Visualizing group II intron catalysis through the stages of splicing (original) (raw)
. Author manuscript; available in PMC: 2013 Apr 26.
SUMMARY
Group II introns are self-splicing ribozymes that share a reaction mechanism and a common ancestor with the eukaryotic spliceosome, thereby providing a model system for understanding the chemistry of pre-mRNA splicing. Here we report fourteen crystal structures of a group II intron at different stages of catalysis. We provide a detailed mechanism for the first step of splicing, we describe a reversible conformational change between the first and the second steps of splicing, and we present the ligand-free intron structure after splicing, in an active state that corresponds to the retrotransposable form of the intron. During each reaction, the reactants are aligned and activated by a heteronuclear four-metal-ion center that contains a metal cluster and obligate monovalent cations, adopting a structural arrangement similar to that of protein endonucleases. Based on our data, we propose a model for the splicing cycle and show that it is applicable to the eukaryotic spliceosome.
INTRODUCTION
Group II introns are self-splicing ribozymes that are essential for gene expression in many organisms. In addition to their impact on modern RNA metabolism, they are important model systems for understanding pre-mRNA splicing, as they share a common ancestor with the spliceosomal machinery that is required for splicing of pre-mRNAs in eukaryotes (Pyle and Lambowitz, 2006). Excised group II introns are important catalysts for genomic diversification because they reverse-splice into DNA and incorporate themselves into new genomic locations through retrotransposition (Dai and Zimmerly, 2002). Given their profound and continuing impact on gene expression and evolution in all domains of life (Mattick, 1994), and their widespread applications in biotechnology (Garcia-Rodriguez et al., 2011), it is important to understand the chemical mechanism of group II intron catalysis.
Fundamental insights into the mechanisms of RNA cleavage and ligation by group II introns have been obtained through biochemical experiments and from crystal structures of the Oceanobacillus iheyensis group IIC intron (Pyle and Lambowitz, 2006; Toor et al., 2008a). These studies have revealed an intricate architectural scaffold that encloses a catalytic pocket within its core. The pocket is formed by three highly conserved motifs that are joined into a triple helix containing nucleotides from the conserved “catalytic triad” of intron domain 5 (D5, C358-G359-C360), the two-nucleotide bulge of D5 (A376-C377), and the J2/3 junction (A287-G288-C289). The resulting motif binds two divalent ions (M1 and M2) that participate directly in the mechanism of chemical catalysis (Gordon and Piccirilli, 2001; Toor et al., 2008a; Toor et al., 2008b). However, the lack of information on other metals near the active site, and on the position of key reactants, had so far precluded a clear understanding of group II intron and, potentially, spliceosomal catalysis.
To address these issues, we present an extensive set of intron crystal structures obtained at different stages of the splicing reaction, in the presence of diverse metal ion cofactors. In this way, we define key structural elements necessary for hydrolytic group II intron splicing, including a heteronuclear metal ion center within the active-site and the position of the reaction nucleophile. We report the structure of the excised intron in an alternative conformation and, on the basis of mutagenesis and in vitro splicing data, we show that it is involved in the second step of splicing. Our results lead us to propose a functional model for group II intron splicing, in which the J2/3 motif toggles between two alternative conformations during the transition from the first to the second step of splicing. This model agrees with functional and structural data available for the nuclear spliceosome, having important implications for understanding the structural basis for all pre-mRNA splicing.
RESULTS
Visualizing the first step of splicing
To determine the structure of the intron at sequential stages of catalysis, we created two constructs that contain O. iheyensis group II intron domains 1–5 with and without a short 5′-exon (Oi5eD1-5 and OiD1-5, respectively). These optimized constructs, which are fully active ribozymes, lack flexible sections that had been included in previous studies (Toor et al., 2008a), thereby facilitating strict control over the binding of reaction substrates and oligonucleotide ligands.
Using the Oi5eD1-5 construct, we solved the intron structure before and after 5′-exon hydrolysis at 3.1 Å and 2.9 Å resolution, respectively. The pre-hydrolytic state (the stage prior to the first step of splicing) was crystallized in the presence of Ca2+ (Figure 1 and Table S1), which is a known inhibitor of Mg2+-dependent enzymes (including group II introns), and which has previously been employed to crystallize endonucleases in complex with their substrates (Erat and Sigel, 2008; Viadiu and Aggarwal, 1998; and this work, Figure 2). The post-hydrolytic state was crystallized in the same condition, but using catalytically active Mg2+ ions that stimulate hydrolysis of the 5′-exon (Figure S1). Overall, both structures are nearly identical to those determined previously (i.e. RMSD values of 0.6 Å with respect to PDB id. 3IGI) and their active sites adopt the characteristic triple helix conformation that is stabilized by a dehydrated metal ion of previously unknown identity, here classified as K1 (vide infra). In the pre-catalytic state, the metal ions M1 and M2 coordinate the pro-R oxygen atom of the scissile phosphate at the 5′-splice junction (Figure 1), as predicted from chemical biology experiments (Padgett et al., 1994; Podar et al., 1995). Additionally, M1 coordinates the 3′-OH leaving group (belonging to the last nucleotide of the exon), while M2, together with C358 (catalytic triad), coordinates a solvent molecule located 2.7 Å from the scissile phosphate. This solvent molecule, identified here for the first time and validated by the calculation of the corresponding simulated-annealing electron density omit-map (Figure S1), is in the precise position that would be occupied by the reaction nucleophile, which consists either of a 2′-OH group when splicing proceeds through the branching pathway, or an H2O in the hydrolytic splicing pathway that is typical for group IIC introns (Pyle, 2010). Finally, the geometry of the 5′-splice junction is constrained by two other interactions. Upstream of the scissile phosphate, the 5′-exon is immobilized by base complementarity to exon-binding site 1 (EBS1), while downstream of the scissile phosphate, the 5′-end of the intron is stabilized by a fourth important yet still uncharacterized metal ion, K2 (vide infra). As a consequence of these interactions, the 5′-splice junction adopts a sharp “kink” (Chan et al., 2012), with an unusually small backbone angle of approximately 50° at the scissile position (the backbone angle is defined here as the angle formed by three consecutive phosphorus atoms, Table S2). Upon hydrolysis, the kink relaxes and the scissile phosphate displaces by ~4 Å, where it is neutralized by electrostatic interactions with K2. By contrast, the 5′-exon does not displace significantly, instead maintaining its coordination to M1 and thus occupying an ideal position to react as the nucleophile at the 3′-splice junction during the second step of splicing.
Figure 1. The mechanism of the first splicing step by hydrolysis.
