tg cells was negligible (fig. S12), suggesting that antigen exposure in the periphery does not favor the development of induced Tregs. Taken together, our results demonstrate that expression of the MJ23 TCR facilitates Treg development in the thymus of male hosts. Thus, T cells of this specificity encounter the antigen(s) driving Treg development during maturation in the thymus, prior to their exposure to the prostate or tumor environment.
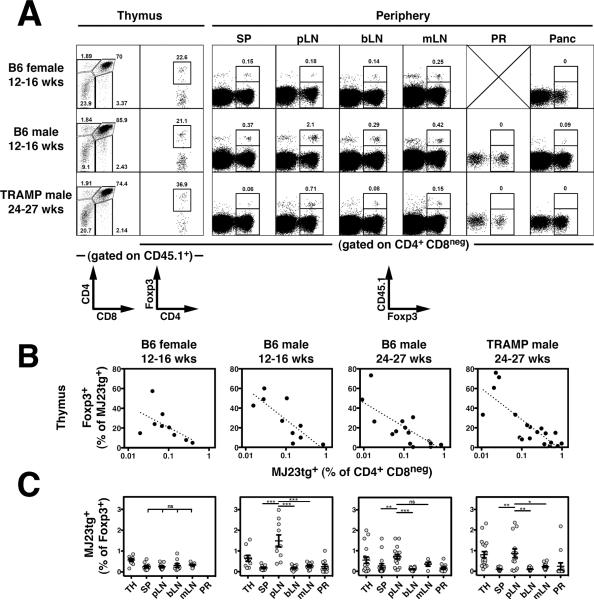
Fig. 3. Thymic development of MJ23 Tregs.
The MJ23 TCR facilitates thymic Treg development at low clonal frequencies. T cell-depleted bone marrow cells from MJ23tg _Rag1_−/− CD45.1+ female donor mice were engrafted, along with polyclonal “filler” cells from B6 females (CD45.2/.2), into sublethally irradiated CD45.2/.2 recipient mice. This approach resulted in seeding of MJ23tg precursors at a low frequency (<1%). 6 weeks post-engraftment, the fate of MJ23tg cells was analyzed. (A) Representative flow cytometric analyses of CD45.1+ MJ23tg T cells from different anatomical sites of male or female mice of the indicated ages and strain. Abbreviations are the same as in Fig. 2. For the thymus, the left column (CD4 vs. CD8) depicts undepleted samples, the right column (Foxp3 vs. CD4) depicts CD8-depleted samples. For the periphery, the percentage of all CD4+Foxp3+ T cells that are CD45.1+ (MJ23tg+) is indicated. (B) Summary plots of the “efficiency” of MJ23tg Treg development in the thymus, in which the percentage of CD4+CD8neg CD45.1+ MJ23tg cells that express Foxp3 is plotted vs. the frequency of CD45.1+ MJ23tg thymocytes (as a percentage of all CD4+CD8neg cells) for cells isolated from host mice of the indicated strain, gender, and age. Dashed lines indicate best-fit semi-log curves. (C) Summary plots of the percentage of CD45.1+ MJ23tg T cells amongst all CD4+Foxp3+ cells isolated from various organs of the indicated hosts. The mean ± SEM is shown. Asterisks indicate p < 0.05. ANOVA was used to compare the secondary lymphoid sites (spleen and lymph nodes) within chimeric hosts of a given type. Data are pooled from at least N = 3 independent experiments.
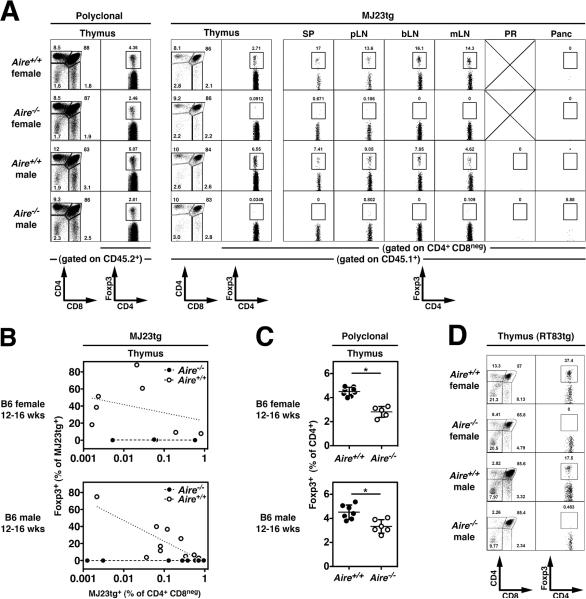
On the basis of our data indicating that MJ23 T cells are reactive to a prostate-associated antigen, we anticipated that Foxp3+ MJ23tg Tregs would not develop in B6 female hosts. However, unexpectedly, Foxp3+ MJ23tg Tregs developed in the thymus of chimeric B6 females (Fig. 3, A and B, and fig. S9). In the periphery of female mice, Foxp3+ MJ23tg T cells were broadly distributed in the spleen and all lymph nodes examined, but selective enrichment in the periaortic lymph nodes was not observed in female hosts (Fig. 3C). In order to elucidate the mechanisms underlying MJ23tg Treg development in both male and female mice, we examined the role of Aire in development. Aire encodes a transcriptional regulator that drives the ectopic expression of peripheral tissue-specific antigens by medullary thymic epithelial cells, and is critical for the maintenance of immune tolerance (15–17). Analysis of MJ23tg development in chimeric mice in which MJ23tg precursors were engrafted into Aire+/+ or _Aire_−/− hosts revealed that Foxp3+ MJ23tg Tregs failed to develop in the thymus and periphery of _Aire_−/− hosts, both male and female (Fig. 4, A and B). In addition, _Aire_−/− hosts exhibited a reduction in the percentage of thymic Tregs relative to Aire+/+ hosts, consistent with previous reports (15, 18), but were not characterized by a complete deficiency of polyclonal Tregs (Fig. 4, A and C). Despite the lack of MJ23tg Treg development, mature CD4+Foxp3neg MJ23tg T cells developed in _Aire_−/− hosts (Fig. 4A and fig. S13), indicating that T