The Oi5eD1-5 ribozyme solved in the presence of K+/Ca2+ at 3.1 Å resolution (left). The inset shows a magnification into the active site (distances are depicted as black dotted lines and are indicated in angstroms). Two divalent ions (M1 and M2, yellow spheres) and two monovalent ions (K1 and K2, purple spheres) tightly coordinate the nucleotides involved in catalysis (sticks, color coded by atom type). The water molecule W1 (cyan sphere) is in an ideal position to perform a nucleophilic attack on the scissile phosphate (black arrows). Upon hydrolysis, the scissile phosphate swings out of the active site (semitransparent sticks and gray dotted arrow) to form an electrostatic interaction with K2 (gray dotted line). See also Figure S1.
Figure 2. Splicing reactions in the presence of different ion analogues.
Splicing is robust in all ion combinations that support the structural integrity of the triple helical, active intron conformation (K+/Mg2+, Rb+/Mg2+, Tl+/Mg2+, and NH4+/Mg2+). By contrast, splicing is impaired in Cs+/Mg2+, likely due to the larger size of Cs+ and to its consequently suboptimal occupation at the K1 site. Mg2+ analogues (i.e. Ca2+, Sr2+, and Ba2+) completely inhibit splicing despite ensuring the overall correct assembly of the intron. Finally, Na+ and Li+ also do not promote catalysis, because they favor the toggled intron conformation (see Figure 4). Upon splicing, the precursor RNA (5e-I-3e) forms intermediate product (I-3e), linear intron (I), ligated exons (5e-3e), and hydrolyzed 5′- and 3′-exons (5e and 3e, respectively). The sizes of the bands are indicated in nucleotides (nt) by the marker lane (M). The reactions were performed at a concentration of 10 mM divalent cation and 150 mM monovalent cation. See also Figure S2.
In conclusion, the structures of the Oi5eD1-5 constructs reveal the position of the key structural elements necessary for catalyzing the first step of splicing as it proceeds through the hydrolytic pathway.
The intron structure in its ligand-free state explains its high reactivity as an infectious retroelement
In addition to structural characterization of 5′-splice site hydrolysis, we also succeeded in solving the intron structure in a completely empty, exon-free state at 3.5 Å resolution. By crystallizing the OiD1-5 construct in the absence of any added or residual oligonucleotides, which were present in all previous studies (Toor et al., 2010; Toor et al., 2008a; Toor et al., 2008b), we show that the free intron possesses a remarkably intact active site. The intron core adopts the typical triple helix conformation and maintains the positions of the four metal ions M1, M2, K1, and K2 (Figure S3). This ligand-free structure represents the intron product at the end of the splicing cycle, after release of ligated exons. In this state, the intron can attack and retrotranspose into host genomes. Upon rebinding ligated exons, it can also catalyze spliced-exon reopening (SER), a reaction that provides a well-established enzymological mimic for the retrotransposition event (Mikheeva et al., 2000; Podar et al., 1995). The empty intron therefore provides a useful starting point for studying the infectious form of this ribozyme as it is poised to recognize substrates.
To visualize the free intron as it moves through a cycle of target binding and cleavage, we co-crystallized OiD1-5 with oligonucleotide substrates that are identical in sequence to native, ligated exons. We obtained two structures at 3.0 Å and 2.7 Å resolution in the presence of Ca2+ and Mg2+, respectively, which describe target recognition both before and after hydrolysis. In the pre-catalytic state, the scissile phosphate of the substrate presents its pro-S oxygen atom to the two catalytic metals M1 and M2 (Figure 3). As in the 5′-exon hydrolysis reaction, M1 coordinates the leaving group, while M2, together with C358, coordinates a solvent molecule that likely represents the reaction nucleophile (Figure S3). Upstream of the scissile position, the 5′-end of the oligonucleotide tightly binds to the EBS1 site, and it does not displace upon hydrolysis. Downstream of the scissile phosphate, the 3′-end of the oligonucleotide forms a well-defined Watson-Crick base pairing between the scissile uridine and the EBS3 adenosine (A223) in the pre-catalytic state, but this contact is released after hydrolysis. Constrained by the interactions with the exon binding sites, the junction of the ligated exons is relatively extended and forms a backbone angle of 137°.
Figure 3. The mechanism of the spliced-exon-reopening (SER) reaction.
Left: structure of OiD1-5 bound to a ligated oligonucleotide in the presence of K+/Ca2+. The scissile phosphate is coordinated by the M1 and M2 ions (Ca2+, yellow spheres), while the reaction nucleophile (W1, cyan sphere) is coordinated by M2 and C358, similar to the 5′-exon hydrolysis reaction (Figure 1). Right: structure of OiD1-5 bound to a hydrolyzed oligonucleotide in the presence of K+/Mg2+. Upon hydrolysis, the portion of the oligonucleotide downstream of the scissile site is released, while the portion upstream of the scissile site remains bound to the intron because of its extensive base complementarity with EBS1 (not shown). In the absence of the scissile phosphate, a water molecule (W2, cyan sphere) bridges M1 and M2. See also Figure S3.
In conclusion, the structures of the OiD1-5 construct with and without a target substrate reveal that the free intron is structurally intact, with a perfectly well-formed core that can readily bind target RNA or DNA molecules without undergoing extensive structural rearrangement.
A heteronuclear metal ion center is required for group II intron splicing reactions
A consistent feature of the structures described above are the four metal ions (M1, M2, K1, and K2) involved in the reaction mechanism. Given their central importance for all the splicing reactions characterized thus far, it was essential to unambiguously establish the identity, location and functional role of these ion cofactors. To this end, we selectively replaced each putative type of ion with alkaline and alkali-earth metals, monitoring the effects of the substitutions crystallographically, using X-ray anomalous scattering, and biochemically, through splicing assays.
Previous crystallographic investigations using the heavy ion Yb3+ established that M1 and M2 correspond to multivalent ion binding sites (Toor et al., 2008a), and here we confirmed this using Ca2+ (3.3 Å resolution) and Ba2+ (4.0 Å resolution, Figure S2). The M1 and M2 sites are also fully occupied in the structures obtained using Mg2+ as the only multivalent cation, and they possess the octahedral coordination geometry typical of Mg2+ (Andreini et al., 2008). It is therefore likely that M1 and M2 are occupied by Mg2+ under physiological conditions. Based on their location relative to the scissile linkage and on the biochemically-established role of their coordinating ligands (Boulanger et al., 1995; Schmidt et al., 1996), it is also likely that M1 and M2 participate directly in the mechanism of chemical catalysis. Not surprisingly, substituting Mg2+ with divalent ion analogues Ca2+, Sr2+, and Ba2+ inhibits splicing almost completely (Figure 2). Such inhibitory effect can be ascribed to the fact that these ions do not allow the correct formation of the trigonal bipyramidal reaction intermediate that is typical of the SN2 nucleophilic reaction performed by the intron, as observed in other systems (McConnell et al., 1997; Viadiu and Aggarwal, 1998).
By contrast, although previous structures revealed electron density peaks at the positions that we define as K1 and K2 (Toor et al., 2010; Toor et al., 2008b), the identity of the species occupying these sites and their precise location were ambiguous and required conclusive characterization. To characterize the K1 and K2 sites, we solved the structure of OiD1-5 in the presence of heavy monovalent ions Tl+ (2.8 Å resolution), Rb+ (3.3 Å resolution) and Cs+ (3.4 Å resolution). All anomalous difference Fourier electron density maps show two pronounced peaks within the intron core at the K1 and K2 positions (and not at the M1 and M2 positions, Figure S2). All structures are intact overall, but Cs+, which is the largest ion (Table S3), induces a distortion in the active site. Additionally, we solved the structure of OiD1-5 in the presence of NH4+ (2.9 Å resolution), which possesses an ionic radius similar to K+, and in Na+ (3.2 Å resolution), which has a smaller radius (Table S3). It is significant that NH4+, but not Na+ (vide infra), can bind at the K1 and K2 positions. Therefore, we conclude that K1 and K2 are specific monovalent ion binding sites with strict size requirements for proper occupancy and it is likely that they are occupied by K+ under physiological conditions. It is significant that the M1, M2 and K1 ions are each joined by a single shared ligand atom (the pro-R oxygen of the scissile phosphate in the 5′-splice junction and the pro-S oxygen of C377, respectively). Given this bonding pattern, these metals can be formally classified as a heteronuclear KMgO cluster (Cotton and Wilkinson, 1980), resembling the clusters observed in metalloproteins (Messerschmidt et al., 2001). However, a metal cluster composed of magnesium and potassium ions has not been reported to date, and it may represent a unique attribute of RNA enzymes. This trinuclear M1-M2-K1 cluster, together with the ion K2, forms a four-metal center that constitutes an essential cofactor for group II intron catalysis.
Complementary functional analysis confirms this hypothesis. Specifically, we examined the effects of different monovalent and divalent cations on the splicing activity of wild type O.i. group II intron. We observed that all ion combinations that produced well-diffracting crystals with well-formed active sites (K+/Mg2+, NH4+/Mg2+, Rb+/Mg2+, and Tl+/Mg2+) also displayed robust splicing (Figure 2). By contrast, splicing is inhibited in the presence of Cs+ and blocked in Na+, two ions that preserve the overall structure of the ribozyme yet distort the local active-site architecture. Thus, enzyme activity perfectly mirrors the results from crystallography.
Taken together, the collection of structures and accompanying biochemistry demonstrate that M1, M2, K1, and K2 form a functional metal center in which each type of ion plays a role in catalysis. It is likely that additional ions also contribute indirectly to activity through their role in supporting the intron structure. Indeed, our collection of high-resolution X-ray anomalous scattering maps obtained for Tl+, Rb+, Cs+, and Ba2+ now reveal the identity of site-bound divalent and monovalent metals throughout the entire OiD1-5 structure (three of which are described in Figure S2), thus providing much-needed information on the role of metals in the architectural fabric of RNA.
The intron active site can adopt two alternative conformations
As reported above, when K+ is replaced by Na+ in the crystallization buffer, the K1 and K2 sites are not occupied and this structural feature induces a loss of electron density at the M1 and M2 sites. Such disruption of the active site is accompanied by the dislocation of the J2/3 junction and D5 bulge residues G288 and C377, which become stabilized by a completely different network of interactions (Figure 4). In the Na+ structure, G288 has rotated by about 90° around an axis connecting its C5′ and C3′ backbone atoms and the guanosine moiety is now located directly underneath A287, extending a stack of nucleobases that is formed by G3, U4, G5, A376 and A287. Additionally, G288 forms two new sets of hydrogen bonds, one with C377 through its O2′ atom and the other with a solvent molecule that is coordinated by A287 and the N2 of G321 (Figure S4). As a consequence of the conformational change in G288, C377 also rotates by about 70° around the glycosidic bond. In this new position, N3 and O2′ of C377 form hydrogen bonds with O2′ of G288 and with the pro-R phosphate oxygen of C360, respectively. In this conformation, G288 and the base moiety of C377 possess higher B-factors than the rest of the ribozyme chain (Bave-overall = 72 Å2, Bave-G288 = 133 Å2, Bave-C377-base = 110 Å2) indicating a higher degree of flexibility for these residues. Perhaps most importantly, upon rotation of G288 and C377, binding of the metal cluster in the active site is abolished, which explains why only 2–5 % of wild type splicing activity is retained under these conditions (Figure S5) and which suggests that the alternative conformation represents an inactive state of the intron.
Figure 4. The toggled conformation of the J2/3 junction.
The structure of the OiD1-5 ribozyme solved in the presence of Na+/Mg2+ at 3.2 Å resolution shows a concerted rotation (gray dotted arrows) of G288 and C377 from the triple helix conformation (semi-transparent sticks, drawn using the coordinates of the K+/Mg2+ structure) to the toggled conformation (pink solid sticks). The toggled conformation is stabilized by a new network of hydrogen bonds (black dotted lines, distances in angstroms), which involves also a solvent molecule (W) and G321 in domain 3. See also Figure S4.
We observed a similar disruption of the active site in the structure of Oi5eD1-5 solved in the presence of Li+ (2.8 Å resolution). Although Li+ does not appear to support splicing (Figure 2), the Li+ structure reveals a post-hydrolytic state in which the 5′-exon has been cleaved over the course of the crystallization experiment. This observation suggests that the active triple helix conformation may form, at least transiently, in Li+ conditions. Indeed, electron density within the active site is compatible with a double conformation for G288 (Figure S4).
Overall, the Na+ and Li+ structures show that the active site of group II introns can exist in two different states: the active triple helical form and the inactive toggled conformation. The presence of residual hydrolytic activity and the high crystallographic B-factors of G288 and C377 suggest that there exists a reversible equilibrium between these two conformations in the core.