cells of this specificity are not dependent on Aire for their positive selection into the CD4+ lineage. This result also suggests that the thymic development of Tregs reactive to an Aire-dependent antigen may require both the recognition of self peptide/MHC ligands for positive selection, and the recognition of an additional Aire-dependent antigen for commitment to the Treg lineage. Importantly, similar experiments utilizing a second naturally occurring tumor-infiltrating Treg TCR, termed “RT83”, demonstrated that Aire is required for the thymic development of RT83tg Tregs (Fig. 4D and fig. S14). Thus, our data demonstrate that Aire is critical for the thymic development of multiple naturally occurring Treg specificities. These findings are consistent with a model in which the Aire-mediated expression of peripheral tissue-restricted antigens by thymic epithelial cells drives the development of a subset of tissue-specific or organ-specific Tregs (19).
Fig. 4. Aire-dependent thymic development of antigen-specific Tregs.
(A–C). The thymic development of MJ23tg Tregs is Aire-dependent. T cell-depleted bone marrow cells from MJ23tg _Rag1_−/− CD45.1+ female donor mice were engrafted, along with “filler” cells from _Aire_−/− females (CD45.2/.2), into sublethally irradiated _Aire_−/− or Aire+/+ recipient mice (CD45.2/.2), both male and female. 6 weeks post-engraftment, the fate of MJ23tg cells was analyzed. (A) Representative flow cytometric analyses of CD45.2+ polyclonal T cells (left) and CD45.1+ MJ23tg T cells from different anatomical sites of 12–16-week-old male or female mice of the indicated Aire genotype. Abbreviations are the same as in Fig. 2. The percentage of CD4+ cells that are Foxp3+ is shown. For the thymus, the left column (CD4 vs. CD8) depicts undepleted samples, the right column (Foxp3 vs. CD4) depicts CD8-depleted samples. (B) Summary plots of the “efficiency” of MJ23tg Treg development, in which the percentage of CD45.1+ MJ23tg cells that express Foxp3 is plotted vs. the frequency of CD45.1+ MJ23tg thymocytes (as a percentage of all CD4+CD8neg cells) for cells isolated from host mice of the indicated sex and genotype. Dashed lines indicate best-fit semi-log curves. (C) Summary plots of the percentage of CD45.2+ polyclonal CD4+CD8neg thymocytes that express Foxp3 in chimeric mice of the indicated sex and genotype. The mean ± SEM is shown. Asterisks indicate p < 0.05 for the comparison of _Aire_−/− vs. Aire+/+ mice (t-test). Data in (A–C) are pooled from N = 3 independent experiments. (D) Aire-dependent thymic development of “RT83tg” Tregs. RT83tg bone marrow chimeras were generated in _Aire_−/− or Aire+/+ hosts, as described above for MJ23tg T cells. Representative flow cytometric analyses of CD45.1+ RT83tg T cells from the thymus of 12–16-week-old male or female mice of the indicated Aire genotype. The percentage of CD4+ cells that are Foxp3+ is shown. For the thymus, the left column (CD4 vs. CD8) depicts undepleted samples, the right column (Foxp3 vs. CD4) depicts CD8-depleted samples. Data in (D) are pooled from N = 2 independent experiments.
While there is substantial evidence demonstrating that Aire plays a critical role in the deletion of autoreactive thymocytes reactive to peripheral tissue antigens (15, 20–22), a definitive role for Aire in the thymic selection of naturally occurring Tregs has not been previously established. Here, we provide direct evidence that Aire is critical for the thymic development of Tregs of naturally arising specificities. Thus, the integration of available evidence suggests a dual role for Aire in the maintenance of immune tolerance, in which Aire drives both the deletion of autoreactive T cells and the development of a subset of Foxp3+ Tregs.