A mutation in the triple helix affects the first and second steps of splicing at different rates
In order to evaluate if the toggled conformation is functionally relevant, we used the Na+ structure to guide the creation of point mutants that would affect the hydrogen bonding networks among the active residues. Specifically, we analyzed the splicing behavior of point mutations at positions G288 and C377, which appear to stabilize the toggled conformation. Based on energy-minimized structural models of such mutations, we expected the G288N mutants to destabilize both the triple helix and the toggled conformations, and the C377N mutants to form different sets of interactions in the two conformations and thus to influence the respective stability of the conformers to different extents. In particular, C377G is likely to have a stabilizing effect on the toggled conformation, in which it can establish an additional hydrogen bond with the N7 atom of G359 (Figure S5). Therefore, we anticipated that mutations on C377 should reveal if and when the toggled conformation forms during splicing.
In line with these expectations, we observed that G288 mutations have a strong impact on both steps of splicing (> 90 % inhibition), while substitutions of C377 affect the two steps differently (Table S4). Most importantly, in the presence of physiological K+ concentrations, the C377G mutant accumulates linear intron/3′-exon intermediate, indicating that it suffers a pronounced defect in the transition to the second step of splicing (40-fold slower second step relative to wild-type, Figure 5).
Figure 5. Effect of the C377G mutation on group II intron splicing.

Wild type O.i. group II intron splices efficiently under physiological K+ concentrations, while the C377G mutant has an evident splicing defect and accumulates intermediate product (I-3e, red and blue boxes, respectively). Specifically, the rate constants of the C377G mutant are reduced by ~11-fold in the first step and by ~37-fold in the second step of splicing with respect to wild type, respectively (Table S4). The graph on the right shows the evolution of the population of intermediate over time. The error bars represent the standard deviation calculated from three independent experiments. Intermediate (I3e) and linear intron (I) migrate as double bands because of cryptic cleavage at nucleotides (−11) and (−10) from the 5′-end of the intron, as explained in Extended Experimental Procedures. See also Figure S5.
Therefore, the behavior of the C377G mutant reveals a differential influence of active site elements on the two splicing steps, suggesting that the network of interactions within the intron active site changes during the splicing reaction. Possibly, such rearrangement involves the transient disruption of the triple helix conformation to adopt the toggled conformation described above for the Na+ and Li+ structures.
DISCUSSION
Visualizing the splicing chemistry of a complex metal ion center
This work provides the spatial description of the structural elements necessary for group II intron catalysis and describes how these elements change position during a cycle of splicing. We visualize the mechanistic interplay of conserved RNA intron motifs, a four-metal-ion heteronuclear center, the scissile phosphate, and the reaction nucleophile.
First, we establish that three conserved ribozyme motifs (D5 catalytic triad, D5 bulge, and J2/3 nucleotides) provide the necessary scaffold for correct formation of the active site and tight binding of the catalytic metal center. Bound oligonucleotide substrate is not required for the integrity of active-site architecture, as demonstrated by the fact that conserved motifs maintain their position, and catalytic metals fully occupy their respective binding sites in all ligand-free structures (Figure S3). This observation is unusual in the context of phosphodiester cleavage enzymes (including protein enzymes), which generally require the binding of an oligonucleotide substrate in order to fix the position of catalytic metal ions (Pingoud et al., 2009). The fact that the group IIC ribozymes have rigid “teeth” is consistent with their biological function, as the free introns are highly reactive, infectious retroelements that attack and integrate throughout host genomes, where they must function with complete autonomy. Having a preorganized active site composed of a rigid metal cluster is a distinct advantage for a genomic predator.
Second, we find that each metal within the heteronuclear center contributes to catalysis, acting as an essential cofactor. The divalent ions M1 and M2 directly activate the reactants for catalysis, as described for other two-metal-ion dependent enzymes (Steitz and Steitz, 1993). But this enzyme is different in that monovalent ions are strictly required as well. K1 and K2 mold the RNA backbone within the conserved core, rigidifying the divalent ion binding sites and stabilizing key reaction intermediates. The exceptionally dehydrated K1 ion acts like a “piton” that anchors the M1 and M2 ligands firmly in place, potentially explaining the stability of these latter sites in the absence of bound oligonucleotide. As such, it is also likely that K1 modulates conformational toggling between the first and the second splicing steps (vide infra). By contrast, K2 plays a dual role both by ensuring correct arrangement of the 5′-splice junction and by stabilizing the post-catalytic state after the first step of splicing. These findings underscore the importance of K+ ions during RNA tertiary structure formation, as observed also in riboswitches (Lambert et al., 2009) and in rRNA fragments (Conn et al., 2002). In a broader perspective, our results show that, by forming complex catalytic metal centers, RNA can be expected to possess the same complex chemistry as proteins. However, we see differences in the bioinorganic chemistry of the two systems: Monovalent ions appear to play a central role in the chemistry of RNA splicing, while protein metalloenzymes are almost exclusively dependent on multivalent ions, and on transition elements in particular (Messerschmidt et al., 2001).
Third, the structural comparison of the 5′-exon hydrolysis and SER reactions provide important information on the geometry of the scissile phosphate and on the reaction nucleophile. On one hand, the structures reveal specific differences between the two reactions. For instance, the stereochemistry of the scissile phosphate is inverted in the two steps (pro-R for 5′-exon hydrolysis and pro-S for SER), as predicted by biochemical experiments (Padgett et al., 1994; Podar et al., 1995, 1998). In addition, the RNA backbone adopts a different conformation at the scissile position during the two steps. It is tightly kinked before 5′-exon hydrolysis but extended prior to SER, suggesting that, although it may favor the reaction (Chan et al., 2012), a kink at the scissile junction is not necessarily required for hydrolysis. However, the two reactions also display common features. Most importantly, they occur at the same active site, sharing even the structural position of the reaction nucleophile. In light of the debate over whether group II introns possess one single or two separate active sites (de Lencastre et al., 2005; Pyle and Lambowitz, 2006), our structural observations are enlightening. Considering that 5′-exon hydrolysis is a pathway by which introns catalyze the first step of splicing, while SER represents the reverse of the second step of splicing, our data are consistent with the single active site hypothesis. This conclusion is supported by the fact that the 5′-exon maintains its coordination to EBS1 and to catalytic ion M1 during both splicing reactions, both in the precatalytic states and after hydrolysis.
In summary, considering the structural organization of the active site elements, we conclude that group II introns possess a significantly different active site from that of group I introns (Cate et al., 1997). Rather, group II introns resemble protein enzymes, like type II endonuclease BamHI (Figure 6; Viadiu and Aggarwal, 1998). The striking similarity between these macromolecules represents an intriguing example of convergent evolution and underscores the fact that catalytic strategy is universal and independent of biopolymer scaffold.
Figure 6. Structural convergence between group II introns and protein endonucleases.
Stereo representation of the intron active site (K+/Ca2+ oligonucleotide-bound structure, blue) superimposed over the R active site of endonuclease BamHI (PDB id.: 2BAM, red). The carboxyl groups of amino acids Glu77 and Asp94 occupy the same location as the phosphates of nucleotides U375 and C377 (D5 bulge), the carbonyl oxygen of Phe112 and other solvent molecules (WA and WB) replace the phosphates of C358-G359 (D5 catalytic triad), Lys126 is analogous to the K2 ion, and Tyr65 plays the same role as the K1 ion. As a consequence, the catalytic ions M1 and M2 and also the reaction nucleophile superimpose precisely.
Toggling of the J2/3 junction guides the transition between the first and second steps of splicing
Although the active-site elements described above maintain a similar structural organization during all of the states that contribute to splicing chemistry, the intron must undergo some conformational rearrangement between the first and the second steps of splicing in order to accommodate the entry and exit of splicing components. The observation of an alternative conformational state sampled by G288 and C377 may help resolve this apparent paradox. G288 and C377 have historically been implicated in transition between the first and second steps of splicing (Boudvillain and Pyle, 1998; de Lencastre et al., 2005; Ho Faix, 1998; Mikheeva et al., 2000), so it is remarkable that these particular nucleotides here emerge as the major structural players involved in core conformational change.
Nucleotide G288 is strictly conserved among all group II introns and is required for both the first and the second splicing steps (de Lencastre and Pyle, 2008; Jacquier and Michel, 1990; Mikheeva et al., 2000), as we confirm in this work (Table S4). In some introns, the G288N mutants were shown to suffer a more serious second step than first step splicing defect, thereby leading to the suggestion that G288 may be a dynamic element that contributes to intron rearrangement between the two steps (Ho Faix, 1998; Mikheeva et al., 2000; and this work, Table S4). The observation that G288 enhances reactivity in constructs where it is attached to D2 but disconnected from D3 (Fedorova et al., 2003) also supports the hypothesis that conformational flexibility of G288 contributes to splicing.
By contrast, C377 is only moderately conserved and one observes that C377 can be phylogenetically substituted by U and (in one case) by A, but it is never replaced by G in any known sequence (Keating et al., 2010). Here we report that the C377G mutant displays a pronounced defect that specifically obstructs the second step of splicing (Table S4). This defect cannot be attributed strictly to a disruption of the catalytic triple helix because we have solved the structure of the C377G mutant at 3.2 Å resolution in the presence of K+ and we show that it is capable of forming the chemically competent active-site form (Figure S5). However, the structural and functional data demonstrate that C377 can establish at least two different interaction networks during the cycle of splicing, of which the triple helix conformation represents only one set. Based on the effects of the mutants, the wealth of past functional data, and the conformational isomerization we have observed crystallographically, we propose that G288 and C377 are flexible and that the triple helix is disrupted during the transition between the first and second steps of splicing. Possibly, the toggled conformation adopted by the intron during this rearrangement resembles that which we observe upon crystallization of the intron in Na+ or Li+.
In addition to the mutagenesis data, two other pieces of evidence suggest that the triple helix is disrupted during the splicing cycle. First, in our structures (which lack D6) we observe that the first intron nucleotide, G1, rotates and occupies the active site at the conclusion of the 5′-exon hydrolysis reaction (Figure S1). During splicing, such rotation would create a steric impediment for the 3′-splice junction to insert into the same position. It is known that an interaction between G1 and the penultimate intron nucleotide is important for the second splicing step (Chanfreau and Jacquier, 1993). The formation of this tertiary interaction, together with the transient disruption of the active site may favor the displacement of G1. Second, D6 is expected to occupy a cavity near the J2/3 junction. D6 was proposed to adopt an “active” conformation in the first step of splicing and a “silent” conformation forming the η-η′ interaction with D2 for the second step of splicing (Pyle, 2010). Modeling shows that the “silent” conformation is likely to involve hydrogen bonds between the branch-site and G288, but only when the latter adopts the toggled conformation, as in the Na+ and Li+ structures (Figure S4). Interestingly, G288 was indeed shown to form short-range crosslinks near the branch site in ai5γ group II intron (de Lencastre et al., 2005). Therefore, the disruption of the triple helix may help orient D6 correctly within η-η′, facilitating the second step of splicing.
A complete model for the splicing reaction and its applicability to the spliceosome
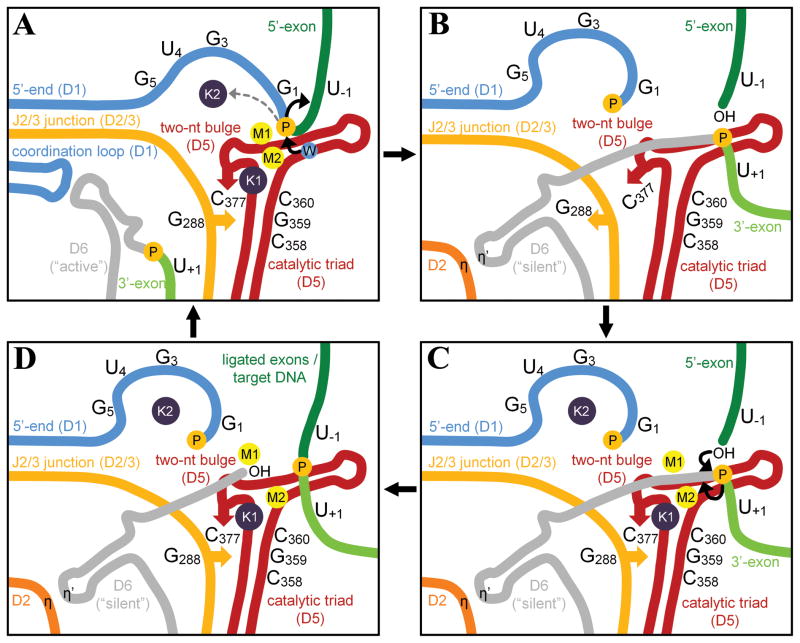
Based on the results of our structural and functional studies, we propose a model for the splicing reaction, which consists of three successive steps (Figure 7). In the precatalytic state, the group II intron adopts the triple helix conformation and catalyzes 5′-exon hydrolysis. Upon hydrolysis, the triple helix is broken by concerted rotation of G288 and C377 and the metal center within the active site is transiently disrupted to favor the release of G1 and the recruitment of the 3′-splice junction. After this rearrangement, the intron undergoes the second splicing step. It is possible that the combination of the 3′-splice junction with the toggled conformation of G288 may create a novel metal binding site for M2 that catalyzes 3′-exon excision. In this case, the geometry of the active site could potentially resemble RNA polymerase, which possesses a similar arrangement of active site elements, except for the location of M2 (Wang et al., 2006). That said, the same triple-helical active-site is observed in group II intron crystal structures that model both steps of splicing (5′-exon hydrolysis and SER reactions), as discussed above. Therefore, it seems more likely that G288 toggles back into the active site after the second-step reactants are brought into place, re-establishing the triple helix and the heteronuclear metal cluster prior to the second chemical step. Such toggling of G288 would explain how the intron can rearrange between the splicing steps despite possessing one single active site (Pyle and Lambowitz, 2006). This hypothesis implies the formation of a transient diffusive intermediate between the splicing steps, in which the active site metals bind diffusively to the toggled conformation of the intron – and are thus invisible in the crystal structure.