In sum, our data support a model in which a tumor does not drive the de novo conversion of tumor-specific CD4+ effector T cells into induced Foxp3+ Tregs, but instead recruits pre-existing thymic-derived Tregs reactive to Aire-dependent self antigens associated with the organ of cancer origin. Thus, a developing neoplasm co-opts endogenous mechanisms that have evolved to preserve the integrity of the host by maintaining organ-specific immune tolerance.
Supplementary Material
Supplementary Material
Table S1
Acknowledgements
We thank A. Bendelac, T. Gajewski, R. Gottschalk, and S. Quezada for critical reading of the manuscript, A. Rudensky for Foxp3_gfp_ reporter mice (6), C. Benoist for cassette vectors for TCRα and TCRβ transgenic mice (23), L. Degenstein for transgenic mouse production, and A. Thomas and N.-L. Yin for deep TCR sequence analysis. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. This work was partially funded by the following sources (to P.A.S.): R01 (#1R01CA160371-01), a Cancer Research Institute Investigator Award, a Cancer Research Foundation Young Investigator Award, an American Cancer Society Institutional Research Grant (#IRG-58-004), and the University of Chicago Comprehensive Cancer Center (including Support Grant #P30 CA14599). N.D.S. was supported by MSKCC Comprehensive Cancer Center (Support Grant #P30 CA008748). J.P.A. was supported by funding from the Howard Hughes Medical Institute.
Footnotes
*
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References and Notes
- 1.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu Rev Immunol. 2011;29:235. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 3.Deleeuw RJ, Kost SE, Kakal JA, Nelson BH. The Prognostic Value of FoxP3+ Tumor-Infiltrating Lymphocytes in Cancer: A Critical Review of the Literature. Clin Cancer Res. 2012 Jun 1;18:3022. doi: 10.1158/1078-0432.CCR-11-3216. [DOI] [PubMed] [Google Scholar]
- 4.Greenberg NM, et al. Prostate cancer in a transgenic mouse. Proc Natl Acad Sci U S A. 1995 Apr 11;92:3439. doi: 10.1073/pnas.92.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gingrich JR, Barrios RJ, Foster BA, Greenberg NM. Pathologic progression of autochthonous prostate cancer in the TRAMP model. Prostate Cancer Prostatic Dis. 1999 Mar;2:70. doi: 10.1038/sj.pcan.4500296. [DOI] [PubMed] [Google Scholar]
- 6.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005 Mar;22:329. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J Exp Med. 2007 Sep 3;204:2039. doi: 10.1084/jem.20070822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong J, et al. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007 Jun 1;178:7032. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 9.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006 Apr;7:401. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 10.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med. 2008 Dec 22;205:3105. doi: 10.1084/jem.20081359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magurran . Ecological Diversity and Its Measurement. Princeton University Press; Princeton, NJ: 1988. [Google Scholar]
- 12.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998 Feb;76:34. doi: 10.1046/j.1440-1711.1998.00709.x. [DOI] [PubMed] [Google Scholar]
- 13.Bautista JL, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol. 2009 Jun;10:610. doi: 10.1038/ni.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung MW, Shen S, Lafaille JJ. TCR-dependent differentiation of thymic Foxp3+ cells is limited to small clonal sizes. J Exp Med. 2009 Sep 28;206:2121. doi: 10.1084/jem.20091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson MS, et al. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005 Aug;23:227. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Anderson MS, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002 Nov 15;298:1395. doi: 10.1126/science.1075958. [DOI] [PubMed] [Google Scholar]
- 17.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 18.Lei Y, et al. Aire-dependent production of XCL1 mediates medullary accumulation of thymic dendritic cells and contributes to regulatory T cell development. J Exp Med. 2011 Feb 14;208:383. doi: 10.1084/jem.20102327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aschenbrenner K, et al. Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat Immunol. 2007 Apr;8:351. doi: 10.1038/ni1444. [DOI] [PubMed] [Google Scholar]
- 20.Liston A, Lesage S, Wilson J, Peltonen L, Goodnow CC. Aire regulates negative selection of organ-specific T cells. Nat Immunol. 2003 Apr;4:350. doi: 10.1038/ni906. [DOI] [PubMed] [Google Scholar]
- 21.Taniguchi RT, et al. Detection of an autoreactive T-cell population within the polyclonal repertoire that undergoes distinct autoimmune regulator (Aire)-mediated selection. Proc Natl Acad Sci U S A. 2012 May 15;109:7847. doi: 10.1073/pnas.1120607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeVoss J, et al. Spontaneous autoimmunity prevented by thymic expression of a single self-antigen. J Exp Med. 2006 Nov 27;203:2727. doi: 10.1084/jem.20061864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kouskoff V, Signorelli K, Benoist C, Mathis D. Cassette vectors directing expression of T cell receptor genes in transgenic mice. J Immunol Methods. 1995 Mar 27;180:273. doi: 10.1016/0022-1759(95)00002-r. [DOI] [PubMed] [Google Scholar]
- 24.Materials and methods are available as supplementary materials in Science Online.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Table S1