Figure 7. A model for group II intron splicing.
A) Precatalytic state. Group II intron adopts the triple helix conformation and catalyzes 5′-exon hydrolysis. “W” is the nucleophilic water molecule. B) Transient intermediate. Upon hydrolysis, the concerted rotation of G288 and C377 disrupts the metal center to favor the release of G1 and the recruitment of the 3′-splice junction, while D6 in the “silent” conformation forms the η-η′ interaction with D2 (Pyle, 2010). G288 interacts with residues near the bulge in D6 (de Lencastre et al., 2005). C) The second step of splicing. After all reactants are rearranged correctly, the metal cluster forms again to release ligated exons and the free intron. The position of M2 in the second step of splicing remains uncertain, as described in the text. D) SER and retrotransposition. The free intron harbors a fully assembled cluster and can rehydrolyze ligated exons or reinsert into genomic DNA. The retrotransposed intron is then transcribed again to start a new splicing cycle. See also Figure S6.
The idea of a rearrangement involving the J2/3 junction and of the formation of a transiently inactive intermediate is not only in agreement with structural and biochemical work on group II introns (Ho Faix, 1998 and this work), but it is strongly supported by data obtained on the analogous eukaryotic splicing machinery, the spliceosome. In the spliceosome, the snRNA subunit U6 possesses a conserved ACAGAGA box in which the last G (G52 in yeast) is likely to correspond with intron G288 (Keating et al., 2010). Interestingly, the box is close in space with the branch site nucleotide (Madhani and Guthrie, 1994), it interacts with the 5′-splice site, and it undergoes a rearrangement between the splicing steps (Konarska et al., 2006). Such rearrangement was proposed to have a double role. On one hand, it would facilitate the release of the 5′-end of spliceosomal introns from the active site after the first splicing step, and on the other hand, it would favor the recruitment of the 3′-splice junction into the active site for the second splicing step (Konarska et al., 2006). Furthermore, it is well accepted that during the splicing cycle the spliceosome “pauses” in transiently inactive states in order to avoid processing non ideal pre-mRNA substrates (Smith et al., 2008). Even if in the spliceosome the conformational rearrangement is primarily modulated by protein subunits (i.e. Prp8, Prp16) (Query and Konarska, 2004; Schwer and Guthrie, 1992), monovalent ions and the RNA elements homologous to functional groups of the intron are also directly involved. For instance, it is known that, similar to group II introns, the spliceosome is specifically dependent on K+ (Hardy et al., 1984). Moreover, it was shown that G52 mutations in the spliceosome have an inhibitory effect on the second step of splicing (Fabrizio and Abelson, 1990; Wolff et al., 1994), similar to what was described for G288 in the group II intron (Ho Faix, 1998; Mikheeva et al., 2000).
In the light of these analogies, it seems plausible that spliceosomal activation may depend on conformational toggling, possibly regulated by monovalent metal ions. We can envisage two different scenarios that differ in the choice of the partner nucleotides involved in the toggling. In the first scenario, we suggest that a toggling of G52 and U80 (corresponding to intron nucleotides G288 and C377, respectively (de Lencastre et al., 2005; Keating and Pyle, 2010)) might regulate spliceosomal catalysis (Figure S6B). Indeed, U80 was shown to bind an essential catalytic Mg2+ ion through its pro-S oxygen atom and thus to be a key residue in catalysis (Yean et al., 2000). However, no direct evidence of a reciprocal conformational change involving G52 and U80 has been reported to date. In group II introns, C377 binds both M1 and M2 metals, while evidence for a second metal coordination by U80 is not yet conclusive (Yean et al., 2000). In the second scenario, we propose that conformational toggling might occur between G52 and A25 (the U2–U6 helix I bulge, Figure S6C). Indeed, these two nucleotides interact before and during the chemical steps of spliceosomal splicing (Brow, 2002; Madhani and Guthrie, 1994). Moreover, a recent NMR structure of a “ground-state inactive” form of the U2/U6 spliceosomal subcomplex indicated that A25 and G52 are oriented in opposite directions (Burke et al., 2012) stimulating the authors to propose that a concerted conformational toggling of G52 and A25 is required during formation of a functional spliceosomal active site (Burke et al., 2012). In this case, rotation of G52 towards A25 may establish a structure similar to the group II intron triple helix. In this scenario, the role of U80 need not be marginal. For instance, it could bind M1, possibly occupying the position of U375. However, experimental support for a direct participation of A25 in the formation of the spliceosomal active site needs to be obtained, for instance determining whether this residue is involved in binding catalytic metal ions.
Regardless of which scenario best describes the still unknown structure of the active spliceosome (and there could be other models as well), our family of group II intron structures reinforces the hypothesis that the two systems share not just a common catalytic core, but a common mechanism for spatially arranging their reactants and switching between the two steps of splicing.
CONCLUDING REMARKS
In conclusion, we present a series of X-ray structures of the O.i. group II intron that provide essential mechanistic details about the complete splicing reaction. On one hand, they show the intron “in action”, and they clarify the mechanism of hydrolytic splicing and the structural basis for retrotransposition. Additionally, they reveal that the intron active site can adopt two alternative conformations and that toggling between them may regulate the transition from the first to the second step of splicing. Our results suggest a model for splicing with direct implications for the human spliceosome, which is expected to follow a reaction mechanism similar to group II introns. Therefore, our data provide a new framework for the design of future experiments aimed at determining the structure of intron domain 6 and of the spliceosomal active site, which are urgently needed to complete our understanding of splicing and pre-mRNA maturation processes.
EXPERIMENTAL PROCEDURES
Cloning and mutagenesis
The constructs of Oceanobacillus iheyensis group II intron used in this work correspond to previously used constructs (Toor et al., 2008a), modified as described in Extended Experimental Procedures.
In vitro transcription, purification and splicing assays
Following restriction with the appropriate endonucleases at 37°C overnight, the intron was transcribed in vitro using T7 polymerase as previously described (Toor et al., 2008a). For crystallization purposes, it was then purified in a native state as previously described (Toor et al., 2008a) omitting the splicing step. Intron RNA was then rebuffered and concentrated to 80 μM in buffer A (10 mM MgCl2 and 5 mM sodium cacodylate pH 6.5). For the splicing studies, the intron was radiolabeled during transcription, purified in a denatured state as previously described (Toor et al., 2008a), and subsequently refolded. Splicing assays were performed at 37°C and in 5 mM MgCl2 if not mentioned otherwise, as described in Extended Experimental Procedures.
Crystallization
Crystals of the purified intron mixed with a 0.5 mM spermine solution in buffer A, and with the crystallization buffer in a 1:1:1 volume ratio (Toor et al., 2008a), were grown at 30°C by the hanging drop vapor diffusion method. Where appropriate, a 1 mM oligonucleotide solution was mixed with all other components in a 1:1:1:1 volume ratio. All crystallization solutions are described in the Extended Experimental Procedure section.
Structure determination
Diffraction data (summarized in Table S1) were collected at beamlines 24ID-C and E (NE-CAT) at the Argonne Photon Source (APS), Argonne, IL and processed with the Rapid Automated Processing of Data (RAPD) software package (https://rapd.nec.aps.anl.gov/rapd/) and with the XDS suite (Kabsch, 1993). The structures were solved by molecular replacement using Phaser [CCP4, (Collaborative computational project number 4, 1994)] and the RNA coordinates of Protein Data Bank (PDB) entry 3IGI (without solvent atoms and bound oligonucleotide) as the initial model (Toor et al., 2010). The models were improved automatically in Phenix (Adams et al., 2010) and Refmac5 using TLS (Collaborative computational project number 4, 1994; Murshudov et al., 1997), and manually in Coot (Emsley and Cowtan, 2004), and finally evaluated by MolProbity (Chen et al., 2010; Davis et al., 2007). Anomalous difference Fourier electron density maps were calculated using the programs SFall and FFT in CCP4 (Agarwal, 1978; Ten Eyck, 1973). Omit maps were calculated in Phenix, excluding the atoms of interest, and avoiding model bias by simulated annealing refinement. The figures depicting the structures were drawn using PyMOL (Schrodinger, 2010).
Supplementary Material
02
03
Table s1
RESEARCH HIGHLIGHTS.
- A heteronuclear, four-metal-ion center catalyzes group II intron splicing
- The functional groups maintain their position during both catalytic steps
- Between the steps, the active site transiently adopts an alternative conformation
- The same conformational change may regulate splicing fidelity in the spliceosome
Acknowledgments
We thank the beamline scientists at 24-ID-C and E, NE-CAT, APS, Argonne (IL), USA for their thorough support during data collection. We acknowledge all members of the Pyle lab, and in particular Dr. Olga Fedorova for synthesizing the oligonucleotide fragment, Dr. Maximilian Bailor for cloning the OiD1-5 construct, Gabriele Drews for cloning the Oi5eD1-5 construct, Dr. Kevin Keating for the analysis of the backbone kink at the 5′-splice junction (Table S2), Dr. Srinivas Somarowthu for calculating the model of intron domain 6 in the silent conformation (Figure S4), Dr. Laura Murray for sharing preliminary unpublished results, Dr. Dahai Luo and Dr. Patrick Lombardi for valuable discussion, and Dr. Isabel Chillón Gázquez for constructive criticism and help preparing the figures. Additionally, we acknowledge Dr. John Abelson, Dr. John Stahley, Dr. Magda Konarska, Dr. Patrick Cramer, Dr. Giovanni Capranico, Dr. Ulrich Ermler and Tanja Hedderich for insightful discussion. This project was supported by the National Institute of Health (RO1GM50313). Prof. Pyle is a Howard Hughes Medical Institute Investigator.
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors were deposited to the Protein Data Bank (PDB) with entry codes: 4E8K, 4E8M, 4E8N, 4E8P, 4E8Q, 4E8R, 4E8T, 4E8V, 4FAQ, 4FAR, 4FAU, 4FAW, 4FAX, and 4FB0.
Supplemental Information includes Extended Experimental Procedures, 6 figures, and 4 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal RC. A new least-squares refinement technique based on the fast Fourier transform algorithm. Acta Crystallogr A Found Crystallogr. 1978;34:791–809. [Google Scholar]
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Boudvillain M, Pyle AM. Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. Embo J. 1998;17:7091–7104. doi: 10.1093/emboj/17.23.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger SC, Belcher SM, Schmidt U, Dib-Hajj SD, Schmidt T, Perlman PS. Studies of point mutants define three essential paired nucleotides in the domain 5 substructure of a group II intron. Mol Cell Biol. 1995;15:4479–4488. doi: 10.1128/mcb.15.8.4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE. Structure of the yeast U2/U6 snRNA complex. RNA. 2012 doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Hanna RL, Doudna JA. A magnesium ion core at the heart of a ribozyme domain. Nat Struct Biol. 1997;4:553–558. doi: 10.1038/nsb0797-553. [DOI] [PubMed] [Google Scholar]
- Chan RT, Robart AR, Rajashankar KR, Pyle AM, Toor N. Crystal structure of a group II intron in the pre-catalytic state. Nat Struct Mol Biol. 2012;19:555–557. doi: 10.1038/nsmb.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A. Interaction of intronic boundaries is required for the 2nd splicing step efficiency of a group-II intron. EMBO J. 1993;12:5173–5180. doi: 10.1002/j.1460-2075.1993.tb06212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative computational project number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Conn GL, Gittis AG, Lattman EE, Misra VK, Draper DE. A compact RNA tertiary structure contains a buried backbone-K+ complex. J Mol Biol. 2002;318:963–973. doi: 10.1016/S0022-2836(02)00147-X. [DOI] [PubMed] [Google Scholar]
- Cotton AF, Wilkinson G. Symmetry and Structure. In Advanced organic chemistry. A comprehensive text. New York, USA: John Wiley and Sons, Inc; 1980. pp. 58–59. [Google Scholar]
- Dai L, Zimmerly S. The dispersal of five group II introns among natural populations of Escherichia coli. Rna. 2002;8:1294–1307. doi: 10.1017/s1355838202023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lencastre A, Hamill S, Pyle AM. A single active-site region for a group II intron. Nat Struct Mol Biol. 2005;12:626–627. doi: 10.1038/nsmb957. [DOI] [PubMed] [Google Scholar]
- de Lencastre A, Pyle AM. Three essential and conserved regions of the group II intron are proximal to the 5′-splice site. RNA. 2008;14:11–24. doi: 10.1261/rna.774008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Erat MC, Sigel RK. Divalent metal ions tune the self-splicing reaction of the yeast mitochondrial group II intron Sc.ai5γ. J Biol Inorg Chem. 2008;13:1025–1036. doi: 10.1007/s00775-008-0390-7. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science. 1990;250:404–409. doi: 10.1126/science.2145630. [DOI] [PubMed] [Google Scholar]
- Fedorova O, Mitros T, Pyle AM. Domains 2 and 3 interact to form critical elements of the group II intron active site. J Mol Biol. 2003;330:197–209. doi: 10.1016/s0022-2836(03)00594-1. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez FM, Barrientos-Duran A, Diaz-Prado V, Fernandez-Lopez M, Toro N. Use of RmInt1, a group IIB intron lacking the intron-encoded protein endonuclease domain, in gene targeting. Applied and Environmental Microbiology. 2011;77:854–861. doi: 10.1128/AEM.02319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PM, Piccirilli JA. Metal ion coordination by the AGC triad in domain 5 contributes to group II intron catalysis. Nat Struct Biol. 2001;8:893–898. doi: 10.1038/nsb1001-893. [DOI] [PubMed] [Google Scholar]
- Hardy SF, Grabowski PJ, Padgett RA, Sharp PA. Cofactor requirements of splicing of purified messenger RNA precursors. Nature. 1984;308:375–377. doi: 10.1038/308375a0. [DOI] [PubMed] [Google Scholar]
- Ho Faix P. PhD thesis. University of Pittsburgh; Pittsburgh: 1998. Conserved nucleotides in the joining segment between domains 2 and 3 are important for group II intron splicing. [Google Scholar]
- Jacquier A, Michel F. Base-pairing interactions involving the 5′ and 3′-terminal nucleotides of group-II self-splicing introns. J Mol Biol. 1990;213:437–447. doi: 10.1016/S0022-2836(05)80206-2. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- Keating KS, Pyle AM. Semiautomated model building for RNA crystallography using a directed rotameric approach. Proc Natl Acad Sci U S A. 2010;107:8177–8182. doi: 10.1073/pnas.0911888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM. A structural analysis of the group II intron active site and implications for the spliceosome. RNA. 2010;16:1–9. doi: 10.1261/rna.1791310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. doi: 10.1016/j.molcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lambert D, Leipply D, Shiman R, Draper DE. The influence of monovalent cation size on the stability of RNA tertiary structures. J Mol Biol. 2009;390:791–804. doi: 10.1016/j.jmb.2009.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Randomization-selection analysis of snRNAs in vivo: evidence for a tertiary interaction in the spliceosome. Genes Dev. 1994;8:1071–1086. doi: 10.1101/gad.8.9.1071. [DOI] [PubMed] [Google Scholar]
- Mattick JS. Introns: evolution and function. Curr Opin Genet Dev. 1994;4:823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- McConnell TS, Herschlag D, Cech TR. Effects of divalent metal ions on individual steps of the Tetrahymena ribozyme reaction. Biochemistry. 1997;36:8293–8303. doi: 10.1021/bi9700678. [DOI] [PubMed] [Google Scholar]
- Messerschmidt A, Huber R, Poulos T, Wieghardt K, editors. Handbook of metalloproteins. Chirchester, UK: Wiley; 2001. [Google Scholar]
- Mikheeva S, Murray HL, Zhou H, Turczyk BM, Jarrell KA. Deletion of a conserved dinucleotide inhibits the second step of group II intron splicing. RNA. 2000;6:1509–1515. doi: 10.1017/s1355838200000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Padgett RA, Podar M, Boulanger SC, Perlman PS. The stereochemical course of group II intron self-splicing. Science. 1994;266:1685–1688. doi: 10.1126/science.7527587. [DOI] [PubMed] [Google Scholar]
- Pingoud V, Wende W, Friedhoff P, Reuter M, Alves J, Jeltsch A, Mones L, Fuxreiter M, Pingoud A. On the divalent metal ion dependence of DNA cleavage by restriction endonucleases of the EcoRI family. J Mol Biol. 2009;393:140–160. doi: 10.1016/j.jmb.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA. Stereochemical selectivity of group II intron splicing, reverse splicing, and hydrolysis reactions. Mol Cell Biol. 1995;15:4466–4478. doi: 10.1128/mcb.15.8.4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M, Perlman PS, Padgett RA. The two steps of group II intron self-splicing are mechanistically distinguishable. RNA. 1998;4:890–900. doi: 10.1017/s1355838298971643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol. 2010;45:215–232. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. Cold Spring Harbor: Cold Spring Harbor Press; 2006. pp. 469–505. [Google Scholar]
- Query CC, Konarska MM. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell. 2004;14:343–354. doi: 10.1016/s1097-2765(04)00217-5. [DOI] [PubMed] [Google Scholar]
- Schmidt U, Podar M, Stahl U, Perlman PS. Mutations of the two-nucleotide bulge of D5 of a group II intron block splicing in vitro and in vivo: phenotypes and suppressor mutations. Rna. 1996;2:1161–1172. [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Query CC, Konarska MM. “Nought may endure but mutability”: spliceosome dynamics and the regulation of splicing. Mol Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Eyck LF. Crystal physics, diffraction, theoretical and general crystallography. Acta Crystallogr A Found Crystallogr. 1973;29:183–191. [Google Scholar]
- Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA. 2010;16:57–69. doi: 10.1261/rna.1844010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science. 2008a;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Rajashankar K, Keating KS, Pyle AM. Structural basis for exon recognition by a group II intron. Nat Struct Mol Biol. 2008b;15:1221–1222. doi: 10.1038/nsmb.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viadiu H, Aggarwal AK. The role of metals in catalysis by the restriction endonuclease BamHI. Nat Struct Biol. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff T, Menssen R, Hammel J, Bindereif A. Splicing function of mammalian U6 small nuclear RNA: conserved positions in central domain and helix I are essential during the first and second step of pre-mRNA splicing. Proc Natl Acad Sci U S A. 1994;91:903–907. doi: 10.1073/pnas.91.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature. 2000;408:881–884. doi: 10.1038/35048617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
02
03
Table s1